Abstract
Viruses rely on the metabolic network of their cellular hosts to provide energy and building blocks for viral replication. We developed a flux measurement approach based on liquid chromatography–tandem mass spectrometry to quantify changes in metabolic activity induced by human cytomegalovirus (HCMV). This approach reliably elucidated fluxes in cultured mammalian cells by monitoring metabolome labeling kinetics after feeding cells 13C-labeled forms of glucose and glutamine. Infection with HCMV markedly upregulated flux through much of the central carbon metabolism, including glycolysis. Particularly notable increases occurred in flux through the tricarboxylic acid cycle and its efflux to the fatty acid biosynthesis pathway. Pharmacological inhibition of fatty acid biosynthesis suppressed the replication of both HCMV and influenza A, another enveloped virus. These results show that fatty acid synthesis is essential for the replication of two divergent enveloped viruses and that systems-level metabolic flux profiling can identify metabolic targets for antiviral therapy.
The capability of mass spectrometry and nuclear magnetic resonance spectroscopy to quantify numerous metabolites simultaneously has given rise to the systems-level examination of metabolites (metabolomics) and their fluxes (fluxomics)1–3. Initial efforts to apply metabolomics to investigate human disease have focused largely on analysis of biofluids in normal versus affected individuals4–7. Although promising, such analysis is complicated by variation between individuals8. Moreover, the complexity of metabolic processes in multicellular organisms renders comprehensive understanding of the underlying biochemistry difficult.
An alternative to studying biofluids of affected individuals involves using cellular models to probe disease-associated metabolic changes in a more controlled experimental setting. One class of human disease with useful cellular models is viral infection, as the core processes of viral replication are recapitulated reliably in cultured mammalian cells.
HCMV is a large, enveloped double-stranded DNA virus of the β-herpes family that latently infects a majority of adults. In healthy individuals, most infections are asymptomatic, although they may have long-term health consequences, such as increased risk of atherosclerosis9,10. More acutely, the virus is a major cause of morbidity and mortality in immunocompromised people11 and is also the major infectious cause of birth defects, most commonly causing hearing loss12.
HCMV replicates in a variety of cell types, including epithelial cells, endothelial cells, smooth muscle cells, macrophages and fibroblasts, which provide a convenient host cell for in vitro cultivation of the virus13. Infection of cultured fibroblasts with HCMV triggers transcription over the first 24 h of viral immediate-early and early genes, as well as modulation of host cell transcription14. This is followed by transcription of a wider set of viral genes, viral DNA replication and extensive viral protein synthesis over the subsequent 48 h, resulting eventually in the production and release of new virions15,16.
It has been known for two decades that HCMV infection in vitro is associated with increased uptake of glucose by infected fibroblasts, suggesting virus-induced upregulation of host cell metabolism17. Recently, we examined the levels of intracellular metabolites in HCMV-infected confluent fibroblasts. Intermediates involved in glycolysis, the tricarboxylic acid (TCA) cycle and pyrimidine nucleotide biosynthesis markedly increased in response to the infection. The extent of metabolite concentration changes far exceeded those found in the switch between the quiescent and growing states of uninfected fibroblasts18.
Although informative, data on metabolite concentrations are inherently incomplete. Elevations in the concentration of a metabolite may reflect either its increased production or decreased consumption. These alternatives lead to fundamentally different understanding of the underlying biology. Moreover, they have distinct practical implications. For example, inhibitors of enzymes catalyzing fluxes that are upregulated by viral infection merit consideration as antiviral agents.
For microbes growing on minimal media, fluxes can be measured based on steady-state labeling patterns of proteinogenic amino acids after feeding with a partially labeled carbon source or mixtures of labeled and unlabeled carbon sources19,20. Unlike common model microorganisms, mammalian cells do not grow on minimal media. Instead, they are bathed in vivo in a plethora of nutrients, including glucose, glutamine and the essential amino acids—all of which are included in common tissue culture media. This increased complexity of metabolic inputs renders the approaches used to deduce metabolic fluxes in microbes inadequate for resolving fluxes in mammalian cells.
The dynamics of assimilation of isotope-labeled nutrients contain a wealth of information beyond that available from steady-state labeling patterns21. Here we capture this information by kinetic flux profiling, in which liquid chromatography–tandem mass spectrometry (LC-MS/MS) is used to measure the passage of an isotope label from nutrients into downstream metabolites22,23 The kinetic data are then computationally integrated with direct measurements of selected metabolite influxes and effluxes and specific steady-state metabolite labeling patterns to determine metabolic fluxes in mammalian cells.
Using this technology, we found that HCMV infection upregulates much of central carbon metabolic flux, as well as efflux to nucleotide and fatty acid biosynthesis. Subsequent experiments showed that this unanticipated upregulation of fatty acid biosynthesis is essential for the replication of HCMV as well as influenza A, another medically important enveloped virus.
RESULTS
Glycolytic, pentose phosphate and nucleotide flux
Flux profiling was conducted 48 h after HCMV infection, when large changes in metabolite concentrations first appear18. A battery of five assays was used: (i) direct measurement of key cellular metabolic influxes (glucose and glutamine) and effluxes (pyruvate, lactate, alanine and glutamate), (ii) kinetic flux profiling with uniformly 13C-labeled glucose, (iii) kinetic flux profiling with uniformly 13C-labeled glutamine, (iv) probing of the branch point between glycolysis and the pentose phosphate pathway (PPP) using [1,2-13C]glucose and (v) probing of the branch point of pyruvate oxidation versus carboxylation using [3-13C]glucose. The kinetic flux profiling experiments involved quick transfer of cells from an unlabeled to a labeled carbon source by aspiration of unlabeled medium and its replacement with otherwise identical medium containing the labeled carbon source. Samples were collected at various time points after the isotope switch for analysis by LC-MS/MS.
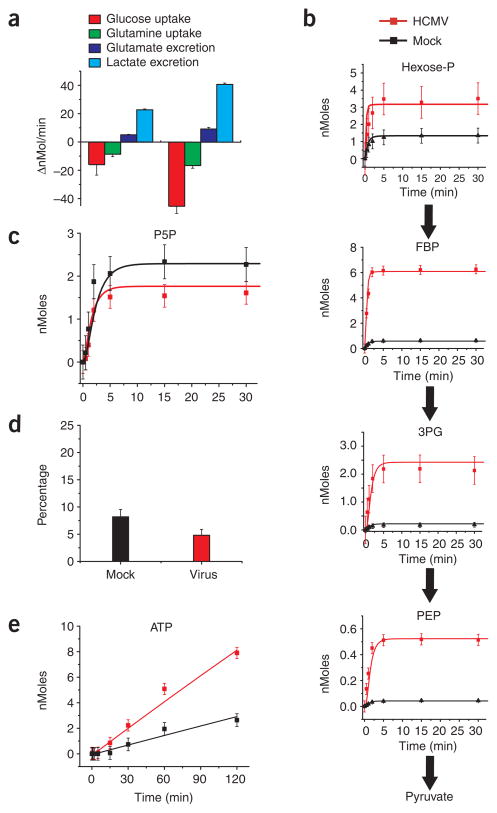
HCMV infection increased the uptake of glucose and glutamine and the excretion of lactate and glutamate (Fig. 1a). Consistent with the increased glucose uptake and lactate excretion, kinetic profiling of glycolysis indicated increased glycolytic flux: labeled glucose was more rapidly converted into labeled glycolytic intermediates in HCMV-infected than in mock-infected cells (Fig. 1b). Labeling of the five carbon species of the PPP was similar in infected and uninfected cells (Fig. 1c). Consistent with viral upregulation of glycolysis relative to the PPP, viral infection reduced the fraction of lactate with a single 13C-labeled carbon after feeding of [1,2-13C]glucose (Fig. 1d). Such singly-labeled lactate is formed when [1,2-13C]glucose is metabolized by the non-oxidative PPP but not by glycolysis. Although PPP flux was not upregulated, incorporation of ribose-5-phosphate into nucleotides was increased (Fig. 1e), consistent with virally induced upregulation of nucleotide biosynthesis16.
Figure 1.
Flux profiling of uninfected and HCMV-infected cells. (a) Measurement of influxes and effluxes of selected metabolites (per 1.5 × 106 cells; mean + 2 s.e.; n ≥ 3). Negative values are influxes, and positive ones effluxes. (b) Intracellular accumulation of 13C-labeled glycolytic metabolites (hexose-P, glucose-6-phosphate and its isomers; FBP, fructose-1,6-bisphosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate) after switching 1.5 × 106 cells into uniformly 13C-labeled glucose medium. Symbols indicate experimental data points ± 2 s.e.; n = 4; lines indicate model output. (c) Labeling dynamics, as in b, for the PPP intermediate pentose-P (ribose-5-phosphate and its isomers). (d) Percentage of labeled lactate containing one 13C atom after feeding of [1,2-13C]glucose. This is indicative of the PPP:glycolytic flux ratio; nonoxidative PPP flux yields lactate containing one 13C atom, whereas glycolytic flux does not. Data are shown as means + 2 s.e.; n = 3). (e) Labeling dynamics, as in b and c, for ATP. The bulk of observed labeling came from the ribose moiety of ATP. Similar results were found for GTP, UTP and CTP.
Tricarboxylic acid cycle and citrate shuttle
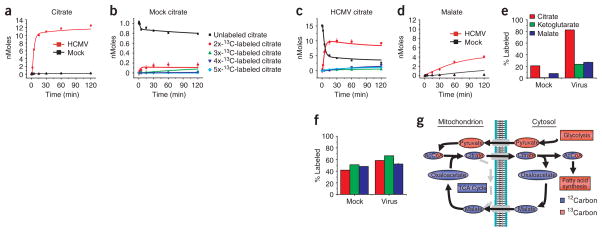
A particularly marked increase in labeling from [13C]glucose occurred for the first compound of the TCA cycle, citrate, which accumulated more than 20-fold faster in infected cells than in mock-infected cells (Fig. 2a). This reflected a larger citrate pool in the infected cells, as well as more rapid and complete labeling of that pool (compare Fig. 2b,c). Although almost all citrate in virally infected cells became labeled with 13C from glucose within 15 min, less than 30% of the malate pool was labeled (Fig. 2d,e). In contrast, [13C]glutamine (but not glucose) labeled citrate, α-ketoglutarate and malate all to a similar extent (Figs. 2e,f). This suggested that some of the carbon passing from glucose to citrate was redirected from the TCA cycle.
Figure 2.
Profiling of TCA cycle fluxes in uninfected and HCMV-infected cells. (a) Intracellular accumulation of 13C-labeled citrate after transfer of 1.5 × 106 cells to uniformly 13C-labeled glucose medium. Symbols indicate experimental data points ± 2 s.e.; n = 4; lines indicate model output. (b) Details of citrate labeling kinetics and patterns after transfer of 1.5 × 106 mock-infected cells to uniformly 13C-labeled glucose medium. Symbols indicate experimental data points ± 2 s.e.; n = 4; lines indicate model output. (c) Comparable data to b, but for HCMV-infected cells. (d) Labeling dynamics, as in a, for malate. (e) Extent of 13C-labeling (partial or complete) of the indicated TCA cycle metabolites after 2 h of exposure to uniformly 13C-labeled glucose in uninfected and virally infected cells (mean + 2 s.e.; n = 4). (f) Comparable data to e, but for labeling with uniformly 13C-labeled glutamine (n = 2). (g) Schematic of labeling patterns induced by citrate shuttle with feeding of uniformly 13C-labeled glucose. The pattern corresponds well to viral labeling data in a–e. The unlabeled portion of acetyl-CoA comes from CoA, which was omitted from the diagram for simplicity.
Citrate, in addition to being a TCA cycle intermediate, also shuttles two carbon units from the mitochondrion to the cytosol, where they are used for fatty acid and cholesterol biosynthesis. The observed TCA cycle labeling patterns (Fig. 2) suggested virus-induced activation of this shuttle. As shown schematically in Figure 2g, after [13C]glucose feeding, the citrate shuttle transfers labeled acetyl units (derived from glucose) from the mitochondrion to the cytosol through citrate with two 13C atoms. Four- and five-carbon compounds of the TCA cycle do not become labeled. Unlabeled cytosolic oxaloacetate, released after cytosolic cleavage of citrate, is reduced by NADH to form malate, which is either decarboxylated to form pyruvate and NADPH or pumped back into the mitochondrion to reenter the TCA cycle24.
Absolute quantitation of metabolite concentration and flux
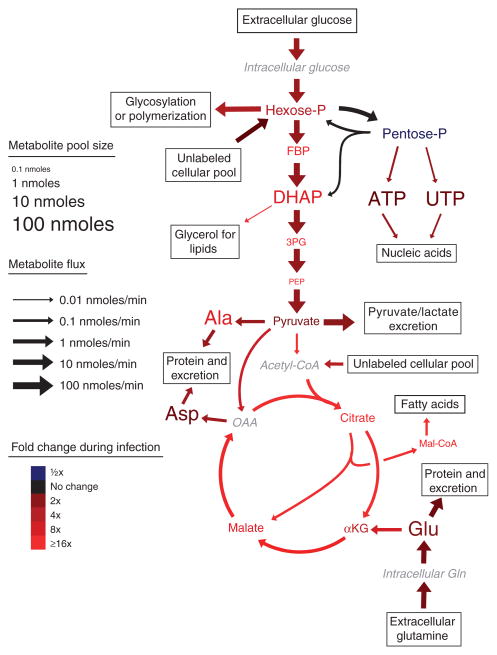
Although manual inspection of labeling data is a useful tool to generate hypotheses, the numerous fluxes that feed to and from the TCA cycle complicate data interpretation. These include formation of oxaloacetate through pyruvate carboxylase, which we probed directly using [3-13C]glucose (Supplementary Fig. 1 online) and fluxes to and from TCA-related amino acids. Of particular importance is the rapid interconversion of α-ketoglutarate and glutamate, an abundant intra-cellular metabolite. The large pool of glutamate can dilute labeled carbon atoms coming from glucose, rendering qualitative interpretation of TCA cycle labeling patterns unreliable25. To achieve a more quantitative understanding of central carbon metabolic flux and its modulation by HMCV infection, we developed an ordinary differential equation model (Fig. 3 and Supplementary Fig. 2 online) that predicts isotope labeling kinetics and patterns under the assumptions of fixed metabolic fluxes and pool sizes—that is, flux balance26.
Figure 3.
Metabolite concentrations and fluxes in uninfected and HCMV-infected confluent human fibroblasts. Font sizes indicate metabolite pool sizes (per 1.5 × 106 cells) in uninfected fibroblasts. Arrow sizes indicate net fluxes (per 1.5 × 106 cells) in uninfected fibroblasts. Colors indicate fold changes in response to HCMV infection. All scales are logarithmic. Fluxes shown are median values (Supplementary Table 5) from the 100 flux sets shown in Supplementary Tables 6 and 7. An exception to the proportionality of font size and pool size is malonyl-CoA, the concentration of which was too small to depict by font size. Metabolites whose levels were not directly measured are shown in gray italics. Amino acids are named by standard three-letter codes. Hexose-P, glucose-6-phosphate and its isomers; FBP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; AKG, α-ketoglutarate; OAA, oxaloacetate.
We began by experimentally determining the absolute pool sizes of metabolites in the model. The approach involved extensive labeling of intracellular metabolites by simultaneously feeding uniformly 13C-labeled glucose and glutamine for 1 week in culture27. We then extracted the labeled intracellular metabolites in the presence of known concentrations of unlabeled internal standard compounds, and determined the ratios of labeled to unlabeled compounds by LC-MS/MS. Corrections were applied to account for incomplete labeling of certain metabolites (Supplementary Table 1 online). The resulting values are shown in Supplementary Table 2 online. The use of internal standards throughout extraction, sample handling and analysis substantially enhanced the reliability of these measurements, compared to measurements based on extracted quantities per se.
We then used the pool size data, kinetic flux profiling data for glucose and glutamine, metabolite influx and efflux data, and specific branch-point data to search for combinations of fluxes consistent with the experimental results. Complete experimental results are shown in Supplementary Tables 2–4 and Supplementary Figures 3 and 4 online. Fluxes consistent with the experimental data were obtained using a global parameter identification algorithm, with a cost assigned to flux sets that did not recapitulate observed experimental data within ± 2 s.e. of their mean28 (Supplementary Methods online). Treating all fluxes that fit the experimental data within the 95% confidence interval as equivalent is a crucial precaution against overfitting, a common problem in flux elucidation29. To further mitigate the risk of overfitting, instead of identifying a single set of fluxes that best recapitulated the experimental results, we identified 100 different flux sets that approximated the observed data. This enabled estimation of confidence limits on the observed fluxes. Median flux values and their confidence limits for uninfected versus virally infected cells are shown in Supplementary Table 5 online.
Absolute pool sizes, absolute fluxes and their fold change after HCMV infection are shown schematically in Figure 3. Pools and fluxes scale exponentially with their diagrammed sizes, with marked differences in absolute pool sizes and fluxes across metabolites and reactions. The most active pathway in uninfected cells was glycolysis, and the majority of pyruvate produced was excreted as lactate. TCA cycle flux was ~1% of glycolytic flux and was driven largely by inflow from glutamine to α-ketoglutarate through glutamate.
Global metabolic changes induced by HCMV
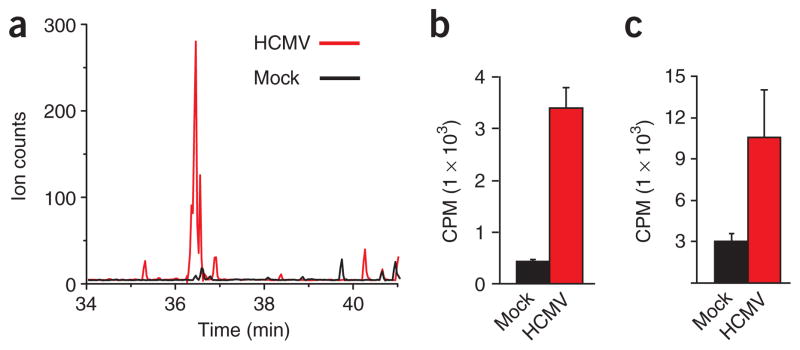
Colors in Figure 3 indicate changes in pool sizes and fluxes induced by HCMV infection, with increases in red and decreases in blue. Notably, almost the entire pathway diagram (with the exception of the PPP) is red, indicating a nearly global upregulation of metabolic flux by HCMV. Glycolytic flux increased approximately two-fold (Fig. 3 and Supplementary Table 5), consistent with the enhanced glucose uptake in virus-infected cells17. Nucleotide biosynthetic flux increased approximately three-fold, consistent with the efficacy of nucleotide-based antimetabolites as HCMV therapeutics30. TCA cycle fluxes increased even more markedly, with substantial upregulation of the core cycle fluxes (for example, ~20-fold from citrate to α-ketoglutarate), the pyruvate dehydrogenase pathway feeding the cycle (~80-fold from pyruvate to acetyl-CoA), the anapleuretic pathway for formation of four-carbon units (~4-fold from pyruvate to oxaloacetate) and the efflux of two-carbon units to fatty acid biosynthesis (~20-fold from citrate to malonyl-CoA). Consistent with upregulation of the fatty acid biosynthetic pathway, malonyl-CoA—the committed intermediate in fatty acid synthesis—increased in concentration from undetectable levels to amounts greater than ten-fold above the detection limit (Fig. 4a).
Figure 4.
HCMV induces lipogenesis. (a) Raw LC-MS/MS chromatograms of malonyl-CoA in uninfected (black) and virally infected (red) cells. (b,c) Production of 14C-labeled lipids from [14C]glucose in uninfected and HCMV-infected fibroblasts. Data are shown separately for the fatty acid (b) and glycerol (c) portions of saponified lipids (mean + s.e.; n = 3).
Fatty acid biosynthesis as an antiviral target
Many of the upregulated fluxes reflect essential processes that are not viable antiviral targets. For example, impairing canonical steps of the TCA cycle would be likely to lead to serious metabolic complications in oxidation-dependent cell types, such as cardiomyocytes and neurons. In contrast, fatty acid synthesis—although important in growth and development—is not acutely essential in mammals31–33. Given the substantial upregulation of fatty acid synthesis, we wished to explore its role in viral infection further. To test more directly whether viral infection increases glucose-driven lipid biosynthesis, we fed uninfected or HCMV-infected fibroblasts 14C-labeled glucose. We then extracted and saponified the lipids and counted radioactivity in the resulting lipid-derived fatty acid and glycerol fractions. The resulting data confirmed that HCMV infection induced a marked increase in glucose assimilation into both the fatty acid and glycerol moieties of lipids (Fig. 4b,c).
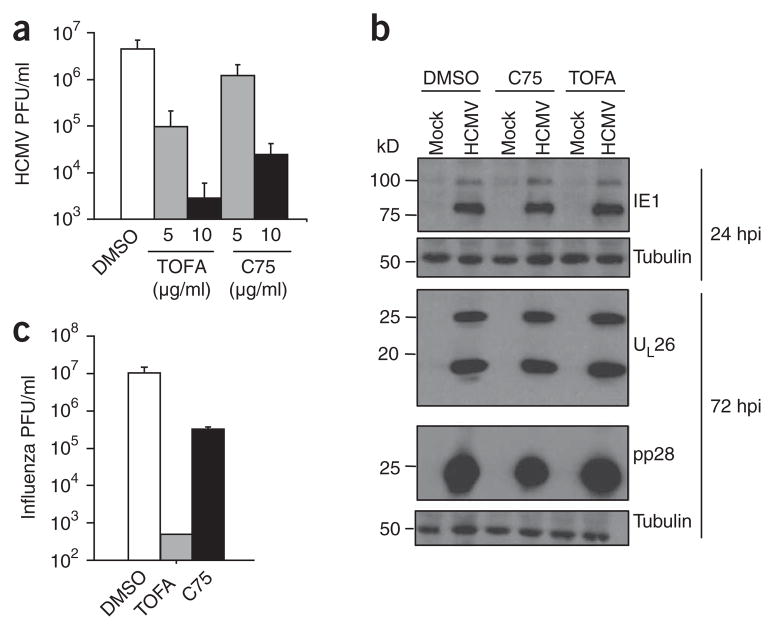
We next used pharmacological inhibitors of the fatty acid biosynthetic enzymes acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) to determine whether HCMV-induced upregulation of fatty acid production is necessary for viral replication (Fig. 5a). Treatment with 5-tetradecyloxy-2-furoic acid (TOFA), an ACC inhibitor34, resulted in a dose-dependent inhibition of HCMV replication with a >1,000-fold effect at 10 μg ml−1 TOFA. Treatment with C75 (trans-4-carboxy-5-octyl-3-methylene-butyrolactone), an inhibitor of FAS35, resulted in similar dose-dependent inhibition, with a >100-fold effect at 10 μg ml−1. Taken together, these results indicate that fatty acid biosynthesis is necessary for normal HCMV replication.
Figure 5.
Effect of pharmacological inhibitors of fatty acid biosynthesis on HCMV and influenza replication. (a) Production of infectious HCMV virions 96 h after infection (MOI, 3.0 PFU per cell) in the presence of carrier (DMSO), the ACC inhibitor TOFA or the FAS inhibitor C75 (mean + s.e., n = 4). (b) Accumulation of the HCMV IE1 protein (the primary isoform is ~75 kDa), UL26 (two isoforms) and pp28 in the presence of carrier, C75 (10 μg ml−1) or TOFA (10 μg ml−1) at an MOI of 3.0 PFU per cell. Tubulin levels are indicated as a control for protein loading. (c) Production of infectious influenza A virions 24 h after infection (MOI, 0.1 PFU per cell) in the presence of carrier, TOFA (50 μg ml−1) or C75 (10 μg ml−1). Data are shown as mean + s.e.; n = 4).
HCMV infection induces a temporally coordinated cascade of viral gene expression and subsequent protein synthesis. To determine whether inhibition of fatty acid biosynthesis affects this cascade, we analyzed the accumulation of the immediate early HCMV protein IE1, the early protein UL26 (which consists of two isoforms) and the late protein pp28 (Fig. 5b). Accumulation of all three proteins was unimpaired by C75 or TOFA treatment. Because the accumulation of pp28 depends on viral DNA synthesis36, these results indicate that the defect in viral growth caused by inhibition of fatty acid biosynthesis occurs subsequent to DNA replication.
Inhibition of FAS as a result of malonyl-CoA accumulation can be toxic and has been shown to induce apoptosis in some transformed cell lines37. The normal accumulation of viral proteins in the presence of C75 and TOFA suggested that the tested drug concentrations were not cytotoxic under the present conditions. To test directly for host cell cytotoxicity, we exposed uninfected fibroblasts to C75 or TOFA for 96 h (the length of the viral growth assays) and analyzed for viability by Trypan blue exclusion and by assessing cellular growth after drug removal. Both endpoints were identical in treated and untreated cells. To test directly for apoptosis induction, we examined the cleavage of the caspase-3 target poly(ADP-ribose) polymerase (PARP). Cleavage of PARP was undetectable in mock- or HCMV-infected cells after 24 or 72 h of TOFA or C75 treatment, whereas induction of apoptosis through osmotic shock with 0.5 M sorbitol resulted in the 85-kDa fragment characteristic of caspase-mediated PARP cleavage (Supplementary Fig. 5 online). Taken together, these results indicate that the tested doses of TOFA and C75 block HCMV replication without inducing host cell toxicity or apoptosis.
The requirement of fatty acid biosynthesis for a late event in HCMV replication may reflect an essential role for de novo synthesized fatty acids in viral envelopment or in lipid modification of viral proteins. To investigate whether this requirement extends to other enveloped viruses, we assessed the growth of influenza A in the presence of the ACC inhibitor TOFA or the FAS inhibitor C75. TOFA inhibited influenza A replication by >1,000-fold, and C75 inhibited replication by >10-fold (Fig. 5c). Thus, inhibitors of fatty acid biosynthesis impair the normal replication of two evolutionarily diverged enveloped viruses.
DISCUSSION
The ability to quantify metabolic fluxes in mammalian cells is crucial to understanding normal metabolic regulation and the pathophysiology of a broad spectrum of diseases. These include overtly metabolic conditions (such as diabetes) and conditions involving secondary metabolic derangements (such as cancer). Although previous studies of metabolic fluxes in mammalian cells have yielded many important results regarding specific pathways and branch points (see, e.g., refs. 38–40), they have not been adequate to produce comprehensive metabolic flux maps. Microbial fluxomic methods based on steady-state isotope labeling patterns of amino acids do not translate readily to mammalian cells, because mammalian cells require diverse nutrient inputs (complicating interpretation of labeling pattern data) and cannot synthesize essential amino acids de novo (limiting the information obtained from analyzing amino acids only).
We used the kinetics of assimilation of isotope-labeled nutrients into downstream metabolites to dissect metabolic fluxes in mammalian cells. Reliable flux determination is enabled by combining kinetic data with selective measurement of metabolite uptake and excretion rates and steady-state labeling patterns. As in microbial flux determination, computational data integration is achieved by a genetic algorithm that searches for flux combinations consistent with the experimental data41. A distinguishing feature of this study is identification of a large set of flux combinations that came close to recapitulating the experimental results within their 95% confidence limits. In contrast to approaches that identify a single flux solution based on mean experimental results, this approach avoids overfitting and provides flux confidence limits. This enables significant flux changes (those cases in which the flux distributions in Supplementary Table 5 are nonoverlapping) to be reliably identified. A next step in flux deconvolution would include acquisition and incorporation of data relevant also to cofactor reactions—for example, oxygen consumption to gain insight into rates of NAD(H) oxidation and reduction.
Application of this flux measurement approach to mock- and HCMV-infected human fibroblasts revealed massive flux upregulation in the infected cells. Of 41 fluxes examined, 28 showed nonoverlapping flux distributions between the uninfected and infected cells, with flux greater in the infected cells in all cases (Supplementary Table 5). Thus, HCMV results in nearly global metabolic upregulation. The mechanisms by which HCMV upregulates metabolic fluxes (including fatty acid biosynthesis) remain largely unknown. Virus-induced transcriptional changes may have a role for some pathways. For example, through microarray and quantitative PCR analysis, we have found that the phosphofructokinase-1 transcript is upregulated throughout infection, potentially contributing to increased glycolytic flux18. In other cases, virally induced metabolic gene transcription does not seem to have a role. ACC transcription, for example, is not changed by HCMV infection14,18. In such cases, post-translational modification of metabolic proteins could have a role in virally induced flux alterations.
Nucleotide biosynthesis, the target of current antimetabolites used in treatment of HCMV infection42, is among the fluxes upregulated by HCMV. Thus, our approach effectively identified an upregulated pathway whose inhibition is known to be clinically relevant for HCMV treatment. Like nucleotide biosynthesis, fatty acid biosynthesis can be pharmacologically inhibited in mammals without severe side effects. Our observation that HCMV increased flux into fatty acid biosynthesis at least as much as nucleotide biosynthesis suggested that inhibitors of fatty acid biosynthesis, developed with the objective of treating hyperlipidemia and obesity, could be used to impair HCMV replication. This proved to be the case with TOFA, an inhibitor of the committed step of fatty acid biosynthesis, which reduced HCMV titers ~1,000-fold. Notably, TOFA also affected replication of influenza A, a virus with little in common with HCMV except for the presence of a lipid envelope.
The specific mechanism by which inhibition of fatty acid biosynthesis targets HCMV and influenza A remains to be determined. Possibilities include precluding changes in membrane composition required for viral budding, impairing synthesis of specialized envelope phospholipids43, or impeding fatty acid modification of proteins44. Notably, hemagglutinin release from influenza A–infected cells is sensitive to the nonspecific lipid synthesis inhibitor cerulenin45. Similarly, hepatitis C virus replication is linked to host cholesterol synthesis through levels of geranylgeranyl pyrophosphate and activities of proteins found in cholesterol-rich membrane domains46. Furthermore, a recent metabolomic analysis identified elevated phospholipase A2 activity in simian immunodeficiency virus–induced encephalitis47, and a recent RNAi-based screen found that several genes involved in glycosphingolipid and inositol metabolism are important for HIV replication48. Taken together, these results suggest that lipid metabolism is a useful therapeutic target to treat infection with various enveloped viruses. Moreover, inhibitors of lipid metabolism might have relatively broad-spectrum antiviral activity, enabling their use in patients with viral syndrome without the need to identify the specific underlying pathogen.
For any potential new therapeutic approach, it is important to weigh anticipated efficacy versus side effects. The clinical success of 3-hydroxy-3-methyl-glutaryl-CoA reductase inhibitors (statins) indicates that inhibitors of lipid metabolism can be safe and effective human therapeutics. For treatment of viral infection by inhibiting fatty acid biosynthesis, targeting of ACC may be more clinically practical than targeting FAS, as FAS inhibition can cause severe anorexia and weight loss49. In mammals, ACC exists as two tissue-specific isoenzymes—ACC1 in adipose tissue and liver, and ACC2 in liver, heart and skeletal muscles. Although ACC1 is essential during embryogenesis32, TOFA (which is not isozyme specific) is well tolerated in rats, including during pregnancy and postnatal development. Oral administration of TOFA (150 mg per kg per day) results in steady-state plasma concentrations (30 μg ml−1) above those required here to block HCMV replication (10 μg ml−1)31. This dose is associated with reductions in plasma cholesterol and fatty acids without obvious signs of toxicity or teratogenicity31. Although extensive clinical testing would be required, this hints at the possibility of a favorable risk-benefit ratio for TOFA or another ACC inhibitor50 in treating HCMV infection or controlling an epidemic of influenza A resistant to current agents.
Links between cancer and viral infection have appeared repeatedly over the past decades. Viruses are important causes of cancer, and cancer and viruses both target specific genes, including tumor suppressors, to override normal control of the cell cycle and DNA replication. Our results extend these similarities to the arena of cellular metabolism. We find substantial virus-induced upregulation of nucleotide biosynthesis, glycolysis and lipid biogenesis. Increased nucleotide biosynthesis has long been known to be a hallmark of cancer, elevated glycolysis is an analog of the Warburg effect51 and, recently, increased flux from glucose into fatty acids has emerged as a feature of oncogenesis52. It is likely that—beyond providing more specific means of targeting viral infection—understanding the mechanisms of virus-induced metabolic flux modulation will also inform cancer research.
METHODS
Biological reagents and cell culture
MRC-5 fibroblasts (ATCC) were cultured in Dulbecco modified Eagle medium (DMEM, Sigma) containing 7.5% fetal calf serum and 4.5 g l−1 glucose. Before infection, fibroblasts were grown to confluence in 10-cm dishes, resulting in ~1.5 × 106 cells per dish. After incubation for 3–5 d at confluence, serum-containing medium was removed and serum-free medium added. Cells were then maintained in serum-free DMEM for 24 h, which has been shown to synchronize cells in the G0 stage of the cell cycle18. Cells were then mock-infected or infected with HCMV at a multiplicity of infection (MOI) of 3.0 plaque-forming units (PFU) per cell. We used the HCMV strain BADwt, which is derived from a bacterial artificial chromosome (BAC) clone of the AD169 strain of HCMV53. The BAC was inserted into the genome of HCMV without deletion of any viral sequence and was excised by a cotransfected Cre recombinase, which mediates recombination at the loxP sites flanking the BAC, leaving only the loxP site in the viral clone. This clone has been tested in various assays and has always shown a wild-type AD169 phenotype. After a 2-h adsorption period, the viral inocula were aspirated and fresh serum-free DMEM was added.
TOFA (Biomol International) and C75 (Calbiochem) were maintained as 10 mg ml−1 stocks in DMSO. To assess HCMV growth in the presence of metabolic inhibitors, densely confluent fibroblasts were infected in the presence of inhibitors at the indicated dose in DMEM containing 10% serum. After 2 h of incubation to allow viral adsorption, unbound virus was inactivated through a sodium citrate wash (40 mM sodium citrate, 10 mM KCl and 135 mM NaCl, pH 3.0) followed by a DMEM wash and then incubated at the indicated inhibitor dose in DMEM containing 10% serum. Forty-eight hours after infection, the cellular medium was changed, and fresh medium and metabolic inhibitors were added. At 96 h, cells were scraped and viral titers were determined by standard plaque assay on MRC-5 cells. To assess influenza A growth, fully confluent Madin-Darby canine kidney epithelial cells (MDCK cells; ATCC) were infected with the A/WSN/33 influenza strain (ATCC) in Flu infection buffer (DMEM containing 0.2% BSA, 0.01% CaCl2, 0.01% MgCl2, 1 μg ml−1 trypsin (Worthington) and 0.1% fetal calf serum). Viral titers were determined by standard plaque assay on MDCK cells.
Potential toxicity of TOFA and C75 was analyzed by treating uninfected cells with various doses of pharmaceutical inhibitors in DMEM (for fibroblasts) or Flu infection buffer (for MDCK cells) for the same duration as the viral growth assay (96 h for fibroblasts and 24 h for MDCK cells). After incubation with inhibitor, treated cells were analyzed for Trypan blue exclusion and found to be >95% viable. Inhibitor-treated cells were also replated in the absence of inhibitor and found to grow with kinetics similar to those of untreated cells. For MDCK cells, 50 μg ml−1 of TOFA was used for the influenza A growth assay, as this dose was found to be well tolerated by MDCK cells. Lower TOFA doses may be effective in nontransformed host cells, as cancer cells generally have strongly upregulated de novo lipid biosynthesis.
Metabolomic experiments
Labeled DMEM medium was prepared from DMEM without glucose or glutamine (Sigma) by addition of 10 mM HEPES and the appropriate forms (labeled or unlabeled) of glucose and glutamine to a final concentration of 4.5 g l−1 glucose and 0.584 g l−1 glutamine (labeled glucose and labeled glutamine from Cambridge Isotope Laboratories), followed by sterile filtration. For kinetic flux profiling experiments, samples were switched to fresh unlabeled medium 1 h before the switch into 13C-labeled medium. This minimized metabolome perturbations at the time of the isotope switch resulting from removal of accumulated metabolic waste products. Metabolome quenching and extraction were conducted as previously described18. Absolute metabolite quantitation involved extended labeling of cellular metabolites with uniformly labeled [13C]glucose and [13C]glutamine and extraction in the presence of known concentrations of unlabeled standards (for details, see refs. 23,27 and Supplementary Methods).
Uptake of glucose and glutamine and excretion of all measured metabolites (excretion of pyruvate, lactate, alanine and glutamate were found to be significant) was determined from medium samples taken over an 8-h time period, centered at 48 h after infection. Glucose was measured by enzyme assay (E00715251, R-Biopharm). The other compounds were measured by LC-MS/MS, with inclusion of isotopic internal standards for glutamine, glutamate, pyruvate, lactate and alanine.
Estimation of the relative carbon flux between glycolysis and the PPP was carried out using [1,2-13C]glucose as described38, with 4 h of incubation in labeled medium and detection of labeled forms of lactate by LC-MS/MS. Estimation of the relative carbon flux between pyruvate dehydrogenase and pyruvate carboxylase was carried out based on the passage of labeled carbon from [3-13C]glucose into malate, aspartate and citrate over 6 h (for details, see Supplementary Methods).
Liquid chromatography–tandem mass spectrometry analysis
Two different LC separations were coupled by electrospray ionization (ESI) to triple-quadrupole mass spectrometers operating in multiple reaction monitoring mode. The LC method coupled to positive-mode ESI was hydrophilic interaction chromatography on an aminopropyl column at basic pH, as described54. The LC method coupled to negative-mode ESI was reversed-phase chromatography using an amine-based ion pairing agent (a variation of the method used in ref. 55). The stationary and mobile phases were identical, but the gradient was altered as follows: t = 0, 0% B; t = 5, 0% B; t = 10, 20% B; t = 20, 20% B; t = 35, 65% B; t = 38, 95% B; t = 42, 95% B, t = 43, 0% B; t = 50, 0% B; where B refers to the methanol-containing mobile phase. For LC, we used an LC-20 AD HPLC system (Shimadzu) with autosampler temperature of 4 °C and injection volume of 10 μl. For MS, we used a TSQ Quantum Ultra or Discovery Max triple-quadrupole mass spectrometer (Thermo Fisher Scientific). Mass spectrometry parameters were as described in ref. 54, with the addition of extra multiple reaction monitoring scans to measure partially labeled compounds. All data were corrected for the natural abundance of 13C (see Supplementary Methods).
Measurement of lipid synthesis
To directly examine the flux of glucose into lipids, we transferred fibroblasts (infected or mock-infected for 48 h) into medium containing radioactive glucose (8 μC ml−1 [14C]glucose, 1 g l−1). After incubation for 4 h, the culture medium was aspirated, cells were washed with PBS and phospholipids were extracted by addition of 500 μl of 60:40 hexane:isopropanol. The culture dishes were then washed with an additional 400 μl of the hexane:isopropanol mixture. The resulting total extract was dried under N2 gas, resuspended in 500 μl of 1 N KOH in 90:10 methanol:water and incubated at 70 °C for 60 min to saponify lipids. Sulfuric acid (100 μl, 2.5 M) was then added, followed by hexane (700 μl) to extract the saponified fatty acids. The organic and aqueous phases were separated by centrifugation and scintillation-counted.
Computational determination of fluxes
An ordinary differential equation model of central carbon metabolism was constructed based on the diagram in Supplementary Figure 2. The model consisted of 69 differential equations, written to maintain flux balance. Equations of the model described the rates of loss of unlabeled forms of metabolites (and the creation of particular labeled forms) after feeding of [U-13C]glucose or [U-13C]glutamine media, and were written in the following manner:
where metabolite B is directly downstream in the model from metabolites A1, A2, …, AN, Ftot is the total flux into (and equivalently out of) the total B pool, BX is the pool size of a specific isotopic form of compound B; Fi is the ith flux into B; is the sum of all isotopic forms of Ai that feed into BX through flux Fi, is the total pool size of Ai (that is, the sum of all labeled and unlabeled forms of Ai), Btot is the total pool size of B and N is the number of fluxes into B. The model did not explicitly include nucleotide oxidation or reduction or phosphate transfer reactions (such as NAD+ reduction or ATP hydrolysis), as the rates of these reactions cannot be deduced using 13C-tracers.
Parameters (fluxes and unmeasured concentrations) were identified by a genetic algorithm implemented in C/C++ that seeks parameter values that minimize the differences between the experimental observations and the computational results. Costs were applied only when the model output fell outside of the 95% confidence limits (mean ± 2 s.e.) of the laboratory data. A series of 20 independent genetic algorithm runs were conducted on both the mock and virally infected data to obtain the distribution of best flux estimates shown in Supplementary Tables 5–7 online. For computational details, see Supplementary Tables 8–10 online, Supplementary Methods and Supplementary Computational Code online.
In creation of the flux-balanced model, amino acid efflux to protein biosynthesis was denoted by the symbol X (as shown in Supplementary Fig. 2). The efflux of each amino acid in the model was written as a fraction of X, where X is the sum of efflux from all amino acids not present in the medium (alanine, aspartate, asparagine, glutamate and proline) and glutamine to protein. These fractions were estimated based on the relative abundance of the amino acid in protein, including in the human proteome, in the HCMV proteome56 and in collagen. The abundance ratios were approximated as 6:4:3:5:5:4 (Ala:Asp:Asn:Glu:Pro:Gln) for both infected and uninfected cells. The fraction shown in Supplementary Figure 2 for glutamate (10/27) accounts for glutamate and its product proline, and that shown for aspartate (7/27) also accounts for asparagine. As shown in Supplementary Figure 2, X is fully constrained by other model fluxes and therefore does not appear in the model as a parameter.
Protein analysis
Protein accumulation was assayed by western blot analysis. Cells were washed with PBS, scraped and solubilized in disruption buffer (50 mM Tris (pH 7.0), 2% SDS, 5% 2-mercaptoethanol and 2.75% sucrose). Resulting extracts were sonicated and centrifuged at 14,000 g for 5 min to pellet insoluble material. Equivalent fractional amounts were subjected to electrophoresis in an SDS-containing 10% polyacrylamide gel and transferred to a nitrocellulose sheet. Blots were then stained with Ponceau S to ensure equivalent protein loading and transfer, blocked by treatment with 5% milk and reacted with primary antibody. Antibodies used were specific for UL99-encoded pp28 (10B4-29; ref. 57), UL123-encoded IE1 (1B12; T.S., unpublished data), UL26 (7H19; ref. 58), PARP (Santa Cruz Biotechnology) or tubulin (Epitomics). Protein bands were visualized through enhanced chemiluminescent detection according to the manufacturer’s instructions (Pierce). Analysis of tubulin levels served as an additional protein loading control.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Metabolomics Roadmap initiative (AI068678), NIH grants CA82396 and CA85786, and the NIH Center for Systems Biology at Princeton University (5 P50 GM071508). Development of the fluxomic technology was supported by the National Science Foundation Faculty Early Career Development award program (MCB-0643859), the Beckman Foundation, the American Heart Association (0635188N) and the National Science Foundation Dynamic Data-Driven Applications Systems program (CNS-0549181). J. Munger was supported by a postdoctoral fellowship from the American Cancer Society.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
J. Munger conducted experiments, analyzed data, conceived ideas and prepared the manuscript; B.D.B. conducted experiments, analyzed data, conceived ideas and prepared the manuscript; A.P. analyzed data; X.-J.F. conceived ideas and analyzed data; J. McArdle conducted experiments and analyzed data; H.A.R. conceived ideas; T.S. conceived ideas and prepared the manuscript; and J.D.R. conceived ideas, analyzed data and prepared the manuscript.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 2.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 3.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikoff WR, Gangoiti JA, Barshop BA, Siuzdak G. Metabolomics identifies perturbations in human disorders of propionate metabolism. Clin Chem. 2007;53:2037–2039. doi: 10.1373/clinchem.2007.089011. [DOI] [PubMed] [Google Scholar]
- 5.Holmes E, et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabatine MS, et al. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 7.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 8.Rezzi S, et al. Human metabolic phenotypes link directly to specific dietary preferences in healthy individuals. J Proteome Res. 2007;6:4469–4477. doi: 10.1021/pr070431h. [DOI] [PubMed] [Google Scholar]
- 9.Bruggeman CA. Does cytomegalovirus play a role in atherosclerosis? Herpes. 2000;7:51–54. [PubMed] [Google Scholar]
- 10.Reinhardt B, et al. Human cytomegalovirus-induced reduction of extracellular matrix proteins in vascular smooth muscle cell cultures: a pathomechanism in vasculopathies? J Gen Virol. 2006;87:2849–2858. doi: 10.1099/vir.0.81955-0. [DOI] [PubMed] [Google Scholar]
- 11.Gerna G, Baldanti F, Revello MG. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum Immunol. 2004;65:381–386. doi: 10.1016/j.humimm.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Burny W, Liesnard C, Donner C, Marchant A. Epidemiology, pathogenesis and prevention of congenital cytomegalovirus infection. Expert Rev Anti Infect Ther. 2004;2:881–894. doi: 10.1586/14789072.2.6.881. [DOI] [PubMed] [Google Scholar]
- 13.Myerson D, Hackman RC, Nelson JA, Ward DC, McDougall JK. Widespread presence of histologically occult cytomegalovirus. Hum Pathol. 1984;15:430–439. doi: 10.1016/s0046-8177(84)80076-3. [DOI] [PubMed] [Google Scholar]
- 14.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa T, Fioretti A, Plotkin S. Growth characteristics of cytomegalovirus in human fibroblasts with demonstration of protein synthesis early in viral replication. J Virol. 1973;11:991–997. doi: 10.1128/jvi.11.6.991-997.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S, Furukawa T, Plotkin SA. Human cytomegalovirus stimulates host cell RNA synthesis. J Virol. 1975;15:297–304. doi: 10.1128/jvi.15.2.297-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landini MP. Early enhanced glucose uptake in human cytomegalovirus-infected cells. J Gen Virol. 1984;65:1229–1232. doi: 10.1099/0022-1317-65-7-1229. [DOI] [PubMed] [Google Scholar]
- 18.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szyperski T. Biosynthetically directed fractional C-13-labeling of proteinogenic aminoacids - an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Baba T, Mori H, Shimizu K. Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl Microbiol Biotechnol. 2004;64:91–98. doi: 10.1007/s00253-003-1458-5. [DOI] [PubMed] [Google Scholar]
- 21.Boros LG, Brackett DJ, Harrigan GG. Metabolic biomarker and kinase drug target discovery in cancer using stable isotope-based dynamic metabolic profiling (SIDMAP) Curr Cancer Drug Targets. 2003;3:445–453. doi: 10.2174/1568009033481769. [DOI] [PubMed] [Google Scholar]
- 22.Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol. 2006;2:529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- 23.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenkel R. Regulation and physiological functions of malic enzymes. Curr Top Cell Regul. 1975;9:157–181. doi: 10.1016/b978-0-12-152809-6.50012-3. [DOI] [PubMed] [Google Scholar]
- 25.Chatham JC, Forder JR, Glickson JD, Chance EM. Calculation of absolute metabolic flux and the elucidation of the pathways of glutamate labeling in perfused rat heart by 13C NMR spectroscopy and nonlinear least squares analysis. J Biol Chem. 1995;270:7999–8008. doi: 10.1074/jbc.270.14.7999. [DOI] [PubMed] [Google Scholar]
- 26.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng XJ, Rabitz H. Optimal identification of biochemical reaction networks. Biophys J. 2004;86:1270–1281. doi: 10.1016/S0006-3495(04)74201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Winden WA, et al. Metabolic-flux analysis of Saccharomyces cerevisiae CEN. PK113–7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEM Yeast Res. 2005;5:559–568. doi: 10.1016/j.femsyr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Fishman JA, et al. Dosing of intravenous ganciclovir for the prophylaxis and treatment of cytomegalovirus infection in solid organ transplant recipients. Transplantation. 2000;69:389–394. doi: 10.1097/00007890-200002150-00014. [DOI] [PubMed] [Google Scholar]
- 31.Gibson JP, et al. Toxicity and teratogenicity studies with the hypolipidemic drug RMI 14,514 in rats. Fundam Appl Toxicol. 1981;1:19–25. doi: 10.1093/toxsci/1.1.19. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Elheiga L, et al. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proc Natl Acad Sci USA. 2005;102:12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 34.Kariya T, Wille LJ. Inhibition of fatty acid synthesis by RMI 14,514 (5-tetradecyloxy-2-furoic acid) Biochem Biophys Res Commun. 1978;80:1022–1024. doi: 10.1016/0006-291x(78)91347-5. [DOI] [PubMed] [Google Scholar]
- 35.Pizer ES, Chrest FJ, DiGiuseppe JA, Han WF. Pharmacological inhibitors of mammalian fatty acid synthase suppress DNA replication and induce apoptosis in tumor cell lines. Cancer Res. 1998;58:4611–4615. [PubMed] [Google Scholar]
- 36.Chambers J, et al. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizer ES, et al. Inhibition of fatty acid synthesis induces programmed cell death in human breast cancer cells. Cancer Res. 1996;56:2745–2747. [PubMed] [Google Scholar]
- 38.Lee WN, et al. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2–13C2]glucose. Am J Physiol. 1998;274:E843–E851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- 39.Zupke C, Sinskey AJ, Stephanopoulos G. Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995;44:27–36. doi: 10.1007/BF00164476. [DOI] [PubMed] [Google Scholar]
- 40.Chance EM, Seeholzer SH, Kobayashi K, Williamson JR. Mathematical analysis of isotope labeling in the citric acid cycle with applications to 13C NMR studies in perfused rat hearts. J Biol Chem. 1983;258:13785–13794. [PubMed] [Google Scholar]
- 41.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 42.Andrei G, De Clercq E, Snoeck R. Novel inhibitors of human CMV. Curr Opin Investig Drugs. 2008;9:132–145. [PubMed] [Google Scholar]
- 43.van Meer G, Simons K. Viruses budding from either the apical or the basolateral plasma membrane domain of MDCK cells have unique phospholipid compositions. EMBO J. 1982;1:847–852. doi: 10.1002/j.1460-2075.1982.tb01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt MF. The transfer of myristic and other fatty acids on lipid and viral protein acceptors in cultured cells infected with Semliki Forest and influenza virus. EMBO J. 1984;3:2295–2300. doi: 10.1002/j.1460-2075.1984.tb02129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conti G, Portincasa P, Chezzi C. Cerulenin inhibits production of mature virion particles in chick embryo fibroblasts infected by influenza A viruses. Res Virol. 1995;146:141–149. doi: 10.1016/0923-2516(96)81083-1. [DOI] [PubMed] [Google Scholar]
- 46.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J Clin Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 49.Loftus TM, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 50.Tong L, Harwood HJ., Jr Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 52.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 53.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 55.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 56.Varnum SM, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munger J, Yu D, Shenk T. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J Virol. 2006;80:3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.