Abstract
Cells can respond to stress in various ways ranging from the activation of survival pathways to the initiation of cell death that eventually eliminates damaged cells. Whether cells mount a protective or destructive stress response depends to a large extent on the nature and duration of the stress as well as the cell type. Also, there is often the interplay between these responses that ultimately determines the fate of the stressed cell. The mechanism by which a cell dies (i.e., apoptosis, necrosis, pyroptosis, or autophagic cell death) depends on various exogenous factors as well as the cell's ability to handle the stress to which it is exposed. The implications of cellular stress responses to human physiology and diseases are manifold and will be discussed in this review in the context of some major world health issues such as diabetes, Parkinson's disease, myocardial infarction, and cancer.
1. Overview of Cellular Stress Responses
Cells respond to stress in a variety of ways ranging from activation of pathways that promote survival to eliciting programmed cell death that eliminates damaged cells. The cell's initial response to a stressful stimulus is geared towards helping the cell to defend against and recover from the insult. However, if the noxious stimulus is unresolved, then cells activate death signaling pathways. The fact that the cell's survival critically depends on the ability to mount an appropriate response towards environmental or intracellular stress stimuli can explain why this reaction is highly conserved in evolution. For example, antioxidant defence mechanisms against oxidative injury and stress proteins such as heat shock proteins occur in lower organisms as well as the mammals.
There are many different types of stress and the response a cell mounts to deal with these conditions will depend on the type and level of the insult. For example, protective responses such as the heat shock response or the unfolded protein response mediate an increase in chaperone protein activity which enhances the protein folding capacity of the cell, thus counteracting the stress and promoting cell survival. The adaptive capacity of a cell ultimately determines its fate.
Therefore, depending on the level and mode of stress, different defense mechanisms and prosurvival strategies are mounted; however, if these are unsuccessful, then the cell death programs are activated to eliminate these damaged cells from the organism. The mechanism by which a cell dies, that is, apoptosis, necrosis, pyroptosis, or autophagic cell death, often depends on its ability to cope with the conditions to which it is exposed. In this review we initially discuss the different forms of cell death that can be activated by adaptive responses because activation of death signaling pathways is the ultimate response to all types of persistent irresolvable stress. In Section 3 we will discuss the many types of stress a cell can encounter and the different responses that are activated to survive adverse conditions. Finally, we will discuss the involvement or contribution of cellular stress responses to disease states.
2. Stress-Induced Cell Death
Cell death has many forms and shapes. Cell death research encompasses not only the study of programmed forms of cell death (both apoptosis and autophagic cell death), necrosis and other modes of cellular demise but also the role these phenomena play in physiological and pathological processes including development, aging, and disease.
The cell death field has attracted much attention in the last two decades, mainly because of its relevance to development, degenerative diseases, and cancer. However, the field of cell death research is by no means new [1]. The concepts of cellular demise and associated terminology have been evolving since the 19th century. The term programmed cell death refers to controlled or regulated forms of death associated with a series of biochemical and morphological changes [2–4]. The realization that some forms of cell death were biologically controlled or programmed has led to exploitation of these processes and has made profound impact in various fields of biology and medicine [5–7].
Nowadays, programmed cell death is synonymous with apoptosis; however, based on the original definition it also refers to autophagic cell death [8]. The term apoptosis was first used to describe a particular morphology of cell death [9] common to the vast majority of physiological cell deaths. This morphology includes shrinkage and blebbing of cells, rounding and fragmentation of nuclei with condensation, and margination of chromatin, shrinkage, and phagocytosis of cell fragments without accompanying inflammatory responses (in most cases) [9–11]. The morphology of cells undergoing apoptosis appeared dissimilar and distinct from the morphology associated with necrosis [9, 10]. Necrosis, a term commonly used by pathologists, refers to any deaths associated with the loss of control of ionic balance, uptake of water, swelling, and cellular lysis [12, 13]. This lysis releases many intracellular constituents, attracting immune cells and provoking an inflammatory response.
2.1. Apoptosis
During the 1980s, apoptosis became the focus of attention, primarily because of the relative ease with which it could be distinguished morphologically from other types of cell death. Within a few years apoptosis and delineation of the underlying biochemical and molecular pathways dominated cell death research. The discoveries of the Bcl-2 family of proteins [14–16], death receptors [17], caspases [18], mitochondrial cytochrome c release [19], and a role for the endoplasmic reticulum [20] in apoptosis were just a few major milestones in the history of the field. Today the morphological and biochemical changes associated with apoptosis are largely explained by activation of caspases, and apoptosis has become generally accepted as caspase-dependent programmed cell death [21].
Of all the forms of cell death apoptosis is the best characterized and its highly regulated nature makes it an attractive target for therapeutic intervention. Apoptosis is highly conserved throughout evolution [22, 23] and plays a major physiological role in both embryonic development and aging [22, 24]. Various types of cellular stress stimuli have been shown to trigger apoptosis, including chemotherapeutic agents, irradiation, oxidative stress, and ER stress. Caspases, a family of cysteine proteases, act as common death effector molecules in various forms of apoptosis [25]. Caspases are synthesized as inactive proenzymes, which upon activation cleave various substrates in the cytoplasm or nucleus. This leads to many of the morphologic features of apoptotic cell death, for example, polynucleosomal DNA fragmentation, loss of overall cell shape, and nuclear shrinking [22, 25–27].
During apoptosis caspases are activated by different mechanisms. Stimulation of death receptors of the tumor necrosis factor (TNF) receptor superfamily such as CD95 (APO-1/Fas) or TNF-related apoptosis inducing ligand (TRAIL) receptors by their respective ligands or agonistic antibodies results in receptor aggregation and recruitment of the adaptor molecule Fas-associated death domain (FADD) and procaspase-8 to form the death inducing signaling complex (DISC) [26]. Upon recruitment caspase-8 becomes activated and initiates apoptosis by direct cleavage of downstream effector caspases [26]. The mitochondrial pathway to caspase activation is initiated by the release from the mitochondrial intermembrane space of apoptogenic factors such as cytochrome c, apoptosis inducing factor (AIF), second mitochondria-derived activator of caspase (Smac)/direct IAP binding protein with low pI (DIABLO) or Omi/high-temperature requirement protein A2 (HtrA2) [28]. The release of cytochrome c into the cytosol results in caspase-3 activation through formation of the cytochrome c/Apaf-1/caspase-9-containing apoptosome complex [29]. Smac/DIABLO or Omi/HtrA2 promotes caspase activation through neutralizing the inhibitory effects of Inhibitor of Apoptosis Proteins (IAPs) [30]. Activation of caspases has to be tightly controlled because of the potential detrimental effects on cell survival if they are inappropriately activated. For example, resistance to apoptosis can be caused by aberrant function or expression of IAPs [30]. IAPs present a group of endogenous inhibitors of caspases with eight members in human cells, that is, XIAP, cIAP1, cIAP2, survivin, livin (ML-IAP), NAIP, Bruce (apollon), and ILP-2 [30]. All IAP proteins harbor one or more baculovirus IAP repeat (BIR) domains that mediate their inhibitory interaction with caspases [30]. Among the IAP family proteins, XIAP is the most potent inhibitor of caspases and blocks apoptosis by binding to active caspase-3 and -7 and by interfering with caspase-9 activation [30].
In addition, the ratio of antiapoptotic versus pro-apoptotic Bcl-2 family proteins regulates apoptosis sensitivity. The Bcl-2 proteins comprise both anti-apoptotic family members, for example, Bcl-2, Bcl-XL, and Mcl-1, and pro-apoptotic molecules such as Bax, Bak, and BH3 domain only molecules [31]. According to the direct activation model of Bcl-2 protein activation, BH3-only proteins that function as direct activators (such as Bim and the cleaved form of Bid (tBid)), directly bind to Bax and Bak to stimulate their activation [32]. In this model, BH3-only proteins that act as sensitizers such as Bad promote apoptosis by binding to the prosurvival Bcl-2 proteins [32]. In contrast, the indirect activation model proposes that BH3-only proteins activate Bax and Bak in an indirect manner by binding to the multiple anti-apoptotic Bcl-2 proteins that inhibit Bax and Bak, which in turn leads to the release of Bax and Bak [33, 34]. Moreover, apoptosis sensitivity may be controlled by IAPs, through the regulation of additional signaling cascades, for example, the NF-κB, JNK, TNFR, and the ubiquitin/proteasome pathway [30, 35]. The anti-apoptotic mechanisms regulating cell death have also been implicated in conferring drug resistance to tumor cells.
2.2. Autophagic Cell Death
Autophagy (self-eating) is a multistep process that is characterized by the vesicular sequestration and degradation of long-lived cytoplasmic proteins and organelles, for example, mitochondria [36]. The resulting double-membrane vesicle is termed an autophagosome [36]. The discovery of autophagy-related (atg) genes, first in yeast and subsequently in humans, has greatly enhanced the molecular understanding of the mechanisms that are involved in the control of autophagy [36]. The protein product of the tumor suppressor gene Beclin 1 is the mammalian homolog of Atg6 and forms a multiprotein complex together with Vps34, a class III phosphatidylinositol 3-kinase, UVRAG (UV irradiation resistance-associated tumor suppressor gene), and a myristylated kinase (Vps15, or p150 in humans) [36, 37]. This complex is required for the initiation of the formation of the autophagosome. Once this complex forms, Vps34 becomes activated and catalyzes the generation of phosphatidylinositol-3-phosphate, which is required for vesicle nucleation.
Two major protein conjugation systems exist that are required for autophagosome formation, that is, the Atg12–Atg5 conjugation and Atg8-phosphatidylethanolamine conjugation systems [38]. Mechanistically, both conjugation systems function in a manner that is closely related to ubiquitin conjugation to proteins, with corresponding conjugation-assisting enzymes that resemble the E1 and E2 enzymes in ubiquitin conjugation [38]. In the Atg12–Atg5 conjugation pathway, Atg12 is covalently conjugated to Atg5 with the help of the E1-like enzyme Atg7 and the E2-like enzyme Atg10 [36]. In the other conjugation pathway, phosphatidylethanolamine (PE) is conjugated to LC3, one of the mammalian homologues of Atg8 [36]. This process involves the sequential action of the protease Atg4, the E1-like enzyme Atg7 and the E2-like enzyme Atg3. Subsequently, lipid conjugation results in the conversion of the soluble form of LC3, that is, LC3-I, to the autophagic-vesicle-associated form that is termed LC3-II [39]. Thus, LC3 is soluble under unstressed conditions and undergoes association with peripheral membranes of autophagosomes during the induction of autophagy. Via the fusion with lysosomes, the content of autophagosomes is degraded by the action of acid-dependent enzymes [36].
Autophagy is typically observed in cells that are exposed to a variety of metabolic and therapeutic stresses, including growth factor deprivation, inhibition of the receptor tyrosine kinase/Akt/ mammalian target of rapamycin (mTOR) signaling, shortage of nutrients, ischemia/reperfusion, inhibition of proteasomal degradation, the accumulation of intracellular calcium, and endoplasmic reticulum (ER) stress [40–43]. Reactive oxygen species (ROS) may provide a common link between cellular stress signals and the initiation of autophagy, as ROS accumulation has been reported to result in inactivation of the cysteine protease ATG4, which in turn causes accumulation of the ATG8-phosphoethanolamine precursor that is required for the initiation of autophagosome formation [44]. The functional relationship between autophagy and cell death is complex in the sense that, under most cellular settings, autophagy functions as a stress adaptation that prevents cell death, whereas in some circumstances, it constitutes an alternative route to cell death. This complex interrelationship between autophagy and cell death implies that these responses are somewhat linked at the molecular level. However, the key molecular events that eventually determine whether autophagy is protective or destructive are still poorly understood.
Although it is still controversial whether autophagy is protective or toxic for the cells, accumulating evidence suggests that it has beneficial roles in the heart under both physiological and pathological conditions [45, 46]. Autophagy was shown to mediate turnover of intracellular proteins and organelles in the heart and protect against hemodynamic stress [45]. Consistent with this, rapamycin, which induces autophagy by inhibiting mTOR, can protect myocardium against ischemia/reperfusion injury [47]. In contrast, recent studies also demonstrated that downregulation of the transcription factors, activating transcription factor 5 or 7 (ATF5 or ATF7), using siRNA prevented stress-induced cell death [48, 49], suggesting that the level or timing of autophagy may be critical for deciding the fate of the cells. Autophagic cell death has mainly been shown during development. However, during recent years accumulating evidences suggest that inhibition of apoptosis induces cell death that is either associated with or dependent on autophagy [48–50]. There is evidence of cross-talk between apoptosis and autophagy at the molecular level, particularly with regard to the Bcl-2 family. In addition to its role in inhibiting apoptosis, Bcl-2 has also been shown to inhibit autophagy [51, 52] and autophagic cell death [53]. This effect is mediated through the ability of Bcl-2 to interact with Beclin 1, a key protein in autophagosome formation [52]. In fact, Beclin 1 has been shown to be a novel BH3-only protein and to interact with a number of anti-apoptotic Bcl-2 family members including Bcl-2, Bcl-xL, Bcl-w, and Mcl-1 [54–57].
2.3. Necrosis
Necrosis has been considered as an accidental mode of cell death for many years, implying that within a multicellular organism it is an unregulated process. However, there is now mounting evidence that the execution of necrotic cell death is also regulated by a set of signaling pathways [58–60]. For instance, death domain receptors, for example, TNFR1, and Toll-like receptors have been reported to trigger necrosis, in particular in the presence of caspase inhibitors [58]. In addition, necrotic cell death has been reported in response to cellular stress stimuli, including ischemia or glutamate excitotoxicity in neurons or cancer cells exposed to alkylating DNA damaging agents [61–63]. Morphologically, necrosis is characterized by a gain in cell volume, swelling of organelles and plasma membrane rupture, which results in the loss of intracellular contents. Several signal transduction cascades have been described that are involved in the propagation of necrotic cell death. There is mounting evidence that the serine/threonine kinase RIP1 is one of the key mediators of necrotic cell death, at least in the case of death receptors or Toll-like receptors [64, 65]. Studies in RIP1-deficient leukemia cells revealed that RIP1 is required for death receptor-induced necrosis [66, 67]. Furthermore, RIP1 has been described to be required for lipopolysaccharide-induced cell death of macrophages [68]. In line with a central role of RIP1 in necrotic cell death, small molecule inhibitors of RIP1 kinase were reported to protect against ischaemic brain injury in an in vivo model of necrosis [69–71]. In addition to RIP1, there is very recent evidence that RIP3 is also critical for necrotic cell death [72–74]. To this end, RIP3 was identified in an RNA interference screen to be essential for necrosis in response to TNFα stimulation and during virus infection [72, 73]. RIP3 interacts with RIP1 and regulates RIP1 phosphorylation and the generation of ROS [72–74].
Moreover, ROS and calcium constitute important mediators that are involved in the propagation of the necrotic signal in various forms of necrosis, for example, upon stimulation with TNFα or exposure to double-stranded DNA [75, 76]. ROS may be generated intracellularly by mitochondria and glycolysis [75, 77]. While the ER is the main intracellular calcium store, mitochondrial calcium has been described to stimulate oxidative phosphorylation, thereby promoting ROS generation [78]. Both ROS and calcium can cause damage to organelles and macromolecules, which contributes to the loss of cell integrity. In addition calcium-mediated activation of calpain can lead to cleavage and inactivation of caspases [79], whereas the ROS can target the active site of caspases and render them inactive [80]. Many stimuli that drive necrosis can inhibit the apoptotic machinery.
3. Cellular Stress Responses
During tissue homeostasis there is an equilibrium between the net growth rate and the net rate of cell death [22]. Upon exposure to cellular stress this physiological homeostasis is in danger. Depending on the type of cellular stress and its severity, the cell's response can be manifold. In essence, if the stress stimulus does not go beyond a certain threshold, the cell can cope with it by mounting an appropriate protective cellular response, which ensures the cell's survival. Conversely, the failure to activate or maintain a protective response, for example, if the stressful agent is too strong, results in activation of stress signaling cascades that eventually fuel into cell death pathways [81, 82].
3.1. The Heat Shock Response
One of the main prosurvival activities of cells, the heat shock response, was originally described as the biochemical response of cells to mild heat stress (i.e., elevations in temperature of 3–5°C above normal) [83, 84]. It has since been recognized that many stimuli can activate this response, including oxidative stress and heavy metals. One of the main cellular consequences of these stresses is protein damage leading to the aggregation of unfolded proteins. In order to counteract this, cells increase the expression of chaperone proteins that help in the refolding of misfolded proteins and alleviate protein aggregation. This confers a transient protection, leading to a state that is known as thermotolerance, whereby cells become more resistant to various toxic insults, including otherwise lethal temperature elevations, oxidative stress, various anticancer drugs, and trophic factor withdrawal [85–88].
During initiation of the heat shock response general protein transcription and translation is halted, presumably to alleviate the burden of misfolded proteins in the cell. However, transcription factors that enhance expression of a specific subset of protective genes are selectively activated under these conditions; these are the heat shock factors (HSFs) [89]. Vertebrate cells have three different HSFs: HSF1 is essential for the heat shock response and is also required for developmental processes, HSF2 and HSF4 are important for differentiation and development, while HSF3 is only found in avian cells and is probably redundant with HSF1 [90, 91]. Cells derived from mice lacking HSF1 are sensitive to stress and are unable to develop thermotolerance or induce heat responsive genes upon heat shock [92–94], which has confirmed that HSF1 in particular is responsible for the heat shock response. More recent work has shown that HSF2 can modulate HSF1-mediated expression of heat-responsive genes [95], suggesting that HSF2 also participates in transcriptional regulation of the heat shock response.
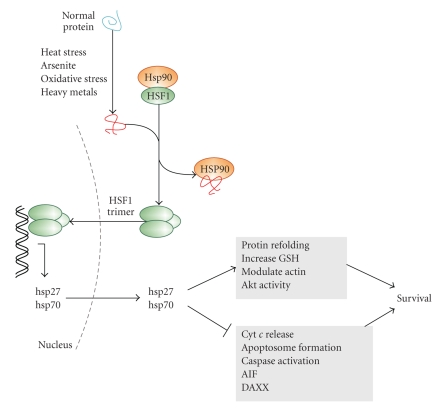
Inactive HSF1 is maintained in a monomeric form in the cytoplasm through interaction with Hsp90 and cochaperones [96, 97] (Figure 1). When the cell is exposed to stressful conditions, there is accumulation of unfolded proteins which compete with HSF1 for Hsp90 binding. Thus, HSF1 is released from the complex stimulating its transition from a monomer to a homotrimer that can translocate to the nucleus and bind to DNA (Figure 1). HSFs bind to upstream sequences (heat shock elements) in the promoters of target genes, leading to the expression of heat shock proteins (Hsps).
Figure 1.
Induction of heat shock proteins inhibits apoptosis and promotes cell survival. Exposure of cells to elevated temperatures, oxidative stress, and heavy metals causes accumulation of unfolded proteins, which through activation of HSF1 leads to induction of Hsp27 and Hsp70. These Hsps inhibit apoptosis and promote survival.
Hsps are a set of evolutionary conserved proteins that are grouped into subfamilies with molecular weights of approximately 110, 90, 70, 60, 40, and 15–30 kDa [85, 98]. Some of these, for example, Hsp90, are constitutively expressed and act intracellularly as molecular chaperones, preventing premature folding of nascent polypeptides [99]. Others, particularly Hsp27 and Hsp70, are usually expressed at low basal levels and increase in response to environmental and physiological stressors, and as such they are termed inducible Hsps and are part of the heat shock response [85]. Hsp27 belongs to a subfamily of stress proteins, the small Hsps, which are detectable in virtually all organisms. Hsp27 is also regulated by phosphorylation and dynamic association/dissociation into multimers ranging from dimers to large oligomers [100]. Hsp70 is the inducible member of the 70 kDa family of Hsps. Both Hsp27 and Hsp70 have been shown to protect cells against the induction of cell death by a variety of stresses and by different modes of cell death, including apoptosis [86, 101] and necrosis [102–104]. They achieve these effects directly, through inhibition of cell death pathways, and indirectly, through general prosurvival activities. For example, in their capacity as molecular chaperones, inducible Hsps bind to and aid the refolding of unfolded proteins, thereby preventing protein aggregation [105]. Hsp27 can interact with actin and is thus important for maintaining the integrity of the cytoskeleton which may play a role in promoting survival [106].
Apart from these indirect mechanisms, Hsp27 and Hsp70 can directly inhibit apoptosis by modulating both the intrinsic and the extrinsic apoptosis pathways and by interfering with caspase activation at several different levels [107–109]. Both Hsp27 and Hsp70 have been reported to directly block release of pro-apoptotic factors, including cytochrome c, from the mitochondria [110–112]. In the cytosol, these Hsps can block apoptosome formation and activation of downstream caspases through their ability to bind to cytochrome c and procaspase-3 (in the case of Hsp27) [107, 108] and procaspases -3, -7 and Apaf-1 (in the case of Hsp70) [101, 113–115]. Hsp70 can also interact with and inhibit apoptosis-inducing factor (AIF) thus inhibiting apoptotic nuclear changes [116, 117]. Hsps can also modulate the death receptor pathway. Hsp27 is reported to inhibit DAXX, an adaptor protein that links the Fas death receptor and the ER stress sensor IRE1 to ASK-1 and downstream JNK pro-apoptotic signaling [118]. Hsp70 also inhibits JNK activity [119–121], although this is not observed in all systems [101]. Hsp27 and 70 can also interact with other proteins that regulate cell survival. For example, Hsp27 can interact with the prosurvival Ser/Thr kinase Akt which is suggested to be important for sustained Akt activity [122–124]. Hsp70 can exist in complex with cochaperones, including DnaJ/Hsp40 and BAG-1 which affect its ability to modulate apoptosis [125, 126]. Overall, Hsps can be activated or induced by a number of stresses and they act to protect the cell by influencing a variety of cellular processes which determine cellular fate. Hsps are, in general, prosurvival and anti-apoptotic molecules.
3.2. The Unfolded Protein Response (UPR)
Secretory and membrane proteins undergo posttranslational processing, including glycosylation, disulfide bond formation, correct folding, and oligomerization, in the ER. In order to effectively produce and secrete mature proteins, cellular mechanisms for monitoring the ER environment are essential. Exposure of cells to conditions such as glucose starvation, inhibition of protein glycosylation, disturbance of Ca2+ homeostasis and oxygen deprivation causes accumulation of unfolded proteins in the ER (ER stress) and results in the activation of a well orchestrated set of pathways during a phenomenon known as the unfolded protein response (UPR) [127, 128] (Figure 2). The UPR is generally transmitted through activation of ER resident proteins, most notably inositol-requiring protein-1 (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK), and activating transcription factor 6 (ATF6). In some cells/tissues, additional ATF6-like bZip type transcription factors such as OASIS, CREB-H, Tisp40, and Luman also transmit the UPR signaling [129–132]. The UPR target genes include molecular chaperones in the ER, folding catalysts, subunits of translocation machinery (Sec61 complex), ER-associated degradation (ERAD) molecules and anti-oxidant genes [127].
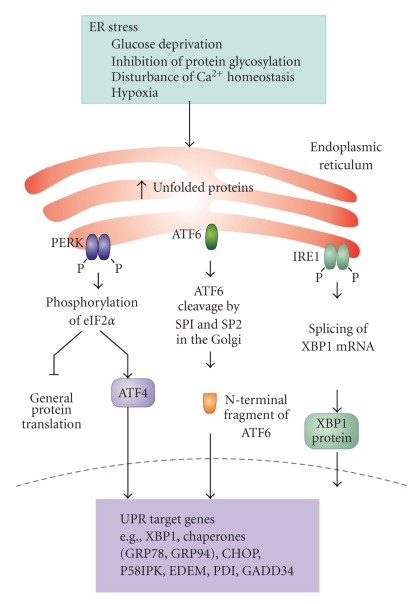
Figure 2.
ER stress and the unfolded protein response. Stress to the ER stimulates the activation of the three endoplasmic reticulum (ER) stress receptors, PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1 (Ire1) that are involved in the unfolded protein response (UPR). PERK phosphorylates eukaryotic initiation factor 2 (eIF2α) which inhibits general protein translation, allowing eIF2α-independent translation of ATF4, which activates transcription of chaperones such as GRP78. ATF6 undergoes specific proteolysis in the Golgi apparatus which leads to activation. One of the ATF6 target genes is XBP1. IRE1 catalyzes the alternative splicing of XBP1 mRNA leading to expression of the active XBP1 transcription factor. Together the three arms of the UPR block protein translation, increase chaperone expression and enhance ER-associated protein degradative pathways.
Among the UPR transmitters so far identified, IRE1 and PERK are both type I transmembrane protein kinases which dimerize to promote autophosphorylation and activation in response to ER stress. Activated IRE1 endonucleolytically cleaves mRNA that encodes a transcription factor named homologous to ATF/CREB1 (Hac1) in yeast [133, 134] and X-box binding protein-1 (XBP1) in higher species [135, 136]. The spliced forms of Hac1 and/or XBP1 in turn activate the transcription of the UPR target genes. In contrast, activated PERK phosphorylates the α-subunit of eukaryotic translation initiation factor-2 (eIF2α) which leads to lower levels of eIF2 and translational suppression [137]. The PERK-eIF2α signaling pathway also activates the transcription of the UPR target genes through CAP-independent upregulation of the translation of a transcription factor ATF4 [138]. PERK can also directly phosphorylate and activate the transcription factor, NF-E2-related factor-2 (Nrf2), which contributes to cellular redox homeostatis by inducing the expression of anti-oxidant genes [139, 140]. ATF6 is a type II transmembrane protein which is cleaved by Golgi apparatus-resident proteases site-1 protease (SP1) and site-2 protease (SP-2) in response to ER stress [141, 142]. The cleaved N-terminal fragment of ATF6 acts as a transcription factor to increase the transcription of the UPR target genes together with XBP1 and ATF4.
UPR signaling generally promotes cell survival by improving the balance between the protein load and the folding capacity in the ER and/or by improving the secretion of trophic factors/growth factors [143, 144]. However, if the protein load in the ER exceeds its folding capacity, or some defects in the UPR exist, cells tend to die, typically, with apoptotic features (ER stress-induced cell death). Although the exact molecular mechanisms that regulate this type of cell death remain to be elucidated, at least three pathways have been identified as being involved: the caspase-12/caspase-4 pathway and CHOP and IRE1-JNK pathways. Caspase-12 [145] in mice and caspase-4 in human [146] have been proposed as caspases that initiate ER stress-induced cell death. Caspase-12 null mice are reported to be relatively resistant to ER stress and amyloid-beta toxicity [145]. Caspase-12 is reported to directly cleave procaspase-9 without involvement of the cytochrome c/Apaf-1 pathway [147]. C/EBP homologous protein (CHOP), a transcription factor that is induced downstream of PERK and ATF6 pathways, induces ER stress-induced cell death at least in part by suppressing the expression of Bcl-2 [148] and inducing Bim expression [149]. IRE1 also participates in ER stress-induced cell death by activating JNK through the binding with ASK1 and Traf2 [150, 151].
Important roles for ER stress and ER stress-induced cell death have also been demonstrated in a broad spectrum of pathophysiological situations, including ischemia, diabetes, atherosclerosis, endocrine defects, development, neurodegenerative disorders, and cancer as described below [143, 144, 152–155].
Among the UPR targets, glucose-regulated proteins (GRPs) are the most studied and best characterized. GRPs were originally identified as proteins induced by glucose starvation [156]. Later, it was found that these molecules were transcriptionally induced by ER stress through the cis-acting element termed ER stress response element (ERSE) [157]. GRPs include molecular chaperones in the ER such as GRP78/Bip, GRP94, ORP150/GRP170, and oxidoreductases in the ER such as PDI, ERp72, and GRP58/ERp57. Accumulating evidence suggests that GRPs promote cell survival when exposed to stresses such as hypoxia/ischemia [143, 158], glutamate excitotoxicity [159], and neurodegeneration [160–162]. GRP78 could be a potential factor to inhibit atherosclerosis by preventing ER stress-induced cell death in endothelial cells [163]. This involves the inhibition of the activation of SREBPs, a molecule that induces cholesterol and triglyceride biosynthesis, or by inhibiting tissue factor procoagulant activity [164–166]. ORP150/GRP170 was found to be associated with insulin sensitivity in both human and mice as described below. Furthermore, GRPs also play important roles in survival during early mammalian development [159, 167–169].
Interestingly, recent studies have revealed that small compounds that mimic the functions of GRPs (chemical chaperones) and those that induce endogenous GRPs (molecular chaperone inducers) can prevent protein aggregation [170], improve protein secretion [171], and protect cells against brain ischemia [172] or neurodegeneration [173]. These results suggest that the regulation of ER stress can be a novel therapeutic target in a variety of diseases.
3.3. The DNA Damage Response
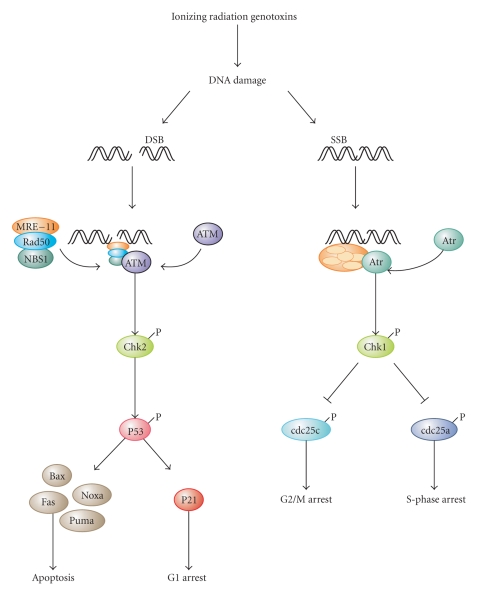
Upon cellular stress conditions that are caused by exposure to chemotherapeutic agents, irradiation, or environmental genotoxic agents such as polycyclic hydrocarbons or ultraviolet (UV) light, damage to DNA is a common initial event [174, 175]. DNA double strand breaks (DSBs) and single strand breaks (SSBs) are considered as key lesions that initiate the activation of the DNA damage response [174]. Since the DNA duplex is more vulnerable to chemical attack or nucleases when it is separated into two single-stranded DNA strands, for example, during DNA replication and transcription, SSBs are preferentially generated under these conditions [176]. Defined SSBs are also generated during distinct pathways of DNA repair, for example, in the course of nucleotide excision repair (NER). After DNA damage recognition, dual incision 5′ to the DNA lesion by ERCC1-XPF and 3′ to the damage by XPG results in the removal of the lesion-containing oligonucleotide [177]. DSBs are produced directly or indirectly by many anticancer drugs, including DNA intercalating, alkylating or crosslinking agents, topoisomerase inhibitors, and nucleotide analogs [174]. Once DSBs are generated, ataxia telangiectasia mutated (ATM) is recruited by the MRE-11-Rad50-NBS1 (MRN) complex to sites of broken DNA and phosphorylates downstream substrates such as checkpoint kinase 2 (Chk2) and p53 [175, 178] (Figure 3). p53 induces transcriptional activation of different functional programs, for example, cell cycle regulatory proteins such as p21 and pro-apoptotic factors such as CD95, PUMA, and BAX [179]. In addition, recent studies have also defined a nontranscriptional pro-apoptotic activity of p53 that regulates the intrinsic mitochondria-mediated pathway of apoptosis [180]. Damage to DNA engages DNA repair processes to ensure the cell's survival in the case of sublethal damage [174]. Alternatively, if the damage is too severe to be repaired—the DNA-damaging insult is transmitted by the cellular stress response to the activation of effector systems to mediate cell death [174]. In the latter case, various stress-inducible molecules, including NF-κB, p53, JNK, or MAPK/ERK, have been implicated in propagating and modulating the cell death signal [81, 82].
Figure 3.
DNA damage responses and cell death. Upon exposure to ionizing radiation or genotoxins, the damage to DNA is a common initial event. DNA double strand breaks (DSBs) or single strand breaks (SSBs) are considered to be key lesions that initiate activation of the DNA damage response. Upon DSBs, ataxia telangiectasia mutated (ATM) is recruited by the MRE-11-Rad50-NBS1 (MRN) complex to sites of broken DNA and phosphorylates downstream substrates such as checkpoint kinase 2 (Chk2), which subsequently phosphorylates p53. Sublethal damage to DNA can engage survival pathways via p21-mediated cell cycle arrest. Alternatively—if the damage is too severe to be repaired—pro-apoptotic p53 target genes are activated including Bax, Puma, Noxa, and Fas, which promote apoptosis. Upon SSBs, it is ataxia telangiectasia and Rad3 related (ATR) that gets activated and phosphorylates Chk1. Chk1 in turn phosphorylates and inhibits cdc25c to mediated G2/M arrest or alternatively cdc25a to promote S-phase arrest.
Depending on the type of lesion, DNA damage initiates one of several mammalian DNA repair pathways, which eventually restore the continuity of the DNA double strand. There are two main pathways for the repair of DSBs, that is, nonhomologous end-joining and homologous recombination [181, 182]. The former constitutes the predominant DNA repair pathway in humans and involves DNA repair proteins such as DNA-PK, Ku70, and Ku80 [181, 182]. Base damage can be repaired either by enzyme-catalyzed reversal or alternatively via excision repair [183]. Mismatch repair is responsible for the removal of incorrectly paired nucleotides [184]. It is important to note that DNA repair can, in principle, be error-free and error-prone. Several proteins have been discovered recently that exert a specific function in error-free repair processes to guarantee high-fidelity reconstitution of the DNA [185]. Faithful genome transmission requires the coordination of this highly complex network of DNA repair pathways and repair surveillance mechanisms linked to cell cycle checkpoints as well as cell death mechanisms [185]. Error-prone repair or complete failure of DNA repair cannot only lead to mutations but can also lead to the initiation of cell death pathways [185].
3.4. The Response to Oxidative Stress
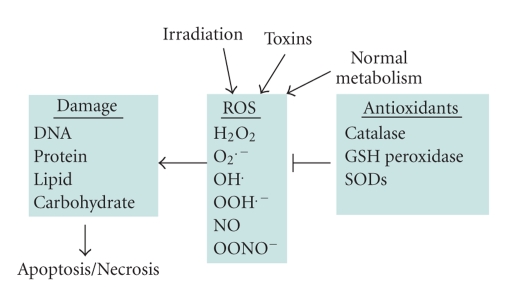
Cell survival requires appropriate proportions of molecular oxygen and various antioxidants. Reactive products of oxygen are amongst the most potent and omnipresent threats faced by cells. These include ROS such as superoxide anion (O2 •−), hydrogen peroxide (H2O2), singlet oxygen, hydroxyl radical (OH•), peroxy radical, as well as the second messenger nitric oxide (NO•) which can react with O2 •− to form peroxynitrite (ONOO−). Normally in cells there exists equilibrium between pro-oxidant species and antioxidant defense mechanisms such as ROS-metabolizing enzymes including catalase, glutathione peroxidase, and superoxide dismutases (SODs) and other antioxidant proteins such as glutathione (GSH) (Figure 4). Oxidative stress occurs when there is a disturbance in this pro-oxidant:antioxidant balance and it has been implicated in several biological and pathological processes [186]. Although most oxidative insults can be overcome by the cell's natural defenses, sustained perturbation of this balance may result in either apoptotic or necrotic cell death [186–190].
Figure 4.
Oxidative stress and cell death. There is a plethora of stimuli that can trigger the generation of reactive oxygen species (ROS), among them irradiation, toxins, and also normal metabolic processes. A range of different ROS species have been identified, which are kept in check by antioxidant defenses. These include several detoxifying enzymes, for example, catalase, GSH peroxidase, and superoxide dismutase (SOD). If these antioxidants defense mechanisms are too weak, ROS-mediated damage to cellular macromolecules will eventually lead to cell death.
ROS can emanate from intracellular or extracellular sources. Auto-oxidation of reduced respiratory components of the mitochondrial electron transport chain causes the production of free radical intermediates, O2 •− and H2O2, which in the presence of iron can produce highly reactive OH• radical via the Fenton reaction. These ROSs are dealt with by SODs, enzymes considered to be the first line of defense against oxygen toxicity. ROS can also be produced in the cytosol. For example, the arachidonic acid cascade, yielding prostaglandins, and leukotrienes may generate ROS when the released lipid is metabolized [191], and some cytochrome P-450 isozymes are well-known ROS producers [192]. Also, the auto-oxidation reactions of ascorbic acid, low molecular weight thiols, adrenalin, and flavin coenzymes can cause ROS production. In many of these cases, cytosolic GSH neutralizes the offenders. In addition to physiological sources of ROS, diverse exogenous agents can contribute to the intracellular production of free radicals. Most of these compounds cause the generation of O2 •− and H2O2 [80, 193, 194]. The mechanism of action of many exogenous agents involves redox cycling whereby an electron is accepted to form a free radical and it is then transferred to oxygen.
Interestingly, there is evidence of cross-talk between oxidative stress and other stress response pathways. For example, oxidative stress is known to cause an increase in the expression of certain inducible Hsps, particularly Hsp27 [195–197]. Hsps have been reported to protect against many stresses apart from heat shock, including heavy metals, radiation, nitric oxide, and other oxidants. In addition, activation of the UPR stimulates upregulation of antioxidant genes through PERK-dependent phosphorylation of the Nrf2 transcription factor, whose target genes include enzymes involved in GSH biosynthesis, and heme oxygenase-1 [198]. Moreover, perturbations in cellular redox status sensitize cells to the harmful effects of ER stress [199]. Similarly, accumulating evidence suggests a role for O2 •− in the activation of autophagy [200].
ROS can cause damage to all of the major classes of biological macromolecules, including nucleic acids, proteins, carbohydrates, and lipids. When the cell's antioxidant defenses are overwhelmed, ROS can induce cell death. Numerous, recent studies have shown that the mode of cell death that occurs depends on the severity of the insult [187–189]. In fact, oxidants and antioxidants not only determine cell fate, but can also modulate the mode of cell death [186, 190].
Many cytotoxic agents induce ROS, including peroxide and O2 •−, which are involved in the induction of apoptotic cell death [201]. H2O2 can cause the release of cytochrome c from mitochondria into the cytosol and H2O2 may also activate nuclear transcription factors, like NF-κB, AP-1, and p53 [202], which may upregulate death proteins or produce inhibitors of survival proteins. One model proposed for H2O2 induction of apoptosis is upregulation of the Fas-FasL system, leading to activation of caspase-8 and downstream caspases [203, 204]. It is also possible that NO• may also inactivate several antioxidant enzymes, including catalase, glutathione peroxidase, and superoxide dismutases [205, 206]. Also, NO• has been reported to induce apoptosis by increasing ceramide generation through caspase-3 activation, induction of mitochondrial permeability transition, and activation of the Fas system [207].
Certain anti-apoptotic proteins have also been reported to have antioxidant roles. An early suggestion regarding the mechanism of action of Bcl-2 was that it inhibited cell death by reducing the generation of reactive oxidants, thus preventing critical intracellular oxidations that are requisite for the completion of the apoptotic program [208]. However, it is now understood that the reduction in ROS observed with Bcl-2 overexpression is probably the result of its ability to prevent loss of cytochrome c from mitochondria. Yet it is interesting to note that separate studies illustrate that Bcl-2-overexpressing cells have higher levels of total cellular GSH [209]. The product of the baculovirus p35 gene, a potent anti-apoptotic protein, is thought to have antioxidant role and is protective against many apoptotic stimuli including growth factor withdrawal, staurosporine, glucocorticoid, and actinomycin-D treatment, and is a broad-spectrum caspase inhibitor [210]. However, caspase inhibition may not be p35's sole mechanism of cytoprotection. Expression of the p35 gene inhibits H2O2-induced apoptosis in insect cells and may be acting as a sink for free radicals [211].
However, ROS are also reported to interfere with the apoptosis death program, compelling cells to adopt an alternative mode of cell death. Apoptotic cell death can be switched to necrosis during oxidative stress by two possible mechanisms: inactivation of caspases or a drop in cellular levels of ATP levels. Caspases contain an active site cysteine nucleophile [212] which is prone to oxidation or thiol alkylation as well as S-nitrosylation [80, 213, 214]. This leads to their inactivation, switching the mode of cell death to necrosis [80, 214]. NO• may act as a molecular switch to control protein function via reactive thiol groups. For example, NO•-mediated inhibition of apoptosis in most cases is due to direct inhibition of caspase activity through S-nitrosylation of the active site cysteine conserved in all caspases although indirect effects on caspases can also be a component of toxicity in certain systems [214]. A switch from apoptosis to necrosis can also occur due to a drop in cellular levels of ATP caused by the failure of mitochondrial energy production by oxidants [215, 216]. As mentioned previously ROS may provide a common link between cellular stress signals and the initiation of autophagy, and ROS accumulation has been reported to result in inactivation of the cysteine protease ATG4, which in turn causes accumulation of the ATG8-phosphoethanolamine precursor that is required for the initiation of autophagosome formation [44]. In most circumstances, the induction of an autophagic response serves as a strategy that should ensure the cell's survival [217]. Under certain conditions, however, it may also bring about cell death, although the molecular determinants that may control the switch from survival to death are still poorly defined. In fact, in response to several anticancer drugs ROS can induce autophagic cell death.
4. Switch from Prosurvival Signaling to Cell Death Signaling
While conditions of stress stimulate cells to mount protective responses to counteract the effect of the stress on cellular processes, if the stress remains unresolved, eventual death of the cell ensues. This raises key questions about the molecular mechanisms involved in this switch from prosurvival signaling to prodeath signaling. For example, is there a particular molecule that acts as a molecular switch? How do the duration and severity of the stress contribute to activation of this switch? As described above, in the face of exposure to cell stress, the cell mounts protective responses such as the heat shock response, or the unfolded protein response, in order to relieve the stress and promote survival. However, it is known that if the stress is very severe or if it is prolonged, the cell will die in spite of the activation of prosurvival signaling.
In the case of the response of cells to heat stress, the induction of Hsps does not occur if the stress is too severe and it has previously been suggested that the induction of thermotolerance, that is, Hsp expression, and of cell death is mutually exclusive events within the same cell [87, 195]. In support of this, we have observed that in a culture exhibiting mixed responses to a stressor, that is, expression of Hsps, induction of apoptosis, and induction of necrosis, the expression of Hsps was mainly observed in the surviving cells [196]. However, a recent report suggests that this may not always be the case, as at least one agent, which induces expression of Hsps through direct activation of HSF1, induces apoptosis rather than being protective [218].
During ER stress, IRE1 may be involved in the switch between the prosurvival UPR and initiation of cell death pathways [219]. Interestingly, the three arms of the UPR are thought to be activated sequentially, with PERK being activated most rapidly, followed by ATF6 and then IRE1. This suggests that time is allowed so that PERK and ATF6 may resolve the stress, and although IRE1 also contributes to the prosurvival UPR, it ultimately terminates it by relieving the translational inhibition by inducing p58IPK [20]. If the stress has been resolved, the cell returns to normal, but if not, then apoptosis is initiated, possibly by IRE1-dependent activation of ASK1 and its downstream target JNK. However, recently it has been shown that attenuation of IRE1 can switch the adaptive UPR to apoptosis and that persistent activation of IRE1 increases cell viability upon ER stress, suggesting that the duration of IRE1 signaling may act as a switch [219].
5. Stress Responses in Disease States
It is currently understood that a pathological stress response is a hallmark of many common human diseases for a number of reasons. Firstly, the stress stimulus may be too strong and/or prolonged, thereby allowing insufficient time for recovery to the normal status. Secondly, a cell's ability to handle even physiological levels of stress may be altered in disease states, similarly resulting in detrimental outcomes. In the following section, we will provide some selected examples of how pathological handling of stress is one of the major underlying causes of the pathophysiological state in very different types of human diseases.
5.1. Diabetes
Loss of function or death of the pancreatic β-cells in the Islets of Langerhans in the pancreas is the major pathological feature of diabetes mellitus. The pancreatic β-cells have a highly developed secretory system, in which the ER has an integral role, enabling a rapid response to glucose stimulation by producing and releasing large amounts of insulin. Both oxidative stress and ER stress are involved in the failure of pancreatic β-cells and development of diabetes.
The reactive species which play an important role in the pathogenesis of pancreatic β-cell loss in diabetes are generated intracellularly when the β-cells are targeted by proinflammatory cytokines in autoimmune Type 1 diabetes or when exposed to a hyperglycaemic and hyperlipidaemic milieu in Type 2 diabetes. There is evidence for the participation of both NO• and ROS in the pathogenesis of β-cell death in Type 1 diabetes, whereas, for β-cell dysfunction in Type 2 diabetes, ROS are the main culprits.
Proinflammatory cytokines, including IL-1β (interleukin 1β), TNFα (tumor necrosis factor α), and IFNγ (interferon γ), released from immune cells infiltrating the pancreas in Type 1 diabetes, target the β-cells via their respective receptors [220]. They activate a multitude of signaling cascades, culminating in apoptosis of β-cells [221]. A number of steps in this chain of events affect the rate of generation of NO• and ROS. It is evident from studies in patients with diabetes and in animal models of Type 1 diabetes, that IL-1β is the key proinflammatory cytokine which significantly contributes to β-cell dysfunction and apoptosis in the pathogenesis of Type 1 diabetes. It does so through activation of the transcription factor NF-κB which is responsible for the induction of iNOS and subsequent production of NO• [155, 221]. The production and release of IFNγ acts synergistically with IL-1β. High concentrations of IFNγ are required to potentiate the effects of IL-1β on iNOS and NO• production [222]. NO• and ROS seem to also cross-talk with ER stress and UPR [223].
IL-1β also induces MnSOD (a manganese-dependent SOD isoenzyme) and this results in an increased rate of conversion of O2 •− into H2O2 in the mitochondria [224]. Cu/ZnSOD, the cytoplasmic isoenzyme, is unaffected by IL-1β. The profile of the effects of TNFα and IFNγ alone, or in combination with IL-1β, on MnSOD is comparable with that of their regulation of iNOS. The effects on the generation of both radicals are not only important in themselves but also affect the balance between NO• and O2 •−, and this can have significant effects on β-cell toxicity. A decrease in O2 •− through MnSOD may present as a protective signal through a reduction of NF-κB activation and other components of the IL-1β signaling pathway [225]. On the other hand, an increased conversion rate of O2 •− into H2O2 by SOD is likely to increase toxicity to the β-cell with its poor enzymatic capacity for H2O2 inactivation [226, 227].
Another major proinflammatory cytokine, TNFα, is released from the infiltrating immune cells speeds up β-cell loss significantly, resulting in an accelerated progression of the disease with rapid loss of the entire pancreatic β-cell population and Islet mass. Ceramide is likely to play a significant role as a mediator of O2 •− formation in TNFα-mediated toxicity [228], thereby explaining the dominance of ROS in the case of TNFα when compared with IL-1β. Thus, with a significant contribution of TNFα produced by the infiltrating immune cells in Type 1 diabetes the resulting greater cytotoxicity is the result of the more pronounced ROS component of TNFα toxicity.
That the ROS-mediated component of cytokine toxicity primarily targets the mitochondria is shown by the fact that exposure of insulin-producing cells to IL-1β, or to a cytokine mixture containing both IFNγ and TNFα, causes mitochondrial damage, while other subcellular structures remain intact. This damage can be prevented by expression of high levels of catalase or GSH in the mitochondria, but not in the cytosol [228]. IL-1β toxicity, mediated through NO• and potentiated by IFNγ and TNFα, is likely to focus its effects in the cytoplasm. This component will presumably contribute to ER stress, which plays a significant role in dysfunction of β-cells under cytokine attack [229].
β-Cell loss in Type 2 diabetes is slower than in Type 1 diabetes, typically with a long phase of β-cell dysfunction, characterized by defective insulin secretion in response to glucose. In Type 2 diabetes, glucolipotoxicity, rather than proinflammatory cytokines, is considered to be an important contributing factor to β-cell dysfunction [230–234]. It is evident from studies on β-cells exposed to a combination of high glucose and a saturated fatty acid that NO• generation through iNOS induction does not contribute to β-cell dysfunction [235].
Increased mitochondrial metabolic flux is required in the β-cell for generation of the ATP signal for glucose-induced insulin secretion [236] and its potentiation through fatty acids [231]. On the other hand, increased metabolic flux through the respiratory chain at high glucose and lipid concentrations should increase O2 •− formation, thereby reducing the mitochondrial membrane potential via uncoupling protein 2 [230, 237]. This should decrease metabolic flux through the respiratory chain and thus reduce O2 •− production, thereby acting in a protective manner against ROS-induced damage, but, at the same time, attenuating nutrient-induced insulin secretion. This casts doubt on the concept that increased intra-mitochondrial generation of ROS crucially contributes to β-cell damage in Type 2 diabetes.
This interpretation is supported by the results of morphological analyses showing that insulin-producing cells exposed to the fatty acid palmitate show no signs of mitochondrial damage, but very pronounced defects of the ER [238], confirming observations of increased ER stress in response to glucolipotoxicity [235]. Thus one of the prominent targets of this free-radical-mediated toxicity might indeed be the ER.
Defects in PERK-eIF2α pathways cause Wolcott-Rallison syndrome, a rare infantile-onset insulin-requiring diabetes [239] and PERK-null mice developed similar phenotypes [240]. Mice with mutated proinsulin (proinsulin-2) that cannot form a disulfide bond (Akita mice) also develop severe diabetes which is associated with the ER stress-induced cell death in pancreatic β-cells [241, 242]. Mice deficient for p58IPK, which suppresses PERK-mediated phosphorylation of eIF2α, exhibit apoptosis of pancreatic β-cells and diabetes [243]. This suggests that the tight regulation of PERK-eIF2α pathway is required for the maintenance of pancreatic β-cells. In contrast, some single nucleotide polymorphisms (SNPs) in the ORP150/GRP170 genome of Pima Indians are associated with insulin sensitivity in peripheral tissues [244]. Accordingly, overexpression of ORP150 enhances insulin sensitivity and suppresses oxidative stress but does not improve insulin secretion [245]. These findings suggest that proper functioning of the ER is important for both insulin synthesis in pancreatic β-cells and insulin sensitivity in peripheral tissues. Consistent with this hypothesis, chemical chaperones such as 4-phenylbutryic acid and tauroursodeoxycholic acid improved both insulin resistance and insulin synthesis [171, 246].
5.2. Parkinson's Disease
Neurodegenerative diseases are characterized by the loss of subsets of neurons. The course of these diseases can last decades, with the accumulation of neuronal loss causing progressively worse symptoms. Postmortem tissue is usually obtained from end-stage patients, at which time many of the evidences regarding the events preceding cell death are long gone. However, there is substantial and growing evidence for the activation of stress responses in neurons in all of the common neurodegenerative diseases. This suggests that when neurons are exposed to stress, they counteract with activation of one or more protective stress responses; however, eventually the neurons are unable to cope and one-by-one they are lost as the disease progresses. There is a growing recognition that protein misfolding and impairment of protein handling play a key role in neuronal cell death in neurodegenerative diseases [153].
As an example of stress responses and stress-induced cell death in neurodegenerative disease, we will describe the evidence pertaining to Parkinson's disease. Parkinson's disease is the second most common neurodegenerative disease, affecting mainly people over 55 years and causing progressively worsening motor impairment. It is characterized pathologically by the degeneration of midbrain dopaminergic neurons in the substantia nigra pars compacta and the presence of proteinaceous intracytoplasmic inclusions (Lewy bodies) within the surviving neurons.
The molecular mechanisms that initiate dopaminergic neuron loss in Parkinson's disease are not known. Evidence from various sources suggest that environmental toxins, genetic predisposition, and aging are important factors in the onset and progression of the disease [247–249]. Insecticides such as rotenone and the mitochondrial toxin 1-Methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP) cause dopaminergic neuronal loss in animal models and have been implicated in Parkinson's disease itself [250, 251]. To date, mutations in at least 13 PARK genes have been linked to the pathogenesis of familial Parkinson's disease which include mutations in genes that encode the proteins α-synuclein, parkin, PTEN-induced kinase 1 (PINK1), DJ-1, leucine-rich repeat kinase2 (LRRK2), Omi/Htra2, and ubiquitin carboxy-terminal hydrolase L1 (UCHL1) [252]. Of these, α-synuclein (along with chaperone proteins and ubiquitin) is a major component of Lewy bodies. Parkin and UCHL1 are linked to the ubiquitin-proteasome system that degrades damaged or misfolded proteins [253]. In addition, several of these genes, including parkin, PINK1, DJ-1, and Omi/Htra2 are linked to the mitochondria and may have roles in mitochondrial function and resistance to oxidative stress [254].
Mutations in PARK genes, as well as toxins that specifically target dopaminergic neurons, have been strongly linked to the activation of stress responses in dopaminergic neurons. For example, mitochondrial dysfunction due to mutations in certain PARK genes or to environmental toxins is linked with impairment of mitochondrial complex I which causes oxidative stress in affected cells. It has long been known that oxidative stress is a feature of Parkinson's disease and it is observed in experimental models of Parkinson's disease and in tissues from individuals with sporadic forms of the disease [255].
Most of the evidence regarding activation of the heat shock response in Parkinson's disease come from models. Targeted overexpression of α-synuclein in mouse substantia nigra causes an increase in the expression of Hsp27, Hsp40, and Hsp70 [256, 257] and elevations in Hsp27 are observed in in vitro models of Parkinson's disease using the neurotoxin 6-hydroxdopamine [196]. Recent findings from Parkinsonian patients have described that DnaJB6 is present in the core of Lewy bodies and is also upregulated in astrocytes [258]. DnaJB6 is one of the Hsp40 chaperones, which stabilizes the interactions of Hsp70s with their substrate proteins. In vitro and in vivo models of Parkinson's disease demonstrate that overexpression of Hsps prevents α-synuclein aggregation as well as dopaminergic neuronal cell death due to α-synuclein and Parkinson mimetic toxins [216, 237, 239–242]. Interestingly, the inducibility of Hsps decreases with aging, which may contribute to the inability of aged neurons to fully protect themselves from stresses such as protein misfolding, aggregation, and oxidative stress [259].
Activation of the UPR has been reported in postmortem brain tissue from patients with Parkinson's disease. Specifically, phosphorylated PERK and phosphorylated eIF2α have been detected in dopaminergic neurons in the substantia nigra of Parkinson's disease patients [260]. Phospho-PERK immunoreactivity was colocalized with increased α-synuclein immunoreactivity in dopaminergic neurons [260]. Supporting evidences from in vitro models of Parkinson's disease show that 6-hydroxydopamine and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPP+) (Parkinson mimetic drugs) trigger ER stress in dopaminergic neurons [261, 262]. Furthermore, neuronal cultures from PERK knockout mice display an increased sensitivity to 6-hydroxydopamine [262], while a null mutation in CHOP results in a reduction in 6-hydroxydopamine-induced apoptosis in vivo [263]. However, protection was not observed in the chronic MPTP model, despite robust expression of CHOP [263].
The information from models, the genetic information, as well as analysis of postmortem tissue, when taken together, strongly connects the induction of stress responses with the loss of dopaminergic neurons in Parkinson's disease. It is likely that the induction of stress responses is the neurons attempts at protection, which eventually fail with neuronal cell death being the inevitable outcome. Interestingly, these observations are mirrored in research findings of other common neurodegenerative diseases, including Alzheimer's disease and Huntington's disease, indicating the important role for protein misfolding, aggregation and formation of protein inclusions in these chronic diseases [153].
5.3. Myocardial Infarction
Cardiovascular disease (CVD), a group of disorders of the heart and the vasculature, includes high blood pressure, coronary heart disease, congestive heart failure, stroke, and congenital heart defects. Apoptotic cell death is a fundamental process in the morphogenesis of the developing heart [264, 265]. Until recently the classical view was that necrosis was the major mode of cardiomyocyte death during CVD. However, accumulating in vitro and in vivo studies provides compelling evidence that terminally differentiated cardiomyocytes, can and do undergo apoptosis [266]. Apoptosis has important pathophysiological consequences, contributing to the loss and functional abnormalities of the myocardium. Cardiomyocyte apoptosis has been reported in a variety of cardiovascular diseases, including myocardial infarction, end-stage heart failure, arrhythmogenic right ventricular dysplasia, and adriamycin-induced cardiomyopathy [267]. Animal models have been instrumental in establishing the occurrence of cardiomyocyte apoptosis and in the elucidation of the apoptotic mechanisms. Features of myocyte apoptosis were first reported in rabbit and rat heart models of MI or ischemia/reperfusion injury [268, 269]. Since these pioneering studies, apoptosis has been repeatedly observed in the injured human heart [270–274]. Due to its sporadic occurrence and the prompt clearance of apoptotic cells by phagocytosis, apoptosis in diseased tissue is grossly underestimated.
Oxidative damage mediated by free radicals is a contributing factor to ischemia/reperfusion-induced injury in cardiomyocytes [275–278]. Plasma and pericardial fluid obtained from patients with end stage heart failure have increased levels of thiobarbituric acid reactive substances, a commonly used marker of ROS production [279, 280]. Reperfusion is associated with a burst of ROS generated via the mitochondrial respiratory chain, where partial reduction of ubiquinone forms ubisemiquinone combine with oxygen to form O2 •− radicals [281]. High levels of ROS can lead to mitochondrial damage and dysfunction [282] and can induce apoptosis in cardiac myocytes [275, 276].
In addition, enhanced levels of the heat shock response and UPR have been demonstrated in animal models of myocardial infarction, and overexpression of either Hsps or GRPs enhanced tolerance against ischemia/reperfusion injury in these models [283, 284]. Although Hsps may work upstream of caspase-3 but downstream of cytochrome c release [285], GRPs likely contribute to the maintenance of intracellular Ca2+ homeostasis [284]. Similarly, overexpression of sarco (endo) plasmic reticulum Ca2+-ATPase (SERCA), which regulates intracellular Ca2+ homeostasis, improved postischemic cardiac function and decreased myocardial infarction [286].
5.4. Cancer
Since tissue homeostasis is the result of a subtle balance between proliferation on one side and cell death on the other side, changes in the rate of cell death can contribute to either the loss or gain of tissue [22]. For example, too little cell death can contribute to tumor formation and is considered to be one of the hallmarks of human cancers [287, 288]. Some oncogenic mutations block cell death pathways creating a permissive environment for genetic instability and resulting in the accumulation of gene mutations leading to tumor initiation and progression [289]. Also, evasion of cell death promotes resistance to immune-based destruction, facilitates growth factor- or hormone-independent survival, and supports anchorage-independent survival during metastasis [288]. In addition, defects in cell death programs may confer resistance to cytotoxic therapies that are currently used in the clinic for the treatment of cancer such as chemotherapy, irradiation, or immunotherapy, since the response of cancer cells to these treatment approaches is, to a large extent, due to their ability to undergo cell death in response to cytotoxic stimuli [290–292].
In principle, the signaling to apoptosis can be blocked in cancers by loss or defective function of proapoptotic molecules, aberrantly high expression of antiapoptotic proteins, and/or by the relative dominance of cell survival signaling pathways. For example, impaired death receptor expression or function has been reported in a variety of human cancers. Reduced expression of CD95 was found in drug-resistant leukemia or neuroblastoma cells, indicating that intact signaling via CD95 is linked to drug response [293, 294]. CD95 mutations have been detected in both hematological malignancies and various solid tumors [295–300]. It is interesting to note that both agonistic TRAIL receptors, that is, TRAIl-R1 and TRAIL-R2, are located on chromosome 8p, a region that is frequently lost in cancers due to heterozygosity [301, 302]. Further, a larger range of antiapoptotic proteins are reported to be expressed at high levels in malignant versus nonmalignant tissue, including death domain-containing proteins that interfere with activation of caspase-8 at the death receptor level such as cellular FLICE-Inhibitory Protein (cFLIP) and phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes-15kDa (PED/PEA-15) [303], anti-apoptotic Bcl-2 family proteins such as Bcl-2, Bcl-XL, and Mcl-1 [31] and IAPs, including XIAP, cIAP1, cIAP2, survivin and livin [304]. Alternatively, apoptosis regulators with proapoptotic functions have been reported to be lost, mutated or epigenetically silenced in cancers. Examples include epigenetic loss or homo- or heterozygous genomic deletions of caspase-8 [305], single nucleotide substitution or frameshift mutations of the bax gene in mismatch repair-deficient colon cancer or hematopoetic malignancies [306, 307], and deletion or epigenetic silencing of the bim gene [308–310].
It is also now generally accepted that the majority of tumors, due to poor vascularisation of the tumor mass, experience stressful conditions in the tumor microenvironment, including low oxygen supply, nutrient deprivation, and pH changes. These conditions activate a range of cellular stress-response pathways, including the UPR. Recent studies have shown that the UPR plays an important role in tumorigenesis [311–315]. Activation of at least one branch of the UPR has been reported in a number of cancers and many ER chaperones and UPR target genes show increased expression in human tumor samples. Although activation of the UPR has been reported in a variety of human cancers, the role of UPR in different forms of cancer is not yet fully characterized.
At present it is unclear how tumor cells adapt to long-term ER stress in vivo—whether the protective elements of the response are enhanced, the destructive components suppressed, or if the compromised apoptotic machinery is sufficient to protect them from UPR-induced apoptosis. Given that the UPR can trigger prosurvival and pro-apoptotic signals, it is important to understand how modulation of the UPR alters the balance between these processes and contributes to carcinogenesis in different cell types. The upregulation of UPR in cancers may be beneficial for the tumor cells by increasing the protein folding capacity and prolonging life.
Moreover, altered redox status can promote tumor initiation and progression by blunting cell death pathways. For example, a pro-oxidant intracellular milieu has been linked to carcinogenesis and tumor promotion. To this end, increased signaling via the PI3K/Akt pathway has been shown to result in enhanced intracellular ROS generation [316]. Similarly, cancer cells that constitutively express oncogenic Ras have been reported to harbor higher intracellular levels of O2 •− and to be resistant to drug-induced apoptosis [317].
Hsps, including Hsp90, Hsp70, and Hsp27, are expressed at increased levels in many solid tumors and haematological malignancies. Since various oncogenic proteins that are critically required for the malignant transformation of cells, for example, Ras, Akt, and HER2, are client proteins of Hsp90, elevated levels of Hsp90 favor tumor initiation and promotion [318]. Similarly, the expression of Hsp27 and Hsp70 is abnormally high in cancers [319]. These chaperones participate in carcinogenesis and in cell death resistance by blocking key effector molecules of the apoptotic machinery at the pre- and post-mitochondrial level [319]. Thus, targeting Hsps, for example, with chemical inhibitors, is currently under investigation as anticancer strategy [318].
Error-prone repair or complete failure to repair DNA damage as well as inherited or acquired defects in maintenance systems of the mammalian genome can lead to mutations [185]. In addition, such deficiencies in the DNA damage response contribute substantially to carcinogenesis and promote the progression and treatment resistance of cancer [185].
6. Summary and Future Perspectives
Cellular stress responses are an integral part of normal physiology to either ensure the cell's survival or alternatively to eliminate damaged or unwanted cells. Several distinct stress responses can be distinguished, among them the heat shock, unfolded protein, DNA damage, and oxidative stress responses. Despite individual signaling components, these different stress responses can eventually fuel into common cell death effector mechanisms, if the cell is unable to cope with the stress. Whether or not cellular stress triggers cell death or cell survival programs is determined by a set of different factors, among them the initial stress stimulus, cell type, and environmental factors. Because aberrant cellular stress responses are tightly linked to many common human diseases, a better understanding of the underlying molecular mechanisms is expected to enable us to interfere with these processes, for example, to switch such response from cell death into survival programs or vice versa, depending on the desired outcome. In addition, new insights into the mechanistic basis of stress responses will open new perspectives for the development of molecular targeted treatment approaches and thus have a great potential for drug discovery.
Acknowledgments
The authors are grateful to Drs. Sandra Healy and Sanjeev Gupta for their suggestions and comments on this manuscript. Fulda's group is supported by grants from the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung, the Deutsche Krebshilfe, the EU (ApopTrain, APO-SYS), the Wilhelm Sander Stiftung, the Else-Kröner-Fresenius Stiftung, the Novartis Stiftung für therapeutische Forschung, and IAP6/18. Research in Samali and Gorman groups is financially supported by Science Foundation Ireland under Grant nos. 09/RFP/BMT2153, 09/RFP/BIC2371, and 05/IN3/B851 as well as grants from Health Research Board of Ireland (HRA/2009/59) and the Breast Cancer Campaign.
References
- 1.Lockshin RA, Zakeri Z. Programmed cell death and apoptosis: origins of the theory. Nature Reviews Molecular Cell Biology. 2001;2(7):545–550. doi: 10.1038/35080097. [DOI] [PubMed] [Google Scholar]
- 2.Lockshin RA, Williams CM. Programmed cell death—I. Cytology of degeneration in the intersegmental muscles of the Pernyi silkmoth. Journal of Insect Physiology. 1965;11(2):123–133. doi: 10.1016/0022-1910(65)90099-5. [DOI] [PubMed] [Google Scholar]
- 3.Lockshin RA, Williams CM. Programmed cell death—IV. The influence of drugs on the breakdown of the intersegmental muscles of silkmoths. Journal of Insect Physiology. 1965;11(6):803–809. doi: 10.1016/0022-1910(65)90159-9. [DOI] [PubMed] [Google Scholar]
- 4.Lockshin RA, Williams CM. Programmed cell death—V. Cytolytic enzymes in relation to the breakdown of the intersegmental muscles of silkmoths. Journal of Insect Physiology. 1965;11(7):831–844. doi: 10.1016/0022-1910(65)90186-1. [DOI] [PubMed] [Google Scholar]
- 5.Pützer BM. E2F1 death pathways as targets for cancer therapy. Journal of Cellular and Molecular Medicine. 2007;11(2):239–251. doi: 10.1111/j.1582-4934.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeve JLV, Duffy AM, O’Brien T, Samali A. Don’t lose heart—therapeutic value of apoptosis prevention in the treatment of cardiovascular disease. Journal of Cellular and Molecular Medicine. 2005;9(3):609–622. doi: 10.1111/j.1582-4934.2005.tb00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samali A. A tribute to professor Richard A. Lockshin on his 70th birthday. Journal of Cellular and Molecular Medicine. 2007;11(6):1210–1211. doi: 10.1111/j.1582-4934.2007.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakeri Z, Bursch W, Tenniswood M, Lockshin RA. Cell death: programmed, apoptosis, necrosis, or other? Cell Death and Differentiation. 1995;2(2):87–96. [PubMed] [Google Scholar]
- 9.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. British Journal of Cancer. 1972;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr JF. Shrinkage necrosis: a distinct mode of cellular death. Journal of Pathology. 1971;105(1):13–20. doi: 10.1002/path.1711050103. [DOI] [PubMed] [Google Scholar]
- 11.Trump BF, Goldblatt PJ, Stowell RE. An electron microscopic study of early cytoplasmic alterations in hepatic parenchymal cells of mouse liver during necrosis in vitro (autolysis) Laboratory Investigation. 1962;11:986–1015. [PubMed] [Google Scholar]
- 12.Berenbom M, Chang PI, Betz HE, Stowell RE. Chemical and enzymatic changes associated with mouse liver necrosis in vitro. Cancer Research. 1955;15(1):1–5. [PubMed] [Google Scholar]
- 13.Trauth BC, Klas C, Peters AMJ, et al. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 1989;245(4915):301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Liu X, Bhalla K, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275(5303):1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 15.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335(6189):440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 16.Hockenbery D, Nunez G, Milliman C, Schreiber RD, Korsmeyer SJ. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348(6299):334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- 17.Itoh N, Yonehara S, Ishii A, et al. The polypeptide encoded by the cDNA for human cell surface antigen fas can mediate apoptosis. Cell. 1991;66(2):233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell. 1993;75(4):641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 19.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl- 2 regulation of apoptosis. Science. 1997;275(5303):1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 20.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Reports. 2006;7(9):880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samali A, Zhivotovsky B, Jones D, Nagata S, Orrenius S. Apoptosis: cell death defined by caspase activation. Cell Death and Differentiation. 1999;6(6):495–496. doi: 10.1038/sj.cdd.4400520. [DOI] [PubMed] [Google Scholar]
- 22.Lockshin RA, Zakeri Z. Cell death in health and disease. Journal of Cellular & Molecular Medicine. 2007;11(6):1214–1224. doi: 10.1111/j.1582-4934.2007.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samali A, Gorman AM, Cotter TG. Apoptosis—the story so far. Experientia. 1996;52(10-11):933–941. doi: 10.1007/BF01920101. [DOI] [PubMed] [Google Scholar]
- 24.Bree RT, Stenson-Cox C, Grealy M, Byrnes L, Gorman AM, Samali A. Cellular longevity: role of apoptosis and replicative senescence. Biogerontology. 2002;3(4):195–206. doi: 10.1023/a:1016299812327. [DOI] [PubMed] [Google Scholar]
- 25.Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22(53):8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 26.Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine and Growth Factor Reviews. 2008;19(3-4):325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Samali A. Apoptosis: a mapped path to cell death. Journal of Cellular and Molecular Medicine. 2007;11(6):1212–1213. doi: 10.1111/j.1582-4934.2007.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiological Reviews. 2007;87(1):99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 29.Zou H, Li Y, Liu X, Wang X. An APAf-1· cytochrome C multimeric complex is a functional apoptosome that activates procaspase-9. Journal of Biological Chemistry. 1999;274(17):11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 30.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27(48):6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 31.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26(9):1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2(3):183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Molecular Cell. 2005;17(3):393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 34.Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315(5813):856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 35.Varfolomeev E, Vucic D. (Un)expected roles of c-IAPs in apoptotic and NFκB signaling pathways. Cell Cycle. 2008;7(11):1511–1521. doi: 10.4161/cc.7.11.5959. [DOI] [PubMed] [Google Scholar]
- 36.Eskelinen E-L. New insights into the mechanisms of macroautophagy in Mammalian cells. International Review of Cell and Molecular Biology. 2008;266:207–247. doi: 10.1016/S1937-6448(07)66005-5. [DOI] [PubMed] [Google Scholar]
- 37.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 38.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nature Reviews Molecular Cell Biology. 2001;2(3):211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 39.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO Journal. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120(2):237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 41.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447(7146):859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 42.Høyer-Hansen M, Bastholm L, Szyniarowski P, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Molecular Cell. 2007;25(2):193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Ogata M, Hino S-I, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Molecular & Cellular Biology. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO Journal. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nature Medicine. 2007;13(5):619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 46.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. American Journal of Pathology. 1980;98(2):425–444. [PMC free article] [PubMed] [Google Scholar]
- 47.Khan S, Salloum F, Das A, Xi L, Vetrovec GW, Kukreja RC. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. Journal of Molecular and Cellular Cardiology. 2006;41(2):256–264. doi: 10.1016/j.yjmcc.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptopic programmed cell death dependent on autophagy genes. Nature Cell Biology. 2004;6(12):1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 49.Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophaglic cell death by caspase-8. Science. 2004;304(5676):1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 50.Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death and Differentiation. 2008;15(2):422–425. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boya P, González-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Molecular & Cellular Biology. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Akar U, Chaves-Reyez A, Barria M, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4(5):669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 54.Erlich S, Mizrachy L, Segev O, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3(6):561–568. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 55.Maiuri MC, Criollo A, Tasdemir E, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-XL. Autophagy. 2007;3(4):374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 56.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO Journal. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-beclin 1 peptide complex: beclin 1 is a novel BH3-only protein. Journal of Biological Chemistry. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 58.Vanlangenakker N, Berghe TV, Krysko DV, Festjens N, Vandenabeele P. Molecular mechanisms and pathophysiology of necrotic cell death. Current Molecular Medicine. 2008;8(3):207–220. doi: 10.2174/156652408784221306. [DOI] [PubMed] [Google Scholar]
- 59.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends in Biochemical Sciences. 2007;32(1):37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135(7):1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes and Development. 2004;18(11):1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ankarcrona M, Dypbukt JM, Bonfoco E, et al. Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. Neuron. 1995;15(4):961–973. doi: 10.1016/0896-6273(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 63.Zhu C, Wang X, Xu F, et al. The influence of age on apoptotic and other mechanisms of cell death after cerebral hypoxia-ischemia. Cell Death and Differentiation. 2005;12(2):162–176. doi: 10.1038/sj.cdd.4401545. [DOI] [PubMed] [Google Scholar]
- 64.Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death and Differentiation. 2007;14(3):400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- 65.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends in Biochemical Sciences. 2005;30(3):151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 66.Holler N, Zaru R, Micheau O, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature Immunology. 2000;1(6):489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 67.Chan FK, Shisler J, Bixby JG, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. Journal of Biological Chemistry. 2003;278(51):51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 68.Ma Y, Temkin V, Liu H, Pope RM. NF-κB protects macrophages from lipopolysaccharide-induced cell death: the role of caspase 8 and receptor-interacting protein. Journal of Biological Chemistry. 2005;280(51):41827–41834. doi: 10.1074/jbc.M510849200. [DOI] [PubMed] [Google Scholar]
- 69.Degterev A, Huang Z, Boyce M, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature Chemical Biology. 2005;1(2):112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 70.Degterev A, Hitomi J, Germscheid M, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature Chemical Biology. 2008;4(5):313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.You Z, Savitz SI, Yang J, et al. Necrostatin-1 reduces histopathology and improves functional outcome after controlled cortical impact in mice. Journal of Cerebral Blood Flow and Metabolism. 2008;28(9):1564–1573. doi: 10.1038/jcbfm.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho YS, Challa S, Moquin D, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137(6):1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He S, Wang L, Miao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α . Cell. 2009;137(6):1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 74.Zhang DW, Shao J, Lin J, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325(5938):332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 75.Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. Journal of Biological Chemistry. 1992;267(8):5317–5323. [PubMed] [Google Scholar]
- 76.Kalai M, Van Loo G, Vanden Berghe T, et al. Tipping the balance between necrosis and apoptosis in human and murine cells treated with interferon and dsRNA. Cell Death and Differentiation. 2002;9(9):981–994. doi: 10.1038/sj.cdd.4401051. [DOI] [PubMed] [Google Scholar]
- 77.Van Herreweghe F, Mao J, Chaplen FWR, et al. Tumor necrosis factor-induced modulation of glyoxalase I activities through phosphorylation by PKA results in cell death and is accompanied by the formation of a specific methylglyoxal-derived AGE. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(2):949–954. doi: 10.1073/pnas.012432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hansford RG, Zorov D. Role of mitochondrial calcium transport in the control of substrate oxidation. Molecular & Cellular Biochemistry. 1998;184(1-2):359–369. [PubMed] [Google Scholar]
- 79.Chua BT, Guo K, Li P. Direct cleavage by the calcium-activated protease calpain can lead to inactivation of caspases. Journal of Biological Chemistry. 2000;275(7):5131–5135. doi: 10.1074/jbc.275.7.5131. [DOI] [PubMed] [Google Scholar]
- 80.Samali A, Nordgren H, Zhivotovsky B, Peterson E, Orrenius S. A comparative study of apoptosis and necrosis in HepG2 cells: oxidant-induced caspase inactivation leads to necrosis. Biochemical and Biophysical Research Communications. 1999;255(1):6–11. doi: 10.1006/bbrc.1998.0139. [DOI] [PubMed] [Google Scholar]
- 81.Weston CR, Davis RJ. The JNK signal transduction pathway. Current Opinion in Cell Biology. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 82.Perkins ND, Gilmore TD. Good cop, bad cop: the different faces of NF-κB. Cell Death and Differentiation. 2006;13(5):759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 83.Lindquist S. The heat-shock response. Annual Review of Biochemistry. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 84.Craig EA. The heat shock response. CRC Critical Reviews in Biochemistry. 1985;18(3):239–280. doi: 10.3109/10409238509085135. [DOI] [PubMed] [Google Scholar]
- 85.Samali A, Orrenius S. Heat shock proteins: regulators of stress response and apoptosis. Cell Stress and Chaperones. 1998;3(4):228–236. doi: 10.1379/1466-1268(1998)003<0228:hspros>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samali A, Cotter TG. Heat shock proteins increase resistance to apoptosis. Experimental Cell Research. 1996;223(1):163–170. doi: 10.1006/excr.1996.0070. [DOI] [PubMed] [Google Scholar]
- 87.Samali A, Holmberg CI, Sistonen L, Orrenius S. Thermotolerance and cell death are distinct cellular responses to stress: dependence on heat shock proteins. FEBS Letters. 1999;461(3):306–310. doi: 10.1016/s0014-5793(99)01486-6. [DOI] [PubMed] [Google Scholar]
- 88.Li GC, Werb Z. Correlation between synthesis of heat shock proteins and development of thermotolerance in Chinese hamster fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(10):3218–3222. doi: 10.1073/pnas.79.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morimoto RI, Kroeger PE, Cotto JJ. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. EXS. 1996;77:139–163. doi: 10.1007/978-3-0348-9088-5_10. [DOI] [PubMed] [Google Scholar]
- 90.Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB Journal. 2001;15(7):1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 91.Shabtay A, Arad Z. Reciprocal activation of HSF1 and HSF3 in brain and blood tissues: is redundancy developmentally related? American Journal of Physiology. 2006;291(3):R566–R572. doi: 10.1152/ajpregu.00685.2005. [DOI] [PubMed] [Google Scholar]
- 92.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. Journal of Biological Chemistry. 1998;273(13):7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 93.Xiao X, Zuo X, Davis AA, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO Journal. 1999;18(21):5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible hsp molecular chaperones. Journal of Cellular Biochemistry. 2002;86(2):376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
- 95.Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat Shock Factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. Journal of Biological Chemistry. 2007;282(10):7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- 96.Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cellular and Molecular Life Sciences. 2008;65(6):855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress and Chaperones. 2004;9(2):122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jäättelä M. Heat shock proteins as cellular lifeguards. Annals of Medicine. 1999;31(4):261–271. doi: 10.3109/07853899908995889. [DOI] [PubMed] [Google Scholar]
- 99.Lindquist S, Craig EA. The heat-shock proteins. Annual Review of Genetics. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 100.Garrido C. Size matters: of the small HSP27 and its large oligomers. Cell Death and Differentiation. 2002;9(5):483–485. doi: 10.1038/sj.cdd.4401005. [DOI] [PubMed] [Google Scholar]
- 101.Jäättelä M, Wissing D, Kokholm K, Kallunki T, Egeblad M. Hsp7O exerts its anti-apoptotic function downstream of caspase-3-like proteases. EMBO Journal. 1998;17(21):6124–6134. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kabakov AE, Gabai VL. Heat-shock-induced accumulation of 70-kDa stress protein (HSP70) can protect ATP-depleted tumor cells from necrosis. Experimental Cell Research. 1995;217(1):15–21. doi: 10.1006/excr.1995.1058. [DOI] [PubMed] [Google Scholar]
- 103.Mehlen P, Preville X, Chareyron P, Briolay J, Klemenz R, Arrigo AP. Constitutive expression of human hsp27, Drosophila hsp27, or human αB- crystallin confers resistance to TNF- and oxidative stress-induced cytotoxicity in stably transfected murine L929 fibroblasts. Journal of Immunology. 1995;154(1):363–374. [PubMed] [Google Scholar]
- 104.Champagne MJ, Dumas P, Orlov SN, Bennett MR, Hamet P, Tremblay J. Protection against necrosis but not apoptosis by heat-stress proteins in vascular smooth muscle cells: evidence for distinct modes of cell death. Hypertension. 1999;33(3):906–913. doi: 10.1161/01.hyp.33.3.906. [DOI] [PubMed] [Google Scholar]
- 105.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 106.Concannon CG, Gorman AM, Samali A. On the role of Hsp27 in regulating apoptosis. Apoptosis. 2003;8(1):61–70. doi: 10.1023/a:1021601103096. [DOI] [PubMed] [Google Scholar]
- 107.Concannon CG, Orrenius S, Samali A. Hsp27 inhibits cytochrome c-mediated caspase activation by sequestering both pro-caspase-3 and cytochrome c. Gene Expression. 2001;9(4-5):195–201. doi: 10.3727/000000001783992605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pandey P, Farber R, Nakazawa A, et al. Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene. 2000;19(16):1975–1981. doi: 10.1038/sj.onc.1203531. [DOI] [PubMed] [Google Scholar]
- 109.Bruey J-M, Ducasse C, Bonniaud P, et al. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nature Cell Biology. 2000;2(9):645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]
- 110.Samali A, Robertson JD, Peterson E, et al. Hsp27 protects mitochondria of thermotolerant cells against apoptotic stimuli. Cell Stress and Chaperones. 2001;6(1):49–58. doi: 10.1379/1466-1268(2001)006<0049:hpmotc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chauhan D, Li G, Hideshima T, et al. Hsp27 inhibits release of mitochondrial protein Smac in multiple myeloma cells and confers dexamethasone resistance. Blood. 2003;102(9):3379–3386. doi: 10.1182/blood-2003-05-1417. [DOI] [PubMed] [Google Scholar]
- 112.Steel R, Doherty JP, Buzzard K, Clemons N, Hawkins CJ, Anderson RL. Hsp72 inhibits apoptosis upstream of the mitochondria and not through interactions with Apaf-1. Journal of Biological Chemistry. 2004;279(49):51490–51499. doi: 10.1074/jbc.M401314200. [DOI] [PubMed] [Google Scholar]
- 113.Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nature Cell Biology. 2000;2(8):476–483. doi: 10.1038/35019510. [DOI] [PubMed] [Google Scholar]
- 114.Beere HM, Wolf BB, Cain K, et al. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nature Cell Biology. 2000;2(8):469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 115.Komarova EYu, Afanasyeva EA, Bulatova MM, Cheetham ME, Margulis BA, Guzhova IV. Downstream caspases are novel targets for the antiapoptotic activity of the molecular chaperone Hsp70. Cell Stress and Chaperones. 2004;9(3):265–275. doi: 10.1379/CSC-27R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Creagh EM, Carmody RJ, Cotter TG. Heat shock protein 70 inhibits caspase-dependent and -independent apoptosis in Jurkat T cells. Experimental Cell Research. 2000;257(1):58–66. doi: 10.1006/excr.2000.4856. [DOI] [PubMed] [Google Scholar]
- 117.Ravagnan L, Gurbuxani S, Susin SA, et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nature Cell Biology. 2001;3(9):839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 118.Charette SJ, Landry J. The interaction of HSP27 with Daxx identifies a potential regulatory role of HSP27 in Fas-induced apoptosis. Annals of the New York Academy of Sciences. 2000;926:126–131. doi: 10.1111/j.1749-6632.2000.tb05606.x. [DOI] [PubMed] [Google Scholar]
- 119.Gabai VL, Yaglom JA, Volloch V, et al. Hsp72-mediated suppression of c-Jun N-terminal kinase is implicated in development of tolerance to caspase-independent cell death. Molecular & Cellular Biology. 2000;20(18):6826–6836. doi: 10.1128/mcb.20.18.6826-6836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Molecular & Cellular Biology. 1997;17(9):5317–5327. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park J, Liu AYC. JNK phosphorylates the HSF1 transcriptional activation domain: role of JNK in the regulation of the heat shock response. Journal of Cellular Biochemistry. 2001;82(2):326–338. doi: 10.1002/jcb.1163. [DOI] [PubMed] [Google Scholar]
- 122.Konishi H, Matsuzaki H, Tanaka M, et al. Activation of protein kinase B (Akt/RAC-protein kinase) by cellular stress and its association with heat shock protein Hsp27. FEBS Letters. 1997;410(2-3):493–498. doi: 10.1016/s0014-5793(97)00541-3. [DOI] [PubMed] [Google Scholar]
- 123.Mearow KM, Dodge ME, Rahimtula M, Yegappan C. Stress-mediated signaling in PC12 cells—the role of the small heat shock protein, Hsp27, and Akt in protecting cells from heat stress and nerve growth factor withdrawal. Journal of Neurochemistry. 2002;83(2):452–462. doi: 10.1046/j.1471-4159.2002.01151.x. [DOI] [PubMed] [Google Scholar]
- 124.Rane MJ, Pan Y, Singh S, et al. Heat shock protein 27 controls apoptosis by regulating Akt activation. Journal of Biological Chemistry. 2003;278(30):27828–27835. doi: 10.1074/jbc.M303417200. [DOI] [PubMed] [Google Scholar]
- 125.Qiu X-B, Shao Y-M, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cellular & Molecular Life Sciences. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takayama S, Bimston DN, Matsuzawa S-I, et al. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO Journal. 1997;16(16):4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annual Review of Biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 128.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 129.Kondo S, Murakami T, Tatsumi K, et al. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nature Cell Biology. 2005;7(2):186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- 130.Zhang K, Shen X, Wu J, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 131.Nagamori I, Yomogida K, Ikawa M, Okabe M, Yabuta N, Nojima H. The testes-specific bZip type transcription factor Tisp40 plays a role in ER stress responses and chromatin packaging during spermiogenesis. Genes to Cells. 2006;11(10):1161–1171. doi: 10.1111/j.1365-2443.2006.01013.x. [DOI] [PubMed] [Google Scholar]
- 132.Liang G, Audas TE, Li Y, et al. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein herp through an ER stress response element. Molecular & Cellular Biology. 2006;26(21):7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 134.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes to Cells. 1996;1(9):803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 135.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 136.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 137.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 138.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. Journal of Cell Biology. 2004;167(1):27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Molecular & Cellular Biology. 2003;23(20):7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. Journal of Biological Chemistry. 2004;279(19):20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 141.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Molecular Biology of the Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ye J, Rawson RB, Komuro R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Molecular Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 143.Tamatani M, Matsuyama T, Yamaguchi A, et al. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nature Medicine. 2001;7(3):317–323. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- 144.Hori O, Miyazaki M, Tamatani T, et al. Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum (ER) translocation sites, leads to ER stress. Molecular & Cellular Biology. 2006;26(11):4257–4267. doi: 10.1128/MCB.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nakagawa T, Zhu H, Morishima N, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β . Nature. 2000;403(6765):98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 146.Hitomi J, Katayama T, Eguchi Y, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. Journal of Cell Biology. 2004;165(3):347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rao RV, Castro-Obregon S, Frankowski H, et al. Coupling endoplasmic reticulum stress to the cell death program. An Apaf-1-independent intrinsic pathway. Journal of Biological Chemistry. 2002;277(24):21836–21842. doi: 10.1074/jbc.M202726200. [DOI] [PubMed] [Google Scholar]
- 148.McCullough KD, Martindale JL, Klotz L-O, Aw T-Y, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bc12 and perturbing the cellular redox state. Molecular & Cellular Biology. 2001;21(4):1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Puthalakath H, O’Reilly LA, Gunn P, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129(7):1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 150.Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 151.Nishitoh H, Matsuzawa A, Tobiume K, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes and Development. 2002;16(11):1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Letters. 2007;581(19):3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gorman AM. Neuronal cell death in neurodegenerative diseases: recurring themes around protein handling. Journal of Cellular & Molecular Medicine. 2008;12(6):2263–2280. doi: 10.1111/j.1582-4934.2008.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mhaille AN, McQuaid S, Windebank A, et al. Increased expression of endoplasmic reticulum stress-related signaling pathway molecules in multiple sclerosis lesions. Journal of Neuropathology & Experimental Neurology. 2008;67(3):200–211. doi: 10.1097/NEN.0b013e318165b239. [DOI] [PubMed] [Google Scholar]
- 155.Holohan C, Szegezdi E, Ritter T, O’Brien T, Samali A. Cytokine-induced β-cell apoptosis is NO-dependent, mitochondria-mediated and inhibited by BCL-XL . Journal of Cellular & Molecular Medicine. 2008;12(2):591–606. doi: 10.1111/j.1582-4934.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shiu RPC, Pouyssegur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(9):3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: involvement of basic leucine zipper transcription factors. Journal of Biological Chemistry. 1998;273(50):33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 158.Tanaka S, Uehara T, Nomura Y. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. Journal of Biological Chemistry. 2000;275(14):10388–10393. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- 159.Kitao Y, Ozawa K, Miyazaki M, et al. Expression of the endoplasmic reticulum molecular chaperone (ORP150) rescues hippocampal neurons from glutamate toxicity. Journal of Clinical Investigation. 2001;108(10):1439–1450. doi: 10.1172/JCI12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Uehara T, Nakamura T, Yao D, et al. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441(7092):513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 161.Kitao Y, Imai Y, Ozawa K, et al. Pael receptor induces death of dopaminergic neurons in the substantia nigra via endoplasmic reticulum stress and dopamine toxicity, which is enhanced under condition of parkin inactivation. Human Molecular Genetics. 2007;16(1):50–60. doi: 10.1093/hmg/ddl439. [DOI] [PubMed] [Google Scholar]
- 162.Hoshino T, Nakaya T, Araki W, Suzuki K, Suzuki T, Mizushima T. Endoplasmic reticulum chaperones inhibit the production of amyloid-β peptides. Biochemical Journal. 2007;402(3):581–589. doi: 10.1042/BJ20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Feaver RE, Hastings NE, Pryor A, Blackman BR. GRP78 upregulation by atheroprone shear stress via p38-, α2β1-dependent mechanism in endothelial cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(8):1534–1541. doi: 10.1161/ATVBAHA.108.167999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang C, Cai Y, Adachi MT, et al. Homocysteine induces programmed cell death in human vascular endothelial cells through activation of the unfolded protein response. Journal of Biological Chemistry. 2001;276(38):35867–35874. doi: 10.1074/jbc.M100747200. [DOI] [PubMed] [Google Scholar]
- 165.Werstuck GH, Lentz SR, Dayal S, et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. Journal of Clinical Investigation. 2001;107(10):1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Watson LM, Chan AKC, Berry LR, et al. Overexpression of the 78-kDa glucose-regulated protein/immunoglobulin-binding protein (GRP78/BiP) inhibits tissue factor procoagulant activity. Journal of Biological Chemistry. 2003;278(19):17438–17447. doi: 10.1074/jbc.M301006200. [DOI] [PubMed] [Google Scholar]
- 167.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Molecular & Cellular Biology. 2006;26(15):5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mesaeli N, Nakamura K, Zvaritch E, et al. Calreticulin is essential for cardiac development. Journal of Cell Biology. 1999;144(5):857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kitao Y, Hashimoto K, Matsuyama T, et al. ORP150/HSP12A regulates purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. Journal of Neuroscience. 2004;24(6):1486–1496. doi: 10.1523/JNEUROSCI.4029-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kubota K, Niinuma Y, Kaneko M, et al. Suppressive effects of 4-phenylbutyrate on the aggregation of Pael receptors and endoplasmic reticulum stress. Journal of Neurochemistry. 2006;97(5):1259–1268. doi: 10.1111/j.1471-4159.2006.03782.x. [DOI] [PubMed] [Google Scholar]
- 171.Choi S-E, Lee Y-J, Jang H-J, et al. A chemical chaperone 4-PBA ameliorates palmitate-induced inhibition of glucose-stimulated insulin secretion (GSIS) Archives of Biochemistry and Biophysics. 2008;475(2):109–114. doi: 10.1016/j.abb.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 172.Kudo T, Kanemoto S, Hara H, et al. A molecular chaperone inducer protects neurons from ER stress. Cell Death and Differentiation. 2008;15(2):364–375. doi: 10.1038/sj.cdd.4402276. [DOI] [PubMed] [Google Scholar]
- 173.Takano K, Tabata Y, Kitao Y, et al. Methoxyflavones protect cells against endoplasmic reticulum stress and neurotoxin. American Journal of Physiology. 2007;292(1):C353–C361. doi: 10.1152/ajpcell.00388.2006. [DOI] [PubMed] [Google Scholar]
- 174.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends in Molecular Medicine. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 175.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193(1-2):3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 176.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Critical Reviews in Biochemistry and Molecular Biology. 2009;44(2-3):98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 177.Staresincic L, Fagbemi AF, Enzlin JH, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO Journal. 2009;28(8):1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 179.Vousden KH, Lane DP. p53 in health and disease. Nature Reviews Molecular Cell Biology. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 180.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochimica et Biophysica Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23(5):687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 182.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22(37):5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 183.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annual Review of Genetics. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 184.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3(8-9):1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 185.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 186.Trachootham D, Lu W, Ogasawara MA, Valle NR-D, Huang P. Redox regulation of cell survival. Antioxidants & Redox Signaling. 2008;10(8):1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Brune B. The intimate relation between nitric oxide and superoxide in apoptosis and cell survival. Antioxidants & Redox Signaling. 2005;7(3-4):497–507. doi: 10.1089/ars.2005.7.497. [DOI] [PubMed] [Google Scholar]
- 188.Niizuma K, Endo H, Chan PH. Oxidative stress and mitochondrial dysfunction as determinants of ischemic neuronal death and survival. Journal of Neurochemistry. 2009;109(supplement 1):133–138. doi: 10.1111/j.1471-4159.2009.05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annual Review of Pharmacology & Toxicology. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 190.Genestra M. Oxyl radicals, redox-sensitive signalling cascades and antioxidants. Cellular Signalling. 2007;19(9):1807–1819. doi: 10.1016/j.cellsig.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 191.Zaccagnino P, Saltarella M, D’Oria S, Corcelli A, Saponetti MS, Lorusso M. N-arachidonylglycine causes ROS production and cytochrome c release in liver mitochondria. Free Radical Biology & Medicine. 2009;47(5):585–592. doi: 10.1016/j.freeradbiomed.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 192.Sapone A, Affatato A, Canistro D, et al. Induction and suppression of cytochrome P450 isoenzymes and generation of oxygen radicals by procymidone in liver, kidney and lung of CD1 mice. Mutation Research. 2003;527(1-2):67–80. doi: 10.1016/s0027-5107(03)00055-1. [DOI] [PubMed] [Google Scholar]
- 193.Gong X, Gutala R, Jaiswal AK. Quinone oxidoreductases and vitamin K metabolism. Vitamins and Hormones. 2008;78:85–101. doi: 10.1016/S0083-6729(07)00005-2. [DOI] [PubMed] [Google Scholar]
- 194.Robertson JD, Chandra J, Gogvadze V, Orrenius S. Biological reactive intermediates and mechanisms of cell death. Advances in Experimental Medicine and Biology. 2001;500:1–10. doi: 10.1007/978-1-4615-0667-6_1. [DOI] [PubMed] [Google Scholar]
- 195.Gorman AM, Heavey B, Creagh E, Cotter TG, Samali A. Antioxidant-mediated inhibition of the heat shock response leads to apoptosis. FEBS Letters. 1999;445(1):98–102. doi: 10.1016/s0014-5793(99)00094-0. [DOI] [PubMed] [Google Scholar]
- 196.Gorman AM, Szegezdi E, Quigney DJ, Samali A. Hsp27 inhibits 6-hydroxydopamine-induced cytochrome c release and apoptosis in PC12 cells. Biochemical & Biophysical Research Communications. 2005;327(3):801–810. doi: 10.1016/j.bbrc.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 197.Swindell WR, Huebner M, Weber AP. Transcriptional profiling of arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8, article 125 doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cullinan SB, Diehl JA. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. International Journal of Biochemistry & Cell Biology. 2006;38(3):317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 199.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. Journal of Biological Chemistry. 2004;279(19):20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 200.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death and Differentiation. 2009;16(7):1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 201.Gorman A, McGowan A, Cotter TG. Role of peroxide and superoxide anion during tumour cell apoptosis. FEBS Letters. 1997;404(1):27–33. doi: 10.1016/s0014-5793(97)00069-0. [DOI] [PubMed] [Google Scholar]
- 202.Meyer M, Schreck R, Baeuerle PA. H2O2 and antioxidants have opposite effects on activation of NF-kappa B and AP-1 in intact cells: AP-1 as secondary antioxidant-responsive factor. EMBO Journal. 1993;12(5):2005–2015. doi: 10.1002/j.1460-2075.1993.tb05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Denning TL, Takaishi H, Crowe SE, Boldogh I, Jevnikar A, Ernst PB. Oxidative stress induces the expression of Fas and Fas ligand and apoptosis in murine intestinal epithelial cells. Free Radical Biology & Medicine. 2002;33(12):1641–1650. doi: 10.1016/s0891-5849(02)01141-3. [DOI] [PubMed] [Google Scholar]
- 204.Hug H, Strand S, Grambihler A, et al. Reactive oxygen intermediates are involved in the induction of CD95 ligand mRNA expression by cytostatic drugs in hepatoma cells. Journal of Biological Chemistry. 1997;272(45):28191–28193. doi: 10.1074/jbc.272.45.28191. [DOI] [PubMed] [Google Scholar]
- 205.Dobashi K, Pahan K, Chahal A, Singh I. Modulation of endogenous antioxidant enzymes by nitric oxide in rat C6 glial cells. Journal of Neurochemistry. 1997;68(5):1896–1903. doi: 10.1046/j.1471-4159.1997.68051896.x. [DOI] [PubMed] [Google Scholar]
- 206.Asahi M, Fujii J, Suzuki K, et al. Inactivation of glutathione peroxidase by nitric oxide. Implication for cytotoxicity. Journal of Biological Chemistry. 1995;270(36):21035–21039. doi: 10.1074/jbc.270.36.21035. [DOI] [PubMed] [Google Scholar]
- 207.Bosca L, Hortelano S. Mechanisms of nitric oxide-dependent apoptosis: involvement of mitochondrial mediators. Cellular Signalling. 1999;11(4):239–244. doi: 10.1016/s0898-6568(98)00064-3. [DOI] [PubMed] [Google Scholar]
- 208.Chen ZX, Pervaiz S. BCL-2: pro-or anti-oxidant? Frontiers in Bioscience. 2009;1:263–268. doi: 10.2741/E25. [DOI] [PubMed] [Google Scholar]
- 209.Mirkovic N, Voehringer DW, Story MD, McConkey DJ, McDonnell TJ, Meyn RE. Resistance to radiation-induced apoptosis in bcl-2-expressing cells is reversed by depleting cellular thiols. Oncogene. 1997;15(12):1461–1470. doi: 10.1038/sj.onc.1201310. [DOI] [PubMed] [Google Scholar]
- 210.Robertson NM, Zangrilli J, Fernandes-Alnemri T, Friesen PD, Litwack G, Alnemri ES. Baculovirus P35 inhibits the glucocorticoid-mediated pathway of cell death. Cancer Research. 1997;57(1):43–47. [PubMed] [Google Scholar]
- 211.Sah NK, Taneja TK, Pathak N, Begum R, Athar M, Hasnain SE. The baculovirus antiapoptotic p35 gene also functions via an oxidant-dependent pathway. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(9):4838–4843. doi: 10.1073/pnas.96.9.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Alnemri ES, Livingston DJ, Nicholson DW, et al. Human ICE/CED-3 protease nomenclature. Cell. 1996;87(2):p. 171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 213.Chandra J, Samali A, Orrenius S. Triggering and modulation of apoptosis by oxidative stress. Free Radical Biology & Medicine. 2000;29(3-4):323–333. doi: 10.1016/s0891-5849(00)00302-6. [DOI] [PubMed] [Google Scholar]
- 214.Melino G, Bernassola F, Knight RA, Corasaniti MT, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388(6641):432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 215.Leist M, Single B, Naumann H, et al. Inhibition of mitochondrial ATP generation by nitric oxide switches apoptosis to necrosis. Experimental Cell Research. 1999;249(2):396–403. doi: 10.1006/excr.1999.4514. [DOI] [PubMed] [Google Scholar]
- 216.Tsujimoto Y, Shimizu S, Eguchi Y, Kamiike W, Matsuda H. BCL-2 and Bcl-xL block apoptosis as well as necrosis: possible involvement of common mediators in apoptotic and necrotic signal transduction pathways. Leukemia. 1997;11(supplement 3):380–382. [PubMed] [Google Scholar]
- 217.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Reviews Molecular Cell Biology. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 218.Kalmar B, Greensmith L. Activation of the heat shock response in a primary cellular model of motoneuron neurodegeneration-evidence for neuroprotective and neurotoxic effects. Cellular & Molecular Biology Letters. 2009;14(2):319–335. doi: 10.2478/s11658-009-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lin JH, Li H, Yasumura D, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jorns A, Gunther A, Hedrich H-J, Wedekind D, Tiedge M, Lenzen S. Immune cell infiltration, cytokine expression, and β-cell apoptosis during the development of type 1 diabetes in the spontaneously diabetic LEW.1AR1/Ztm-iddm rat. Diabetes. 2005;54(7):2041–2052. doi: 10.2337/diabetes.54.7.2041. [DOI] [PubMed] [Google Scholar]
- 221.Eizirik DL, Mandrup-Poulsen T. A choice of death—the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia. 2001;44(12):2115–2133. doi: 10.1007/s001250100021. [DOI] [PubMed] [Google Scholar]
- 222.Souza KLA, Gurgul-Convey E, Elsner M, Lenzen S. Interaction between pro-inflammatory and anti-inflammatory cytokines in insulin-producing cells. Journal of Endocrinology. 2008;197(1):139–150. doi: 10.1677/JOE-07-0638. [DOI] [PubMed] [Google Scholar]
- 223.Pirot P, Eizirik DL, Cardozo AK. Interferon-γ potentiates endoplasmic reticulum stress-induced death by reducing pancreatic beta cell defence mechanisms. Diabetologia. 2006;49(6):1229–1236. doi: 10.1007/s00125-006-0214-7. [DOI] [PubMed] [Google Scholar]
- 224.Lortz S, Gurgul-Convey E, Lenzen S, Tiedge M. Importance of mitochondrial superoxide dismutase expression in insulin-producing cells for the toxicity of reactive oxygen species and proinflammatory cytokines. Diabetologia. 2005;48(8):1541–1548. doi: 10.1007/s00125-005-1822-3. [DOI] [PubMed] [Google Scholar]
- 225.Azevedo-Martins AK, Lortz S, Lenzen S, Curi R, Eizirik DL, Tiedge M. Improvement of the mitochondrial antioxidant defense status prevents cytokine-induced nuclear factor-κB activation in insulin-producing cells. Diabetes. 2003;52(1):93–101. doi: 10.2337/diabetes.52.1.93. [DOI] [PubMed] [Google Scholar]
- 226.Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radical Biology & Medicine. 1996;20(3):463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 227.Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46(11):1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- 228.Gurgul E, Lortz S, Tiedge M, Jorns A, Lenzen S. Mitochondrial catalase overexpression protects insulin-producing cells against toxicity of reactive oxygen species and proinflammatory cytokines. Diabetes. 2004;53(9):2271–2280. doi: 10.2337/diabetes.53.9.2271. [DOI] [PubMed] [Google Scholar]
- 229.Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocrine Reviews. 2008;29(1):42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 230.Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Are oxidative stress-activated signaling pathways mediators of insulin resistance and β-cell dysfunction? Diabetes. 2003;52(1):1–8. doi: 10.2337/diabetes.52.1.1. [DOI] [PubMed] [Google Scholar]
- 231.Diakogiannaki E, Dhayal S, Childs CE, Calder PC, Welters HJ, Morgan NG. Mechanisms involved in the cytotoxic and cytoprotective actions of saturated versus monounsaturated long-chain fatty acids in pancreatic β-cells. Journal of Endocrinology. 2007;194(2):283–291. doi: 10.1677/JOE-07-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Robertson R, Zhou H, Zhang T, Harmon JS. Chronic oxidative stress as a mechanism for glucose toxicity of the beta cell in type 2 diabetes. Cell Biochemistry & Biophysics. 2007;48(2-3):139–146. doi: 10.1007/s12013-007-0026-5. [DOI] [PubMed] [Google Scholar]
- 233.Cnop M, Welsh N, Jonas J-C, Jorns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic β-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(supplement 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 234.Jorns A, Rath KJ, Bock O, Lenzen S. Beta cell death in hyperglycaemic Psammomys obesus is not cytokine-mediated. Diabetologia. 2006;49(11):2704–2712. doi: 10.1007/s00125-006-0413-2. [DOI] [PubMed] [Google Scholar]
- 235.Kharroubi I, Ladriere L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Free fatty acids and cytokines induce pancreatic β-cell apoptosis by different mechanisms: role of nuclear factor-κB and endoplasmic reticulum stress. Endocrinology. 2004;145(11):5087–5096. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]
- 236.Ashcroft FM. KATP channels and insulin secretion: a key role in health and disease. Biochemical Society Transactions. 2006;34, part 2:243–246. doi: 10.1042/BST20060243. [DOI] [PubMed] [Google Scholar]
- 237.Saleh MC, Wheeler MB, Chan CB. Uncoupling protein-2: evidence for its function as a metabolic regulator. Diabetologia. 2002;45(2):174–187. doi: 10.1007/s00125-001-0737-x. [DOI] [PubMed] [Google Scholar]
- 238.Moffitt JH, Fielding BA, Evershed R, Berstan R, Currie JM, Clark A. Adverse physicochemical properties of tripalmitin in beta cells lead to morphological changes and lipotoxicity in vitro. Diabetologia. 2005;48(9):1819–1829. doi: 10.1007/s00125-005-1861-9. [DOI] [PubMed] [Google Scholar]
- 239.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nature Genetics. 2000;25(4):406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 240.Harding HP, Zeng H, Zhang Y, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Molecular Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 241.Wang J, Takeuchi T, Tanaka S, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic β-cell dysfunction in the Mody mouse. Journal of Clinical Investigation. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Oyadomari S, Koizumi A, Takeda K, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. Journal of Clinical Investigation. 2002;109(4):525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ladiges WC, Knoblaugh SE, Morton JF, et al. Pancreatic β-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54(4):1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 244.Kovacs P, Yang X, Permana PA, Bogardus C, Baier LJ. Polymorphisms in the oxygen-regulated protein 150 gene (ORP150) are associated with insulin resistance in Pima Indians. Diabetes. 2002;51(5):1618–1621. doi: 10.2337/diabetes.51.5.1618. [DOI] [PubMed] [Google Scholar]
- 245.Ozawa K, Miyazaki M, Matsuhisa M, et al. The endoplasmic reticuluin chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54(3):657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 246.Ozcan U, Yilmaz E, Ozcan L, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gandhi S, Wood NW. Molecular pathogenesis of Parkinson’s disease. Human Molecular Genetics. 2005;14(18):2749–2755. doi: 10.1093/hmg/ddi308. [DOI] [PubMed] [Google Scholar]
- 248.Przedborski S. Pathogenesis of nigral cell death in Parkinson’s disease. Parkinsonism and Related Disorders. 2005;11(supplement 1):S3–S7. doi: 10.1016/j.parkreldis.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 249.Sulzer D. Multiple hit hypotheses for dopamine neuron loss in Parkinson’s disease. Trends in Neurosciences. 2007;30(5):244–250. doi: 10.1016/j.tins.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 250.Steece-Collier K, Maries E, Kordower JH. Etiology of Parkinson’s disease: genetics and environment revisited. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(22):13972–13974. doi: 10.1073/pnas.242594999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hatcher JM, Pennell KD, Miller GW. Parkinson’s disease and pesticides: a toxicological perspective. Trends in Pharmacological Sciences. 2008;29(6):322–329. doi: 10.1016/j.tips.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mochizuki H. Gene therapy for Parkinson’s disease. Expert Review of Neurotherapeutics. 2007;7(8):957–960. doi: 10.1586/14737175.7.8.957. [DOI] [PubMed] [Google Scholar]
- 253.Farrer MJ. Genetics of Parkinson’s disease: paradigm shifts and future prospects. Nature Reviews Genetics. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 254.Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Current Opinion in Neurobiology. 2007;17(3):331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 255.Banerjee R, Starkov AA, Beal MF, Thomas B. Mitochondrial dysfunction in the limelight of Parkinson’s disease pathogenesis. Biochimica et Biophysica Acta. 2009;1792(7):651–663. doi: 10.1016/j.bbadis.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.St Martin JL, Klucken J, Outeiro TF, et al. Dopaminergic neuron loss and up-regulation of chaperone protein mRNA induced by targeted over-expression of alpha-synuclein in mouse substantia nigra . Journal of Neurochemistry. 2007;100(6):1449–1457. doi: 10.1111/j.1471-4159.2006.04310.x. [DOI] [PubMed] [Google Scholar]
- 257.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces α-synuclein aggregation and toxicity. Journal of Biological Chemistry. 2004;279(24):25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 258.Durrenberger PF, Filiou MD, Moran LB, et al. DnaJB6 is present in the core of Lewy bodies and is highly up-regulated in Parkinsonian astrocytes. Journal of Neuroscience Research. 2009;87(1):238–245. doi: 10.1002/jnr.21819. [DOI] [PubMed] [Google Scholar]
- 259.Fargnoli J, Kunisada T, Fornace AJ, Jr., Schneider EL, Holbrook NJ. Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(2):846–850. doi: 10.1073/pnas.87.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hoozemans JJM, van Haastert ES, Eikelenboom P, de Vos RAI, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochemical & Biophysical Research Communications. 2007;354(3):707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 261.Holtz WA, O’Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. Journal of Biological Chemistry. 2003;278(21):19367–19377. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- 262.Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. Journal of Neuroscience. 2002;22(24):10690–10698. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Silva RM, Ries V, Oo TF, et al. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. Journal of Neurochemistry. 2005;95(4):974–986. doi: 10.1111/j.1471-4159.2005.03428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fisher SA, Langille BL, Srivastava D. Apoptosis during cardiovascular development. Circulation Research. 2000;87(10):856–864. doi: 10.1161/01.res.87.10.856. [DOI] [PubMed] [Google Scholar]
- 265.James TN. Normal and abnormal consequences of apoptosis in the human heart: from postnatal morphogenesis to paroxysmal arrhythmias. Circulation. 1994;90(1):556–573. [PubMed] [Google Scholar]
- 266.Gill C, Mestril R, Samali A. Losing heart: the role of apoptosis in heart disease—a novel therapeutic target? FASEB Journal. 2002;16(2):135–146. doi: 10.1096/fj.01-0629com. [DOI] [PubMed] [Google Scholar]
- 267.Gustafsson AB, Gottlieb RA. Mechanisms of apoptosis in the heart. Journal of Clinical Immunology. 2003;23(6):447–459. doi: 10.1023/b:joci.0000010421.56035.60. [DOI] [PubMed] [Google Scholar]
- 268.Fliss H, Gattinger D. Apoptosis in ischemic and reperfused rat myocardium. Circulation Research. 1996;79(5):949–956. doi: 10.1161/01.res.79.5.949. [DOI] [PubMed] [Google Scholar]
- 269.Gottlieb RA, Burleson KO, Kloner RA, Babior BM, Engler RL. Reperfusion injury induces apoptosis in rabbit cardiomyocytes. Journal of Clinical Investigation. 1994;94(4):1621–1628. doi: 10.1172/JCI117504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Misao J, Hayakawa Y, Ohno M, Kato S, Fujiwara T, Fujiwara H. Expression of bcl-2 protein, an inhibitor of apoptosis, and Bax, an accelerator of apoptosis, in ventricular myocytes of human hearts with myocardial infarction. Circulation. 1996;94(7):1506–1512. doi: 10.1161/01.cir.94.7.1506. [DOI] [PubMed] [Google Scholar]
- 271.Narula J, Haider N, Virmani R, et al. Apoptosis in myocytes in end-stage heart failure. New England Journal of Medicine. 1996;335(16):1182–1189. doi: 10.1056/NEJM199610173351603. [DOI] [PubMed] [Google Scholar]
- 272.Olivetti G, Abbi R, Quaini F, et al. Apoptosis in the failing human heart. New England Journal of Medicine. 1997;336(16):1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 273.Olivetti G, Quaini F, Sala R, et al. Acute myocardial infarction in humans is associated with activation of programmed myocyte cell death in the surviving portion of the heart. Journal of Molecular & Cellular Cardiology. 1996;28(9):2005–2016. doi: 10.1006/jmcc.1996.0193. [DOI] [PubMed] [Google Scholar]
- 274.Saraste A, Pulkki K, Kallajoki M, Henriksen K, Parvinen M, Voipio-Pulkki L-M. Apoptosis in human acute myocardial infarction. Circulation. 1997;95(2):320–323. doi: 10.1161/01.cir.95.2.320. [DOI] [PubMed] [Google Scholar]
- 275.Cook SA, Sugden PH, Clerk A. Regulation of bcl-2 family proteins during development and in response to oxidative stress in cardiac myocytes association with changes in mitochondrial membrane potential. Circulation Research. 1999;85(10):940–949. doi: 10.1161/01.res.85.10.940. [DOI] [PubMed] [Google Scholar]
- 276.von Harsdorf R, Li P-F, Dietz R. Signaling pathways in reactive oxygen species-induced cardiomyocyte apoptosis. Circulation. 1999;99(22):2934–2941. doi: 10.1161/01.cir.99.22.2934. [DOI] [PubMed] [Google Scholar]
- 277.Neuss M, Monticone R, Lundberg MS, Chesley AT, Fleck E, Crow MT. The apoptotic regulatory protein ARC (apoptosis repressor with caspase recruitment domain) prevents oxidant stress-mediated cell death by preserving mitochondrial function. Journal of Biological Chemistry. 2001;276(36):33915–33922. doi: 10.1074/jbc.M104080200. [DOI] [PubMed] [Google Scholar]
- 278.Fleury C, Mignotte B, Vayssiere J-L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84(2-3):131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 279.McMurray J, McLay J, Chopra M, Bridges A, Belch JJF. Evidence for enhanced free radical activity in chronic congestive heart failure secondary to coronary artery disease. American Journal of Cardiology. 1990;65(18):1261–1262. doi: 10.1016/0002-9149(90)90985-a. [DOI] [PubMed] [Google Scholar]
- 280.Singh N, Aggarwal S. The effect of active oxygen generated by xanthine/xanthine oxidase on genes and signal transduction in mouse epidermal JB6 cells. International Journal of Cancer. 1995;62(1):107–114. doi: 10.1002/ijc.2910620120. [DOI] [PubMed] [Google Scholar]
- 281.Zweier JL, Flaherty JT, Weisfeldt ML. Direct measurement of free radical generation following reperfusion of ischemic myocardium. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(5):1404–1407. doi: 10.1073/pnas.84.5.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Logue SE, Gustafsson AB, Samali A, Gottlieb RA. Ischemia/reperfusion injury at the intersection with cell death. Journal of Molecular & Cellular Cardiology. 2005;38(1):21–33. doi: 10.1016/j.yjmcc.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 283.Suzuki K, Sawa Y, Kaneda Y, Ichikawa H, Shirakura R, Matsuda H. In vivo gene transfection with heat shock protein 70 enhances myocardial tolerance to ischemia-reperfusion injury in rat. Journal of Clinical Investigation. 1997;99(7):1645–1650. doi: 10.1172/JCI119327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Aleshin AN, Sawa Y, Kitagawa-Sakakida S, et al. 150-kDa oxygen-regulated protein attenuates myocardial ischemia-reperfusion injury in rat heart. Journal of Molecular & Cellular Cardiology. 2005;38(3):517–525. doi: 10.1016/j.yjmcc.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 285.Li C-Y, Lee J-S, Ko Y-G, Kim J-I, Seo J-S. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. Journal of Biological Chemistry. 2000;275(33):25665–25671. doi: 10.1074/jbc.M906383199. [DOI] [PubMed] [Google Scholar]
- 286.Talukder MAH, Kalyanasundaram A, Zhao X, et al. Expression of SERCA isoform with faster Ca2+ transport properties improves postischemic cardiac function and Ca2+ handling and decreases myocardial infarction. American Journal of Physiology. 2007;293(4):H2418–H2428. doi: 10.1152/ajpheart.00663.2007. [DOI] [PubMed] [Google Scholar]
- 287.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21(3):485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 288.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 289.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 290.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108(2):153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 291.Fulda S, Debatin K-M. Targeting apoptosis pathways in cancer therapy. Current Cancer Drug Targets. 2004;4(7):569–576. doi: 10.2174/1568009043332763. [DOI] [PubMed] [Google Scholar]
- 292.Makin G, Dive C. Apoptosis and cancer chemotherapy. Trends in Cell Biology. 2001;11(11):S22–S26. doi: 10.1016/s0962-8924(01)02124-9. [DOI] [PubMed] [Google Scholar]
- 293.Friesen C, Fulda S, Debatin K-M. Deficient activation of the CD95 (APO-1/Fas) system in drug-resistant cells. Leukemia. 1997;11(11):1833–1841. doi: 10.1038/sj.leu.2400827. [DOI] [PubMed] [Google Scholar]
- 294.Fulda S, Los M, Friesen C, Debatin K-M. Chemosensitivity of solid tumor cells in vitro is related to activation of the CD95 system. International Journal of Cancer. 1998;76(1):105–114. doi: 10.1002/(sici)1097-0215(19980330)76:1<105::aid-ijc17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 295.Maeda T, Yamada Y, Moriuchi R, et al. Fas gene mutation in the progression of adult T cell leukemia. Journal of Experimental Medicine. 1999;189(7):1063–1071. doi: 10.1084/jem.189.7.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Landowski TH, Qu N, Buyuksal I, Painter JS, Dalton WS. Mutations in the Fas antigen in patients with multiple myeloma. Blood. 1997;90(11):4266–4270. [PubMed] [Google Scholar]
- 297.Gronbaek K, Straten PT, Ralfkiaer E, et al. Somatic fas mutations in non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood. 1998;92(9):3018–3024. [PubMed] [Google Scholar]
- 298.Lee SH, Shin MS, Park WS, et al. Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene. 1999;18(25):3754–3760. doi: 10.1038/sj.onc.1202769. [DOI] [PubMed] [Google Scholar]
- 299.Lee SH, Shin MS, Park WS, et al. Alterations of Fas (APO-1/CD95) gene in transitional cell carcinomas of urinary bladder. Cancer Research. 1999;59(13):3068–3072. [PubMed] [Google Scholar]
- 300.Park WS, Oh RR, Kim YS, et al. Somatic mutations in the death domain of the Fas (Apo-I/CD95) gene in gastric cancer. Journal of Pathology. 2001;193(2):162–168. doi: 10.1002/1096-9896(2000)9999:9999<::aid-path759>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 301.Wistuba II, Behrens C, Virmani AK, et al. Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Research. 1999;59(8):1973–1979. [PubMed] [Google Scholar]
- 302.Emi M, Fujiwara Y, Nakajima T, et al. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Research. 1992;52(19):5368–5372. [PubMed] [Google Scholar]
- 303.Micheau O. Cellular FLICE-inhibitory protein: an attractive therapeutic target? Expert Opinion on Therapeutic Targets. 2003;7(4):559–573. doi: 10.1517/14728222.7.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12(9):1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 305.Fulda S. Caspase-8 in cancer biology and therapy. Cancer Letters. 2009;281(2):128–133. doi: 10.1016/j.canlet.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 306.Kitada S, Pedersen IM, Schimmer AD, Reed JC. Dysregulation of apoptosis genes in hematopoietic malignancies. Oncogene. 2002;21(21):3459–3474. doi: 10.1038/sj.onc.1205327. [DOI] [PubMed] [Google Scholar]
- 307.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275(5302):967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 308.Zinkel SS, Ong CC, Ferguson DO, et al. Proapoptotic BID is required for myeloid homeostasis and tumor suppression. Genes and Development. 2003;17(2):229–239. doi: 10.1101/gad.1045603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tagawa H, Karnan S, Suzuki R, et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24(8):1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 310.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109(1):271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 311.Fels DR, Koumenis C. The PERK/eIF2α/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biology & Therapy. 2006;5(7):723–728. doi: 10.4161/cbt.5.7.2967. [DOI] [PubMed] [Google Scholar]
- 312.Koumenis C, Wouters BG. “Translating” tumor hypoxia: unfolded protein response (UPR)-dependent and UPR-independent pathways. Molecular Cancer Research. 2006;4(7):423–436. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- 313.Zheng H-C, Takahashi H, Li X-H, et al. Overexpression of GRP78 and GRP94 are markers for aggressive behavior and poor prognosis in gastric carcinomas. Human Pathology. 2008;39(7):1042–1049. doi: 10.1016/j.humpath.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 314.Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Research. 2007;67(20):9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 315.Koong AC, Chauhan V, Romero-Ramirez L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biology & Therapy. 2006;5(7):756–759. doi: 10.4161/cbt.5.7.2973. [DOI] [PubMed] [Google Scholar]
- 316.Qin S, Chock PB. Implication of phosphatidylinositol 3-kinase membrane recruitment in hydrogen peroxide-induced activation of PI3K and Akt. Biochemistry. 2003;42(10):2995–3003. doi: 10.1021/bi0205911. [DOI] [PubMed] [Google Scholar]
- 317.Pervaiz S, Cao J, Chao OSP, Chin YY, Clement M-V. Activation of the RacGTPase inhibits apoptosis in human tumor cells. Oncogene. 2001;20(43):6263–6268. doi: 10.1038/sj.onc.1204840. [DOI] [PubMed] [Google Scholar]
- 318.Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Targeting HSP90 for cancer therapy. British Journal of Cancer. 2009;100(10):1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5(22):2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]