Abstract
Prions are proteins that access self-templating amyloid forms, which confer phenotypic changes that can spread from individual to individual within or between species. These infectious phenotypes can be beneficial, as with yeast prions, or deleterious, as with mammalian prions that transmit spongiform encephalopathies. However, the ability to form self-templating amyloid is not unique to prion proteins. Diverse polypeptides that tend to populate intrinsically unfolded states also form self-templating amyloid conformers that are associated with devastating neurodegenerative disorders. Moreover, two RNA-binding proteins, FUS and TDP-43, which form cytoplasmic aggregates in amyotrophic lateral sclerosis, harbor a ‘prion domain’ similar to those found in several yeast prion proteins. Can these proteins and the neurodegenerative diseases to which they are linked become ‘infectious’ too? Here, we highlight advances that define the transmissibility of amyloid forms connected with Alzheimer's disease, Parkinson's disease and Huntington's disease. Collectively, these findings suggest that amyloid conformers can spread from cell to cell within the brains of afflicted individuals, thereby spreading the specific neurodegenerative phenotypes distinctive to the protein being converted to amyloid. Importantly, this transmissibility mandates a re-evaluation of emerging neuronal graft and stem-cell therapies. In this Commentary, we suggest how these treatments might be optimized to overcome the transmissible conformers that confer neurodegeneration.
Keywords: Amyloid, Infectivity, Prion, Stem cell, Therapy, Transmissibility
Introduction
Prion diseases are a heterogeneous collection of fatal, infectious neurodegenerative disorders that afflict mammals, including: scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in deer and elk, and Creutzfeldt-Jakob Disease (CJD), Gerstmann-Sträussler-Scheinker syndrome (GSS) and fatal familial insomnia (FFI) in humans. These currently untreatable disorders are unusual because they can be inherited, acquired by infection or arise spontaneously. They are caused by pure proteinaceous infectious particles known as prions (Prusiner, 1982; Prusiner, 1998). Prions are proteins that exist in several alternative but functionally distinct conformations, at least one of which is self templating (Shorter and Lindquist, 2005; Soto and Castilla, 2004). Typically, the self-templating form is a ‘cross-β’ fibrous structure termed amyloid, in which the strands of the β-sheets align orthogonal to the fiber axis. These fibers elongate at both ends. Indeed, the fiber ends are the active sites of the prion. Fiber ends capture and convert natively folded proteins to the cross-β form (Fig. 1A). This self-templating or ‘seeding’ activity forms the basis of infectivity. Owing to the remarkable stability of the cross-β prion form, which resists detergents, proteases and heat denaturation (Dobson, 2003; Knowles et al., 2007; Prusiner, 1982; Prusiner et al., 1983; Smith et al., 2006), transmission between individuals (e.g. BSE is transmitted from cow to cow) and occasionally even between species (e.g. variant CJD is transmitted from cow to human) becomes possible. Because of their resistance to harsh environments, prions can be found in saliva and blood (Mathiason et al., 2006; Saa et al., 2006), and can even survive the normally denaturing environment of the digestive system (Beekes and McBride, 2007). Thus, even before presenting with symptoms of CWD, deer excrete prions in feces that persist in the environment and enable rapid horizontal transmission of CWD via a fecal-oral route within deer populations (Tamguney et al., 2009).
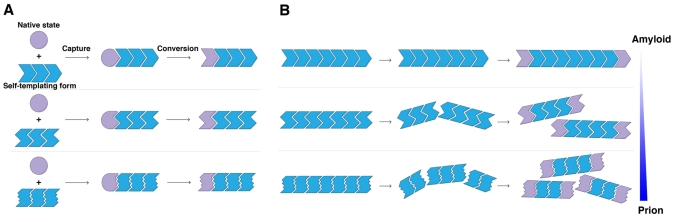
Fig. 1.
Prion seeding, strain and fragmentation phenomena. (A) Prions are proteins that exist in several alternative conformations. The first is the native state, depicted by a purple circle. Prion proteins can also exist in numerous distinct self-templating amyloid forms or ‘strains’, depicted by distinct blue polymers. Amyloid forms capture and convert native conformers to the self-templating form at their ends. Typically, this capture and conversion process requires native conformers to exist in a transiently unfolded state or to possess an intrinsically unfolded domain. (B) Amplification of conformational replication is achieved by the fragmentation of amyloid forms to liberate new ends. Fragmentation also allows the dissemination of infectious material. Fibers can fragment spontaneously (Smith et al., 2006), or fragmentation can be catalyzed by cellular factors, such as Hsp104 in yeast (Shorter and Lindquist, 2004). Typically, different strains fragment at different rates. More frangible strains tend to be more potent prions because they expose more fiber ends (the active sites of conformational replication) per unit mass and therefore convert monomers more rapidly (Colby et al., 2009; Tanaka et al., 2006). Indeed, one possible explanation for the reason all self-templating amyloid conformers are not prions is that some strains do not fragment readily enough to sufficiently amplify conformational replication (Salnikova et al., 2005). Thus, a gradient of forms with increasing frangibility can be envisioned, with prions and amyloids existing at opposite ends of the spectrum.
It is now clear that these devastating transmissible spongiform encephalopathies (TSEs) are all due to misfolding of one specific protein: mammalian prion protein (PrP) (Prusiner, 1998), a glycosylphosphatidylinositol (GPI)-anchored plasma membrane protein of uncertain function (Bremer et al., 2010; Le Pichon et al., 2009; Steele et al., 2007). For decades, however, the identity of the infectious agent was puzzling and controversial because it is virtually devoid of nucleic acid, the canonical disease agent at the time (Alper et al., 1967; Bruce and Dickinson, 1987). Rather, it was gradually comprehended that infectious amyloid forms of a host-encoded protein, PrP, were causative (Bolton et al., 1982; Oesch et al., 1985; Pan et al., 1993; Prusiner et al., 1983). Strong support for the prion hypothesis has come from experiments showing that amyloid forms of PrP generated from solely recombinant protein can eventually induce transmissible neurodegenerative disease upon inoculation into transgenic mice that overexpress PrP (Colby et al., 2009; Legname et al., 2004) or wild-type hamsters (Castilla et al., 2005). Importantly, Ma and colleagues have recently provided even stronger support (Wang et al., 2010). Using a clever strategy that combined specific lipid- and RNA-facilitating factors, a potent prion was generated from recombinant PrP that induced the rapid appearance of a classic prion disease in wild-type mice (Wang et al., 2010).
Compelling genetic evidence has also linked PrP to TSE pathogenesis. Missense mutations in the PrP gene are tightly linked to familial forms of GSS (Hsiao et al., 1989), FFI (Medori et al., 1992) and CJD (Goldgaber et al., 1989). Moreover, in mice, an FFI-linked mutation in PrP can induce neurodegenerative disease and spontaneous generation of infectious material (Jackson et al., 2009). At the other extreme, missense mutations in PrP can confer resistance to prion disease (Mead et al., 2009). Importantly, PrP-knockout mice resist infection by exogenous TSE-inducing prions (Bueler et al., 1993). This resistance arises because the infectious form of PrP must recruit and convert endogenous PrP to transmit disease. Indeed, if PrP-expressing neurons are grafted into PrP-knockout mice, then only the grafts become infected upon prion exposure, whereas surrounding tissue is unperturbed (Brandner et al., 1996). This experimental observation might prove to be pivotal for devising strategies to mitigate the transmissibility of other human neurodegenerative disease proteins.
The cascade of amyloid seeding incited by prions, however, is not always problematic and is certainly not restricted to mammals. In yeast, several proteins form prions that confer specific heritable phenotypes, which are either benign or advantageous under diverse environmental conditions (Alberti et al., 2009; Shorter and Lindquist, 2005; True and Lindquist, 2000; Tyedmers et al., 2008). These specific heritable phenotypes can be induced de novo by the infection of prion-free cells with pure infectious amyloid forms of a specific prion protein; for example, Sup35, Ure2, Rnq1 or Mot3 (Alberti et al., 2009; Brachmann et al., 2005; King and Diaz-Avalos, 2004; Patel and Liebman, 2007; Shorter and Lindquist, 2006; Tanaka et al., 2004). Typically, a loss-of-function phenotype specific to the prion protein in question arises because steric effects of the amyloid form often preclude functionality (Baxa et al., 2002). Thus, prion forms of Sup35, a translation-termination factor, cause heritable reductions in the fidelity of translation termination (Tanaka et al., 2006). However, in other cases, a gain of function occurs (Derkatch et al., 2001; Derkatch et al., 2004; Si et al., 2003). Many other yeast prions have been defined by compelling genetic criteria, but have yet to be confirmed by infection with pure prion conformers (Alberti et al., 2009; Brown and Lindquist, 2009; Du et al., 2008; Patel et al., 2009). Lessons learned from yeast prion biology will undoubtedly shed light onto mammalian prion pathogenesis.
Typically, prionogenic proteins fold into several structurally distinct amyloid forms or ‘strains’ (Fig. 1). Each distinct strain confers a distinct phenotype (Brachmann et al., 2005; Colby et al., 2009; King and Diaz-Avalos, 2004; Legname et al., 2004; Prusiner, 1998; Safar et al., 1998; Tanaka et al., 2004; Tanaka et al., 2006). Beyond the cross-β form, which is common to each of the distinct strains, little is known about their underlying atomic structures. Pioneering structural work on short amyloidogenic peptides has provided an initial atomic glimpse of the numerous ways a single sequence can generate cross-β diversity (Sawaya et al., 2007; Wiltzius et al., 2009). However, how structural polymorphism can encode distinct phenotypes or disease states remains unclear. One principle is beginning to emerge from pure protein studies of both mammalian and yeast prions. Prions with an amyloid structure that is more readily fragmented (i.e. more frangible), but not so fragile that it is easily eradicated by the cellular machinery, tend to induce a more severe phenotype. Thus, more frangible Sup35 prions confer a stronger phenotype in yeast (Tanaka et al., 2006) and more labile PrP strains amplify and kill the host more rapidly (Colby et al., 2009; Legname et al., 2006). Increased frangibility leads to more fiber ends (the active sites of prion replication) per unit mass and, consequently, more rapid conversion of available monomers and contingent phenotypic change (Fig. 1B).
This ability to access self-templating amyloid forms is not, however, unique to prion proteins. Several fatal neurodegenerative diseases are also associated with the accumulation of self-templating amyloid forms of specific proteins. For example, β-amyloid (Aβ) and tau misfold in Alzheimer's disease (AD), α-synuclein misfolds in Parkinson's disease (PD), and huntingtin misfolds in Huntington's disease (HD). Beyond providing minor symptomatic relief, there are no effective treatments for these conditions. Curiously, neither these diseases nor the amyloid forms involved are generally considered infectious (Forman et al., 2004; Soto, 2003). In this Commentary, we highlight several advances that support the alarming possibility that these disorders and their associated amyloid forms are considerably more transmissible and prion-like than previously suspected. These findings have several important therapeutic implications and blur the distinctions between amyloid and prion, as well as between transmissibility and infectivity.
Alzheimer's disease and β-amyloid
AD is the most common fatal neurodegenerative disorder, afflicting ~35 million people worldwide (Prince et al., 2009). Extracellular neuritic plaques composed primarily of Aβ peptides are the defining pathological lesions of AD (Duyckaerts et al., 2009). Aβ is a cleavage peptide of the amyloid precursor protein (APP), which is rapidly cleared from brains of healthy individuals (Miners et al., 2008). Aβ varies in size from 39 to 43 amino acids, but the major species observed in AD neuritic plaques is the highly amyloidogenic Aβ42 (Glenner and Wong, 1984; Iwatsubo et al., 1994; Masters et al., 1985). Similar to prionogenic proteins, in different environments pure Aβ40 forms fibers with different molecular structures, which are maintained when the fibers are used to seed further assembly (Petkova et al., 2005). Aβ fibers are toxic to neurons in culture (Kayed et al., 2003) and different strains confer different toxicities (Petkova et al., 2005). It will be important to determine whether different strains are associated with differing severity of neurodegeneration in the brains of patients with AD. Notably, Aβ plaques can assemble extremely rapidly and be toxic to neighboring neurons (Meyer-Luehmann et al., 2008). Seeding Aβ40 assembly in vitro with extracts from the brains of patients with AD yields a strain that is distinct to those accessed by purely synthetic Aβ40 (Paravastu et al., 2009). However, the significance of this strain difference and how it contributes to disease remains unclear.
Similarly to PrP, amyloid forms of Aβ accumulate in the extracellular space, and thus the prospect of transmissible pathology through infectious material becomes a concern. Indeed, when brain extracts from patients with AD were directly introduced into the brains of transgenic mice producing human APP (APP23), the material induced plaque formation and extensive deposition of Aβ, whereas the introduction of brain extracts from age-matched patients without AD showed minimal Aβ accumulation (Meyer-Luehmann et al., 2006). Consistent with the property of permissive templating (self seeding), plaques did not form in mice that did not produce human Aβ (Meyer-Luehmann et al., 2006). Furthermore, transmissibility was eliminated when extracts were immunodepleted of Aβ, treated with formic acid (to dissolve fibers) or blocked with Aβ-specific antibodies. Moreover, mice that were passively immunized with an Aβ-specific antibody were resistant to infection. Thus, transmission from extract to host brain probably occurs through pure protein templating (Meyer-Luehmann et al., 2006). Unlike mammalian prions, systemic or peripheral inoculation of dilute Aβ-containing brain extract failed to induce any phenotype in the central nervous system (Eisele et al., 2009). Moreover, unlike mammalian PrP, it has not yet been possible to generate synthetic Aβ conformers that induce a disease phenotype (Meyer-Luehmann et al., 2006). Thus, definitive evidence of ‘protein-only’ transmission by Aβ is lacking.
Another remarkable convergence of Aβ and PrP in neurodegeneration has recently become clear. Before its assembly into fibers, Aβ accesses an oligomeric form that possesses a generic conformation that is distinct to fibers and is extremely cytotoxic (Kayed et al., 2003; Lesne et al., 2006). Indeed, Aβ oligomers might be more closely connected with neurodegeneration in AD than the fibers themselves (Lesne et al., 2006; Walsh et al., 2002). Remarkably, PrPC (the native plasma membrane form of PrP) can function as a cell-surface receptor for toxic Aβ oligomers, and this interaction seems to be crucial for Aβ oligomer-mediated disruption of synaptic function (Lauren et al., 2009). Deletion of PrP renders neurons resistant to the toxic effects of Aβ oligomers (Lauren et al., 2009). This finding is superficially reminiscent of the resistance of PrP-null neurons to mammalian prions (Bueler et al., 1993). However, it is unclear how the PrP-Aβ oligomer interaction leads to synaptic impairment (Balducci et al., 2010; Lauren et al., 2009). Given that mutations in PrP are also associated with AD (Riemenschneider et al., 2004), it is important to unravel the downstream sequelae of the PrP-Aβ interaction. This convergence of PrP and Aβ seems to be specific to the pathology of AD, because deletion of PrP does not ameliorate mouse models of PD, HD or tauopathy (Steele et al., 2009). Perhaps other relevant cell-surface receptors will be discovered for the proteins associated with these diseases.
Alzheimer's disease and tau
Aβ is not the only protein that accesses an amyloid form in AD. Tau is a microtubule-associated protein that normally binds to and stabilizes microtubules, thereby promoting their elongation and enabling intracellular transport (Skovronsky et al., 2006). However, in AD and other tauopathies, tau dissociates from microtubules and forms amyloid accumulations throughout the cell (Duyckaerts et al., 2009; Forman et al., 2004). During the course of AD, these pathological forms of tau spread in a stepwise, orderly manner throughout the brain (Braak and Braak, 1997), but the mode of transmission between cells remains unknown. Thus, it is unclear whether tau in adjacent cells is susceptible to an unknown stressor or whether a self-templating prion-like process underlies the propagation of the amyloid form. Consistent with a potential prion-like mode of transmission, extracellular fibers (known as ‘ghost tangles’) that consist of a fibrillogenic tau fragment encompassing the microtubule-binding region (MTBR) are observed in the brains of patients with AD (Endoh et al., 1993).
Cell culture experiments are beginning to explain how tau might spread between cells. When amyloid forms of recombinant MTBR were applied externally to cultured neural progenitor cells, they entered the cells and induce the formation of intracellular tau aggregates (Frost et al., 2009a). Endogenous full-length tau colocalized with the exogenous MTBR in intracellular, insoluble aggregates, indicating seeded assembly (Frost et al., 2009a). Furthermore, co-culture experiments revealed that intracellular tau aggregates can be transferred from cell to cell, although the mode of transmission remains uncertain (Frost et al., 2009a). Infectious forms of PrP transfer between cells via tunneling nanotubes (TNTs), which are exceedingly thin membrane protrusions of up to several micrometers long that can connect cells that are several cell diameters apart (Gousset et al., 2009). Cells can exchange components of the plasma membrane and cytoplasm (e.g. small vesicles and mitochondria) using this mechanism (Belting and Wittrup, 2008). It is important to investigate whether self-templating tau conformers also spread from cell to cell via this route.
A disease-associated tau missense mutant, P301S, promotes tau fibrillization in mouse models, whereas mice expressing wild-type human tau do not develop aggregates (Clavaguera et al., 2009). When insoluble tau isolated from mice expressing P301S tau was injected into brains of mice expressing wild-type human tau, filamentous tau pathology resulted and these filaments were composed of wild-type human tau (Clavaguera et al., 2009). The aggregates appear over time and spread to anatomically connected regions (Clavaguera et al., 2009). Despite the spread of wild-type tau fibers, there was no accompanying neurodegeneration. Thus, transmissible tau conformers might be distinct from neurotoxic tau conformers (Clavaguera et al., 2009). Alternatively, P301S fibers might seed a non-toxic conformational strain when wild-type tau is the available substrate. Short tau peptides can form polymorphic amyloid conformers (Wiltzius et al., 2009). Moreover, full-length tau seems to be able to access different strains (Crowther, 1991; Frost et al., 2009b), but these remain poorly characterized in terms of molecular structure and the phenotypes they confer. Infectivity from brain extract to host, and the gradual spreading of tau pathology through cell-to-cell transmission, are very reminiscent of TSEs and warrant further investigation. As with Aβ, however, it has not yet been possible to induce tau pathology or a transmissible neurodegenerative disease by injecting pure self-templating tau conformers. The progressive, stepwise spread of tau pathology, however, is also reminiscent of events that occur in PD.
Parkinson's disease and α-synuclein
PD is the most common neurodegenerative movement disorder, primarily affecting individuals over the age of 65, and is caused by a selective devastation of dopaminergic neurons in the substantia nigra (Skovronsky et al., 2006). Although PD is mostly sporadic, mutations in several genes have been linked with familial forms of the disease. These include point mutations and gene multiplications in the SNCA gene, which encodes α-synuclein, a small, unstructured presynaptic protein of uncertain function (Farrer et al., 2004; Kruger et al., 1998; Polymeropoulos et al., 1997; Singleton et al., 2003; Zarranz et al., 2004). Signature lesions of PD are cytoplasmic aggregates in dopaminergic neurons known as Lewy bodies (LBs) and Lewy neurites (LNs), which consist of amyloid forms of ubiquitylated and hyperphosphorylated α-synuclein (Braak and Del Tredici, 2008; Braak et al., 2003).
Curiously, α-synuclein pathology also hints at a prion-like mechanism of propagation through the brains of patients with PD. LB and LN pathology is observed in discrete areas of the brain at early stages of the disease, but exhibits an ascending and highly predictable pattern of progression through anatomically connected brain regions as the disease develops, suggestive of a transmissible agent (Braak and Del Tredici, 2008). In 2008, three groups examined fetal mesencephalic dopaminergic neuron grafts at autopsy from eight patients with PD (Kordower et al., 2008; Li et al., 2008; Mendez et al., 2008). Although the grafts seemed to remain functional and clinically efficacious to some extent, three patients developed α-synuclein-positive LB pathology after more than a decade, despite the young age of the grafted cells (fetal donor tissue) relative to the surrounding host tissue (Kordower et al., 2008; Li et al., 2008). The spread was gradual: grafts that were less than 10 years old exhibited no abnormal α-synuclein pathology, whereas after 10 years, increasing numbers of tyrosine hydroxylase-positive cells containing LB pathology were identified with increasing age (Li et al., 2008). Thus, the aged host brain can propagate LB pathology to young, genetically unrelated neurons, implying that α-synuclein aggregation and progression in the brain can be non-cell-autonomous (Brundin et al., 2008). One study reported reduced tyrosine hydroxylase levels in the grafted neurons, which is an indicator of dysfunction (Kordower et al., 2008). Thus, it seems likely that the grafted neurons were beginning to fail.
These observations raise the question of how amyloid forms of α-synuclein propagate among neurons. Degenerating neurons probably release large quantities of α-synuclein fibers upon cell death in a non-specific manner. Furthermore, α-synuclein monomers and aggregates are incorporated into vesicles and secreted from neurons via a non-classical mechanism (Lee et al., 2005). This expulsion might ensure that a single neuron does not become overwhelmed by an aggregation-prone protein, which might then be cleared by extracellular proteases or by lysosomal degradation following endocytosis by neighboring cells (Lee et al., 2008). Indeed, cultured stem and neuronal cells endocytose extracellular α-synuclein oligomers and fibers (Desplats et al., 2009; Lee et al., 2008). However, subsequent lysosomal degradation seems error-prone, because endocytosed α-synuclein can gain access to the cytoplasm and form ubiquitylated amyloid aggregates that are similar to LBs (Desplats et al., 2009). Moreover, when neural stem cells are grafted into the brains of transgenic mice overexpressing human α-synuclein, they develop LB-like aggregates, whereas in wild-type mice they do not (Desplats et al., 2009).
It is not known how α-synuclein escapes a membrane-bound vesicle to form cytoplasmic aggregates. Inhibition of lysosomal function stimulates deposition of transmitted α-synuclein in cell culture (Desplats et al., 2009). Curiously, mutations in a putative lysosomal transmembrane ATPase, ATP13A2 (also known as PARK9), are connected with familial parkinsonism (Ramirez et al., 2006), and overexpression of PARK9 counters α-synuclein toxicity in models ranging from yeast to rat midbrain dopaminergic neurons (Gitler et al., 2009). A natural decline in lysosomal function with aging might contribute to α-synuclein pathogenesis by facilitating intercellular transmission of α-synuclein and reduced clearance of α-synuclein in the lysosome by chaperone-mediated autophagy (Cuervo et al., 2004). Thus, restoring lysosomal function might be a promising therapeutic strategy for PD.
Another study, however, found that monomeric or fibrillar forms of α-synuclein failed to enter cells in culture unless they were transduced with cationic liposomes (Luk et al., 2009). Consistent with seeding activity, α-synuclein fibers, but not monomers, induced the formation of LB-like aggregates that were ubiquitylated, hyperphosphorylated and composed of amyloid (Luk et al., 2009). These aggregates perturbed Golgi integrity and tethered numerous small vesicles (Luk et al., 2009), consistent with the idea that misfolding of α-synuclein perturbs membrane-traffic events (Cooper et al., 2006; Gitler et al., 2008; Gitler and Shorter, 2007; Larsen et al., 2006; Soper et al., 2008). Despite these advances, there are no reports that intracranial injection of pure misfolded α-synuclein conformers induces a PD-like phenotype in mice. Nonetheless, these studies provide proof-of-principle that the seeding activity of α-synuclein fibers can induce LB formation (Luk et al., 2009).
Tau and α-synuclein cross seeding
Typically, amyloidogenesis is a specific process in which the amyloid form of a protein only converts other copies of the same protein and not proteins with a different primary sequence. This selectivity is known as self seeding or permissive templating, and arises because of the energy-minimized, in-register configurations of primary sequences within amyloid structures (Sawaya et al., 2007; Wiltzius et al., 2009). Thus, too many primary sequence mismatches will preclude access to the requisite set of steric configurations that achieves net stability (Sawaya et al., 2007; Wiltzius et al., 2009). Rarely, however, ‘cross seeding’ can occur, when an amyloid form of one protein catalyzes the assembly of another. Usually, cross seeding is not as effective as self seeding, but can have an important role in the initiation of fiber assembly. For example, prion forms of Rnq1 can initiate assembly of Sup35 prions in S. cerevisiae (Derkatch et al., 2004). Cross-seeding events might also have an important role in neurodegenerative disorders. For example, pure α-synuclein and tau synergize to promote the fibrillization of each other (Giasson et al., 2003a). When viewed by immunolabeling and electron microscopy, the resulting fibers remained separate, i.e. composed singly of tau or α-synuclein, except for a few instances in which spatially segregated portions of a fiber were labeled positive for either tau or α-synuclein (Giasson et al., 2003a). Indeed, this interplay is apparent in the brains of patients with PD, in which tau can colocalize with α-synuclein in portions of LBs (Kotzbauer et al., 2004). Moreover, even when controlling for age, the probability of developing concurrent AD and PD is greater than the product of the probabilities of developing each (Giasson et al., 2003b). Recent genome-wide association studies also indicate that variation in the tau gene (MAPT) can confer genetic risk for PD (Edwards et al., 2010; Satake et al., 2009; Simon-Sanchez et al., 2009). Thus, misfolding of one neurodegenerative-disease-associated protein might trigger the misfolding and transmissibility of a protein associated with another disease.
Huntington's disease and polyglutamine
HD is a progressive, autosomal dominant inherited neurodegenerative disorder, which is caused by the expansion of the CAG trinucleotide repeat (>36) in the huntingtin gene, leading to an expanded polyglutamine (polyQ) tract that confers increased amyloidogenicity (MacDonald et al., 1993; Scherzinger et al., 1997). HD is mainly characterized by the selective loss of striatal neurons in the basal ganglia, although pathology can spread to other areas of the brain (Walker, 2007). Intriguingly, pure polyQ can spontaneously access distinct fiber strains that confer distinct toxicities (Nekooki-Machida et al., 2009). In a mouse model of HD, brain regions with the most severe neurodegeneration contained predominantly toxic strains, whereas more benign strains were found in less-affected regions (Nekooki-Machida et al., 2009). Similarly to PD, fetal grafts of striatal tissue have also been tested in the clinic as a potential treatment for HD, and have provided some marginal and transient therapeutic benefits (Cicchetti et al., 2009). However, autopsy results revealed that these grafts were susceptible to disease-like neurodegeneration (Cicchetti et al., 2009). Although grafted cells displayed increased caspase-3 activation, vacuolization and decreased structural integrity, abnormal huntingtin aggregation was not observed (Cicchetti et al., 2009). Thus, polyQ fibers do not seem to invade these neuronal grafts. By contrast, polyQ fibers seem able to pierce the cell membrane of cells in culture, gain access to the cytoplasm and seed the amyloidogenesis of soluble polyQ (Ren et al., 2009). These assemblies persist despite their dilution when cells divide, suggesting a self-sustaining seeding and fragmentation process (Ren et al., 2009). As with Aβ, tau and α-synuclein, but in contrast to PrP, it has not yet been possible to induce a transmissible neurodegenerative disease by intracranial injection of pure polyQ conformers.
Prions or prionoids?
Several reasons for this general failure to generate infectious forms of Aβ, tau, polyQ and α-synuclein from pure protein are conceivable. For example, perhaps the correct strain with the optimal fragility has not yet been generated in vitro (Fig. 1B), such that there are too few fiber ends to drive infectivity in experimental systems. Auxiliary factors might be more crucial for these amyloid forms to survive in the extracellular space or otherwise transfer from cell to cell and gain access to native substrate. PrP is a GPI-anchored plasma membrane protein, which probably increases its accessibility to infectious conformers (Chesebro et al., 2005; Kanu et al., 2002; Speare et al., 2009). Until protein-only induction of infectious disease is achieved, however, it seems difficult to refer to the amyloid conformers involved (Aβ, tau, α-synuclein, polyQ) as prions. By contrast, it has been possible, albeit difficult, to generate synthetic PrP conformers that induce transmissible disease (Colby et al., 2009; Legname et al., 2004; Wang et al., 2010). Similarly, yeast prion conformers are readily generated from pure protein (Alberti et al., 2009; Brachmann et al., 2005; King and Diaz-Avalos, 2004; Patel and Liebman, 2007; Shorter and Lindquist, 2006; Tanaka et al., 2004). Thus, the amyloid forms involved in AD, PD and HD are perhaps only ‘prion-like’ because they have never been shown to move naturally from one individual to another and have never been connected with infectious disease epidemics. To make this important distinction clearer, the term ‘prionoid’ has been introduced to distinguish experimentally transmissible amyloid templates from bona fide prions (Aguzzi, 2009; Aguzzi and Rajendran, 2009).
Amyloid A amyloidosis
There is one amyloid disorder, however, that might be even more similar to prion diseases than those discussed thus far. The systemic amyloidosis, amyloid A (AA) amyloidosis, occurs in response to chronic inflammation when AA, a ~76 amino acid N-terminal fragment of serum amyloid A protein, forms amyloid conformers that are deposited in various tissues including the spleen, liver and kidneys (Westermark and Westermark, 2009). Parenteral or oral administration of AA fibers purified from the liver of mice with AA amyloidosis is sufficient to accelerate AA amyloidosis in response to inflammatory stress (Lundmark et al., 2002). AA fibers isolated from commercially available foie gras also accelerate AA amyloidosis in mice (Solomon et al., 2007). However, amyloid forms of various proteins, including Sup35, amylin and transthyretin have the same effect, which suggests that AA amyloidogenesis is readily elicited by cross-seeding events (Johan et al., 1998; Lundmark et al., 2005). Curiously, AA amyloidosis is common in captive cheetahs and might move naturally between individuals by a fecal-oral route (Zhang et al., 2008). Moreover, AA amyloidosis can also be transmitted between individuals by peripheral blood monocytes, highlighting a potential risk for human-to-human transmission through blood transfusions (Sponarova et al., 2008). These potentially natural routes of infection by AA fibers are reminiscent of prions.
Amyotrophic lateral sclerosis: are TDP-43 and FUS similar to yeast prions?
A unifying feature of the major yeast prions defined so far is the presence of a prion domain that is enriched in uncharged polar amino acids (particularly asparagine, glutamine and tyrosine) and glycine. This domain can switch between an intrinsically unfolded conformation (non-prion form) and an infectious cross-β conformation (prion form) (Alberti et al., 2009; Shorter and Lindquist, 2005). A Hidden Markov Model has been developed to identify these prion domains, and a proteome-wide survey of S. cerevisiae yielded 19 novel yeast prions that have been confirmed experimentally (Alberti et al., 2009). However, PrP and HET-s, a prion protein from Podospora anserina (Saupe, 2007), do not contain this type of domain, raising the possibility that other primary sequences can access prion states. Nonetheless, this unique tool presents an opportunity to identify potential metazoan prions. Indeed, specific CPEB proteins possess this type of domain, which is crucial for their function in long-term memory formation in Aplysia and Drosophila and might involve prionogenesis (Keleman et al., 2007; Si et al., 2010; Si et al., 2003).
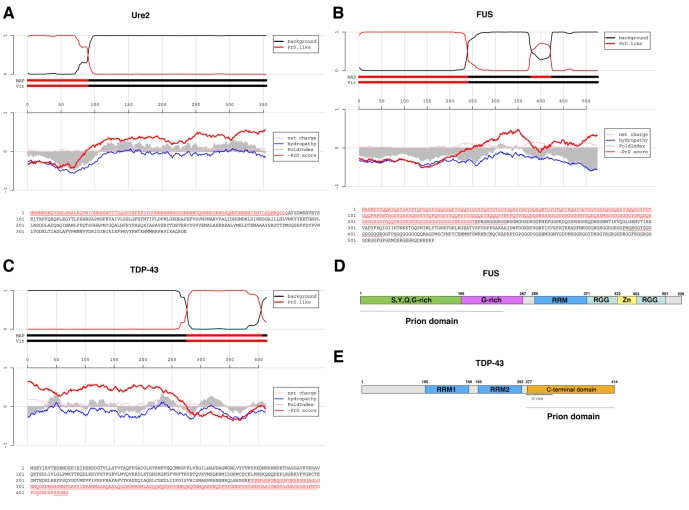
Intriguingly, when the yeast prion-domain algorithm (Alberti et al., 2009) is used to scan the human genome, two nuclear RNA-binding proteins involved in transcriptional regulation and RNA splicing, FUS and TDP-43, rank very highly – 15th and 69th, respectively (out of 27,879 proteins) (Fig. 2A-C). By comparison, in the yeast genome, two known yeast prion proteins, Sup35 and Ure2 (Fig. 2A), rank 15th and 65th, respectively (out of 5808 proteins). The putative prion domain in FUS also satisfies the criteria for predicted prion formation based on ‘prion propensities’ (derived from random mutagenesis experiments in yeast) (Toombs et al., 2010); namely, they contain a region with predicted disorder and with ‘prion propensity’ >0.05. The putative prion domain of TDP-43 is close to this cut-off, with a ‘prion propensity’ of 0.04.
Fig. 2.
Putative prion domains in FUS and TDP-43. (A) Prion domain prediction for Ure2, a known yeast prion protein. The lower part of the panel shows the primary sequence of Ure2, with the predicted prion domain highlighted in red. In accord with experimental data (Masison and Wickner, 1995), the algorithm successfully identifies amino acids 1-89 as the prion domain. The top panel shows the probability of each residue belonging to the Hidden Markov Model state prion domain or ‘background’; the tracks ‘MAP’ and ‘Vit’ illustrate the Maximum a Posteriori and the Viterbi parses of the protein into these two states (for details, see Alberti et al., 2009; this article contains similar plots for 179 yeast proteins in the supplement). The middle panel shows sliding averages over a window of width 51 of net charge (pink), hydropathy (blue) and predicted disorder (gray) (Prilusky et al., 2005), along with a sliding average based on the prion domain amino acid propensities (red). (B) Prion domain prediction for FUS. (C) Prion domain prediction for TDP-43. (D) Domain architecture of FUS. FUS harbors an N-terminal S,Y,Q,G-rich domain (green), followed by a G-rich domain (purple), an RNA-recognition motif (RRM; blue) and two RGG-rich domains (cyan) that surround a zinc-finger domain (yellow). The predicted prion domain encompasses the S,Y,Q,G-rich domain and a portion of the G-rich domain. (E) Domain architecture of TDP-43. TDP-43 harbors two RNA-recognition motifs, RRM1 and 2 (blue), and a C-terminal domain that has a G-rich N-terminal portion. The predicted prion domain spans this C-terminal domain.
Both FUS and TDP-43 are genetically linked with amyotrophic lateral sclerosis (ALS) (Kabashi et al., 2008; Kwiatkowski et al., 2009; Pesiridis et al., 2009; Sreedharan et al., 2008; Van Deerlin et al., 2008; Vance et al., 2009). ALS is a common neurodegenerative motor neuron disease characterized by an unrelenting devastation of both upper and lower motor neurons, leading to progressive weakness, muscle wasting and spasticity, which culminate in paralysis and death within 3-5 years. ALS is characterized by ubiquitylated, cytoplasmic aggregates, that are non-amyloid in nature, in afflicted neurons. In most ALS cases, the major aggregated protein is TDP-43, which is simultaneously depleted from the nucleus (Neumann et al., 2006). Indeed, widespread TDP-43 neuropathology is observed in ALS patients beyond stereotyped upper and lower motor neurons (Geser et al., 2008). In ALS cases connected with FUS mutations, however, TDP-43 does not appear to be affected. Rather, in the motor neurons of these patients, FUS itself is depleted from the nucleus and aggregated in the cytoplasm (Kwiatkowski et al., 2009; Vance et al., 2009). Thus, neuronal defects that give rise to ALS are probably caused by the loss of function of these RNA-binding proteins, as well as the potential toxic properties gained by the aggregated forms (Pesiridis et al., 2009).
The putative prion domain of FUS comprises amino acids ~1-239 (Fig. 2B,D). Several mutations connected with ALS lie within this domain, although the majority of mutations are found at the extreme C-terminal region of FUS (Fig. 2D) (Kwiatkowski et al., 2009; Vance et al., 2009). It is not yet clear which domain of FUS drives its aggregation.
By contrast, the putative prion domain of TDP-43 comprises amino acids ~277-414 (Fig. 2C,E) and has a key role in TDP-43 pathogenesis. Aggregated C-terminal fragments of TDP-43 containing this region are biochemical signatures of ALS (Neumann et al., 2006). This domain is the minimal region that can induce TDP-43 aggregation in yeast and neuroblastoma culture models of TDP-43 proteinopathies (Johnson et al., 2008; Nonaka et al., 2009; Zhang et al., 2009). Curiously, however, the prion domain of TDP-43 plus portions of the neighboring RNA-recognition motif (Fig. 2E) are required for toxicity in both yeast and neuroblastoma cell lines (Johnson et al., 2008; Zhang et al., 2009). In isolation, pure TDP-43 is intrinsically aggregation-prone and rapidly assembles into aggregated species that are morphologically very similar to the aggregates that accumulate in the degenerating neurons of ALS patients (Johnson et al., 2009; Lin and Dickson, 2008; Mori et al., 2008; Nishihira et al., 2008). Here, too, the C-terminal prion domain is crucial: deletion of this domain precludes aggregation (Johnson et al., 2009). Furthermore, all but one of over 30 missense mutations connected with ALS fall in the prion domain of TDP-43 (Pesiridis et al., 2009). The ALS-linked TDP-43 mutants Q331K and M337V accelerate the aggregation of pure TDP-43 and promote aggregation and toxicity in yeast (Johnson et al., 2009). Several ALS-linked mutations in this region also promote aggregation in neuroblastoma cell lines (Nonaka et al., 2009), can be toxic to neurons in culture (Barmada et al., 2010) and confer neurodegeneration in Drosophila (Li et al., 2010) and mice (Wegorzewska et al., 2009). Collectively, these data suggest that the prion domain of TDP-43 directs the protein into toxic misfolding trajectories that cause ALS.
Despite the presence of this domain, and in contrast to yeast prions, TDP-43 does not access amyloid forms in isolation or in ALS (Johnson et al., 2009; Kerman et al., 2010; Kwong et al., 2008). Rather, the aggregates formed by this protein are non-amyloid in nature. However, the presence of this type of domain also predisposes proteins to access pre-amyloid, oligomeric forms (Shorter and Lindquist, 2004; Shorter and Lindquist, 2006; Vitrenko et al., 2007), which can be extremely toxic (Kayed et al., 2003). Indeed, TDP-43 rapidly forms pore-shaped oligomers (Johnson et al., 2009), which are strikingly reminiscent of toxic oligomers formed by Aβ42 and α-synuclein (Lashuel et al., 2002). These TDP-43 oligomers tend to clump together to form large masses that might be highly toxic to motor neurons. TDP-43 is likely to become trapped in this toxic pre-amyloid state and is unable to access the amyloid form. Surprisingly, TDP-43 is also found to be aggregated in numerous other neurodegenerative disorders, including AD and HD. Whether this reflects perturbed proteostasis leading indirectly to the misfolding of the intrinsically aggregation-prone TDP-43 (Gidalevitz et al., 2006; Gidalevitz et al., 2009) or to a more direct cross-seeding event remains unclear.
Implications for neuronal-graft and stem-cell-based therapies
Whether the amyloid templates involved in AD, PD and HD are prions or not, their transmissibility within the brain of an individual has important repercussions for potential therapies. Patient-derived, induced pluripotent stem cells (iPSCs) and the neural progenitor cells that can be derived from them hold great therapeutic potential because they will not elicit a host immunological response following transplantation (Kiskinis and Eggan, 2010; Yamanaka, 2007). Such cells have been generated from fibroblasts of patients with PD, and can be differentiated into dopaminergic neurons (Soldner et al., 2009). Moreover, iPSC-derived dopaminergic neuronal grafts relieved parkinsonian-like symptoms in rats that had previously been treated with 6-hydroxydopamine (6-OH-DA), which selectively kills dopaminergic neurons (Wernig et al., 2008). Yet it is difficult to assess the utility of this stem-cell-based therapy for all types of PD because 6-OH-DA treatment does not generate amyloid forms of α-synuclein. Indeed, such therapies face the same challenge as any other neuronal graft: transmissible conformers will inexorably penetrate these cells and elicit the chain reaction of protein misfolding events that culminate in neurodegeneration. Hence, these approaches might only offer short-term improvements, rather than a long-term solution.
Therefore, steps must be taken to improve the quality of transplanted neurons such that they resist phenotypic conversion by self-templating species. One approach would be to tailor the cells to be grafted for the disease in question. Here, inspiration can be drawn from early experiments with PrP. Neurons lacking PrP are resistant to infection by mammalian prions (Brandner et al., 1996). Thus, deletion of PrP or depletion of PrP by small-interfering RNAs in grafted neurons might be advantageous in CJD and related disorders. Furthermore, because PrP can act as a receptor for toxic Aβ oligomers (Lauren et al., 2009), such neurons might also be useful for the treatment of AD because, in principle, these grafted neurons would resist Aβ oligomer invasion and toxicity. By extension, depletion of tau or α-synuclein from grafted neurons might be beneficial in tauopathies and PD, respectively. By removing the substrate of the transmissible conformer, it might be possible to preclude phenotypic conversion. Indeed, depletion of PrP, tau or huntingtin, even after the onset of disease symptoms and pathology, has proven to be an effective treatment in mouse models of prion disease, AD and HD (DiFiglia et al., 2007; Mallucci et al., 2003; Mallucci et al., 2007; Roberson et al., 2007; Yamamoto et al., 2000).
Of course, it might simply not be possible to deplete a protein from the graft if it is essential for the functionality of the grafted neurons, which might be a concern with tau. Alternatively, one might envision arming the cells to be grafted with a reinforced arsenal of defenses against transmissible conformers. Thus, for the treatment of PD, grafts might be generated that express (perhaps inducibly) high levels of Rab1 or PARK9, which protect against α-synuclein toxicity (Cooper et al., 2006; Gitler et al., 2009). It might also be useful to express (or inducibly express) high levels of molecular chaperones, such as Hsp70, that antagonize the toxicity of α-synuclein and polyQ aggregates (Auluck et al., 2002; Warrick et al., 1999), or even protein disaggregases, such as Hsp104, which dissolve transmissible amyloid conformers as well as toxic oligomers, and protect dopaminergic neurons from α-synuclein toxicity (Lo Bianco et al., 2008; Shorter, 2008; Vashist et al., 2010). Again, it will be essential to determine that these alterations do not in themselves alter graft functionality and to explore these options in rodent models of PD.
Other methods of directly antagonizing transmissible conformers might also be used to protect grafted neurons, including small-molecule therapies. Although several small-molecule antagonists of amyloidogenesis have been described (Crowe et al., 2009; Gestwicki et al., 2004; Wang et al., 2008), there are many issues surrounding delivery of the small molecules to the appropriate site, including their passage across the blood-brain barrier (Brunden et al., 2009). Furthermore, the conformational diversity of amyloid strains severely complicates the development of potential small-molecule therapies. For example, prion strains can vary in susceptibility to small-molecule antagonists and drug-resistant prion strains can arise (Ghaemmaghami et al., 2009; Li et al., 2009; Roberts et al., 2009). However, synergistic small-molecule combinations can be isolated that antagonize several prion strains (Roberts et al., 2009). Finally, detailed knowledge of the mechanisms of non-cell-autonomous propagation of transmissible conformers is urgently needed (Aguzzi and Rajendran, 2009). Small molecules or protein therapeutics that can target and inhibit the intercellular transfer process might slow or halt clinical symptoms in mild-to-moderate cases because more neurons and glia are spared, or extend the time of disease onset in susceptible patients. Such approaches coupled with optimized neuronal graft or stem cell technologies offer great hope in defeating these currently untreatable and increasingly prevalent disorders.
Acknowledgments
This work was supported by an NIH/NIA training grant (AG00255) (M.C.); an NIH Director's New Innovator Award (1DP2OD004417-01), an NINDS grant (1R01NS065317-01) (A.D.G.); A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts; an NIH Director's New Innovator Award (1DP2OD002177-01), an Ellison Medical Foundation New Scholar in Aging Award, and an NINDS grant (1R21NS067354-0110) (J.S.). Grants for PMC depositing: 1DP2OD004417, 1R01NS065317, AG00255, 1DP2OD002177, 1R21NS067354. Deposited in PMC for release after 12 months.
References
- Aguzzi A. (2009). Cell biology: beyond the prion principle. Nature 459, 924-925 [DOI] [PubMed] [Google Scholar]
- Aguzzi A., Rajendran L. (2009). The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64, 783-790 [DOI] [PubMed] [Google Scholar]
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper T., Cramp W. A., Haig D. A., Clarke M. C. (1967). Does the agent of scrapie replicate without nucleic acid? Nature 214, 764-766 [DOI] [PubMed] [Google Scholar]
- Auluck P. K., Chan H. Y., Trojanowski J. Q., Lee V. M., Bonini N. M. (2002). Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295, 865-868 [DOI] [PubMed] [Google Scholar]
- Balducci C., Beeg M., Stravalaci M., Bastone A., Sclip A., Biasini E., Tapella L., Colombo L., Manzoni C., Borsello T., et al. (2010). Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc. Natl. Acad. Sci. USA 107, 2295-2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada S. J., Skibinski G., Korb E., Rao E. J., Wu J. Y., Finkbeiner S. (2010). Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 30, 639-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U., Speransky V., Steven A. C., Wickner R. B. (2002). Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc. Natl. Acad. Sci. USA 99, 5253-5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekes M., McBride P. A. (2007). The spread of prions through the body in naturally acquired transmissible spongiform encephalopathies. FEBS J. 274, 588-605 [DOI] [PubMed] [Google Scholar]
- Belting M., Wittrup A. (2008). Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J. Cell Biol. 183, 1187-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton D. C., McKinley M. P., Prusiner S. B. (1982). Identification of a protein that purifies with the scrapie prion. Science 218, 1309-1311 [DOI] [PubMed] [Google Scholar]
- Braak H., Braak E. (1997). Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiol. Aging 18, 351-357 [DOI] [PubMed] [Google Scholar]
- Braak H., Del Tredici K. (2008). Nervous system pathology in sporadic Parkinson disease. Neurology 70, 1916-1925 [DOI] [PubMed] [Google Scholar]
- Braak H., Del Tredici K., Rub U., de Vos R. A., Jansen Steur E. N., Braak E. (2003). Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging 24, 197-211 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B. (2005). Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. (1996). Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature 379, 339-343 [DOI] [PubMed] [Google Scholar]
- Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A. D., Toyka K. V., Nave K. A., Weis J., et al. (2010). Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 13, 310-318 [DOI] [PubMed] [Google Scholar]
- Brown J. C., Lindquist S. (2009). A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 23, 2320-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce M. E., Dickinson A. G. (1987). Biological evidence that scrapie agent has an independent genome. J. Gen. Virol. 68, 79-89 [DOI] [PubMed] [Google Scholar]
- Brunden K. R., Trojanowski J. Q., Lee V. M. (2009). Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat. Rev. Drug Discov. 8, 783-793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundin P., Li J. Y., Holton J. L., Lindvall O., Revesz T. (2008). Research in motion: the enigma of Parkinson's disease pathology spread. Nat. Rev. Neurosci. 9, 741-745 [DOI] [PubMed] [Google Scholar]
- Bueler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., Weissmann C. (1993). Mice devoid of PrP are resistant to scrapie. Cell 73, 1339-1347 [DOI] [PubMed] [Google Scholar]
- Castilla J., Saa P., Hetz C., Soto C. (2005). In vitro generation of infectious scrapie prions. Cell 121, 195-206 [DOI] [PubMed] [Google Scholar]
- Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., et al. (2005). Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435-1439 [DOI] [PubMed] [Google Scholar]
- Cicchetti F., Saporta S., Hauser R. A., Parent M., Saint-Pierre M., Sanberg P. R., Li X. J., Parker J. R., Chu Y., Mufson E. J., et al. (2009). Neural transplants in patients with Huntington's disease undergo disease-like neuronal degeneration. Proc. Natl. Acad. Sci. USA 106, 12483-12488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F., Bolmont T., Crowther R. A., Abramowski D., Frank S., Probst A., Fraser G., Stalder A. K., Beibel M., Staufenbiel M., et al. (2009). Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby D. W., Giles K., Legname G., Wille H., Baskakov I. V., Dearmond S. J., Prusiner S. B. (2009). Design and construction of diverse mammalian prion strains. Proc. Natl. Acad. Sci. USA 106, 20417-20422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F., et al. (2006). Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313, 324-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe A., Huang W., Ballatore C., Johnson R. L., Hogan A. M., Huang R., Wichterman J., McCoy J., Huryn D., Auld D. S., et al. (2009). Identification of aminothienopyridazine inhibitors of tau assembly by quantitative high-throughput screening. Biochemistry 48, 7732-7745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R. A. (1991). Straight and paired helical filaments in Alzheimer disease have a common structural unit. Proc. Natl. Acad. Sci. USA 88, 2288-2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004). Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292-1295 [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. (2001). Prions affect the appearance of other prions: the story of [PIN+]. Cell 106, 171-182 [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Uptain S. M., Outeiro T. F., Krishnan R., Lindquist S. L., Liebman S. W. (2004). Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 101, 12934-12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P., Lee H. J., Bae E. J., Patrick C., Rockenstein E., Crews L., Spencer B., Masliah E., Lee S. J. (2009). Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc. Natl. Acad. Sci. USA 106, 13010-13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R. K., Rajeev K. G., et al. (2007). Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA 104, 17204-17209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. M. (2003). Protein folding and misfolding. Nature 426, 884-890 [DOI] [PubMed] [Google Scholar]
- Du Z., Park K. W., Yu H., Fan Q., Li L. (2008). Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40, 460-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyckaerts C., Delatour B., Potier M. C. (2009). Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 118, 5-36 [DOI] [PubMed] [Google Scholar]
- Edwards T. L., Scott W. K., Almonte C., Burt A., Powell E. H., Beecham G. W., Wang L., Zuchner S., Konidari I., Wang G., et al. (2010). Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. PMID: 20070850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele Y. S., Bolmont T., Heikenwalder M., Langer F., Jacobson L. H., Yan Z. X., Roth K., Aguzzi A., Staufenbiel M., Walker L. C., et al. (2009). Induction of cerebral beta-amyloidosis: intracerebral versus systemic Abeta inoculation. Proc. Natl. Acad. Sci. USA 106, 12926-12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh R., Ogawara M., Iwatsubo T., Nakano I., Mori H. (1993). Lack of the carboxyl terminal sequence of tau in ghost tangles of Alzheimer's disease. Brain Res. 601, 164-172 [DOI] [PubMed] [Google Scholar]
- Farrer M., Kachergus J., Forno L., Lincoln S., Wang D. S., Hulihan M., Maraganore D., Gwinn-Hardy K., Wszolek Z., Dickson D., et al. (2004). Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol. 55, 174-179 [DOI] [PubMed] [Google Scholar]
- Forman M. S., Trojanowski J. Q., Lee V. M. (2004). Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10, 1055-1063 [DOI] [PubMed] [Google Scholar]
- Frost B., Jacks R. L., Diamond M. I. (2009a). Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845-12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Ollesch J., Wille H., Diamond M. I. (2009b). Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J. Biol. Chem. 284, 3546-3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F., Brandmeir N. J., Kwong L. K., Martinez-Lage M., Elman L., McCluskey L., Xie S. X., Lee V. M., Trojanowski J. Q. (2008). Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch. Neurol. 65, 636-641 [DOI] [PubMed] [Google Scholar]
- Gestwicki J. E., Crabtree G. R., Graef I. A. (2004). Harnessing chaperones to generate small-molecule inhibitors of amyloid beta aggregation. Science 306, 865-869 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Ahn M., Lessard P., Giles K., Legname G., DeArmond S. J., Prusiner S. B. (2009). Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 5, e1000673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson B. I., Forman M. S., Higuchi M., Golbe L. I., Graves C. L., Kotzbauer P. T., Trojanowski J. Q., Lee V. M. (2003a). Initiation and synergistic fibrillization of tau and alpha-synuclein. Science 300, 636-640 [DOI] [PubMed] [Google Scholar]
- Giasson B. I., Lee V. M., Trojanowski J. Q. (2003b). Interactions of amyloidogenic proteins. Neuromolecular Med. 4, 49-58 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. (2006). Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471-1474 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Krupinski T., Garcia S., Morimoto R. I. (2009). Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 5, e1000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A. D., Shorter J. (2007). Prime time for alpha-synuclein. J. Neurosci. 27, 2433-2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J. C., McCaffery J. M., et al. (2008). The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. USA 105, 145-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C., et al. (2009). Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 41, 308-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner G. G., Wong C. W. (1984). Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885-890 [DOI] [PubMed] [Google Scholar]
- Goldgaber D., Goldfarb L. G., Brown P., Asher D. M., Brown W. T., Lin S., Teener J. W., Feinstone S. M., Rubenstein R., Kascsak R. J., et al. (1989). Mutations in familial Creutzfeldt-Jakob disease and Gerstmann-Straussler-Scheinker's syndrome. Exp. Neurol. 106, 204-206 [DOI] [PubMed] [Google Scholar]
- Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D. T., Chenouard N., de Chaumont F., Martino A., Enninga J., et al. (2009). Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 11, 328-336 [DOI] [PubMed] [Google Scholar]
- Hsiao K., Baker H. F., Crow T. J., Poulter M., Owen F., Terwilliger J. D., Westaway D., Ott J., Prusiner S. B. (1989). Linkage of a prion protein missense variant to Gerstmann-Straussler syndrome. Nature 338, 342-345 [DOI] [PubMed] [Google Scholar]
- Iwatsubo T., Odaka A., Suzuki N., Mizusawa H., Nukina N., Ihara Y. (1994). Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43). Neuron 13, 45-53 [DOI] [PubMed] [Google Scholar]
- Jackson W. S., Borkowski A. W., Faas H., Steele A. D., King O. D., Watson N., Jasanoff A., Lindquist S. (2009). Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron 63, 438-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johan K., Westermark G., Engstrom U., Gustavsson A., Hultman P., Westermark P. (1998). Acceleration of amyloid protein A amyloidosis by amyloid-like synthetic fibrils. Proc. Natl. Acad. Sci. USA 95, 2558-2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. S., McCaffery J. M., Lindquist S., Gitler A. D. (2008). A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 105, 6439-6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009). TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329-20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., et al. (2008). TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572-574 [DOI] [PubMed] [Google Scholar]
- Kanu N., Imokawa Y., Drechsel D. N., Williamson R. A., Birkett C. R., Bostock C. J., Brockes J. P. (2002). Transfer of scrapie prion infectivity by cell contact in culture. Curr. Biol. 12, 523-530 [DOI] [PubMed] [Google Scholar]
- Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003). Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486-489 [DOI] [PubMed] [Google Scholar]
- Keleman K., Kruttner S., Alenius M., Dickson B. J. (2007). Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 10, 1587-1593 [DOI] [PubMed] [Google Scholar]
- Kerman A., Liu H. N., Croul S., Bilbao J., Rogaeva E., Zinman L., Robertson J., Chakrabartty A. (2010). Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta Neuropathol. PMID: 20111867 [DOI] [PubMed] [Google Scholar]
- King C. Y., Diaz-Avalos R. (2004). Protein-only transmission of three yeast prion strains. Nature 428, 319-323 [DOI] [PubMed] [Google Scholar]
- Kiskinis E., Eggan K. (2010). Progress toward the clinical application of patient-specific pluripotent stem cells. J. Clin. Invest. 120, 51-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. P., Fitzpatrick A. W., Meehan S., Mott H. R., Vendruscolo M., Dobson C. M., Welland M. E. (2007). Role of intermolecular forces in defining material properties of protein nanofibrils. Science 318, 1900-1903 [DOI] [PubMed] [Google Scholar]
- Kordower J. H., Chu Y., Hauser R. A., Freeman T. B., Olanow C. W. (2008). Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson's disease. Nat. Med. 14, 504-506 [DOI] [PubMed] [Google Scholar]
- Kotzbauer P. T., Giasson B. I., Kravitz A. V., Golbe L. I., Mark M. H., Trojanowski J. Q., Lee V. M. (2004). Fibrillization of alpha-synuclein and tau in familial Parkinson's disease caused by the A53T alpha-synuclein mutation. Exp. Neurol. 187, 279-288 [DOI] [PubMed] [Google Scholar]
- Kruger R., Kuhn W., Muller T., Woitalla D., Graeber M., Kosel S., Przuntek H., Epplen J. T., Schols L., Riess O. (1998). Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat. Genet. 18, 106-108 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T. J., Jr, Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., et al. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205-1208 [DOI] [PubMed] [Google Scholar]
- Kwong L. K., Uryu K., Trojanowski J. Q., Lee V. M. (2008). TDP-43 proteinopathies: neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals 16, 41-51 [DOI] [PubMed] [Google Scholar]
- Larsen K. E., Schmitz Y., Troyer M. D., Mosharov E., Dietrich P., Quazi A. Z., Savalle M., Nemani V., Chaudhry F. A., Edwards R. H., et al. (2006). Alpha-synuclein overexpression in PC12 and chromaffin cells impairs catecholamine release by interfering with a late step in exocytosis. J. Neurosci. 26, 11915-11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel H. A., Hartley D., Petre B. M., Walz T., Lansbury P. T., Jr (2002). Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291 [DOI] [PubMed] [Google Scholar]
- Lauren J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009). Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature 457, 1128-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pichon C. E., Valley M. T., Polymenidou M., Chesler A. T., Sagdullaev B. T., Aguzzi A., Firestein S. (2009). Olfactory behavior and physiology are disrupted in prion protein knockout mice. Nat. Neurosci. 12, 60-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Patel S., Lee S. J. (2005). Intravesicular localization and exocytosis of alpha-synuclein and its aggregates. J. Neurosci. 25, 6016-6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Suk J. E., Bae E. J., Lee J. H., Paik S. R., Lee S. J. (2008). Assembly-dependent endocytosis and clearance of extracellular alpha-synuclein. Int. J. Biochem. Cell Biol. 40, 1835-1849 [DOI] [PubMed] [Google Scholar]
- Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2004). Synthetic mammalian prions. Science 305, 673-676 [DOI] [PubMed] [Google Scholar]
- Legname G., Nguyen H. O., Peretz D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2006). Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA 103, 19105-19110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesne S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006). A specific amyloid-beta protein assembly in the brain impairs memory. Nature 440, 352-357 [DOI] [PubMed] [Google Scholar]
- Li J. Y., Englund E., Holton J. L., Soulet D., Hagell P., Lees A. J., Lashley T., Quinn N. P., Rehncrona S., Bjorklund A., et al. (2008). Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat. Med. 14, 501-503 [DOI] [PubMed] [Google Scholar]
- Li J., Browning S., Mahal S. P., Oelschlegel A. M., Weissmann C. (2009). Darwinian evolution of prions in cell culture. Science 327, 8869-8872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ray P., Rao E. J., Shi C., Guo W., Chen X., Woodruff E. A., 3rd, Fushimi K., Wu J. Y. (2010). A Drosophila model for TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA 107, 3169-3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. L., Dickson D. W. (2008). Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol. 116, 205-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C., Shorter J., Regulier E., Lashuel H., Iwatsubo T., Lindquist S., Aebischer P. (2008). Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J. Clin. Invest. 118, 3087-3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk K. C., Song C., O'Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M. (2009). Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. USA 106, 20051-20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark K., Westermark G. T., Nystrom S., Murphy C. L., Solomon A., Westermark P. (2002). Transmissibility of systemic amyloidosis by a prion-like mechanism. Proc. Natl. Acad. Sci. USA 99, 6979-6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark K., Westermark G. T., Olsen A., Westermark P. (2005). Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: Cross-seeding as a disease mechanism. Proc. Natl. Acad. Sci. USA 102, 6098-6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M. E., Ambrose C. M., Duyao M. P., Myers R. H., Lin C., Srinidhi L., Barnes G., Taylor S. A., James M., Groot N., et al. (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971-983 [DOI] [PubMed] [Google Scholar]
- Mallucci G., Dickinson A., Linehan J., Klohn P. C., Brandner S., Collinge J. (2003). Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302, 871-874 [DOI] [PubMed] [Google Scholar]
- Mallucci G. R., White M. D., Farmer M., Dickinson A., Khatun H., Powell A. D., Brandner S., Jefferys J. G., Collinge J. (2007). Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron 53, 325-335 [DOI] [PubMed] [Google Scholar]
- Masison D. C., Wickner R. B. (1995). Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93-95 [DOI] [PubMed] [Google Scholar]
- Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. (1985). Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 82, 4245-4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiason C. K., Powers J. G., Dahmes S. J., Osborn D. A., Miller K. V., Warren R. J., Mason G. L., Hays S. A., Hayes-Klug J., Seelig D. M., et al. (2006). Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 314, 133-136 [DOI] [PubMed] [Google Scholar]
- Mead S., Whitfield J., Poulter M., Shah P., Uphill J., Campbell T., Al-Dujaily H., Hummerich H., Beck J., Mein C. A., et al. (2009). A novel protective prion protein variant that colocalizes with kuru exposure. N. Engl. J. Med. 361, 2056-2065 [DOI] [PubMed] [Google Scholar]
- Medori R., Tritschler H. J., LeBlanc A., Villare F., Manetto V., Chen H. Y., Xue R., Leal S., Montagna P., Cortelli P., et al. (1992). Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene. N. Engl. J. Med. 326, 444-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I., Vinuela A., Astradsson A., Mukhida K., Hallett P., Robertson H., Tierney T., Holness R., Dagher A., Trojanowski J. Q., et al. (2008). Dopamine neurons implanted into people with Parkinson's disease survive without pathology for 14 years. Nat. Med. 14, 507-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M., Coomaraswamy J., Bolmont T., Kaeser S., Schaefer C., Kilger E., Neuenschwander A., Abramowski D., Frey P., Jaton A. L., et al. (2006). Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 313, 1781-1784 [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M., Spires-Jones T. L., Prada C., Garcia-Alloza M., de Calignon A., Rozkalne A., Koenigsknecht-Talboo J., Holtzman D. M., Bacskai B. J., Hyman B. T. (2008). Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease. Nature 451, 720-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners J. S., Baig S., Palmer J., Palmer L. E., Kehoe P. G., Love S. (2008). Abeta-degrading enzymes in Alzheimer's disease. Brain Pathol. 18, 240-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F., Tanji K., Zhang H. X., Nishihira Y., Tan C. F., Takahashi H., Wakabayashi K. (2008). Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta Neuropathol. 116, 193-203 [DOI] [PubMed] [Google Scholar]
- Nekooki-Machida Y., Kurosawa M., Nukina N., Ito K., Oda T., Tanaka M. (2009). Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl. Acad. Sci. USA 106, 9679-9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130-133 [DOI] [PubMed] [Google Scholar]
- Nishihira Y., Tan C. F., Onodera O., Toyoshima Y., Yamada M., Morita T., Nishizawa M., Kakita A., Takahashi H. (2008). Sporadic amyotrophic lateral sclerosis: two pathological patterns shown by analysis of distribution of TDP-43-immunoreactive neuronal and glial cytoplasmic inclusions. Acta Neuropathol. 116, 169-182 [DOI] [PubMed] [Google Scholar]
- Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009). Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353-3364 [DOI] [PubMed] [Google Scholar]
- Oesch B., Westaway D., Walchli M., McKinley M. P., Kent S. B., Aebersold R., Barry R. A., Tempst P., Teplow D. B., Hood L. E., et al. (1985). A cellular gene encodes scrapie PrP 27-30 protein. Cell 40, 735-746 [DOI] [PubMed] [Google Scholar]
- Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., et al. (1993). Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 90, 10962-10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu A. K., Qahwash I., Leapman R. D., Meredith S. C., Tycko R. (2009). Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. USA 106, 7443-7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B. K., Liebman S. W. (2007). “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+]. J. Mol. Biol. 365, 773-782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B. K., Gavin-Smyth J., Liebman S. W. (2009). The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11, 344-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesiridis G. S., Lee V. M., Trojanowski J. Q. (2009). Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 18, R156-R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. (2005). Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science 307, 262-265 [DOI] [PubMed] [Google Scholar]
- Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276, 2045-2047 [DOI] [PubMed] [Google Scholar]
- Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L. (2005). FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435-3438 [DOI] [PubMed] [Google Scholar]
- Prince M., Jackson J., Ferri C. P., Sousa R., Albanese E., Ribeiro W. S., Honyashiki M. (2009). Alzheimer's disease international world alzheimer report. In In International AsD (ed), pp. 1-96
- Prusiner S. B. (1982). Novel proteinaceous infectious particles cause scrapie. Science 216, 136-144 [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. (1998). Prions. Proc. Natl. Acad. Sci. USA 95, 13363-13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B., McKinley M. P., Bowman K. A., Bolton D. C., Bendheim P. E., Groth D. F., Glenner G. G. (1983). Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35, 349-358 [DOI] [PubMed] [Google Scholar]
- Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L. P., Goebel I., Mubaidin A. F., Wriekat A. L., Roeper J., et al. (2006). Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 38, 1184-1191 [DOI] [PubMed] [Google Scholar]
- Ren P. H., Lauckner J. E., Kachirskaia I., Heuser J. E., Melki R., Kopito R. R. (2009). Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat. Cell Biol. 11, 219-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider M., Klopp N., Xiang W., Wagenpfeil S., Vollmert C., Muller U., Forstl H., Illig T., Kretzschmar H., Kurz A. (2004). Prion protein codon 129 polymorphism and risk of Alzheimer disease. Neurology 63, 364-366 [DOI] [PubMed] [Google Scholar]
- Roberson E. D., Scearce-Levie K., Palop J. J., Yan F., Cheng I. H., Wu T., Gerstein H., Yu G. Q., Mucke L. (2007). Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science 316, 750-754 [DOI] [PubMed] [Google Scholar]
- Roberts B. E., Duennwald M. L., Wang H., Chung C., Lopreiato N. P., Sweeny E. A., Knight M. N., Shorter J. (2009). A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat. Chem. Biol. 5, 936-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saa P., Castilla J., Soto C. (2006). Presymptomatic detection of prions in blood. Science 313, 92-94 [DOI] [PubMed] [Google Scholar]
- Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. (1998). Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4, 1157-1165 [DOI] [PubMed] [Google Scholar]
- Salnikova A. B., Kryndushkin D. S., Smirnov V. N., Kushnirov V. V., Ter-Avanesyan M. D. (2005). Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 280, 8808-8812 [DOI] [PubMed] [Google Scholar]
- Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. (2009). Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 41, 1303-1307 [DOI] [PubMed] [Google Scholar]
- Saupe S. J. (2007). A short history of small s: a prion of the fungus Podospora anserina. Prion 1, 110-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., et al. (2007). Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453-457 [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997). Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549-558 [DOI] [PubMed] [Google Scholar]
- Shorter J. (2008). Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals 16, 63-74 [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. (2004). Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793-1797 [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. (2005). Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435-450 [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. (2006). Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23, 425-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K., Lindquist S., Kandel E. R. (2003). A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 115, 879-891 [DOI] [PubMed] [Google Scholar]
- Si K., Choi Y. B., White-Grindley E., Majumdar A., Kandel E. R. (2010). Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140, 421-435 [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J., Schulte C., Bras J. M., Sharma M., Gibbs J. R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S. W., Hernandez D. G., et al. (2009). Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 41, 1308-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. (2003). alpha-Synuclein locus triplication causes Parkinson's disease. Science 302, 841 [DOI] [PubMed] [Google Scholar]
- Skovronsky D. M., Lee V. M., Trojanowski J. Q. (2006). Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 1, 151-170 [DOI] [PubMed] [Google Scholar]
- Smith J. F., Knowles T. P., Dobson C. M., Macphee C. E., Welland M. E. (2006). Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA 103, 15806-15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G. W., Cook E. G., Hargus G., Blak A., Cooper O., Mitalipova M., et al. (2009). Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon A., Richey T., Murphy C. L., Weiss D. T., Wall J. S., Westermark G. T., Westermark P. (2007). Amyloidogenic potential of foie gras. Proc. Natl. Acad. Sci. USA 104, 10998-11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper J. H., Roy S., Stieber A., Lee E., Wilson R. B., Trojanowski J. Q., Burd C. G., Lee V. M. (2008). Alpha-synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 1093-1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. (2003). Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4, 49-60 [DOI] [PubMed] [Google Scholar]
- Soto C., Castilla J. (2004). The controversial protein-only hypothesis of prion propagation. Nat. Med. 10, S63-S67 [DOI] [PubMed] [Google Scholar]
- Speare J. O., Offerdahl D. K., Hasenkrug A., Carmody A. B., Baron G. S. (2009). GPI anchoring facilitates propagation and spread of misfolded Sup35 aggregates in mammalian cells. EMBO J. 29, 782-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponarova J., Nystrom S. N., Westermark G. T. (2008). AA-amyloidosis can be transferred by peripheral blood monocytes. PLoS One 3, e3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. D., Lindquist S., Aguzzi A. (2007). The prion protein knockout mouse: a phenotype under challenge. Prion 1, 83-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. D., Zhou Z., Jackson W. S., Zhu C., Auluck P., Moskowitz M. A., Chesselet M. F., Lindquist S. (2009). Context dependent neuroprotective properties of prion protein (PrP). Prion 3, 240-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamguney G., Miller M. W., Wolfe L. L., Sirochman T. M., Glidden D. V., Palmer C., Lemus A., DeArmond S. J., Prusiner S. B. (2009). Asymptomatic deer excrete infectious prions in faeces. Nature 461, 529-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004). Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323-328 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006). The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585-589 [DOI] [PubMed] [Google Scholar]
- Toombs J. A., McCarty B. R., Ross E. D. (2010). Compositional Determinants of Prion Formation in Yeast. Mol. Cell. Biol. 30, 319-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H. L., Lindquist S. L. (2000). A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477-483 [DOI] [PubMed] [Google Scholar]
- Tyedmers J., Madariaga M. L., Lindquist S. (2008). Prion switching in response to environmental stress. PLoS Biol. 6, e294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., et al. (2008). TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7, 409-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobagyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208-1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Cushman M., Shorter J. (2010). Applying Hsp104 to protein-misfolding disorders. Biochem. Cell Biol. 88, 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrenko Y. A., Gracheva E. O., Richmond J. E., Liebman S. W. (2007). Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J. Biol. Chem. 282, 1779-1787 [DOI] [PubMed] [Google Scholar]
- Walker F. O. (2007). Huntington's disease. Lancet 369, 218-228 [DOI] [PubMed] [Google Scholar]
- Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002). Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535-539 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang X., Yuan C. G., Ma J. (2010). Generating a prion with bacterially expressed recombinant prion protein. Science 327, 1132-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Duennwald M. L., Roberts B. E., Rozeboom L. M., Zhang Y. L., Steele A. D., Krishnan R., Su L. J., Griffin D., Mukhopadhyay S., et al. (2008). Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc. Natl. Acad. Sci. USA 105, 7159-7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J. M., Chan H. Y., Gray-Board G. L., Chai Y., Paulson H. L., Bonini N. M. (1999). Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23, 425-428 [DOI] [PubMed] [Google Scholar]
- Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009). TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA 106, 18809-18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Zhao J. P., Pruszak J., Hedlund E., Fu D., Soldner F., Broccoli V., Constantine-Paton M., Isacson O., Jaenisch R. (2008). Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc. Natl. Acad. Sci. USA 105, 5856-5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark G. T., Westermark P. (2009). Serum amyloid A and protein AA: molecular mechanisms of a transmissible amyloidosis. FEBS Lett. 583, 2685-2690 [DOI] [PubMed] [Google Scholar]
- Wiltzius J. J., Landau M., Nelson R., Sawaya M. R., Apostol M. I., Goldschmidt L., Soriaga A. B., Cascio D., Rajashankar K., Eisenberg D. (2009). Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973-978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Lucas J. J., Hen R. (2000). Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101, 57-66 [DOI] [PubMed] [Google Scholar]
- Yamanaka S. (2007). Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell 1, 39-49 [DOI] [PubMed] [Google Scholar]
- Zarranz J. J., Alegre J., Gomez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atares B., et al. (2004). The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164-173 [DOI] [PubMed] [Google Scholar]
- Zhang B., Une Y., Fu X., Yan J., Ge F., Yao J., Sawashita J., Mori M., Tomozawa H., Kametani F., et al. (2008). Fecal transmission of AA amyloidosis in the cheetah contributes to high incidence of disease. Proc. Natl. Acad. Sci. USA 105, 7263-7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., et al. (2009). Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 106, 7607-7612 [DOI] [PMC free article] [PubMed] [Google Scholar]