Abstract
Hsp33, a molecular chaperone specifically activated by oxidative stress conditions that lead to protein unfolding, protects cells against oxidative protein aggregation. Stress sensing in Hsp33 occurs via its C-terminal redox switch domain, which consists of a zinc center that responds to the presence of oxidants and an adjacent metastable linker region, which responds to unfolding conditions. Here we show that single mutations in the N terminus of Hsp33 are sufficient to either partially (Hsp33-M172S) or completely (Hsp33-Y12E) abolish this post-translational regulation of Hsp33 chaperone function. Both mutations appear to work predominantly via the destabilization of the Hsp33 linker region without affecting zinc coordination, redox sensitivity, or substrate binding of Hsp33. We found that the M172S substitution causes moderate destabilization of the Hsp33 linker region, which seems sufficient to convert the redox-regulated Hsp33 into a temperature-controlled chaperone. The Y12E mutation leads to the constitutive unfolding of the Hsp33 linker region thereby turning Hsp33 into a constitutively active chaperone. These results demonstrate that the redox-controlled unfolding of the Hsp33 linker region plays the central role in the activation process of Hsp33. The zinc center of Hsp33 appears to act as the redox-sensitive toggle that adjusts the thermostability of the linker region to the cell redox status. In vivo studies confirmed that even mild overexpression of the Hsp33-Y12E mutant protein inhibits bacterial growth, providing important evidence that the tight functional regulation of Hsp33 chaperone activity plays a vital role in bacterial survival.
Keywords: Chaperone Chaperonin, Oxidative Stress, Protein Folding, Protein Stability, Zinc, Natively Disordered, Redox Regulation
Introduction
The heat shock protein Hsp33 is a highly conserved molecular chaperone, which appears to act by specifically protecting bacteria against oxidative stress conditions that cause protein unfolding (1–3). These stress conditions are the result of either very fast-acting oxidants, like hypochlorous acid (i.e. bleach), which directly unfold proteins or the combined action of kinetically slow, non-denaturing oxidants, such as H2O2, with protein unfolding conditions (i.e. oxidative heat shock) (1, 4). In both cases, widespread protein aggregation occurs in vitro and in vivo. Moreover, under these specific stress conditions, ATP-dependent chaperone foldases, such as the DnaK system, seem unable to effectively prevent protein aggregation, presumably because of the oxidative stress-mediated drop in cellular ATP levels (2, 5). Specific activation of the ATP-independent chaperone holdase Hsp33 appears, therefore, to serve as a compensatory mechanism to minimize protein aggregation and to increase bacterial stress resistance.
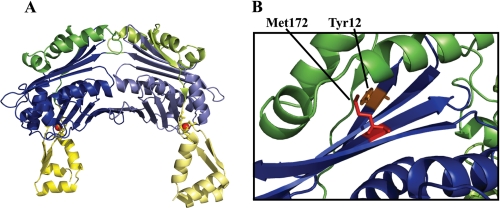
The mechanistic features that allow Hsp33 to sense oxidizing stress conditions that lead to the protein unfolding center around a C-terminal cysteine-rich domain (Fig. 1A, yellow), which harbors four absolutely conserved cysteines arranged in a Cys232-X-Cys234—X—Cys265-X-X-Cys268 motif. Under reducing, non-stress conditions, all four cysteines are engaged in the tetrahedral coordination of one Zn2+ ion (Fig. 1A, red sphere), whose high affinity binding provides significant stability to the Hsp33 C terminus (6). The zinc binding domain connects to the Hsp33 N-terminal domain (Fig. 1A, blue) via a highly flexible ∼52-amino acid linker region (Fig. 1A, green). In the crystal structure of reduced, inactive Hsp33, the linker region is compactly folded and makes extensive contacts with a largely hydrophobic, four-stranded β-sheet platform of the Hsp33 N-terminal domain (7) (Fig. 1A and supplemental Fig. S1). Activation of the Hsp33 chaperone function is triggered by intramolecular disulfide bond formation, which leads to the unfolding of the Hsp33 zinc binding domain and unfolding of the linker region. The latter has been shown to lead to the exposure of hydrophobic surface areas, the proposed binding sites of Hsp33 for unfolded proteins (supplemental Fig. S1 and Refs. 8–10).
FIGURE 1.
Location of the Hsp33 single site mutations. A, domain structure of full-length reduced Hsp33 from B. subtilis Hsp33 (PDB code: 1VZY) (7). Hsp33 is a two-domain protein with an N-terminal domain (blue), a highly flexible linker region (green), and a zinc binding domain (yellow) that harbors the four absolutely conserved cysteines, which coordinate one zinc ion (red sphere). The linker region appears to be stably folded on top of a large hydrophobic four-stranded β-sheet, which has been postulated to serve as a substrate binding site upon linker unfolding and activation of Hsp33 (see supplemental Fig. S1). Although reduced, inactive Hsp33 is monomeric in solution, it forms dimers in the crystal structure. B, to alter the hydrophobic character of the linker binding interface, either Tyr12 in β-sheet 1 was replaced with glutamate (Hsp33-Y12E) or Met172 in β-sheet 9 was substituted with serine (Hsp33-M172S). To illustrate the location of the targeted residues in E. coli Hsp33, the x-ray structure of the truncated E. coli Hsp331–255 (PDB code:1HW7) was used.
Recent studies suggested a mechanistic model for the activation of Hsp33 in which unfolding of the linker region simultaneously depends on and controls disulfide bond formation (4). In this model, incubation of Hsp33 with H2O2 leads to rapid formation of the Cys265–Cys268 disulfide bond, zinc release, and unfolding of the zinc binding domain. This unfolding appears to destabilize the adjacent linker region, which is now in a dynamic equilibrium between a folded state in which the second pair of active site cysteines is not accessible for oxidation and an unfolded state in which the critical second disulfide bond between Cys232 and Cys234 can be formed (11). Kinetically slow oxidants, such as H2O2, induce the formation of the second disulfide bond only in the presence of additional unfolding conditions, which shift the equilibrium to the unfolded conformation. Kinetically fast oxidants, such as HOCl, rapidly form the second disulfide bond. Once the second disulfide bond is formed, the linker region appears to remain in an unfolded conformation, and two oxidized monomers associate to form the highly active Hsp33 dimer (4).
The activation model of Hsp33 implies that the N-terminal hydrophobic platform serves a dual role: as interaction site for the linker region under non-stress conditions and as a binding site for unfolded substrate proteins under stress conditions. We decided to test this model by individually replacing distinct conserved hydrophobic amino acids in the N-terminal domain of Hsp33 with polar (M172S) or charged (Y12E) amino acids, thereby substantially altering the hydrophobic character of this interaction site. Surprisingly, we found that neither of the mutations affected the apparent affinity of Hsp33 for substrate proteins or its ability to sense oxidants. In contrast, the single mutations dramatically altered the regulation of Hsp33, converting the protein into a temperature-regulated (Hsp33-M172S) or constitutively active (Hsp33-Y12E) chaperone, respectively. Circular dichroism (CD) measurements revealed a close correlation between the thermostability of the linker region and the functional regulation of the mutant proteins, providing experimental evidence that unfolding of the linker region is indeed the crucial event in the Hsp33 activation process. The zinc center in Hsp33 appears to act as the switch, which adjusts the thermostability of the linker region according to the oxidation status of the cell. This control mechanism seems to be critical for the Hsp33 in vivo function because expression of the constitutively active Hsp33-Y12E mutant is deleterious to Escherichia coli growth.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
The Hsp33-M172S and Hsp33-Y12E mutants were generated by introducing single site mutations into the wild-type Hsp33 gene (hslO) using pUJ30 (pET11a-hslO) (3) as a template. The plasmids were transformed into JH13 (BL21, ΔhslO) (12), generating the expression strains JHa21 (JH13, pET11a-hslO-M172S) and CC4 (JH13, pET11a-hslO-Y12E). Additional strains used in this study are UJ83, a BL21 strain expressing wild-type Hsp33 from a pET11a vector, and CC20, a JH13 strain carrying the empty pET11a plasmid.
Purification of Wild-type and Hsp33 Mutant Proteins
Cultivation of the Hsp33-overexpressing strains for protein purification was conducted as previously described (12). For the induction of the Hsp33-M172S mutant, the protocol for wild-type Hsp33 was followed (3). To overexpress large amounts of soluble Hsp33-Y12E protein, cells were grown to an A600 of 0.6–0.8 at 37 °C and then shifted to 18 °C. Once the temperature was reached, Hsp33-Y12E expression was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside (IPTG)4 for 24 h. Afterward, the standard protocol for the Hsp33 purification was followed (12). All proteins were stored in 40 mm potassium phosphate buffer (KH2PO4) (pH 7.5) at −20 °C. Reduced, zinc reconstituted Hsp33red was prepared as previously described (12). To prepare oxidized Hsp33, 50 μm Hsp33red was incubated in 2 mm H2O2 at either 30 °C (Hsp33ox30 °C) or 43 °C (Hsp33ox43 °C) for 3 h. Oxidants were removed using NAP-5 columns.
Hsp33 Chaperone Activity Assay
The chaperone activity of wild-type Hsp33 and its variants was measured as previously described (13). In short, 12 μm citrate synthase (CS, Roche) was denatured with 4.5 m Gdn-HCl in 40 mm HEPES-KOH (pH 7.5) overnight at room temperature. To initiate protein aggregation, denatured CS was diluted to a final concentration of 75 nm in 40 mm HEPES-KOH, pH 7.5 at 30 °C under continuous stirring in the absence or presence of a 4× molar excess of wild-type Hsp33 or Hsp33 variants. To assess the activity of reduced wild-type Hsp33 and the variants at heat shock temperatures, CS (final concentration 150 nm) was incubated in the absence or presence of a 4× molar excess of Hsp33 in 40 mm HEPES-KOH, pH 7.5 at 43 °C. Light scattering was monitored at λex/λem at 360 nm using a Hitachi F4500 fluorescence spectrophotometer equipped with a temperature-controlled cuvette holder and stirrer.
Far-UV CD Spectroscopy
To determine changes in the secondary structure of wild-type Hsp33 and its variants, far-UV CD spectra were recorded in 20 mm KH2PO4, pH 7.5 at 20 °C using a Jasco-J810 spectropolarimeter as previously described (6). To determine the thermostability of Hsp33 and its derivates, the CD signal at either 195 nm or 222 nm was recorded at temperatures from 20 to 50 °C or 20 to 80 °C, respectively. The temperature was increased at a rate of 1 °C per minute. The temperature was controlled with a Jasco Peltier device.
ANS Fluorescence Measurements
The fluorescence probe bis-ANS (Molecular Probes) was used to test for the presence of hydrophobic surfaces in wild-type Hsp33 and the variants as previously described (6).
Thiol Redox State of Hsp33 and Its Variants
To assess whether oxidation of Hsp33 occurred during the chaperone activity assay, thiol trapping with the 490-Da thiol-reactive probe AMS was performed immediately after the activity measurement (3). An aliquot (815 μl) of the activity assay mixture was supplemented with 100% (v/v) trichloroacetic acid to a final concentration of 10% (v/v) trichloroacetic acid and incubated on ice for 30 min. The solution was centrifuged (30 min, 16,000 × g, 4 °C), and the protein pellet was resuspended in 20 μl of AMS-DAB solution (15 mm AMS, 6 m urea, 200 mm Tris-HCl, pH 8.5, 50 mm EDTA, 7.5% w/v SDS). After a 1-h incubation in the dark with shaking (1,300 rpm), 5 μl of 5× non-reducing Laemmli buffer was added, and the sample was loaded onto a non-reducing SDS-PAGE gel. Proteins were visualized by Western blot or Coomassie Blue staining of the gels.
Phenotype, Hsp33 Expression Levels, and Aggregate Formation in Strains Expressing Wild-type Hsp33 and Mutants
Overnight cultures of strains expressing either wild-type Hsp33 (UJ83), the two Hsp33 variants (CC4, JHa21), or no protein (CC20) from a pET11a vector in MOPS minimal medium (10× MOPS modified buffer (TEKnovo), 0.2 (w/v) glucose, 132 mm K2HPO4 pH 7, 20 μm thiamine) were prepared. Cell cultures were diluted 1:100 into MOPS minimal medium, supplemented with 50 μm IPTG, and cultivated at 37 °C. Growth was monitored every hour with absorbance measurements at A600. To compare soluble and insoluble proteins in these strains, 5 ml of the culture were removed, and cells were harvested by centrifugation (5,000 × g, 4 °C, 10 min) after 5.5 h of cultivation. Cells were resuspended in 650 μl of sucrose buffer (50 mm Tris-HCl pH 8.0, 25% w/v sucrose, 1 mm NaEDTA, 10 mm DTT) and homogenized by sonication (Kontes, Micro Ultrasonic cell disrupter, 2 × 10–15 s with the output control set to 70). 5 μl of lysozyme (50 mg/ml), 12.5 μl of DNase l (2 mg/ml), and 2.5 μl of MgCl2 (0.5 m) were added, followed by the addition of 650 μl of lysis buffer (50 mm Tris-HCl pH 8.0, 1% Triton X-100, 100 mm NaCl, 0.1% sodium azide, 0.1% sodium deoxycholate, 10 mm DTT). Samples were incubated at room temperature for 15 min, frozen at −80 °C for 20 min, and thawed at 37 °C for 15 min. Then, 2.5 μl of MgCl2 (0.5 m) were added, and the solution was incubated at room temperature until the viscosity decreased. NaEDTA was added, and the lysate was centrifuged (11,000 × g, 4 °C, 20 min). The supernatant was removed, and the pellet was resuspended in 500 μl of washing buffer A (50 mm Tris-HCl, pH 8.0, 0.5% Triton X-100, 100 mm NaCl, 1 mm NaEDTA, 1 mm DTT). The solution was again homogenized by sonication as before. Protein aggregates were pelleted by centrifugation (11,000 × g, 4 °C, 20 min). The supernatant was discarded, and the pellet was resuspended in 500 μl of washing buffer B (50 mm Tris-HCl pH 8.0, 100 mm NaCl, 1 mm NaEDTA, 1 mm DTT), and again homogenized by sonication. The washing steps were repeated three times. The final pellet was resuspended in 2× reducing Laemmli buffer and boiled for 5 min. Aggregate formation was visualized by SDS-PAGE followed by Coomassie Blue staining of the gels.
Homology Modeling and Calculation of the Connectivity Map
The homology modeling of reduced E. coli Hsp33 was performed using the MODELLER software through an interface of the MODWEB modeling server. The crystal structure of reduced Bacillus subtilis Hsp33 (PDB code:1VZY) was used as a template (7). The E. coli Hsp33 sequence was aligned to the B. subtilis Hsp33 structure revealing a sequence identity of 24%. To validate the model, the highly conserved core domain of Hsp33 (residues 7–178) was aligned (RMSD = 0.3 Å). A connectivity map was derived using the AQUAPROT server, which calculates interatomic interactions between residues in two chains (14). For this purpose, the E. coli Hsp33 sequence was divided into N-terminal (amino acids 7–178) and C-terminal (amino acids 179–288) domains. The results were confirmed by a standalone version of the same algorithm, which computes interactions within one chain. The definition of interactions between residues was based on the “all-atom contact” method (15), calculating the detailed atomic contacts including hydrogen bonds, van der Waals, aromatic and electrostatic interactions (15). The van der Waals interactions also reflect side chain packing between protein residues. The residue interaction network was plotted using the Cytoscape V2.6.3 program (16).
RESULTS
Site-specific Mutations to Alter the Linker Binding Interface
The surface of the Hsp33 N-terminal β-sheet, which is masked by the folded linker in the crystal structure of reduced, inactive Hsp33 (Fig. 1A), has been postulated to serve as a substrate binding site for unfolded proteins when Hsp33 is oxidized and active (8–10). Analysis of the Hsp33 x-ray structure reveals a cluster of highly conserved hydrophobic amino acids whose side chains face toward the linker region under reducing conditions and might interact with unfolded substrate proteins under oxidizing conditions. We focused on two conserved residues that mapped to the hydrophobic platform, Tyr12 on β-strand 1 and Met172 on β-strand 9 (Fig. 1B, supplemental Fig. S1). We predicted that alteration of these residues would affect the surface features of the four-stranded β-sheet but not the stability or fold of the Hsp33 N-terminal domain. To efficiently disrupt potential hydrophobic interactions, we individually replaced Tyr12 with glutamate (Hsp33-Y12E) or Met172 with the polar residue serine (Hsp33-M172S). We then overexpressed both mutant proteins in BL21 strains lacking the endogenous Hsp33 gene and determined their solubilities. Hsp33-M172S was fully soluble upon IPTG-induced overexpression at 37 °C. In contrast, the Hsp33-Y12E variant was overexpressed but largely insoluble under these conditions. A 24-h induction with IPTG at 18 °C was required to accumulate significant amounts of soluble Hsp33-Y12E. We purified the two variants according to the wild-type Hsp33 protocol and prepared fully reduced, zinc-reconstituted proteins (Hsp33-Y12Ered, Hsp33-M172Sred) for subsequent functional studies.
Mutations in the Hsp33 N-terminal Domain Dramatically Alter Functional Regulation
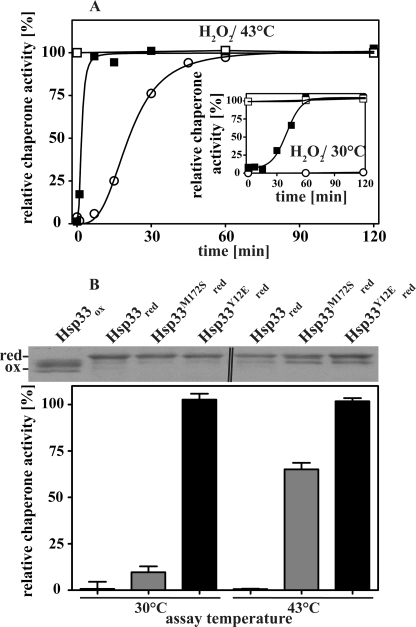
Hsp33 activation by fast-acting oxidants such as HOCl occurs within the mixing time of the experiment, making evaluation of activation kinetics and determination of oxidation intermediates technically challenging. In contrast, H2O2-mediated activation of Hsp33 is significantly slower (t½ of ∼20 min) and depends on the additional presence of unfolding conditions (43 °C) (Fig. 2A, open circles). In the absence of unfolding conditions (H2O2, 30 °C), an oxidation intermediate accumulates, which appears to lack the second disulfide bond (11) and is inactive as a chaperone (Fig. 2A, inset). To determine how the introduced mutations affect the redox regulation and chaperone function of our Hsp33 variants, we monitored their activity upon incubation in 2 mm H2O2 at either 30 or 43 °C. As shown in Fig. 2A, both variants displayed chaperone activity very similar to that of activated wild-type Hsp33 when incubated with H2O2 at 43 °C for 2 h. All three oxidized variants suppressed the aggregation of chemically denatured CS to the same extent. These results clearly indicate that the introduced mutations did not substantially affect the substrate binding affinity of Hsp33. Similar results were observed when the activity of the proteins was tested using thermally unfolding malate dehydrogenase as an in vitro chaperone substrate (supplemental Fig. S2A).
FIGURE 2.
In vitro chaperone activity of Hsp33 and variants. A, activation kinetics of wild-type Hsp33 and its mutant proteins. Reduced wild-type Hsp33 (open circles), Hsp33-M172S (closed squares), or Hsp33-Y12E (open squares) were incubated in 2 mm H2O2 at either 43 or at 30 °C (inset). At the time points indicated, aliquots of Hsp33 (final concentration 300 nm) were tested for their ability to suppress the aggregation of chemically denatured CS (final concentration 75 nm) at 30 °C. B, chaperone activity of reduced, zinc-reconstituted wild-type Hsp33 and its variants. The influence of a 4-fold molar excess of the reduced Hsp33 variants on the aggregation of chemically unfolded CS (75 nm) at 30 °C or thermally unfolded CS (150 nm) at 43 °C was determined. 0% activity is defined as the light scattering signal 4 min after addition of CS in the absence of chaperones, and 100% activity corresponds to the light scattering signal of CS in the presence of a 4-fold molar excess of wild-type Hsp33 that had been activated for 180 min in 2 mm H2O2 at 43 °C (Hsp33ox43 °C). To detect any potential changes in the thiol oxidation status of wild-type Hsp33 and the variants during the activity assay at 30 °C or 43 °C, aliquots were taken immediately after the end of the activity measurement (i.e. 4 min) and labeled with the thiol alkylating reagent AMS. The 490-Da AMS interacts irreversibly with any free thiol group in the protein and leads to a mass increase corresponding to the number of reduced cysteines. The change in mass can be visualized using non-reducing SDS-PAGE (top of panel). The double line (‖) in the top panel indicates the position at which a second Hsp33ox control was removed from the image.
In contrast to the chaperone function, which seems unchanged, the regulation of Hsp33 chaperone function appears to be dramatically affected by the mutations. Activation of Hsp33-M172S was significantly faster at 43 °C and lacked the lag phase characteristic of wild-type Hsp33 (Fig. 2A, compare Hsp33-M172S, closed squares and wild-type Hsp33, open circles). This lag phase has been shown to correlate to the formation of the critical second disulfide bond in Hsp33 and is thought to represent the rate-limiting step in its activation (11). Moreover, activation of Hsp33-M172S no longer required the simultaneous presence of oxidizing and unfolding conditions. Incubation of Hsp33-M172S in 2 mm H2O2 at 30 °C was fully sufficient for its activation (Fig. 2A, inset, closed squares) whereas wild-type Hsp33 (Fig. 2A, inset, open circles) remained inactive. Likewise, simple incubation of reduced, zinc-reconstituted Hsp33-M172S at elevated temperatures (43 °C) caused activation of its chaperone function (Fig. 2B). In contrast to the oxidative activation of Hsp33, however, which is irreversible unless reducing agents are added, thermal activation of Hsp33-M172S was fully reversible and required the activity tests to be performed at 43 °C (Fig. 2B).
Whereas Hsp33-M172S maintained at least some requirement for post-translational regulation, the freshly reduced and zinc-reconstituted Hsp33-Y12E mutant appeared to be fully active even without incubation in H2O2 or exposure to elevated temperatures (Fig. 2, A and B). Additional exposure to H2O2 at either 30 or 43 °C did not further increase its chaperone activity, suggesting that Hsp33-Y12E is constitutively active (Fig. 2A, open squares). To exclude the possibility that high oxidation sensitivity of the active site cysteines in the two Hsp33 variants leads to their uncontrolled oxidation and subsequent activation during the chaperone assays, we performed thiol trapping experiments on the reduced proteins immediately after the activity assays to assess their cysteine oxidation status (Fig. 2B). We made use of the 490-Da thiol-alkylating reagent AMS, which irreversibly reacts with reduced cysteines and causes a distinct mass increase that is easily discernable on SDS-PAGE. We found that the cysteines in both mutant proteins are reduced and remain reduced even during incubation at 43 °C. This result agrees well with zinc binding studies, which revealed that both mutant proteins remain fully zinc-reconstituted even in the presence of a 20-fold excess of the zinc chelator PAR (Kα = 2 × 1012 m−1 at pH 7) (17) (supplemental Fig. S2B).
To test the oligomerization status of the reduced Hsp33 variants, we performed analytical ultracentrifugation and confirmed that both variants are monomeric, as is reduced wild-type Hsp33 (data not shown) (12, 18). These results are in contrast to previous studies, which suggested that dimerization of Hsp33 is required for the full activation of its chaperone function (6, 18). Some of these conclusions were based on a single Hsp33 mutation designed to render Hsp33 constitutively monomeric (Hsp33-E150R) (6). Our results with the Hsp33-Y12E mutant suggest that Hsp33 does not require dimerization for its chaperone function in vitro. It is therefore conceivable that the E150R mutation not only interferes with Hsp33 dimerization but, more importantly, with substrate binding. This possibility remains to be investigated. Nevertheless, we conclude from our results that single amino acid mutations at the linker binding interface of the Hsp33 N terminus are sufficient to generate Hsp33 variants that function as highly effective chaperone holdases despite their reduced, zinc-reconstituted, and monomeric conformation.
Hsp33 Variants Show a Decrease in Surface Hydrophobicity
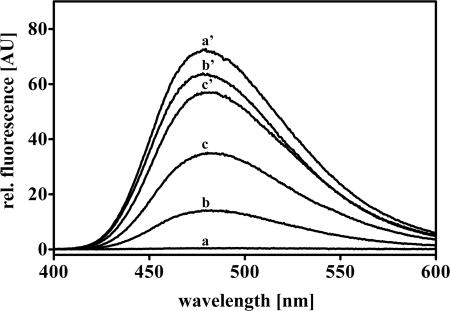
Our results indicated that mutations designed to alter the hydrophobic character of the N-terminal β-sheet platform affect the redox regulation but not the chaperone function of Hsp33. To assess whether the mutations actually caused a detectable change in hydrophobicity, particularly in those surfaces that specifically become exposed upon Hsp33 activation, we recorded the fluorescence spectra of reduced and oxidized wild-type and mutant Hsp33 proteins using the hydrophobic probe bis-ANS. As previously shown, wild-type Hsp33 undergoes substantial conformational rearrangements upon oxidative activation, which lead to the exposure of extensive hydrophobic surfaces that interact with bis-ANS (10) (Fig. 3, compare inactive Hsp33red, trace a and activated Hsp33ox43 °C, trace a′). When both mutant proteins were exposed to the same oxidizing conditions (2 mm H2O2, 43 °C, 3 h), which fully activated wild-type Hsp33, their extent of surface hydrophobicity was significantly reduced (Fig. 3, compare traces b′ and c′ with a′). These results confirmed that the mutations did indeed affect the hydrophobicity of surface areas that become exposed during the activation process. Furthermore, analysis of the surface hydrophobicity of the reduced, constitutively active Hsp33-Y12E mutant revealed that not all of the hydrophobic surface areas exposed upon oxidative activation of Hsp33 are necessary for its chaperone function. This mutant protein is as active as oxidized wild-type Hsp33 when reduced and zinc-coordinated but shows significantly less overall surface hydrophobicity (Fig. 3, trace c).
FIGURE 3.
Hydrophobicity of Hsp33 and variants. 3 μm freshly reduced wild-type Hsp33red (trace a), Hsp33-M172S red (trace b), Hsp33-Y12Ered (trace c), or active wild-type Hsp33ox43 °C (trace a′), Hsp33-M172S ox43 °C (trace b′), Hsp33-Y12E ox43 °C (trace c′) were incubated with 10 μm of the hydrophobic probe bis-ANS in 40 mm Hepes buffer, pH 7.5. Fluorescence spectra were recorded and buffer corrected.
Close Correlation between Linker Destabilization and Hsp33 Activity
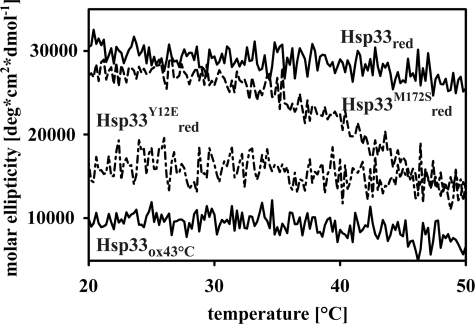
Based on the proposed activation model, Hsp33 chaperone function depends on the formation of the critical disulfide bond between Cys232 and Cys234, which is both kinetically and thermodynamically linked to the folding status of the linker region (4). Mutations that destabilize the linker region should therefore enhance the rate of disulfide bond formation and activation. To investigate whether the M172S and Y12E mutations indeed altered the thermostability of the linker region in the Hsp33 variants, we performed far-UV circular dichroism (CD) spectroscopy to monitor changes in Hsp33 secondary structure as a function of temperature. As previously shown, when fully reduced and zinc-reconstituted, wild-type Hsp33 begins to irreversibly unfold at temperatures above ∼50 °C (supplemental Fig. S3A) (1). When fully oxidized, zinc-depleted, and activated (Hsp33ox43 °C), the linker region and zinc binding domain of wild-type Hsp33 remain unfolded at temperatures as low as 20 °C (Fig. 4), and the N-terminal domain begins to unfold at a temperature that is similar to the onset of unfolding of reduced Hsp33 (supplemental Fig. S3A). In stark contrast, the reduced, zinc-reconstituted Hsp33-M172S variant begins to unfold at temperatures as low as 30 °C (Fig. 4). This unfolding (Tm ∼ 40 °C), which is fully reversible up to 50 °C (supplemental Fig. S3B), is highly reminiscent of the oxidized, inactive Hsp33ox30 °C intermediate, which lacks zinc coordination and has a substantially destabilized linker region (Tm < 43 °C) (1). A further increase in the incubation temperature of reduced Hsp33-M172S led to the irreversible unfolding of the remaining protein with the onset of unfolding at a temperature that was again similar to reduced wild-type Hsp33 (supplemental Fig. S3A). These results suggest that the M172S mutation does not globally destabilize Hsp33. Our finding that the linker region of Hsp33-M172S reversibly unfolds upon exposure to heat shock temperatures (supplemental Fig. S3B) agrees well with our previous observation that the reduced Hsp33-M172S acts as a molecular chaperone when tested at 43 °C but not at 30 °C (Fig. 2B). We concluded from these results that unfolding of the linker region is a critical element for the chaperone function of Hsp33.
FIGURE 4.
Thermal stability of wild-type Hsp33 and variants. Preparations of freshly reduced wild-type Hsp33, Hsp33-Y12E, Hsp33-M172S, or fully active Hsp33ox43 °C were heated to 50 °C (1 °C per minute) and changes in the molar ellipticity were recorded at 195 nm using a circular dichroism spectropolarimeter.
This conclusion was further supported by the CD analysis of the reduced, constitutively active Hsp33-Y12E mutant, which showed dramatically decreased secondary structure content, suggestive of a constitutively unfolded linker region (Fig. 4). Subsequent incubation with H2O2 caused some additional loss of secondary structure as monitored by CD spectroscopy, which likely corresponds to the oxidative zinc release and the unfolding of the zinc binding domain (supplemental Fig. S3C). Unfolding of the remaining protein structure started at temperatures similar to the wild-type Hsp33red and Hsp33-M172Sred mutant protein, suggesting that also this substitution did not globally destabilize Hsp33 (supplemental Fig. S3A).
These results provide good evidence that destabilization of the linker region by single mutations in the Hsp33 N-terminal linker binding domain is sufficient to eliminate Hsp33 post-translational regulation. They further imply that the stability of the linker region plays a central role in the chaperone function of Hsp33. The Hsp33 redox-sensing zinc binding domain appears to primarily serve as the molecular toggle whose redox status controls the thermostability of the linker region and ultimately the activity of the protein.
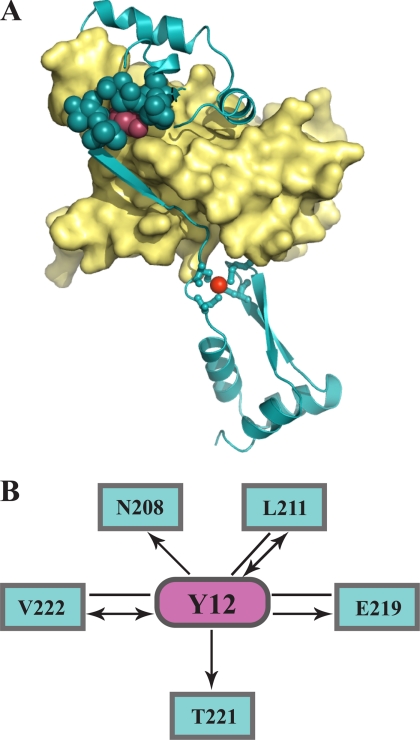
Tyrosine 12-a Central Interaction Hub between the Hsp33 N and C Terminus
To better understand how substitution at either Met172 or Tyr12 can exert such dramatic effects on Hsp33 functional regulation, we decided to analyze the interatomic connectivity of each of these residues with their surroundings. Unfortunately, the structure of reduced E. coli, Hsp33 is not known, and the only two available crystal structures of E. coli Hsp33 are domain-swapped and truncated protein variants (9, 19). We therefore chose to model the structure of reduced E. coli Hsp33 using the x-ray structure of reduced B. subtilis Hsp33 (1VZY) as a template (sequence identity 24%) (7). The quality of the obtained model, which we calculated using the software MODELLER (20), appeared to be very high, as reflected by the maximum GA341 score of 1 (21). When we analyzed the location of Tyr12 in this model (Fig. 5A, magenta spheres), we found that this residue makes extensive contacts with a large number of linker residues (Fig. 5A, cyan spheres). To characterize the architecture and connectivity of these interacting residues in more detail, we computed a contact network using AQUAPROT (14). This type of presentation reveals the environmental importance of specific residues and unveils potential cooperative interactions between residues (22). The contact map further confirmed the high connectivity of Tyr12 with residues located in the Hsp33 linker region (Fig. 5B). Tyr12 was found to interact with at least five residues in the linker region predominantly by side chain interactions, with one of the strongest contacts formed between Tyr12 and Glu219. Therefore, substitution of Tyr12 with a glutamate as in our Hsp33-Y12E variant should not only cause a decrease in residue packing but also strong repulsive interactions between the Hsp33 N and C terminus. This would explain how substitution of a single residue in the Hsp33 N terminus could lead to separation of the linker region from the N-terminal β-sheet platform and subsequent linker unfolding. The side chain atoms of Met172 are involved in interactions with three residues, only one of which, Leu214, is located in the linker region (data not shown). This result might explain why the M172S mutation leads to a more moderate destabilization of the linker region.
FIGURE 5.
Tyrosine 12: a central interaction hub in Hsp33. A, homology modeling of reduced E. coli Hsp33 was conducted with MODELLER using the crystal structure of reduced B. subtilis Hsp33 (PDB code: 1VZY) as a template. The N-terminal core domain is shown in the yellow surface representation, and the C-terminal redox switch domain is shown in cyan ribbon presentation. The zinc ion is depicted as red sphere. The N-terminal Tyr12 (magenta spheres) appears to interact tightly with several linker residues (cyan spheres). A high GA341 score of 1 provided by the MODWEB modeling server indicates that the model is accurate. B, connectivity map of Tyr12. Interactions between Tyr12 and residues in the Hsp33 C terminus (amino acids 179–288) are depicted as magenta and cyan nodes, respectively. The interaction types shown are van der Waals interactions. Arrows point toward the residues that contribute a backbone atom to the interaction.
Lack of Post-translational Regulation of Hsp33 Affects E. coli Growth
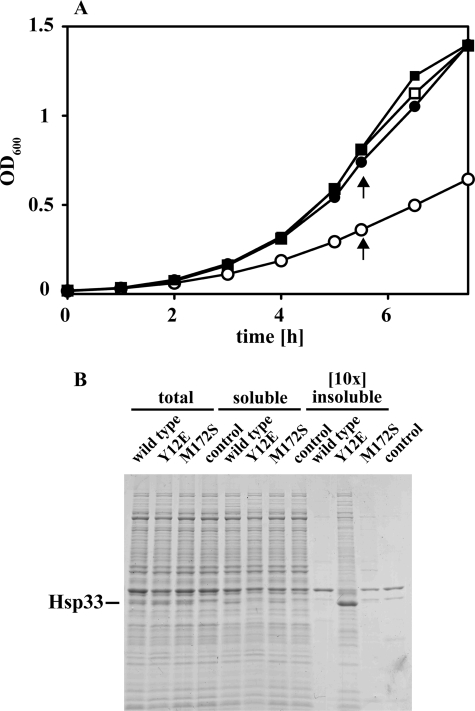
Our results suggested that we generated two Hsp33 mutants that lack their proper post-translational redox regulation in vitro. These mutants provided us now with the opportunity to investigate what role the post-translational regulation of Hsp33 plays in the cell. We cultivated BL21ΔhslO strains expressing either wild-type Hsp33 or the variants from an IPTG-inducible pET11a vector in the presence of increasing concentrations of IPTG at 37 °C (supplemental Fig. S4A). We observed a significant growth defect in strains expressing the constitutively active Hsp33-Y12E mutant at IPTG concentrations as low as 50 μm (Fig. 6A, open circles). In contrast, no growth defect was detected in strains expressing wild-type Hsp33 or the temperature-controlled Hsp33-M172S mutant protein under the same growth conditions. Increasing the cultivation temperature to 43 °C, however, made expression of Hsp33-M172S disadvantageous for E. coli as well (supplemental Fig. S4B). These results provided preliminary evidence that expression of Hsp33 variants lacking their tight post-translational redox control might be harmful to bacteria. Analysis of the respective cell lysates using SDS-PAGE revealed that all three Hsp33-expressing strains contained similar steady-state levels of Hsp33 (supplemental Fig. S4C). The Hsp33 expression levels in the presence of 50 μm IPTG were estimated by Western blot analysis to be ∼20-fold higher than the expression level of chromosomally encoded Hsp33 under non-stress conditions (data not shown). No obvious change in the general protein expression pattern (i.e. induction of the heat shock response) was observed (supplemental Fig. 4C). These results argued against the possibility that the growth defect observed in Hsp33-Y12E expressing strains is due to massive overexpression of a partially unfolded protein. When we analyzed and compared the soluble and insoluble protein population in these three strains, however, we detected a significant difference; a substantial proportion of Hsp33-Y12E protein was found to be insoluble and co-aggregated with a large number of different E. coli proteins (Fig. 6B). This co-aggregation led to a small but reproducible decrease in the overall amount soluble E. coli proteins. In contrast, both wild-type Hsp33 or Hsp33-M172S proteins remained mostly soluble at 37 °C and did not cause any protein aggregation. Cultivation of the Hsp33-M172S expressing strain at temperatures that activated the mutant protein in vitro (Fig. 2B) and caused a growth defect in vivo (supplemental Fig. S4B), however, also sequestered a large number of E. coli proteins into the insoluble protein pellet (supplemental Fig. S4D). These results suggest that in the absence of any post-translational regulation, activated Hsp33 binds to either newly synthesized or pre-existing cellular proteins, which may cause Hsp33 to co-aggregate with these proteins. It remains to be determined whether the formation of aggregates per se, or the sequestration of specific E. coli proteins necessary for optimal bacterial growth is responsible for the observed growth defect. In either case, these results provide a first clue as to why chaperone holdases, such as Hsp33, require tight functional regulation.
FIGURE 6.
Expression of constitutively active Hsp33-Y12E mutant causes growth defects in vivo. A, E. coli strains JH21 (BL21 ΔhslO) expressing either (closed circles) wild-type Hsp33, (closed squares) Hsp33-M172S, (open circles) Hsp33-Y12E or (open squares) no protein from a pET11a plasmid were cultivated in the presence of 50 μm IPTG in MOPS minimal medium at 37 °C. Growth was monitored at A600. Aliquots of the four cell cultures were harvested by centrifugation as indicated by arrows. B, total cell lysates were prepared and separated into soluble proteins and insoluble aggregates. The amount of total cell lysate and supernatant loaded was adjusted to the number of cells harvested. A 10-fold higher amount of aggregates was loaded onto the SDS-PAGE to visualize cellular proteins that precipitate.
DISCUSSION
Hsp33 belongs to a growing group of ATP-independent chaperone holdases, including small heat shock proteins (sHsps) and HdeA, that are inactive under non-stress conditions and use sophisticated post-translational mechanisms to synchronize their activation with specific cellular stress conditions (1, 23–25). For their activation, each of these chaperones appears to undergo the same type of stress-specific conformational rearrangements and unfolding events that cause the widespread aggregation of countless proteins in stress-treated organisms. The Hsp33 chaperone function is activated by the oxidative unfolding of its C-terminal redox switch domain, and it protects cells against oxidative stress-induced protein aggregation (1, 2). Activation of sHsps is triggered by temperature-induced structural rearrangements, which allows them to protect other cellular proteins against heat-induced aggregation (26, 27). Finally, the small HdeA dimer is specifically activated by acid-induced dissociation and unfolding, and it prevents aggregation of a wide range of acid-dissociated and denatured proteins (24, 25). The common denominator for the activation of these chaperone holdases appears to be local protein unfolding accompanied by a substantial increase in surface hydrophobicity (6, 10, 25, 27–29). Because the few known substrate binding sites on chaperones are hydrophobic in nature, these observations led to the very plausible hypothesis that local unfolding of the chaperones exposes previously buried substrate binding sites in the chaperones; hence their activation (23, 28, 30). Investigation of this hypothesis was straightforward in Hsp33 because crystal structures are available that reveal the precise location of the Hsp33 folding-sensitive linker in the reduced, inactive form (7). The linker region covers an extended ∼3800 Å2 N-terminal surface area, 75% of which is rich in hydrophobic residues (supplemental Fig. S1). To test the model that this area serves as a linker binding surface under non-active conditions and as a substrate binding site upon linker unfolding and activation, we substituted two of the most centrally located hydrophobic residues with either polar (M172S) or charged (Y172E) residues. We confirmed that substitution of either one of these residues significantly decreased the hydrophobicity of those surfaces that apparently become exposed upon Hsp33 oxidative activation. We were unable, however, to detect any apparent reduction in Hsp33 ability to prevent protein aggregation in vitro using different substrate proteins. These results suggested that the hydrophobic surface area directly beneath the linker binding site might not be directly involved in substrate binding. Where do unfolded substrate proteins then bind in Hsp33? Given the extended unfolding that occurs upon activation of Hsp33, it is tempting to speculate that the natively unfolded regions in Hsp33 themselves might be involved in substrate binding. Natively unfolded regions are, by definition, highly flexible and charged (31–33). Whereas their high flexibility would foster binding of a large variety of substrate proteins, their highly charged nature would increase the solubility of unfolding proteins and effectively prevent protein aggregation. Supported by the facts that disordered regions in RNA chaperones very often map with the respective RNA binding sites, and that many protein chaperones have extensive disordered regions, these observations led to the hypothesis that natively disordered regions might be directly involved in substrate interaction (34). This hypothesis found experimental support with a very recent study by Vierling and co-workers (35), who used a cross-linking approach to demonstrate that a flexible, intrinsically disordered N-terminal arm of the small heat shock protein PsHsp18.1 cross-links with unfolded substrate proteins in vitro, suggesting that it plays a crucial role in substrate interactions. Because of the hydrophobic nature of the substrate proteins, it is however likely that many of these intrinsically disordered chaperones use a combination of hydrophobic and hydrophilic regions to bind and solubilize their substrate proteins. This possibility has been recently suggested for the acid-activated HdeA, where mutations in the amphiphilic regions were found to reduce its chaperone function (28). Simultaneous mutations within the flexible linker region of Hsp33 and the hydrophobic surface areas might therefore be necessary to interfere with substrate binding in Hsp33 and to locate the substrate binding site of Hsp33.
Although the introduced mutations did not affect the chaperone function of Hsp33, they dramatically altered its redox regulation. Mutation of Met172 to serine turned Hsp33 into a purely heat or oxidative stress-regulated chaperone, whereas mutation of Tyr12 to glutamate caused Hsp33 to be constitutively active. Neither mutation significantly altered the stability of Hsp33 N-terminal or zinc binding domains. However, both substantially lowered the thermostability of the Hsp33 linker region, thereby apparently eradicating the thermodynamic control exerted by the Hsp33 oxidation-sensitive zinc binding domain. In wild-type Hsp33, stress sensing occurs via an interdependent sensing mechanism in which the zinc binding domain senses oxidative stress and the metastable linker region senses the folding status in the cell. Our mutant studies now demonstrate that a simple mutation that destabilizes the linker region is sufficient to abrogate this regulation. Our results provide the experimental evidence that the folding status of the linker region plays a central role for the Hsp33 chaperone function. Furthermore, they imply that the Hsp33 zinc center functions primarily as a redox sensitive pivot whose oxidation status controls the thermodynamic stability of the adjacent linker region and therefore the activity of Hsp33. Based on these results, it is therefore conceivable that stress conditions that act mainly on destabilizing the Hsp33 linker region, might be sufficient to activate Hsp33 independent of oxidative stress conditions. This conclusion would not only explain recent findings, which showed that Hsp33 expression significantly increases bacterial resistance toward solvent stress (36) but would imply that Hsp33 might be a much more versatile chaperone than previously anticipated.
Finally, our studies provide the first evidence that lack of the Hsp33 stringent post-translational control becomes highly disadvantageous to E. coli cells. Expression of the constitutively active Hsp33-Y12E variant caused a large number of cellular proteins to co-precipitate with Hsp33. These results suggest that the ability of active Hsp33 to interact with a wide variety of proteins in vivo (2) generates a clear need to switch Hsp33 off under non-stress conditions. The fact that the constitutively active Hsp33-Y12E mutant can no longer regulate substrate binding and release might be sufficient to target Hsp33 into the insoluble aggregates. Whether it is the newly synthesized proteins that fall victim to Hsp33-Y12E constitutive chaperone activity or mature proteins that are potentially unfolded by Hsp33 remains to be investigated. In either case, these results open the door to future investigations as to why chaperone holdases, such as Hsp33, require such exquisitely tight functional regulation.
Supplementary Material
Acknowledgments
We thank Dr. Zhaohui Xu for helpful discussions regarding Hsp33 structure and Dr. James Bardwell for critically reading the manuscript. We thank Dr. Janine Maddock for providing us with protocols for aggregate preparation and Dr. Titus Franzmann for help with the ultracentrifugation experiments.
This work was supported, in whole or in part, by a National Institutes of Health Grant GM065318 (to U. J.) and a Human Frontiers postdoctoral fellowship (to D. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Methods.
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- AMS
- 4-acetamido-4-maleimidyl-stilbene-2′-disulfonate
- MOPS
- 4-morpholinepropanesulfonic acid
- DTT
- dithiothreitol
- bis-ANS
- 3,3′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid
- CS
- citrate synthase.
REFERENCES
- 1.Ilbert M., Horst J., Ahrens S., Winter J., Graf P. C., Lilie H., Jakob U. (2007) Nat. Struct. Mol. Biol. 14, 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winter J., Linke K., Jatzek A., Jakob U. (2005) Mol. Cell 17, 381–392 [DOI] [PubMed] [Google Scholar]
- 3.Jakob U., Muse W., Eser M., Bardwell J. C. (1999) Cell 96, 341–352 [DOI] [PubMed] [Google Scholar]
- 4.Winter J., Ilbert M., Graf P. C., Ozcelik D., Jakob U. (2008) Cell 135, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrette W. C., Jr., Hannum D. M., Wheeler W. D., Hurst J. K. (1989) Biochemistry 28, 9172–9178 [DOI] [PubMed] [Google Scholar]
- 6.Graf P. C., Martinez-Yamout M., VanHaerents S., Lilie H., Dyson H. J., Jakob U. (2004) J. Biol. Chem. 279, 20529–20538 [DOI] [PubMed] [Google Scholar]
- 7.Janda I., Devedjiev Y., Derewenda U., Dauter Z., Bielnicki J., Cooper D. R., Graf P. C., Joachimiak A., Jakob U., Derewenda Z. S. (2004) Structure 12, 1901–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graf P. C., Jakob U. (2002) Cell Mol. Life Sci. 59, 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijayalakshmi J., Mukhergee M. K., Graumann J., Jakob U., Saper M. A. (2001) Structure 9, 367–375 [DOI] [PubMed] [Google Scholar]
- 10.Raman B., Siva Kumar L. V., Ramakrishna T., Mohan Rao C. (2001) FEBS Lett. 489, 19–24 [DOI] [PubMed] [Google Scholar]
- 11.Leichert L. I., Gehrke F., Gudiseva H. V., Blackwell T., Ilbert M., Walker A. K., Strahler J. R., Andrews P. C., Jakob U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graumann J., Lilie H., Tang X., Tucker K. A., Hoffmann J. H., Vijayalakshmi J., Saper M., Bardwell J. C., Jakob U. (2001) Structure 9, 377–387 [DOI] [PubMed] [Google Scholar]
- 13.Beissinger M., Buchner J. (1998) Biol. Chem. 379, 245–259 [PubMed] [Google Scholar]
- 14.Reichmann D., Phillip Y., Carmi A., Schreiber G. (2008) Biochemistry 47, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 15.Reichmann D., Cohen M., Abramovich R., Dym O., Lim D., Strynadka N. C., Schreiber G. (2007) J. Mol. Biol. 365, 663–679 [DOI] [PubMed] [Google Scholar]
- 16.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Genome Res. 13, 2498– 2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z. S., Peariso K., Penner-Hahn J. E., Matthews R. G. (1999) Biochemistry 38, 15915–15926 [DOI] [PubMed] [Google Scholar]
- 18.Akhtar M. W., Srinivas V., Raman B., Ramakrishna T., Inobe T., Maki K., Arai M., Kuwajima K., Rao Ch. M. (2004) J. Biol. Chem. 279, 55760–55769 [DOI] [PubMed] [Google Scholar]
- 19.Kim S. J., Jeong D. G., Chi S. W., Lee J. S., Ryu S. E. (2001) Nat. Struct. Biol. 8, 459–466 [DOI] [PubMed] [Google Scholar]
- 20.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 21.Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., Stuart A. C., Marti-Renom M. A., Madhusudhan M. S., Yerkovich B., Sali A. (2003) Nucleic Acids Res. 31, 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichmann D., Rahat O., Cohen M., Neuvirth H., Schreiber G. (2007) Curr. Opin. Struct. Biol. 17, 67–76 [DOI] [PubMed] [Google Scholar]
- 23.Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 24.Hong W., Jiao W., Hu J., Zhang J., Liu C., Fu X., Shen D., Xia B., Chang Z. (2005) J. Biol. Chem. 280, 27029–27034 [DOI] [PubMed] [Google Scholar]
- 25.Tapley T. L., Körner J. L., Barge M. T., Hupfeld J., Schauerte J. A., Gafni A., Jakob U., Bardwell J. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzmann T. M., Menhorn P., Walter S., Buchner J. (2008) Mol. Cell 29, 207–216 [DOI] [PubMed] [Google Scholar]
- 27.Giese K. C., Vierling E. (2002) J. Biol. Chem. 277, 46310–46318 [DOI] [PubMed] [Google Scholar]
- 28.Wu Y. E., Hong W., Liu C., Zhang L., Chang Z. (2008) Biochem. J. 412, 389–397 [DOI] [PubMed] [Google Scholar]
- 29.Yang H., Huang S., Dai H., Gong Y., Zheng C., Chang Z. (1999) Protein Sci. 8, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter J., Jakob U. (2004) Crit. Rev. Biochem. Mol. Biol. 39, 297–317 [DOI] [PubMed] [Google Scholar]
- 31.Tompa P. (2009) FEBS J. 276, 5406–5415 [DOI] [PubMed] [Google Scholar]
- 32.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) J. Mol. Graphics Modelling 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 33.Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 34.Tompa P., Csermely P. (2004) Faseb J. 18, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 35.Jaya N., Garcia V., Vierling E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15604–15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang H. J., Heo D. H., Choi S. W., Kim K. N., Shim J., Kim C. W., Sung H. C., Yun C. W. (2007) Biochem. Biophys. Res. Commun. 358, 743–750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.