Abstract
In general, double-stranded RNA (dsRNA)-binding proteins (dsRBPs) are not sequence-specific. A dsRNA molecule in a cell will interact with any dsRBP it comes in contact with, suggesting that different dsRNA-mediated pathways intersect and affect each other. This paper analyzes evidence that the ADAR RNA editing enzymes, which act on dsRNA, affect dsRNA-mediated gene silencing pathways. Examples of how ADARs alter gene silencing pathways such as RNA interference, as well as mechanisms that allow the pathways to coexist and maintain their unique functions, are discussed.
Most of the functional groups in a double helix that allow a protein to recognize a specific base pair are in the major groove (Seeman et al. 1976), and there are many examples of proteins that enter the major groove of B-form DNA to make sequence-specific interactions (see, e.g., Wolberger 1999). However, double-stranded RNA (dsRNA) adopts an A-form helical structure with a major groove that is more narrow than that of DNA. It is difficult for a protein to enter the major groove of dsRNA to make sequence-specific contacts, and in fact, all characterized dsRNA-binding proteins (dsRBPs) bind to dsRNA of any sequence. Consistent with this, structural analyses of protein–dsRNA complexes show interactions that are predominantly in the minor groove, with major groove interactions limited to those on the exterior, involving the phosphodiester backbone (Ryter and Schultz 1998; Ramos et al. 2000; Wu et al. 2004).
Because dsRBPs are not sequence-specific, a dsRNA molecule in a cell will bind to any dsRBP it comes in contact with, and consequently, different dsRNA-mediated pathways affect each other. Researchers involved in early studies of RNA interference (RNAi) encountered this principle when they applied the RNAi protocols that worked well in flies and worms to mammalian cells (Elbashir et al. 2001). dsRNA introduced into mammalian cells encounters not only dsRBPs involved in RNAi, but also a dsRBP called PKR (Williams 2001). Upon binding to dsRNA, PKR becomes an active kinase and phosphorylates the translation initiation factor eIF2α, leading to a global shutdown of protein synthesis (Hunter et al. 1975). Introducing long dsRNA into a mammalian cell reduces expression of all mRNAs, not just the one targeted by RNAi.
Viruses capitalize on the fact that dsRBPs are not sequence-specific by synthesizing their own dsRBPs to antagonize cellular dsRNA-mediated pathways. For example, viral infection is sometimes accompanied by production of dsRNA from the viral genome (Boone et al. 1979; Maran and Mathews 1988), and to avoid the activation of PKR, viruses such as vaccinia and reovirus encode a dsRBP to compete with PKR for dsRNA (for review, see Stark et al. 1998). Not surprisingly, virus-encoded dsRBPs and RNAs are also reported to antagonize the RNAi pathway (Li et al. 2004; Andersson et al. 2005).
The realization that dsRNA-mediated gene silencing is central to many biological processes invites the question as to how cellular dsRBPs affect this pathway. Studies are only beginning, but it seems possible that there is a finely tuned interplay between pathways involving dsRNA. This paper analyzes evidence that the dsRBPs known as adenosine deaminases that act on RNA (ADARs) affect dsRNA-mediated gene silencing.
ADAR Basics
ADARs are RNA editing enzymes that convert adenosine (A) to inosine (I) in dsRNA (for review, see Bass 2002; Keegan et al. 2004; Valente and Nishikura 2005). The enzymes are found in all animals, where they are usually in the nucleus, and are most highly expressed in the nervous system. The A to I conversion involves a hydrolytic deamination, and, depending on the extent of base-pairing in an RNA substrate, the enzyme can selectively deaminate a specific adenosine, or nonselectively deaminate up to 50–60% of the adenosines within a double-stranded region. These two modes of selectivity can be observed in vitro with synthetic RNA, or in vivo, in the endogenous targets of ADARs (for review, see Bass 2002).
Selective deamination occurs in RNA that, although largely double-stranded, is frequently interrupted by mismatches, bulges, or loops. It is this “imperfect” helical structure that is associated with editing in codons, and, as shown in Figure 1A, often forms by pairing between introns and exons. Inosine prefers to pair with cytidine, and thus, an A to I conversion can change the amino acid specified by a codon. Editing in codons results in the synthesis of multiple protein isoforms from a single encoded mRNA and serves as a mechanism of diversifying and increasing an organism's proteome. Deamination of codons typically involves the selective type of editing, which is not surprising, since nonselective editing would result in many amino acid changes and likely produce a nonfunctional protein.
Figure 1.
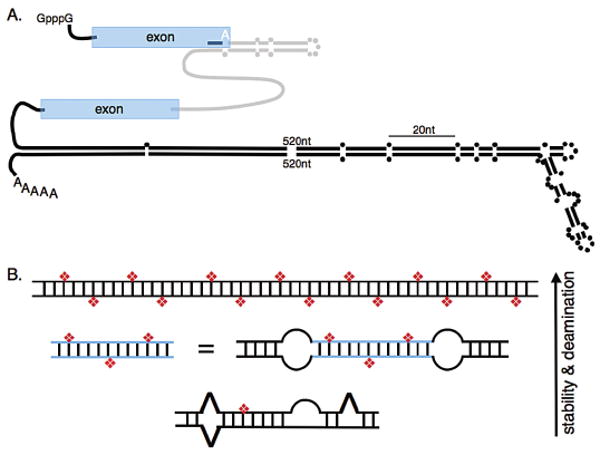
(A) The illustration depicts a pre-mRNA that contains examples of structures that are selectively (upper) and nonselectively (lower) edited. Coding exons are depicted in blue, an intron in gray, and 5′ and 3′ UTRs as black lines. Parallel lines represent base-paired regions with dots representing unpaired nucleotides. The selectively edited hairpin involves pairing between exon and intron sequences and is patterned after the R/G editing site, shown as a white A, found in certain mammalian glutamate receptor pre-mRNAs; the hairpin contains 28 bps, a loop, and mismatches as indicated (Aruscavage and Bass 2000). The nonselectively edited structure is patterned after the 3′UTR of the C. elegans gene, C35E7.6 (Morse et al. 2002). The edited structures are scaled relative to their actual lengths (20 nucleotides indicated), but 520 nucleotides of each strand of the 3′UTR structure were omitted as indicated. (B) The cartoon illustrates that the number of adenosines deaminated by an ADAR at reaction completion, or the selectivity of the enzyme, increases with the thermodynamic stability of the RNA structure. The top structure represents a long, completely base-paired dsRNA ≥ 50 bp which is deaminated nonselectively, showing 50–60% of its adenosines converted to inosines (red diamonds) at reaction completion. As indicated, structures that are less stable because they are shorter, or interrupted by mismatches, bulges, or loops, contain fewer inosines at the end of the reaction. Blue lines represent a specific sequence that exists as a separate molecule or between two internal loops of a longer structure.
The R/G hairpin that is edited at a specific adenosine in certain mammalian glutamate receptor pre-mRNAs is used to represent the selectively edited structure in Figure 1A (upper structure; R/G indicates that editing at the labeled A changes an arginine codon to a glycine codon). This structure is short, with a predicted free energy of −35 kcal/mole (mfold at 37°C; Mathews et al. 1999; Zuker 2003). Although selective editing usually occurs in structures that are less stable than those promoting nonselective editing, this is not always the case, as exemplified by editing of the antigenomic RNA of hepatitis delta virus (HDV; for review, see Casey 2006). The ∼1700-nucleotide HDV RNA folds into a “rod-like” helical structure with a predicted free energy of −930 kcal/mol (mfold at 37°C). Despite its length and stability, the HDV RNA is edited at a specific adenosine to change an amber stop codon to a tryptophan codon (Polson et al. 1996, 1998). Importantly, although the HDV antigenomic RNA is highly base-paired, it is rare to find a complete helical turn within its structure that does not contain a mismatch, bulge, or loop. As discussed in the next section, this is key to its selective deamination.
Nonselective deamination occurs in RNA that is completely, or nearly completely, base-paired and often involves long helices containing hundreds of base pairs. In Figure 1A (lower hairpin), this type of structure is represented by a Caenorhabditis elegans 3′UTR which involves 1423 nucleotides that fold into a structure with a predicted free energy of −1201 kcal/mole (mfold at 20°C). This structure is heavily edited, with 42% of its 467 adenosines appearing as inosines in at least a fraction of the steady-state mRNA isolated from wild-type worms, and ∼18% showing editing at all of these sites (Morse et al. 2002). As illustrated, although the nonselectively edited structures do contain mismatches, bulges, and loops, such disruptions are less frequent, and there are typically long stretches of uninterrupted, contiguous base pairs. In vivo, these types of substrates are usually found in noncoding regions of mRNAs, such as introns and untranslated regions, and are often formed by pairing between repeat elements (Morse and Bass 1999; Morse et al. 2002). Recent bioinformatic studies indicate that these types of substrates are remarkably abundant, occurring in an estimated 5% of the mRNAs encoded by our genomes (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004). As yet, the function of these structures, and the inosines within them, is unknown.
A Model for ADAR Selectivity
The principles that control ADAR selectivity have been summarized previously (Bass 2002) and are briefly reviewed here using Figure 1B. As discussed above, a completely, or largely, base-paired duplex, greater than ∼50 bp, is deaminated nonselectively, showing 50–60% of its adenosines deaminated at reaction completion. However, the reaction does stop, and the model is based on this observation. When AU base pairs are changed to IU mismatches, the RNA structure becomes less stable, and the idea is that the reaction stops when there are so many IU mismatches that the RNA is too single-stranded in character to be acted on by an ADAR. Assuming there is some critical thermodynamic stability after which no further reaction can occur, it is easy to see that structures which are less stable to begin with will accommodate fewer deaminations before the reaction stops. Decreasing the length of an RNA helix, or disrupting it with mismatches, bulges, or loops, will decrease the number of deamination sites at reaction completion. At the extreme, internal loops can uncouple a long helix into a series of short helices that are each deaminated selectively (Lehmann and Bass 1999). In fact, as shown in Figure 1B, a short helix bounded by two internal loops will be deaminated at the same sites targeted when the helix exists as a separate molecule. The ability of loops to uncouple helices explains how a very long helix such as the HDV antigenomic RNA can be selectively deaminated.
Do ADARs Affect RNA Interference?
Given that dsRBPs are not sequence-specific, it was proposed that ADARs might antagonize RNAi (Bass 2000), and support for this idea came from the observation that mutant C. elegans strains that lack ADARs exhibit aberrant gene silencing (Knight and Bass 2002). The mutant animals respond as expected when dsRNA is introduced by injection or feeding; that is, expression of the cognate mRNA is silenced at levels indistinguishable from wild type. Possibly this dsRNA never reaches the nucleus where ADARs usually reside. However, as illustrated in Figure 2, transgenic DNA, which is transcribed in the nucleus and normally expressed in a wild-type animal, is silenced in animals lacking ADARs. The silencing is dependent on factors important for the RNAi pathway, such as Dicer (Knight and Bass 2001), and is reminiscent of the cosuppression first observed in plants (for review, see Matzke and Matzke 2004). Although cosuppression of transgenic DNA is frequently observed in the germ line of C. elegans (Kelly et al. 1997; for review, see Seydoux and Schedl 2001), the cosuppression observed in ADAR mutant animals is distinct because it is observed with transgenes expressed in the soma.
Figure 2.
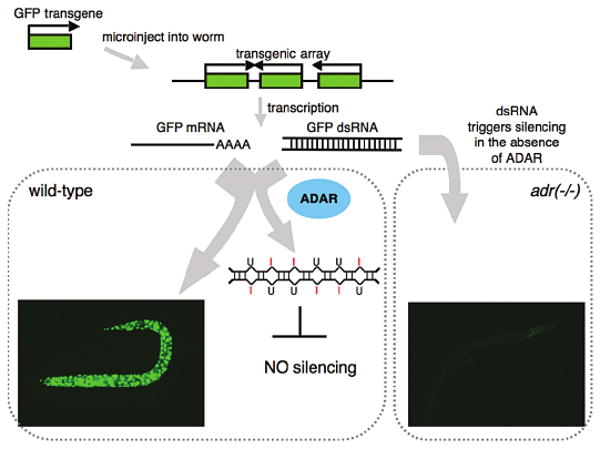
The cartoon illustrates the proposed pathway leading to transgene silencing in C. elegans strains that lack ADARs (adr(−/−)). Starting at the upper left corner, DNA encoding GFP (green rectangle) is injected into a worm, whereby it is covalently linked to form repetitive arrays that are maintained as extrachromosomal elements. In wild-type animals, GFP mRNA is expressed, as evidenced by strong GFP fluorescence (bottom left corner). Black arrows indicate the direction of transcription, and as shown, readthrough transcription of the repetitive array also gives rise to sense and antisense RNAs that hybridize to form dsRNA. In a wild-type animal, this dsRNA is deaminated by ADARs and thus does not lead to silencing. In animals lacking ADARs (adr(-/-)), the dsRNA enters the RNAi pathway, leading to silencing. Photographs of worms were taken by Jeff Habig, and strains are as described (Knight and Bass 2002).
What is the mechanism of the somatic cosuppression observed in the ADAR mutants? In this regard, as shown in Figure 2, it is important to note that DNA introduced into C. elegans is covalently linked to form repetitive arrays, with genes in tandem, as well as inverted, orientation. Although in most cases transcription terminates normally, in the illustrated example giving rise to GFP mRNA, in some cases readthrough transcription of genes in an inverted orientation gives rise to dsRNA. Analysis of the wild-type animal illustrated in Figure 2 confirmed that both sense and antisense RNA were synthesized from the transgene. Because it prefers to pair with cytidine, inosine appears as guanosine in cDNA, and cDNA derived from the transgenic RNA showed the typical A to G changes indicative of editing (Knight and Bass 2002). Consistent with the fact that the sense and antisense RNAs would be completely base-paired, the RNA was nonselectively edited and contained many inosines. In contrast, but in line with the observed silencing, neither sense nor antisense RNA corresponding to the transgene sequence was detected in the ADAR mutant.
There are two obvious mechanisms by which ADARs could prevent an RNA from entering the RNAi pathway, and both may contribute to the observed silencing. First, a simple competition between ADARs and dsRBPs of the RNAi pathway might exist. Dicer and Drosha are both dsRBPs and contain the dsRNA-binding motif found in many dsRBPs (for review, see Doyle and Jantsch 2002). Furthermore, in many organisms, additional dsRBPs have been identified that facilitate the action of these enzymes (Tabara et al. 2002; Gregory et al. 2004; Han et al. 2004, 2006; Haase et al. 2005; Liu et al. 2006). In this scenario, ADARs would antagonize the RNAi pathway simply by binding dsRNA and sequestering it so that it is unavailable to dsRBPs involved in gene silencing. Of course, such a competition could occur between any dsRBP, not just ADARs, and indeed, there may be a fine balance between the concentration of dsRNA and various dsRBPs.
Alternatively, the silencing phenotype of ADAR mutants may relate to the lack of editing, rather than, or in addition to, dsRNA binding. As shown in Figure 2, and discussed earlier, when ADARs edit dsRNA, AU base pairs are changed to IU mismatches, and the RNA becomes less double-stranded. Although it has not yet been demonstrated directly, by definition one would predict a dsRBP would bind less well to RNA after it has been modified by ADARs. According to this scenario, if ADARs deaminate a dsRNA before it enters the RNAi pathway, dsRBPs such as Dicer cannot bind the RNA and gene silencing cannot occur. Studies of RNA interference in vitro, using Drosophila extracts, support this idea. For example, if dsRNA is reacted with an ADAR before adding it to a Drosophila extract, it is less effective at triggering an RNAi response (Scadden and Smith 2001); production of siRNAs is reduced, and consequently, mRNA degradation is also reduced. When the dsRNA is highly edited (∼50%), siRNAs are completely absent, but at intermediate amounts of editing, siRNAs are present. These authors, and others (Zamore et al. 2000), isolated siRNAs produced from edited dsRNA and found that inosine is present in the siRNA, albeit at levels lower than in the starting material, i.e., the unreacted, edited dsRNA. The latter data indicate that at intermediate amounts of editing Dicer can cleave edited dsRNA, possibly in regions that have fewer deamination sites and thus retain their overall double-stranded character.
Of course, the most interesting question in regard to the silencing observed in ADAR mutant animals is whether ADARs are involved in regulating dsRNA-mediated gene silencing of endogenous genes in vivo. So far, a specific example of this does not exist, which is not surprising, given that few genes that are natural targets of dsRNA-mediated gene silencing have been identified. Here, of course, I am not referring to genes whose expression is modulated by miRNA, of which there are many, but to systems more analogous to silencing that involves an antisense transcript, such as the Stellate genes in Drosophila melanogaster (Aravin et al. 2001, 2004), or cyclin E in C. elegans (Grishok and Sharp 2005). However, in support of the idea that ADARs do modulate silencing of endogenous genes, the chemotaxis defects of C. elegans that lack ADARs are rescued in strains that also lack components of the RNAi pathway (Tonkin and Bass 2003). Recent studies show that antisense transcription is abundant in mammalian genomes (Lehner et al. 2002; Yelin et al. 2003; Cawley et al. 2004; Chen et al. 2004; for review, see Chen et al. 2005), and it is intriguing to imagine that ADARs play a role in attenuating silencing that might result from dsRNA formed with this antisense.
When considering how ADARs might regulate silencing, it is important to note that the enzymes are very sensitive to substrate inhibition (Hough and Bass 1994). Thus, for a case where ADARs were allowing expression of a gene by deaminating its cognate dsRNA, simply increasing the levels of dsRNA would inhibit ADARs and allow silencing to take over. The inhibiting dsRNA could be generated in cis from the gene being silenced. Alternatively, since ADARs bind dsRNA of any sequence, a dsRNA synthesized in trans, at a different locus, could also inhibit ADARs and provide another way to regulate dsRNA-mediated silencing.
Do ADARs Target siRNA and miRNA?
As mentioned above, long dsRNA that contains intermediate amounts of inosine is cleaved by Dicer to produce siRNAs that contain low amounts of inosine. However, because it is less stable than long dsRNA, very short dsRNA is a poor substrate for an ADAR (see prior discussion of selectivity). Thus, it seems probable that siRNA and miRNA duplexes are only rarely acted on directly by ADARs. However, it is intriguing to imagine scenarios by which ADARs might regulate the function of the slightly longer precursors of miRNAs. Again, regulation could occur because of a simple competition between ADARs and the dsRBPs required for the biogenesis and functions of small RNAs, or as a direct consequence of editing. In the latter case, an A to I change within the mature miRNA sequence would change its base-pairing properties and, thus, could actually change the mRNA that is targeted by the miRNA.
Although several studies provide “proof of principle” results, that is, they show that ADARs could in theory regulate the function of miRNAs, as yet, a definitive example of an ADAR regulating the function of an miRNA in vivo has not been found. However, it is increasingly clear that endogenous miRNA precursors are targeted by ADARs and contain inosine. Initial studies focused on specific pri-miRNAs, and although some of these were edited to a significant extent when incubated with ADARs in vitro (Yang et al. 2006) or in cells overexpressing ADARs (Luciano et al. 2004; Yang et al. 2006), only very low levels of editing were detected within endogenous miRNA precursors. For example, in 3–7% of the pri-miR-22 molecules isolated from human brain, lung, and testes, and mouse brain, at least one of the adenosines in the miRNA was edited, and in some cases, the inosines were within the mature miRNA sequence (Luciano et al. 2004). However, when considering a given adenosine within the population of pri-miR-22 molecules, the amount of editing at any particular site was extremely low, and furthermore, the editing sites were not conserved between human and mouse. Similarly, low levels of editing were observed in pri-miR-142 in mouse spleen (Yang et al. 2006).
Recently, a more systematic analysis of pri-miRNA sequences from a variety of human tissues was conducted. Six of 99 pri-miRNA sequences analyzed contained editing sites, and in contrast to the previous studies, for these 6 pri-miRNAs, specific adenosines were edited to significant levels (>10%) in a tissue-specific manner, in some cases approaching 100% editing at a single site (Blow et al. 2006). Whereas the lower levels of editing observed in the earlier studies may well represent RNA that accidentally got caught up with ADAR, the higher levels of editing in this study are more suggestive of functional editing.
Given the diverse ways metazoa regulate biological processes, it seems likely that, because ADARs could regulate the function of small RNAs, they probably do, and eventually a bona fide example will be discovered. In this regard, the proof-of-principle experiments are interesting because they illustrate potential mechanisms.
One idea is that editing would interfere with pri-mRNA or pre-mRNA processing, by Drosha and Dicer, respectively. Consistent with this idea, mature miR-142 levels were elevated two- to threefold in the spleen of ADAR null mice (Yang et al. 2006). This same study showed that, in vitro, pri-miRNAs containing inosine were less efficiently processed to pre-miRNAs by recombinant Drosha. This may explain why, despite the large number of mature miRNAs sequenced, no one has yet reported editing in a processed, mature miRNA sequence. Another possibility is that editing of pri- and pre-miRNAs leads to their degradation, possibly by the staphyloccoccal nuclease, Tudor-SN, a component of RISC (Caudy et al. 2003). Whereas some reports indicate that this enzyme is a non-sequence-specific nuclease that targets single-stranded RNA (Caudy et al. 2003), others observe that the enzyme stimulates cleavage of dsRNA that has three or four contiguous IU (or UI) base pairs (Scadden 2005). In fact, when HEK293 cells are transfected with a plasmid encoding pri-miR142, along with an inhibitor of Tudor-SN, an increase in edited pri-miRNA is observed (Yang et al. 2006).
Finally, another study suggests that certain ADARs may antagonize RNAi by binding to siRNA in the cytoplasm (Yang et al. 2005). Using recombinant mammalian ADARs, binding to siRNA in vitro was studied using gel-shift analyses as well as filter-binding. The interferon-inducible ADAR1p150, the only ADAR known to be present in the cytoplasm, had the highest affinity for siRNA, but ADAR1p110 and ADAR2 also bound tightly. Consistent with the idea that the ADAR1p150 antagonizes RNAi in vivo, the efficacy of RNAi in mouse embryonic fibroblasts null for ADAR1 was threefold higher than in wild-type cells or cells null for ADAR2.
Yet, dsRNA-Mediated Pathways Coexist
Although this paper has focused on the antagonism between dsRNA-mediated pathways, it is important to note that these pathways have also evolved ways to minimize this interplay and protect their unique functions. Examples of this have been mentioned throughout this paper, and in closing, the topic will be specifically addressed.
As for all pathways in eukaryotes, one way to minimize antagonism between dsRNA-mediated pathways is to simply sequester the relevant enzymes into different cellular compartments. In the presence of interferon, a cytoplasmic form of an ADAR is induced (George and Samuel 1999), but under normal conditions, these enzymes are sequestered in the nucleus, away from PKR and Dicer, which are largely cytoplasmic (Jimenez-Garcia et al. 1993; Billy et al. 2001). As mentioned earlier, this differential compartmentalization explains why C. elegans that lack ADARs exhibit aberrant dsRNA-mediated silencing only when dsRNA is synthesized in the nucleus. However, like ADARs, Drosha is in the nucleus (Lee et al. 2002), where it processes pri-miRNAs to pre-miRNAs. As discussed above, some miRNA precursors contain inosines indicating that, albeit infrequently, these molecules do interact with ADARs. But why don't more miRNA precursors contain inosine? Why aren't ADAR substrates such as the mRNAs with long, structured UTRs cleaved by Drosha in the nucleus, or Dicer in the cytoplasm, where they must be translated? Or are they?
Definitive answers to these questions await future research. However, predictions can be made by considering that, although dsRBPs are not sequence-specific, they exhibit what is sometimes called a structural specificity. Structural specificity likely plays an important role in allowing dsRBPs to coexist and carry out their unique functions. As gleaned from the earlier discussion of ADAR selectivity, an understanding of dsRBP specificity is based in the obvious: dsRBPs are just that—double-stranded RNA-binding proteins. They prefer to bind dsRNA, and although they don't recognize sequence, they recognize any structural alteration to the A-form helix. The length of a helix, whether the terminus of the helix is blunt or frayed, and whether it contains mismatches, bulges, or loops, all affect how a dsRBP recognizes a substrate. We currently have only limited information linking these observations to the actual interactions that occur between a dsRBP and its substrate. It seems likely that in some cases the structural features of the RNA will affect the physical interactions it has with a dsRBP, but it is also important to consider that anything that makes an RNA substrate more single-stranded in character will shift the double-stranded ⇆ single-stranded equilibrium toward the single-stranded state and affect binding.
An example of structural specificity is illustrated by returning to the discussion started at the beginning of this paper, where it was noted that long dsRNA introduced into mammalian cells not only triggers RNAi, but also activates PKR, a dsRBP that phosphorylates eIF2α to shut down all translation. A solution to the problem of how to apply RNAi techniques to mammalian cells without activating PKR came when it was determined that a specific response could be incurred using a very short RNA: the siRNA that is an intermediate of the RNAi process (Elbashir et al. 2001). Presumably, specificity is achieved because PKR binds less well to short dsRNA than at least some proteins required for RNAi (Hunter et al. 1975; Minks et al. 1979; Manche et al. 1992). The C. elegans dsRBP, RDE-4, which acts with Dicer in dsRNA cleavage, is an RNAi factor that shows a preference for longer dsRNA (Parker et al. 2006), and in this case, cooperativity is proposed to allow the protein to preferentially bind long dsRNA. Like PKR, ADARs do not act efficiently with very short dsRNA, consistent with the idea that mature siRNA and miRNA duplexes are not direct targets of ADARs. Thus, length appears to be one way dsRBPs gain specificity.
miRNA precursors are slightly longer, and indeed, some of these molecules are edited by ADARs. However, many are not. Even under in vitro conditions optimized for the ADAR enzyme, only 4 of 8 pri-miRNAs tested showed inosine after incubation with recombinant ADAR (Yang et al. 2006). This may indicate that, in general, these hairpin structures have evolved so as to avoid deamination by ADARs. Figure 3 compares the predicted structures of 4 pri-mRNAs tested for their ability to be deaminated in vitro by recombinant ADAR (Yang et al. 2006). Consistent with the model of ADAR selectivity presented earlier, the predicted structures of the unedited pri-miRNAs contain more unpaired nucleotides and are less stable than those that are edited.
Figure 3.

The lowest free-energy secondary structure is shown for four miRNA precursors, as predicted by mfold (Mathews et al. 1999; Zuker 2003). Nucleotides shown in red represent mature miRNA sequences as experimentally determined (Lagos-Quintana et al. 2002; Houbaviy et al. 2003; Poy et al. 2004). When incubated with recombinant ADAR, inosine can be detected within the two pri-miRNA sequences above the dotted line (edited), whereas inosine cannot be detected in those below the line (unedited; as reported in Yang et al. 2006).
Although studies of the specificity of dsRBPs involved in RNAi are just beginning, it is already apparent that these dsRBPs will follow similar principles. Dicer requires a helix with a blunt terminus (Zhang et al. 2002; Vermeulen et al. 2005), whereas Drosha utilizes a frayed terminus (Han et al. 2006). Given this, it is predicted that the long structured UTRs of ADAR substrates would not be substrates for Dicer, but could be cleaved by Drosha. Similar to the ADARs, Dicer's activity is influenced by the extent of base-pairing in an RNA: Completely base-paired dsRNA is cleaved at many sites, whereas pre-miRNAs are specifically cleaved to generate the functional, mature miRNA. As evidenced from the data reviewed in this paper, dsRNA-mediated pathways clearly affect each other, but we have a long way to go in order to understand the subtleties of this interplay. Extending our current knowledge of how dsRBPs achieve specificity to a more molecular level, as well as to newly discovered dsRBPs, will no doubt lead to a more complete understanding of how dsRNA-mediated pathways intersect, yet maintain their unique functions.
Acknowledgments
Thanks to members of the Bass lab for their hard work, constant questions, and excellent insight. A special thanks to lab members: to Jeff Habig, for photographs of C. elegans; and to Taraka Dale, Sabine Hellwig, and Heather Hundley for critically reading this manuscript. This work was supported by National Institutes of Health grants (GM067106 and GM44073). B.L.B. is an Investigator of the Howard Hughes Medical Institute.
References
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aruscavage PJ, Bass BL. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA. 2000;6:257. doi: 10.1017/s1355838200991921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci. 2001;98:14428. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone RF, Parr RP, Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J Virol. 1979;30:365. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JL. RNA editing in hepatitis delta virus. Curr Top Microbiol Immunol. 2006;307:67. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J Struct Biol. 2002;140:147. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci. 1999;96:4621. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A, Sharp PA. Negative regulation of nuclear divisions in Caenorhabditis elegans by retinoblastoma and RNA interference-related genes. Proc Natl Acad Sci. 2005;102:17360. doi: 10.1073/pnas.0508989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Hough RF, Bass BL. Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J Biol Chem. 1994;269:9933. [PubMed] [Google Scholar]
- Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975;250:409. [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Green SR, Mathews MB, Spector DL. Organization of the double-stranded RNA-activated protein kinase DAI and virus-associated VA RNAI in adenovirus-2-infected HeLa cells. J Cell Sci. 1993;106:11. doi: 10.1242/jcs.106.1.11. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Leroy A, Sproul D, O'Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci. 2004;101:1350. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran A, Mathews MB. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology. 1988;164:106. doi: 10.1016/0042-6822(88)90625-3. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJ. Planting the seeds of a new paradigm. PLoS Biol. 2004;2:E133. doi: 10.1371/journal.pbio.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J Biol Chem. 1979;254:10180. [PubMed] [Google Scholar]
- Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc Natl Acad Sci. 1999;96:6048. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci. 2002;99:7906. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- Polson AG, Ley HL, III, Bass BL, Casey JL. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol Cell Biol. 1998;18:1919. doi: 10.1128/mcb.18.4.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Scadden AD, Smith CW. RNAi is antagonized by A→I hyper-editing. EMBO Rep. 2001;2:1107. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci. 1976;73:804. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Schedl T. The germline in C. elegans: Origins, proliferation, and silencing. Int Rev Cytol. 2001;203:139. doi: 10.1016/s0074-7696(01)03006-6. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci STKE. 2001;89:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Wolberger C. Multiprotein-DNA complexes in transcriptional regulation. Annu Rev Biophys Biomol Struct. 1999;28:29. doi: 10.1146/annurev.biophys.28.1.29. [DOI] [PubMed] [Google Scholar]
- Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci. 2004;101:8307. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


