Abstract
LSm1-7 complexes promote cellular mRNA degradation, in addition to translation and replication of positive-strand RNA viruses such as the Brome mosaic virus (BMV). Yet, how LSm1-7 complexes act on their targets remains elusive. Here, we report that reconstituted recombinant LSm1-7 complexes directly bind to two distinct RNA-target sequences in the BMV genome, a tRNA-like structure at the 3′-untranslated region and two internal A-rich single-stranded regions. Importantly, in vivo analysis shows that these sequences regulate the translation and replication of the BMV genome. Furthermore, both RNA-target sequences resemble those found for Hfq, the LSm counterpart in bacteria, suggesting conservation through evolution. Our results provide the first evidence that LSm1-7 complexes interact directly with viral RNA genomes and open new perspectives in the understanding of LSm1-7 functions.
Keywords: BMV, LSm1-7, RNA virus, Sm proteins, translation, replication
INTRODUCTION
The Sm and Sm-like (LSm) proteins constitute a conserved family whose members function in multiple aspects of RNA metabolism (for review, see Beggs 2005; Khusial et al. 2005; Wilusz and Wilusz 2005). They are characterized by the presence of a so-called Sm fold that enables their association into homomeric (in eubacteria) or heteromeric (in eukaryotes) ring-shaped complexes. Sm proteins form a stable complex on U-rich small nuclear RNA (U snRNA), and as part of the spliceosome, they function in pre-mRNA processing (Chari et al. 2008, and references therein). LSm complexes, in contrast, bind transiently to a broad spectrum of RNAs and influence their fate. Eight of the LSm proteins (LSm1–LSm8) are conserved from yeast to humans and form two distinct heteroheptameric rings that differ only by the exchange of the LSm1 and LSm8 subunits. The LSm2-8 complex is localized in the nucleus and functions in pre-mRNA maturation (as part of the U6 and U6ATAC snRNPs), pre-mRNA decay, and processing of pre-tRNAs, pre-small nucleolar RNA (snoRNA), and pre-rRNAs. Conversely, the LSm1-7 complex is localized in the cytoplasm and plays a key role in mRNA decay (for review, see Beggs 2005; Khusial et al. 2005; Wilusz and Wilusz 2005).
In eukaryotes, there are two major pathways of mRNA decay that are both initiated by the shortening of the poly(A) tail (Garneau et al. 2007). In the 3′-to-5′ decay pathway, deadenylated mRNAs are degraded by a large complex of 3′–5′ exonucleases, the exosome. Conversely, in the 5′-to-3′ decay pathway, deadenylation triggers decapping and subsequent 5′–3′ exonucleolytic decay by Xrn1. In Saccharomyces cerevisiae it has been shown that LSm1-7 complexes act as activators of decapping in the 5′–3′ decay pathway (Bouveret et al. 2000; Tharun et al. 2000) and as inhibitors of the 3′–5′ exonucleolytic decay (He and Parker 2001; Tharun et al. 2005). In mammalian cells, LSm1-7 complexes are also required for decapping of mRNAs. In particular, they have been shown to participate in the decay of mRNAs containing AU-rich elements (ARE) and histone mRNAs, whose 3′ untranslated regions (UTRs) end in a conserved stem–loop instead of a poly(A) tail (Mukherjee et al. 2002; Stoecklin et al. 2006; Mullen and Marzluff 2008). The mechanism by which the LSm1-7 complexes promote mRNA decapping and their mode of interaction with the target RNA remain elusive. However, several in vivo and in vitro binding studies in yeast suggest a direct interaction with deadenylated 3′ UTRs (He and Parker 2001; Tharun and Parker 2001; Tharun et al. 2005; Chowdhury et al. 2007).
Viruses are obligatory intracellular parasites that depend on the host machinery to multiply. As such, they are useful tools to provide functional insights into cellular regulatory pathways. We have previously observed that the LSm1-7 complex plays a fundamental role in the replication of positive-strand RNA [(+)RNA] viruses (Diez et al. 2000; Noueiry et al. 2003; Mas et al. 2006). This viral group includes serious plant, animal, and human pathogens, such as the hepatitis C virus and the SARS coronavirus. Their genomes are single-stranded RNA molecules that are replicated in the cytoplasm of the host. Early in infection, (+)RNA viral genomes perform two essential functions. They act as mRNAs for expression of viral replicases and as templates for replication. As these two functions are mutually exclusive, a key step in the replication of all (+)RNA viruses is the regulated exit of their genomic RNAs from the cellular translation machinery to a membrane-associated viral replication complex, a process referred to as recruitment. The molecular features underlying such transition are poorly understood.
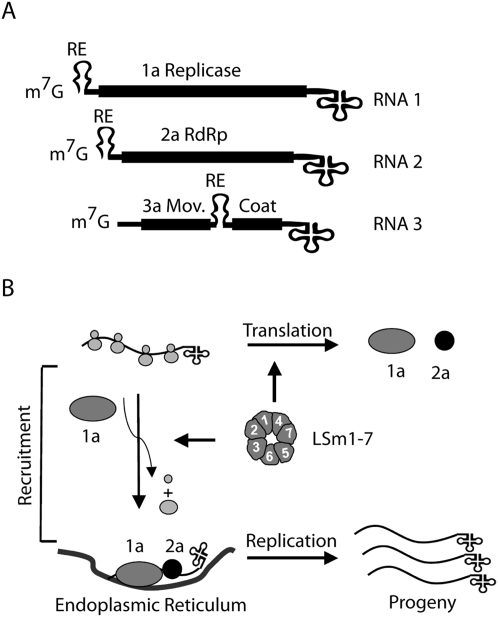
The propagation of the plant Brome mosaic virus in the yeast Saccharomyces cerevisiae has proven to be an excellent model system for studying common and fundamental steps of (+)RNA virus biology in a relatively simple genetic background (Alves-Rodrigues et al. 2006; Galão et al. 2007). The genome of BMV consists of three RNAs with 5′-terminal m7G-caps and a tRNA-like structure (TLS) located at the end of the 3′ UTRs (Fig. 1A; Ahlquist 1992). The cis-acting signals in BMV RNA translation and replication have been extensively characterized (for review, see Noueiry and Ahlquist 2003). In brief, both the 5′ and 3′ UTRs of all BMV genomic RNAs contain partly overlapping sequences that control translation and initiation of negative-strand (in the 3′ UTR) or positive-strand (in the 5′ UTR) RNA synthesis. Moreover, an element, the recruitment element (RE), at the 5′ terminal ends of RNA1 and RNA2 and in the intergenic region (IR) of RNA3 is necessary and sufficient for proper selection and recruitment of the viral RNAs. RNA1 and RNA2 encode the helicase 1a, and the polymerase 2a, respectively. The only BMV protein required for the recruitment of the BMV genome is 1a (Fig. 1B; Janda and Ahlquist 1998). RNA3 encodes the movement protein 3a and, through a subgenomic RNA generated during replication, the coat protein. Both proteins are required for systemic infection of plants, but not for viral replication.
FIGURE 1.
The BMV genome and the initial BMV replication steps. (A) Schematic diagram of the BMV tripartite genome showing the ORFs (solid black boxes) and the untranslated regions (UTRs) (single lines). The three BMV RNAs are capped (m7G) and end in a tRNA-like structure (cloverleaf structure). The location of the recruitment element (RE) is shown for the three RNAs. (B) After translation of the viral proteins, the 1a helicase recognizes the RE sequence to specifically recruit the viral genomes from the cellular translation machinery to viral-induced invaginations in the endoplasmic reticulum where replication occurs. The LSm1-7 complex plays a key role in the regulation of these processes.
With the BMV/yeast system we previously reported that LSm1-7 complexes were required for both the translation and the recruitment of the BMV genome (Fig. 1; Diez et al. 2000; Noueiry et al. 2003; Mas et al. 2006). Yet how these complexes act mechanistically on the BMV genome is unknown. With the availability of recombinant LSm1-7 complexes, we were now able to address this issue experimentally. Using an in vitro binding approach we provide evidence that LSm1-7 directly interacts with BMV RNAs via two specific RNA-targeting elements, namely, a tRNA-like structure located in the 3′ UTR and two internal A-rich single-stranded regions. Functional in vivo analysis showed that these LSm1-7 RNA-target sequences act as regulators of translation and recruitment of the BMV genome, suggesting that the intrinsic RNA-binding characteristics of the LSm1-7 complexes determine their function. Moreover, the newly defined LSm1-7/BMV RNA interactions resemble those found for the Hfq complex, the bacterial counterpart of the LSm proteins. Hfq was first identified as a factor required for the replication of the (+)RNA bacteriophage Qβ. Together our results have important implications for the understanding of the function of LSm1-7 complexes and their conservation through evolution.
RESULTS
The dependence of RNA3 translation on LSm1-7 complexes is functionally linked to the 3′ UTR of the BMV genome
From the three BMV genomic RNAs, it is RNA3 whose regulation of the translation/recruitment steps is characterized best. With the BMV/yeast system we have shown previously that the substitution of the 3′ UTR of BMV RNA3 by a poly(A) tail suppressed the requirement of LSm1-7 complexes for RNA3 recruitment. This indicates a functional link between the presence of the natural 3′ UTR and the LSm1-7 complex (Diez et al. 2000; Mas et al. 2006). In contrast, the dependence of the Lsm1-7 complex on RNA2 translation was not solely linked to the 3′ UTR but required both 5′ and 3′ UTR and the 2a polymerase open reading frame (ORF) (Noueiry et al. 2003). However, parallel experiments with RNA3 resulted in conflicting results. To examine if, as in RNA3 recruitment, the dependence on LSm1-7 complex for RNA3 translation was exclusively linked to the 3′ UTR, we transformed wild-type (WT) yeast and the deletion strain lsm1Δ with plasmids expressing the BMV RNA3 or a RNA3 derivative, in which the 3′ UTR was substituted by the yeast ADH1 polyadenylation signal (Fig. 2A). Since neither the helicase 1a nor the polymerase 2a was expressed, these RNAs will be translated but not recruited and replicated. We focused on lsm1Δ because LSm1 is the subunit that defines the function of the LSm1-7 complex. After induction of expression, total protein and RNA were extracted and analyzed. RNA3 translation efficiency was obtained by normalizing the 3a protein levels detected via Western blot by the respective RNA3 levels detected via Northern blot (see the histogram in Fig. 2B). As expected, RNA3 translation was strongly inhibited in the lsm1Δ deletion strain relative to WT (Fig. 2B, lanes 1,3; Noueiry et al. 2003). However, upon replacement of the 3′ UTR by a poly(A) tail, similar RNA3 translation levels were observed in WT and lsm1Δ cells (Fig. 2B, lanes 2,4). In WT yeast, RNA3 was more efficiently translated than its RNA3p(A) derivative (Fig. 2B, lanes 1,2), highlighting the relevance of the natural BMV 3′ UTR in translation. Thus, as previously observed for RNA3 recruitment, the 3′ UTR of BMV RNA3 confers dependence on LSm1-7 for RNA3 translation.
FIGURE 2.
A 3′poly(A) tail suppresses the requirement of LSm1-7 complexes for RNA3 translation. (A) Schematic representation of the RNA3 and the RNA3-poly(A) derivative [RNA3p(A)] in which the 3′ UTR was replaced by the yeast ADH1 polyadenylation site. (B) WT and lsm1Δ yeast strains were transformed with a plasmid harboring RNA3 or RNA3p(A). (Upper panels) Western blot analysis of the 3a protein expression. As a control for equal loading of total protein, expression of the phosphoglycerate kinase protein (PGK) was also analyzed. (Lower panels) Northern blot analysis of RNA3 accumulation. Detection of the 18S ribosomal RNA was used to assure equal loading and sample quality. Histograms show the average and standard error of the mean of the accumulation of 3a protein relative to the amount of RNA3 for at least three independent colonies. The average value obtained for RNA3 in WT cells was set to 100.
Reconstituted LSm1-7 complexes bind specifically to the 3′ UTR of BMV RNAs
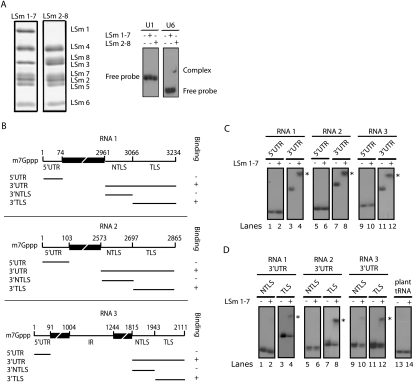
LSm1-7 complexes are believed to influence the metabolism of cellular mRNAs by interacting directly with their 3′ UTR (Chowdhury et al. 2007). Hence, we considered the possibility that LSm1-7 complexes regulate translation and replication of BMV through a direct interaction with the viral genome. To test this possibility, we carried out in vitro RNA-binding experiments with reconstituted recombinant human LSm1-7 complexes (Zaric et al. 2005). We focused our binding studies on the 3′ and 5′ UTRs of the three genomic BMV RNAs because they contain conserved and structured sequences that are involved in the regulation of BMV RNA translation and replication. The reconstituted LSm1-7 complexes were incubated with the respective radiolabeled RNA molecules, and binding was analyzed by gel-shift assays in native polyacrylamide gels. First, we determined the specificity of the LSm complexes used in this study (Fig. 3A). The RNA-binding specificity of LSm1-7 complexes is not known, whereas binding of LSm2-8 complexes to U6 snRNA has been documented. Thus, we used the binding pattern of LSm2-8 as a control to evaluate the ability of the reconstituted complexes to discriminate between cognate and noncognate RNAs. Consistently, we observed specific binding of the LSm2-8 to U6, but not to U1 snRNA, while the LSm1-7 complex did not bind to any of the two (Fig. 3A). We next tested the binding of the LSm1-7 complex to radiolabeled BMV RNA derivatives using identical conditions as determined for the LSm2-8 complex (Fig. 3B–D). Interestingly, a robust binding to the 3′ UTR of all three genomic RNAs was observed. The 5′ UTR, in contrast, failed to bind under the same conditions irrespective of whether they were m7G-capped or uncapped (Fig. 3C; data not shown). Thus, these experiments reveal a direct binding of the LSm1-7 complexes to the 3′ UTR of the three BMV RNAs, i.e., those sequences that our in vivo data have linked to the dependence on LSm1-7 for BMV RNA translation and recruitment.
FIGURE 3.
Binding of LSm1-7 complexes to the 3′ UTRs of the three BMV RNAs depends on a tRNA-like structure. (A) Coomassie blue-stained SDS-polyacrylamide gel of purified reconstituted LSm1-7 and LSm2-8 complexes (left). Binding specificity of LSm1-7 and LSm2-8 complexes were determined by gel-shift analysis using recombinant rings and radiolabeled gel-purified U1 and U6 snRNAs (right). After complex formation, samples were loaded on a nondenaturating polyacrylamide gel and visualized by autoradiography. (B) Schematic representation of generated transcripts. The numeration refers to the position of the corresponding nucleotides in the complete BMV RNA genomes. (C,D) Gel-shift assays were performed as described above. LSm1-7 complexes were incubated with radiolabeled transcribed RNAs corresponding to the 5′ and 3′ UTRs (C) or with the 3′ UTR TLS and NTLS sequences of the three genomic BMV RNAs (D). The plant tRNA corresponds to the Tyr-tRNA of Nicotania bentamiana. Asterisks indicate the position of the gel-shifted RNAs.
LSm1-7 complexes bind to the tRNA-like structure within the 3′ UTR of BMV RNAs
All 3′ UTRs of the three BMV RNAs contain two sequence elements, one with a nondefined structure, referred to as a non-tRNA-like structure (NTLS), and the other with a highly conserved TLS. The latter is 98% identical among the three RNAs and fulfills crucial functions in the BMV life cycle. It is required for efficient translation, initiation of replication, and encapsidation of the BMV RNA genome (Choi and Rao 2003; Noueiry and Ahlquist 2003; Barends et al. 2004). Gel-shift assays were conducted to test which of the sequence elements interacts with the LSm1-7 complex. As shown in Figure 3D, lanes 4, 8, and 12, interaction of the LSm1-7 complex with the TLS of all three RNAs was observed. This interaction was highly specific as no binding was observed to a structurally related plant tRNA (tRNA-tyr) (Fig. 3D, lane 14; Zerfass and Beier 1992). In contrast, the NTLS regions either failed to interact completely (RNA1 and RNA2) or bound LSm1-7 only very weakly (RNA3) (Fig. 3D, lanes 2,6,10).
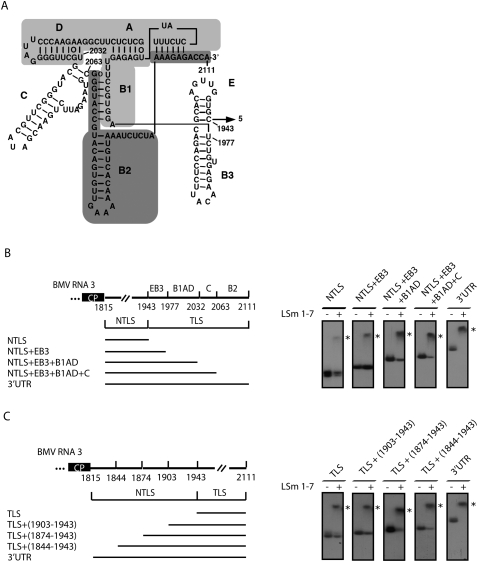
Binding of the LSm1-7 complex to the TLS sequence was weaker as compared with the complete 3′ UTR, suggesting that additional sequence elements contribute to this interaction. To determine the minimal 3′ UTR sequence required to obtain a complete band shift, we designed a set of RNA3 constructs starting either from the NTLS sequence and inserting consecutive domains from the TLS region or from the TLS sequence and inserting equivalent segments of the NTLS region (Fig. 4). In both cases the addition of consecutive sequence elements gradually increased LSm1-7 binding capability. Similar results were obtained under conditions of saturating LSm1-7 concentrations (i.e., ≥20 pmol LSm1-7 complex) (Fig. 4B,C) and in titration experiments (Supplemental Fig. 1). Binding of LSm proteins to all RNA fragments tested was highly reproducible. However, slightly variable results were obtained with RNA3 NTLS+EB3+B1AD+C. Depending on the components used, the binding efficiency varied between 90% and 100%.
FIGURE 4.
The complete 3′ UTR is required for optimal interaction with the LSm1-7 complexes. (A) Secondary structure of the TLS from the 3′ UTR of BMV RNA3. Numbers refer to corresponding nucleotide positions in the BMV RNA3 genome. The various stem–loop structures are indicated (A–E). To facilitate their identification some of them are highlighted with different shadowing (data adapted from Barends et al. 2004). (B) RNA substrates corresponding to the NTLS region of the RNA3 3′ UTR and to the same sequence plus consecutive TLS domains or (C) RNA substrates corresponding to the TLS region of the RNA3 3′ UTR, and to the same sequence plus additional stretches from the NTLS region were incubated with purified LSm1-7 complexes and analyzed by gel-shift assays as described in Figure 3.
Thus, even though the TLS alone is sufficient to bind the LSm1-7 complex, the flanking sequences strongly increase its affinity. The need for this 5′ flanking sequence might be sequence specific, or alternatively, length related. As LSm1-7 and TLS are both required for BMV RNA translation and replication, our data suggest that the LSm1-7 effect on the BMV genome is linked to its ability to bind to the TLS region.
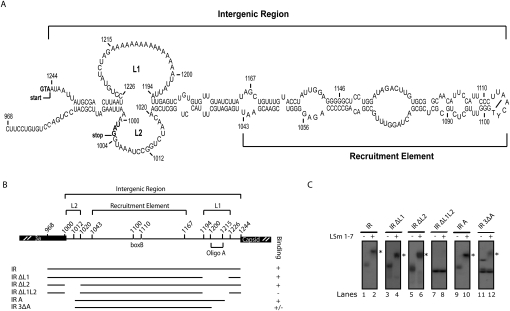
LSm1-7 complexes bind to the intergenic region of BMV RNA3
BMV RNA3 contains an IR between its ORFs, which includes the RE, and probably other unidentified regulatory sequences. The RE is required for 1a-mediated recruitment of the viral RNA to replication complexes (Sullivan and Ahlquist 1999). Chemical mapping together with computer predictions and phylogenetic studies have shown that the IR folds into a long stem–loop structure (Fig. 5A; Baumstark and Ahlquist 2001). Interestingly, the LSm1-7 complex also interacts with this important regulatory region, as determined by gel-shift assays (Fig. 5B,C, lane 2). Two sequences, termed loops L1 and L2, were identified to be essential for LSm1-7 binding. L1 contains an A-rich region with a 16-nucleotide oligo(A) stretch followed by a UUAUUA sequence, while L2 contains a shorter A-rich region (Fig. 5A). Both of them are only present in RNA3 and no regulatory function has been ascribed to them yet. In silico RNA folding analysis suggested that removal of the loops had no effect on the overall structure of the IR. LSm1-7 binding was not affected by removal of either L1 or L2; however, the simultaneous deletion of both entire loops totally abolished binding under conditions of saturating LSm1-7 concentrations (i.e., ≥20 pmol Lsm1-7 complex) (Fig. 5C, cf. lanes 4 and 6 and 8). Deletion of the oligo(A) stretch in L1, plus the AU stretch in L2, strongly inhibited binding, indicating the importance of these sequences for the interaction with LSm1-7 (Fig. 5C, lane 12). The two bands observed for the IR3ΔA RNA reflect two conformations under native conditions, and only the slower migrating one is able to interact with LSm1-7. The RE element did not interact with LSm1-7 complexes (Fig. 5C, lane 8). This is consistent with the finding that neither the 5′ UTR of RNA1 nor the 5′ UTR of RNA2, which both contain RE elements, interacted with LSm1-7 (Fig. 3C, lanes 2,6). Taken together, the results show that the LSm1-7 rings are able to bind A-rich sequences located internally in RNA3.
FIGURE 5.
Binding of LSm1-7 complexes to the intergenic region (IR) of RNA3 depends on the A-rich loops L1 and L2. (A) Secondary structure of the IR of BMV RNA3. Numbers refer to the corresponding nucleotide positions in the BMV RNA3 genome. The recruitment element and the loops L1 and L2 are shown. In bold are indicated the stop codon of the 3a ORF and the start codon of the coat protein that limit the IR. (B) Schematic diagram of the generated RNA transcripts used to map the sequences in the IR that mediate the interaction with the LSm1-7 complexes. (C) The corresponding radiolabeled RNAs were used to perform gel-shift assays with LSm1-7 complexes as described in Figure 3.
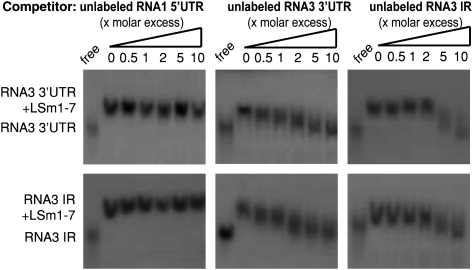
LSm1-7 complexes specifically bind to the 3′ UTRs and the intergenic region through the same RNA binding site
To determine the specificity of the observed LSm1-7 binding interactions, we performed competition assays. The interaction of LSm1-7 with the 3′ UTRs and the IRs was specific after addition of excess unlabeled 3′ UTRs and IRs, respectively, resulted in binding competition, while addition of excess nonbinding unlabeled 5′ UTRs did not (Fig. 6).
FIGURE 6.
LSm1-7 complexes bind to the 3′ UTR and the IR specifically and through a common binding site. Radiolabeled RNA3 3′ UTR (upper panels) and RNA3 IR (lower panels) were incubated with reconstituted LSm1-7 complexes in the presence of increasing amounts of unlabeled RNA1 5′ UTR, RNA3 3′ UTR, or RNA3 IR transcripts as competitors.
To explore whether the two determined RNA-target sequences interact with the same or distinct RNA-binding sites in the LSm1-7 complex, we perform cross-competition assays. Radiolabeled RNA substrates derived from the RNA3 3′ UTR or IR were incubated with LSm1-7 rings in the presence of increasing amounts of an unlabeled IR or 3′ UTR, respectively (Fig. 6). In both cases the binding was efficiently competed and this competition was comparable to the one obtained with the self-unlabeled RNA. These results strongly suggest that both the 3′ UTR and the IR interact with the same RNA-binding site of the LSm1-7 complex.
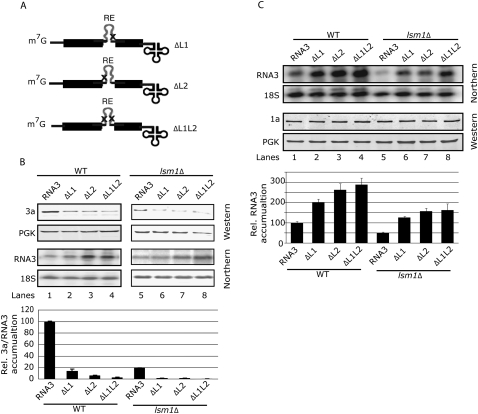
Loops L1 and L2 in the intergenic region of RNA3 regulate translation and recruitment to replication in vivo
The putative function of loops L1 and L2 in the IR on the regulation of the BMV RNA3 translation and recruitment has not been explored. With this aim, we generated yeast plasmids that express BMV RNA3 derivatives in which L1, L2, or both, were deleted (ΔL1, ΔL2, and ΔL1L2, respectively) (see Fig. 7A). To study putative effects on translation, WT and lsm1Δ yeast strains were transformed with the corresponding plasmids and the levels of 3a protein and RNA3 derivatives were analyzed. If the defined in vitro binding of LSm1-7 to these loops has a functional meaning, one would predict that the lack of the LSm1-7 complex would have a comparable effect as the deletion of the loops. Indeed, similar to the effect observed for the lsm1Δ strain in RNA3 translation (Fig. 7B, cf. lanes 1 and 5), the single deletion of L1, L2, or both, simultaneously in RNA3 resulted in a strong inhibition of RNA3 translation in WT yeast (Fig. 7B, lanes 2–4). In the lsm1Δ strain, deletion of the loops resulted in an even higher inhibition, suggesting that additional regulatory factors might be binding to the loops (Fig. 7B, lanes 6–8). Therefore, the obtained results indicate that L1 and L2 loops are translation regulatory sites.
FIGURE 7.
Deletion of loops L1 and L2 in the IR inhibits translation and favors recruitment of BMV RNA3. (A) Generated BMV RNA3 derivatives in which the internal loop1 (ΔL1), loop2 (ΔL2), or both (ΔL1L2), were deleted. (B) WT and lsm1Δ strains were transformed with a plasmid harboring the RNA3 or the RNA3 derivatives (ΔL1, ΔL2, or ΔL1L2) and the accumulations of the 3a protein and RNA3 were analyzed by Western and Northern blotting, respectively, as in Figure 2. Histograms show the average and standard error of the mean of the accumulation of 3a protein relative to the amount of RNA3 from at least three independent colonies. The average 3a/RNA3 value in WT yeast was set to 100. (C) WT and lsm1Δ yeast strains were transformed with a plasmid expressing the 1a protein plus a plasmid transcribing the WT RNA3 or the RNA3 derivatives (ΔL1, ΔL2, or ΔL1L2). RNA3 accumulation was analyzed by Northern blotting and 1a protein expression by Western blotting. Histograms represent relative accumulation of RNA3 and the RNA3 derivatives in WT and lsm1Δ cells of three independent colonies. The average accumulation of RNA3 in WT yeast was set to 100.
Next, we analyzed the effect of the deletion of the internal loops on RNA3 recruitment to the sites of replication. This 1a-dependent recruitment process protects the viral RNAs from degradation, and thus dramatically increases their stability and subsequent accumulation (Janda and Ahlquist 1993; Sullivan and Ahlquist 1999; Schwartz et al. 2002; Mas et al. 2006). Accordingly, when RNA3 and 1a protein are co-expressed, differences in RNA3 accumulation reflects differences in recruitment. WT and lsm1Δ yeast strains were transformed with plasmids transcribing WT RNA3 or the corresponding deletion derivatives plus or minus another plasmid expressing the BMV 1a protein. In the absence of 1a, a similar steady-state level of RNA3 was obtained for all the constructs in WT and lsm1Δ yeast (data not shown). When 1a was co-expressed, the recruitment of RNA3 was inhibited in lsm1Δ when compared with WT yeast (Fig. 7C, upper panel, histogram, lanes 1,5; Diez et al. 2000). However, deletion of the loops resulted in a two- to threefold increase of recruitment when compared with WT RNA3 in both WT and lsm1Δ yeast strains. (Fig. 7C, cf. histograms corresponding to lane 1 and lanes 2–4 and to lane 5 and lanes 6–8). These effects were not related to differences in 1a expression levels (Fig. 7C, lower panel). Therefore, deletion of loops L1 and L2 enhance recruitment independently of the presence of LSm1-7 complexes. Taken together, our experiments indicate that L1 and L2, the LSm1-7 recognition signals in the IR, play an essential role in the regulation of BMV RNA3 translation, as well as a secondary role in its recruitment.
DISCUSSION
The LSm1-7 complex, which functions in cellular mRNA decay, specifically binds to BMV genomes, and instead of decay, promotes their translation and replication. By using in vitro binding assays with reconstituted LSm1-7 complexes and BMV RNA sequences, we have identified two specific LSm1-7 interaction sites in the BMV genome. Importantly, in vivo analysis showed that these sites are required for the regulation of translation and recruitment of BMV RNA. These results are consistent with the role of LSm1-7 complexes in the BMV life cycle and suggest that the intrinsic RNA-binding characteristics of these complexes determine their function. Our data open new perspectives in the mode of action of these versatile complexes in viral and cellular RNA biology.
Reconstituted recombinant human LSm1-7 complexes specifically recognized two translation/replication regulatory signals in the BMV genome, the TLS region in the 3′ UTR of all three genomic RNAs and two internal single-stranded A-rich sequences (L1 and L2) in the IR of RNA3 that are located ∼1 kilobases (kb) upstream of the 3′ end. These are the first defined LSm1-7 binding sites in viral genomes and represent novel LSm1-7 interaction sites. Only two previous studies have addressed the binding specificity of LSm1-7 to RNA sequences other than homopolymers. Binding assays with purified yeast LSm1-7/Pat1 complexes have shown a preferential binding to oligoadenylated versus polyadenylated RNAs and a direct interaction near or at the 3′ ends that contain oligo(U) stretches (Chowdhury et al. 2007). However, since the protein Pat1 was present in the complex, its contribution to the identified RNA-binding properties remains unclear. The other study was performed with reconstituted human LSm1-7 rings and showed their direct interaction and subsequent stabilization of reporter RNAs containing 5′ poly(A) tracts. Although these studies were performed with nonnatural RNA sequences, a function of LSm1-7 in the stabilization of poxviridae messenger RNA, which contain 5′ poly(A) tracts, was suggested via a 5′–3′ circularization by binding of the LSm1-7 complexes to the 3′ and 5′ends (Bergman et al. 2007).
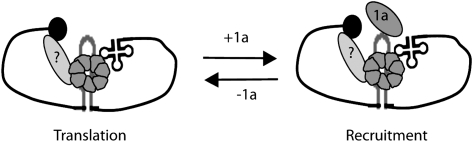
For BMV RNA3, substitution of the TLS region by a poly(A) tail suppressed the requirement of LSm1-7 complexes for RNA3 translation and recruitment. Since poly(A) tails in cellular mRNAs mediate 5′–3′ circularization via the binding of the poly(A) binding protein, the LSm1-7/TLS binding may establish such 5′–3′ interactions in BMV RNAs. Proteins containing Sm folds act as chaperones facilitating a variety of RNA–RNA and RNA–protein interactions (Wilusz and Wilusz 2005). Accordingly, the binding of LSm1-7 to TLS and the L1 and L2 internal loops might facilitate rearrangements in the viral ribonucleoprotein (RNP) structure required for the 5′–3′ interaction. This circularization would then stabilize the RNA permitting both efficient translation and 1a-dependent recruitment via recognition of the RE element (Fig. 8). The fact that the lack of LSm1-7 complexes inhibits both BMV RNA3 translation and recruitment suggests that binding of LSm1-7 is required for both processes. Since translation and recruitment are two antagonist processes, changes such as the presence of 1a protein expression or the absence of some of the Lsm1-7 interaction sites in the viral genomes would result in favoring one over the other. In line with this, we observed that deletion of the L1 and L2 loops inhibits translation but favors recruitment. This may be an indirect effect due to the absence of the competing translation function. Alternatively, deletion of L1 and L2 could affect the RNP reorganization in such a way that the RE signals would be more accessible to the 1a protein. On the other hand, the observed effects also may be due to the binding to loops L1 and L2 of additional host factors not yet identified.
FIGURE 8.
Model of LSm1-7 function on BMV RNA3. Binding of LSm1-7 complexes to the tRNA-like structure in the 3′ UTR and the internal loops in the IR would regulate translation and recruitment to replication of BMV RNA3. Given that the LSm1-7 ring does not bind simultaneously to both RNA sequences, the interaction with two rings might be considered.
Deletion of both L1 and L2 loops in the IR of the RNA3 was required to abolish LSm1-7 binding under saturating LSm1-7 concentrations (Fig. 5). However, deletion of either L1 or L2 inhibited translation and enhanced recruitment (Fig. 7). These findings are corroborated by titration band-shift experiments using subsaturating concentrations of LSm1-7 complexes (Supplemental Fig. 2). Under these conditions we find that LSm1-7 complexes bind both IRΔL1 and IRΔL2 with lower affinity than the WT IR sequence. Thus, in the in vivo setting, where the local concentration of viral RNA and Lsm proteins are low, the presence of both L1 and L2 sites are important to promote efficient Lsm1-7 binding. Hence, under in vivo conditions, a suboptimal binding site would not suffice to allow LSm binding and function.
The newly defined RNA-target sites of LSm1-7 rings resemble those found for Hfq, the bacterial counterpart of Sm/LSm proteins (Wilusz and Wilusz 2005; Lee and Feig 2008). Hfq forms a homohexameric ring that functions in degradation and also in stabilization and translation of cellular RNAs by facilitating RNA–RNA and RNA–protein interactions (Wilusz and Wilusz 2005). Interestingly, Hfq was first identified as a factor required for the replication of the (+)RNA bacteriophage Qß (Franze de Fernandez et al. 1968). By promoting changes in the 3′ secondary structure of the Qß RNA genome, it allows the association of the viral polymerase with the viral genome and the subsequent initiation of replication (Schuppli et al. 1997). In contrast to the LSm1-7 complex, a crystal structure of the Hfq complex is available (Schumacher et al. 2002). Two faces of the ring have been shown to bind RNA substrates with different specificities. The distal face binds to poly(A) tails, while the proximal face binds to internal AU-rich regions located close to stem–loop structures in noncoding RNAs (ncRNAs), or to tRNAs (Mikulecky et al. 2004; Lee and Feig 2008). As for Hfq, LSm1-7 complexes bound to internal regions, in this case A-rich stretches, that flank a stem–loop RNA structure and to a TLS in the BMV 3′ UTR. In addition, our cross-competition binding analysis showed that both sequences bound through the same surface in the LSm1-7 complex (Fig. 6). Mapping of the minimal BMV 3′ UTR sequence necessary for optimal binding suggests that, as described for the Hfq/tRNA interaction, it is not a sequence motif per se, but a structural motif, that promotes binding (Fig. 4). The TLS is tyrosylated in vivo by host enzymes (Kohl and Hall 1974); however, the LSm1-7 complex did not bind the related tRNAtyr (Fig. 3). Thus, the observed LSm1-7/TLS interactions are not solely dependent on the structural mimicry of tRNA, but instead might involve additional residues or structures absent in canonical tRNAs. The fact that the described binding properties and functions of LSm1-7 complexes are similar to those observed for the bacterial Hfq suggests a strong conservation through evolution.
Viruses require the cellular machinery to multiply. In order to express their genomes and regulate the viral life cycle, a complex interplay between the cellular translation and degradation machinery is starting to emerge (Sokoloski et al. 2006). Our data indicate that BMV genomes usurp the LSm1-7 complex, which functions in cellular mRNA decay, to regulate their translation and replication. Given the common strategies of (+)RNA virus replication and that at least three viruses from this group, the bacterial phage Qß, the plant BMV, and the human HCV virus (Scheller et al. 2009), require this complex to regulate their replication, it is likely that LSm1-7 is broadly used within this viral group. The obtained results open new perspectives on the role and way of function of LSm1-7 complexes, not only on viral genomes, but possibly also on specific cellular RNAs. For example, LSm1 protein has been found to be overexpressed in several human cancers (Kelley et al. 2003; Fraser et al. 2005; Streicher et al. 2007). Recent findings show that the long ncRNA MALAT1, which is overexpressed in many human carcinomas, generates a small ncRNA that adopts a tRNA-like structure and localizes to the cytoplasm (Wilusz et al. 2008). The putative role of LSm1-7 complexes in the stability and function of such ncRNAs and other cellular RNAs with special features remains to be explored further and might uncover new functionalities of this versatile complex.
MATERIALS AND METHODS
Yeast cells
Standard yeast genetics and media were used (Guthrie and Fink 1991). For recruitment experiments, WT Saccharomyces cerevisiae strain YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3-Δ200 leu2-Δ1) and the derivative lsm1Δ deletion strain were used (Diez et al. 2000). For translation experiments, WT BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and the derivative lsm1Δ deletion strain were used (Winzeler et al. 1999), and purchased from EUROSCARF.
Plasmids
Plasmids for in vivo experiments
For recruitment assays, plasmid pJDSal1 was used to transcribe WT BMV RNA3 by using a CUP1 promoter (Diez et al. 2000). Plasmids pIR1, pIR2, and pIR3 were derived from pJDSal1 by using the QuickChange XL II site-directed mutagenesis kit (Stratagene) to delete the loop L1 (nucleotides 1194–1226), loop L2 (nucleotides 1000–1020), or both loops, simultaneously. An additional mutation A998T was inserted into pIR2 and pIR3 to introduce a stop codon at the end of the 3a ORF. The primers used are listed in Supplemental Table 1. Plasmids pIR1, pIR2, and pIR3 were then used to transcribe the BMV RNA3 derivatives ΔL1, ΔL2, and ΔL1L2, respectively. To achieve similar 1a expression levels in WT and Δlsm1 yeast, the 1a helicase was expressed from either pB1CT19 or pB1YT3H under the ADH1 or GAL1 promoter, respectively, as described previously (Mas et al. 2006). In these plasmids the 5′ UTR and the 3′ UTR of the BMV RNA1 were replaced by cellular leader sequences and the ADH1-polyadenylation signals (Janda and Ahlquist 1993; Ahola et al. 2000;). All the generated plasmids used to test effects on 3a protein translation were derived from pB3RQ39-URA, a yeast plasmid transcribing WT BMV RNA3 under the GAL1 promoter (Beckham et al. 2007). pB3RQ39JDURA was constructed by cloning the SalI/EcoRI fragment from pB3RQ39JD (Diez et al. 2000) into pB3RQ39-URA and used to transcribe a BMV RNA3 derivative [RNA3p(A)] with the yeast ADH1 polyadenylation site substituting the original viral 3′ end. Plasmids pIR7, pIR8, and pIR9 transcribe from the GAL1 promoter the BMV RNA3 derivatives ΔL1, ΔL2, and ΔL1L2, respectively. To construct these plasmids, pIR1, pIR2, and pIR3 were ClaI/StuI digested and the resulting ∼1.2-kb fragments cloned into pB3RQ39 (Ishikawa et al. 1997), Following digestion with EcoRI/MscI resulted in ∼2-kb fragments that were cloned into the same sites in pB3RQ39-URA.
Constructs used in gel-shift assays
Unless otherwise mentioned, constructs were generated by PCR amplification of the fragments of interest from BMV RNA1, RNA2, and RNA3 genomes cloned, respectively, in the plasmids pB1tp3, pB2tp5, and pB3tp8 (Janda et al. 1987; French and Ahlquist 1988). Forward primers carried a T7 promoter sequence at their 5′ end. Amplified fragments were cloned into SmaI digested pUC18 (New England Biolabs) or pGEM-T vectors (Promega). Fragments IR A and IR 3ΔA were synthetically produced by Geneart and cloned into plasmid pPCR-Script-ampR (Stratagene) using KpnI and SacI restriction sites. Finally, constructs used to generate the transcripts IRΔL1, IRΔL2, and IRΔL12, were obtained by PCR amplification of the relevant parts of pIR1, pIR2, and pIR3, respectively, and cloned into pGEM-T vector (Promega). Standard procedures were used for DNA manipulations and all generated constructs were verified by sequencing. Sequences of the primers used for each construct are given in Supplemental Table 2. BMV sequence numbering refers to the position of the corresponding nucleotides in the complete BMV RNA genomes (GenBank acession numbers NC_002026, NC_002027, and NC_002028). Plasmids used for expression and purification of individual human LSm proteins, and for transcription of the Xenopus U6 and U1 snRNA and the tRNA-tyr from Nicotania have been described previously (Hamm et al. 1990; Zerfass and Beier 1992; Zaric et al. 2005).
Recruitment and translation assays
To analyze recruitment, yeast cells were transformed with the corresponding plasmids and grown in 0.5 mM CuSO4, 2% dextrose minimal media at 30°C until the midlog phase, and then induced in 2% galactose for 72 h. Total RNA from yeast cells was isolated by the hot-phenol method and analyzed by Northern blot as previously described (Janda and Ahlquist 1993). Specific probes to detect positive-strand BMV RNA3 and 18S rRNA has been described previously (Ishikawa et al. 1997; Alves-Rodrigues et al. 2007) and were generated using the MAXIscript In Vitro Transcription Kit (Ambion). Northern blots were imaged on a Typhoon 8600 Instrument (Amersham Biosciences) and band intensities were quantified using the ImageQuant Software (Molecular Dynamics).
To evaluate BMV 3a protein translation, yeast cells were transformed with the corresponding plasmids and grown in 2% galactose at 30°C until the midlog phase. Total protein was extracted from yeast cells as previously described (Ishikawa et al. 1997), separated on 10% SDS-polyacrylamide gels, and immunoblotted. Antibodies against BMV 1a (Restrepo-Hartwig and Ahlquist 1999), BMV 3a (Noueiry et al. 2003), and phophoglycerate kinase (PGK) (Molecular Probes) were used for Western blot. Protein detection and quantification were made using the infrared imaging system Odyssey (LI-COR Biosciences). Total RNA was extracted and RNA3 accumulation was measured as explained above.
LSm recombinant proteins and RNA gel-shift assays
Expression and purification of individual LSm proteins and subcomplexes, as well as the reconstitution and purification of LSm1-7 and LSm2-8 rings, were performed essentially as described previously (Zaric et al. 2005), with the exception of LSm8, which was expressed singly and purified as described for LSm1. Transcripts used to perform gel-shift assays were transcribed in vitro and labeled with [α-32P]UTP. In a typical binding experiment, 50 fmol of labeled RNA were incubated with 20 pmol of reconstituted LSm complexes in a buffer containing 20 mM HEPES-NaOH at pH 7.5, 200 mM NaCl, 2 mM MgCl2, 0.1 U/μL RNasin, and 0.1 μg/μL yeast tRNA in a 5 μL assay at 30°C for 1 h. Samples were loaded on previously prerun 5% native polyacrylamide gels, and run at 4°C for 2 h, 30 mA as described (Chari et al. 2008). Gels were autoradiographed at −80°C on maximum sensivity Kodak Biomax Films (Sigma-Aldrich). For gel-shift assays that included RNA competition, the indicated amount of RNA competitor was added to the reactions.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank P. Ahlquist and F. Gebauer for helpful discussions. This work was supported by grants from the Spanish Ministerio de Educación y Ciencia (BFU2007-66933/BMC) and the German Research Foundation (DFG-FOR855). I.A.-R. and D.L. were supported by Fundação para a Ciência e Tecnología (SARH/BD/9630/2002; SFRH/BD/37047/2007).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1712910.
NOTE ADDED IN PROOF
While this paper was under revision a direct binding of LSm1-7 complexes to translation and replication regulatory sequences in the HCV genome was shown by Scheller et al. (2009).
REFERENCES
- Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- Ahola T, den Boon JA, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J Virol. 2000;74:8803–8811. doi: 10.1128/jvi.74.19.8803-8811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Rodrigues I, Galao RP, Meyerhans A, Diez J. Saccharomyces cerevisiae: A useful model host to study fundamental biology of viral replication. Virus Res. 2006;120:49–56. doi: 10.1016/j.virusres.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Rodrigues I, Mas A, Diez J. Xenopus Xp54 and human RCK/p54 helicases functionally replace yeast Dhh1p in brome mosaic virus RNA replication. J Virol. 2007;81:4378–4380. doi: 10.1128/JVI.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Rudinger-Thirion J, Florentz C, Giege R, Pleij CW, Kraal B. tRNA-like structure regulates translation of brome mosaic virus RNA. J Virol. 2004;78:4003–4010. doi: 10.1128/JVI.78.8.4003-4010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark T, Ahlquist P. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem–loop and is modified in vivo. RNA. 2001;7:1652–1670. [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Light HR, Nissan TA, Ahlquist P, Parker R, Noueiry A. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- Bergman N, Moraes KC, Anderson JR, Zaric B, Kambach C, Schneider RJ, Wilusz CJ, Wilusz J. Lsm proteins bind and stabilize RNAs containing 5′ poly(A) tracts. Nat Struct Mol Biol. 2007;14:824–831. doi: 10.1038/nsmb1287. [DOI] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhäger M, Neuenkirchen N, Sander B, Engelbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Choi YG, Rao AL. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Watson PM, Fraig MM, Kelley JR, Nelson PS, Boylan AM, Cole DJ, Watson DK. CaSm-mediated cellular transformation is associated with altered gene expression and messenger RNA stability. Cancer Res. 2005;65:6228–6236. doi: 10.1158/0008-5472.CAN-05-0650. [DOI] [PubMed] [Google Scholar]
- French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galão RP, Scheller N, Alves-Rodrigues I, Breinig T, Meyerhans A, Diez J. Saccharomyces cerevisiae: A versatile eukaryotic system in virology. Microb Cell Fact. 2007;6:32. doi: 10.1186/1475-2859-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to yeast genetics and molecular biology. Academic; San Diego, CA: 1991. [Google Scholar]
- Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Janda M, Krol MA, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- Kelley JR, Fraser MM, Hubbard JM, Watson DK, Cole DJ. CaSm antisense gene therapy: A novel approach for the treatment of pancreatic cancer. Anticancer Res. 2003;23:2007–2013. [PubMed] [Google Scholar]
- Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Kohl RJ, Hall TC. Aminoacylation of RNA from several viruses: Amino acid specificity and differential activity of plant, yeast and bacterial synthetases. J Gen Virol. 1974;25:257–261. doi: 10.1099/0022-1317-25-2-257. [DOI] [PubMed] [Google Scholar]
- Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Diez J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80:246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS, and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′-to-3′ and 3′ -to-5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- Noueiry AO, Diez J, Falk SP, Chen J, Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galão RP, Chari A, Gímenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppli D, Miranda G, Tsui HC, Winkler ME, Sogo JM, Weber H. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc Natl Acad Sci. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- Sokoloski KJ, Wilusz CJ, Wilusz J. Viruses: Overturning RNA turnover. RNA Biol. 2006;3:140–144. doi: 10.4161/rna.3.4.4076. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104–2114. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zaric B, Chami M, Remigy H, Engel A, Ballmer-Hofer K, Winkler FK, Kambach C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem. 2005;280:16066–16075. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]
- Zerfass K, Beier H. Pseudouridine in the anticodon G psi A of plant cytoplasmic tRNATyr is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res. 1992;20:5911–5918. doi: 10.1093/nar/20.22.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]