Abstract
Bone morphogenetic proteins (BMPs) regulate many processes in embryonic development as well as in the maintenance of normal tissue function later in adult life. However, the role of this family of proteins in formation of adipose tissue has been underappreciated in the field of developmental biology. With the growing epidemic of obesity, improved knowledge of adipocyte development and function is urgently needed. Recently, there have been significant advances in understanding the role of different members of BMP superfamily in control of adipocyte differentiation and systemic energy homeostasis. This review summarizes recent progress in understanding how BMPs specify adipose cell fate in stem/progenitor cells and their potential role in energy metabolism. We propose that BMPs provide instructive signals for adipose cell fate determination and regulate adipocyte function. These findings have opened up exciting opportunities for developing new therapeutic approaches for the treatment of obesity and its many associated metabolic disorders.
1. Introduction
According to the World Health Organization, the continuing surge in the obesity pandemic creates a substantial increase in incidences of metabolic diseases, such as type 2 diabetes mellitus, cardiovascular dysfunction, liver steatosis and cirrhosis, as well as the neurodegenerative Alzheimer’s disease and even some cancers (1–4). Treatment of obesity-related morbidities has imposed a huge economic burden on societies, with 147 billion per year estimated to be the annual medical cost of obesity in the US to date (5). Increasing body adiposity is the defining characteristic of obesity. The past two decades have shed considerable light on the understanding of adipocyte biology and function. Originally considered as an inert mass for energy storage, adipose tissue is now seen as an endocrine organ that actively participates in the regulation of whole body energy metabolism (6). Adipokines produced by fat cells, such as leptin and adiponectin, are key mediators of physiological processes in distant organs, such as brain, liver and muscle, where they control appetite, digestion of nutrients, energy expenditure and storage, glucose and lipid metabolism and insulin sensitivity (7–9). Therefore, improved knowledge on the mechanisms underlying the formation of adipose tissue and its role in energy homeostasis is urgently needed to counter the growing epidemic of obesity.
While research on transcriptional regulation of adipocyte differentiation has been a central focus in studies of adipocyte biology, emerging evidence suggests that secreted factors, such as cytokines or developmental regulators, play a crucial role in controlling the differentiation of mesenchymal progenitor cells into adipocytes and in regulating adipocyte function and energy metabolism. These cytokines and developmental regulators can modulate expression and activities of specific adipogenic transcriptional regulators. This review will focus on the current understanding of how a group of prominent morphogens, the bone morphogenetic proteins (BMPs), regulates adipose cell fate, white versus brown adipocyte formation, and systemic energy metabolism, as well as their potential use for anti-obesity therapies.
2. Development of adipose tissue
2.1. Function and distribution of adipose tissue
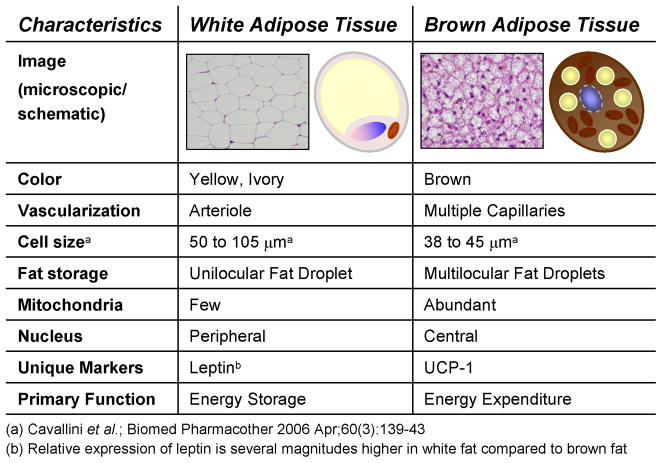
The capacity to store fat as a reservoir of readily available energy in times of scarce nutrient supply is found in most animal species and is conserved throughout evolution. While invertebrates such as the nematode Caenorhabditis elegans store lipids in the intestine, and the fruit fly Drosophila melanogaster stores excess energy in the “fat body”, only higher organisms have developed a specialized tissue for lipid storage -- the adipose tissue/organ (10). There are two functionally and morphologically different types of adipose tissue in mammals: white adipose tissue is the primary site of triglyceride storage; and brown adipose tissue is specialized in energy expenditure (Figure 1). The white fat cell is characterized by a single large lipid droplet, a nucleus located in close proximity to the cell membrane and low mitochondrial density, while the brown adipocyte features multi-locular lipid inclusions, numerous well-developed mitochondria with the unique expression of uncoupling protein-1 (UCP-1) and resides in rich vasculatures (11). UCP-1 is a 32-kDa protein exclusively expressed in the inner membrane of the mitochondria of brown fat and allows the dissipation of the proton electrochemical gradient generated by respiration as the form of heat. UCP-1 is generally regarded as the defining marker of brown fat, whereas leptin is more highly expressed in white fat than brown fat (Figure 1).
Figure 1.
Characteristics of brown versus white adipocytes
The distribution of fat varies among different species. In humans, white fat is dispersed throughout the body with the subcutaneous and intra-abdominal depots as two major compartments for fat storage. Distribution of these two white fat depots is highly associated with the risk of developing metabolic syndrome (6). Increased accumulation of visceral fat is associated with higher risk for metabolic complication of obesity, while no association is found with increased subcutaneous adiposity (12). Brown fat is primarily a thermogenic tissue that burns fat to generate heat in order to maintain body temperature in cold environment and dissipate excess energy in response to overfeeding (11). In rodents, induction of brown fat promotes energy expenditure, reduces adiposity and protects from diet-induced obesity (13;14). Conversely, targeted ablation of brown fat results in reduced energy expenditure and increased obesity (15). In newborn humans, significant amounts of brown fat are found in interscapular, axillary, cervical, perirenal, and periadrenal regions (16). The interscapular brown fat disappears shortly after birth, and thus it has traditionally been assumed that there is no functional brown fat present in adult humans. However, this concept has been radically revised during the past few months. In the spring of 2009, five independent teams reported studies using PET-CT (positron emission tomography- computed tomography) imaging to prove conclusively that adult humans have metabolically active brown fat (17–21). The most common location for brown fat in adults is the cervical-supraclavicular depot, and in a small subset of patients, brown fat is also found in the thoracic and paraspinal regions. More importantly, these brown fat depots appear to correlate inversely with body mass index in older people (17), suggesting a critical role of brown fat in human adult energy metabolism and the potential of using brown fat-mediated energy expenditure as an anti-obesity therapy.
2.2. The origin of adipose tissue
Adipose tissue, like muscle and bone, is considered to be of mesodermal origin, although precise lineage tracing studies have not yet been performed (6). Adipocytes develop from mesenchymal stem/progenitor cells, which derive from embryonic stem cells. When triggered by appropriate developmental cues, these cells become committed to adipocyte lineages, i.e. the preadipocytes (Figure 2). In adolescents, both fat cell hypertrophy and hyperplasia occur with the development of obesity (22). The turnover of adipocytes is tightly maintained in a steady state in adults (23). Thus, the adipocyte stem/progenitor cells residing within the stromo-vascular fraction constantly replenish adipose tissues with newly formed adipocytes. These progenitor cells are characterized by the capacity for self-renewal, and commitment to the adipogenic lineage, which is marked by expression of the transcription factor PPARγ, an early marker of adipogenesis, but do not accumulate lipids (24). Moreover, a subset of vasculature resident adipose precursor cells possesses the ability to regenerate an entire fat depot and induce de novo vascularization. These cells express the surface antigens CD29, CD34, Sca-1, and CD24, and are negative for markers of the hematopoietic lineage (25). As for the development of brown adipose tissue, recent evidence suggests that brown fat and skeletal muscle may share a common early developmental program (26). More recently, Seale et al., used a myogenic marker, myf5, to perform cell fate mapping in the mouse and found that both skeletal muscle and interscapular brown fat, but not white fat, arise from progenitors expressing myf5 (27). In addition to these discrete interscapular brown fat cells, UCP-1-positive brown adipocytes are also found systemically distributed in the body, especially within white fat depots (28) and between muscle bundles (29). Interestingly, these “systemic” brown adipocytes, such as those present in white fat and muscle, are not derived from myf5-expressing precursors (27), suggesting different developmental origins for these different pools of brown fat. Thus, an important unsolved issue in adipocyte biology is the identification of brown fat progenitor cells. Interplay between the progenitor cells and the inductive signal specifies the developmental fate of the precursors into specific adipose cell lineages.
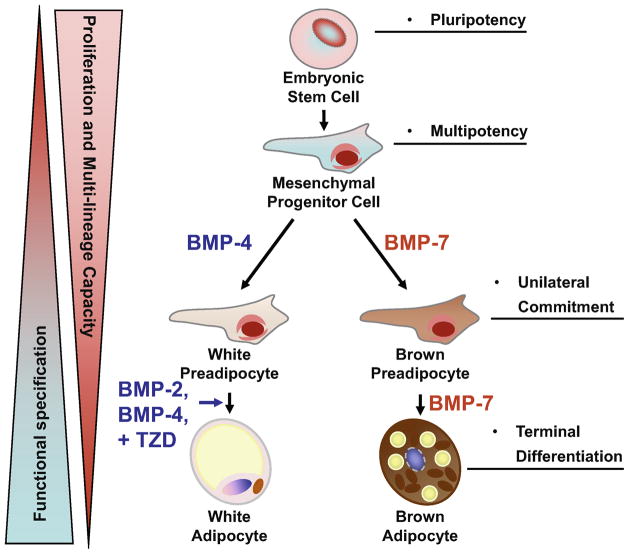
Figure 2. Role of BMPs in adipocyte development.
Adipocytes arise from multipotent mesenchymal progenitor cells which originally derived from the pluripotent embryonic stem cells. When triggered by specific instructive signals, these progenitors become committed to the adipocyte lineage. While BMP-2 and BMP-4 induces commitment and subsequent differentiation into white adipocyte lineage, BMP-7 drives brown fat cell fate in both mesenchymal progenitor cells and committed brown preadipocytes.
2.3. Molecular controls of adipocyte differentiation
Several developmental signaling molecules implicated in the evolution of mesodermal tissues have been shown to impact the development of adipose tissue. These include nodal, wingless, fibroblast growth factors, members of the transforming growth factor (TGF)-β family, BMPs, and others. These factors are often produced by the microenvironment or niche and provide instructive cues to guide differentiation and maturation of the progenitors (30).
Once the multipotent progenitor cells become committed to the adipocyte lineage, these cells are then referred to as preadipocytes. It is believed that white and brown preadipocytes are pre-determined towards differentiation into either one or the other adipose cell type (31). Decisive markers that allow a clear distinction between mesenchymal progenitor and committed preadipocyte are currently unavailable; therefore, the only common phenotypic characteristic of cell culture models of preadipocytes is that they do not undergo differentiation into cell types other than adipocytes. Over the past two decades, considerable progress has been made on defining the transcriptional events controlling differentiation of preadipocytes into mature adipocytes (32;33). Prior to adipogenic transcriptional cascade initiation, both brown and white preadipocytes need to be released from suppression and become committed to terminal differentiation. The known inhibitors of this early adipogenic event include the notch family of epidermal growth factor-like-repeat-containing protein preadipocyte factor-1 (Pref-1) (34), the wingless (Wnt) family of developmental regulators (35), proteins of the retinoblastoma (Rb) family (36;37) and a member of the melanoma-associated antigen family of proteins, functionally resembling RB, named necdin (38). Interestingly, the Rb family of proteins and necdin appear to selectively suppress brown preadipocyte differentiation at the early stage. After release from suppression, the committed preadipocytes then initiate a transcriptional cascade involving transcription factors CCAAT/enhancer-binding proteins (C/EBPs) and peroxisome proliferator-activated receptor (PPAR)γ to turn on lipid synthesis and other adipocyte-specific programs.
Some factors that underlie brown versus white adipocyte differentiation have been identified, including the zinc-finger binding protein PRDM16 (39), nuclear coactivator PPARγ coactivator-1 (PGC-1) α (40), members of the pRb protein family, members of the p160 family of coactivators (41;42), the nuclear corepressor RIP140 (43), and others (44). While these transcriptional regulators may indeed play an important role in the determination of adipose cell fate between BAT and WAT, upstream secreted factors that modulate expression and activities of these transcriptional regulators have just begun to be elucidated. One protein family of great interest in the control of brown versus white adipose fate determination is the BMP family. The role of BMPs in adipocyte development is detailed below.
3. BMPs, BMP receptors and signal transduction
3.1. Discovery and members of BMPs
BMPs, although originally named for their ability to induce bone formation, are a group of pleiotropic proteins that regulate processes as diverse as cell fate determination, proliferation, apoptosis, and differentiation during both embryogenesis and adulthood. The discovery of the BMP protein family was initiated in 1945, when Pierre Lacroix hypothesized that a bone derived substance, which he called osteogenin, could initiate bone formation and growth (45). This hypothesis was later confirmed by Marshall Urist’s seminal work, which demonstrated that intramuscular implants of cell-free, lyophilized bone extracts could induce de novo formation of bone at the site of implantation (46). However, it was not until in the late 1980s that the first individual BMP proteins, BMP-2, -3, and -4, isolated and characterized by Wozney and colleagues (47). BMPs belong to the superfamily of TGF-β proteins, where they form a large subfamily including the currently known 14 BMP proteins, and the growth and differentiation factors (GDFs) (Table 1). BMP homologues are also found in many species, including daf-4 and daf-7 in the nematode (Caenorhabditis elegans), Univin in the sea urchin (class: Echinoidea), decapentaplegic, glass bottom boat 60A, and screw in fruit fly (Drosophila melanogaster), VG1 in the African clawed frog (Xenopus laevis), as well as Dorsalin-1 in chicken (Gallus gallus) (48–50). BMPs are secreted as precursor protein dimers which are cleaved by pro-protein convertases to yield the mature active form of the protein (51;52). Although bone is an important site producing BMPs, most members of this family are expressed in other tissues where they regulate the formation and function of many other organ systems (53–55).
Table 1.
Members of the mammalian BMP-subfamily of the TGF-β superfamily and their functional role in development, disease progression and physiology
| BMP Ligand Subfamily | Alternative Names | Function |
|---|---|---|
| BMP-4 | ZYME; BMP2B; OFC11; BMP2B1; MCOPS6 | mesenchymal progenitor differentiation, (white) adipogenesis, osteogenesis, chondrogenesis |
| BMP-2 | BMP2a; XBMP2; xBMP-2; MGC114605 | mesenchymal progenitor differentiation, (white) adipogenesis, osteogenesis, chondrogenesis |
| BMP-5 | MGC34244 | development of trabecular meshwork and optic nerve head; potential role in glaucoma pathogenesis and cartilage development |
| BMP-6 | Vgr-1; DVR-6 | early embryonic development; joint integrity |
| BMP-7 | OP-1 | brown adipogenesis; early embryonic development and bone formation |
| BMP-8a | OP-2; FLJ14351; FLJ45264 | |
| BMP-8b | OP-3; PC-8; MGC131757 | early embryonic development, possibly bone inductive activity |
| BMP-12 | GDF-7; CDMP-3 | specification of neuronal identity in the dorsal spinal cord |
| BMP-13 | KFS; KFSL; SGM1; CDMP2; MGC158100; MGC158101; GDF6 | formation of some bones and joints in the limbs, skull, and axial skeleton; mutations lead to colobomata and Klippel-Feil syndrome (KFS) |
| BMP-14 | OS5; LAP4; CDMP1; SYNS2; MP52 | cell growth and differentiation in embryonic and adult tissues; mutations associated with acromesomelic dysplasia, Hunter-Thompson type, brachydactyly type C, and chondrodysplasia Grebe type |
| GDF-1 | establishment of left-right asymmetry and in neural development during embryogenesis | |
| GDF-3 | Vgr-2 | cell growth and differentiation in embryonic and adult tissues |
| BMP-9 | GDF-2 | cell growth and differentiation in embryonic and adult tissues; plays a role in adult liver and in differentiation of cholinergic central nervous system neurons |
| BMP-10 | MGC126783 | trabeculation of the embryonic heart |
| BMP-11 | GDF-11 | mesodermal formation and neurogenesis during embryonic development |
| BMP-15 | ODG2; POF4; GDF-9B | oocyte maturation and follicular development |
| BMP-3 | Osteogenin, BMP-3A | induces bone formation; possible role in energy metabolism |
| BMP-3b | GDF-10; Sumitomo-BIP | skeletal morphogenesis |
3.2. BMP receptors
While specific receptors for BMPs exist, the binding specificity of these proteins to their receptors is very complex. BMPs can also bind to some receptors of other members of the TGF-β superfamily, namely the activin receptors, with similar affinity. The activation of the BMP signaling cascade requires binding to two receptor types (BMPRs), which then form a hetero-oligomeric complex that relays the signal to downstream targets. Three type 1 receptors are known to bind BMPs, which include the activin receptor like kinases (ALK)-2, ALK-3 (also known as BMPR1A), and ALK-6 (BMPR1B). Similarly, three type 2 receptors possess binding affinity for BMPs, including BMPR2, activin type 2 A receptor (ActR2A), and ActR2B (56). Upon ligand binding, the type 2 receptor, a serine threonine kinase, trans-phosphorylates the type 1 receptor. Once activated, the serine threonine kinase type 1 receptor further activates downstream targets to transduce signals. The specificity of the BMP signal is believed to be regulated by at least four mechanisms: (a) binding affinity of the BMP ligand to the receptor, (b) the stoichiometric composition of the individual receptors in a given cell type, (c) accessory proteins such as co-receptors (57), and (d) the order of ligand-receptor-complex formation. A BMP ligand can either bind to a single receptor type subunit which then recruits the second subunit, or alternatively bind to a pre-existing loose complex of both receptor types which are activated following association with the BMP ligand. Both possibilities can lead to activation of different downstream signaling pathways, with the Smad (mammalian homologues of the Drosophila melanogaster mothers against decapentaplegic) proteins and the p38 mitogen activated protein kinase (p38MAPK) pathways as the two major signaling cascades activated by most BMP ligands (58).
3.3. Signal transduction
While assembly of the hetero-oligomeric receptor complex prior to ligand binding entails activation of the Smad pathway, ligand binding followed by recruitment of the type 2 receptor activates p38MAPK signaling (58). These two pathways represent the canonical signaling cascades that relay ligand-binding to elicit physiological responses in most cell types, although the differential responses largely depend on cell type and other interacting factors within the cell. While the TGF-β proteins and activins can phosphorylate Smad 2 and Smad 3, the proteins of the BMP subfamily are known to phosphorylate Smad 1, 5 and 8, the so called R-Smads (receptor-activated Smads). The specificity of this interaction depends on three-dimensional interaction of the L45 loop of the type 1 BMP receptor kinases which interacts with the compatible L3 loop on the Smad 1, 5, and 8 proteins only (59). Phosphorylated R-Smad then binds to the universal co-Smad, Smad 4, which in turn facilitates the migration into the nucleus and transcriptional activity of the Smad protein. Smad phosphorylation can be antagonized by the inhibitory Smad 6 and Smad 7 proteins, which interfere with the receptor substrate interaction and can thus contribute another layer of signal specificity from BMP ligand to intercellular response (60–63).
As discussed above, BMPs can also activate the p38MAPK signaling cascade, an alternative signaling pathway which has been characterized as an important regulator of energy metabolism directing both mitochondrial biogenesis and insulin-dependent glucose uptake (64;65). The cascade begins with BMP-2 and BMP-4, whose binding leads to phosphorylation of the respective BMP receptor kinase and the activation of MAPK kinase kinase (MAPKKK) TAK1. TAK1 in turn phosphorylates the MAPK kinase MKK6, which can directly phosphorylate p38MAPK (66). Signal transduction from receptor to MAPKKK is mediated by two accessory proteins, TAB1 and XIAP1 which have been shown to modulate signaling downstream of BMP ligand binding (67;68). Although Smad and p38 MAPK pathways represent the main transmitters for BMP binding signals to the nucleus, it should be noted that other signaling cascades have also been implicated in mediating BMPs’ signals in different cell types. These alternative pathways include activation of the extracellular signal-related kinase (ERK), the c-Jun N-terminal kinase (JNK), the protein kinase C, the phosphoinositide 3-kinase (PI3K), and the p70S6 kinase (69).
4. Effects of BMPs on development of adipocyte tissue
4.1. Determination of adipose cell fate in mesenchymal stem/progenitor cells
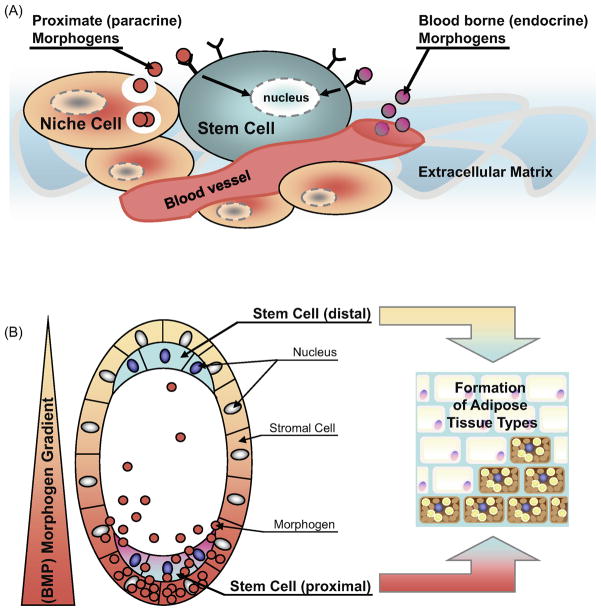
Cell fate determination in the pluripotent stem/progenitor cells is controlled by the integration of cell intrinsic factors with extrinsic cues supplied by the surrounding microenvironment, known as the niche. The concept of a stem cell niche was introduced in 1978, which postulates that stem cells are believed to reside within a microenvironment of defined anatomical structure that helps sustain the typical characteristics of these cells (70). The surrounding cells, forming the niche, not only provide an extracellular matrix as an anchoring point for adhesion of stem cells but also determine stem cell proliferation (i.e. self-renewal) and the differentiation fate of daughter cells. Daughter cells then can either undergo a committing step and terminal differentiation, or can retain their ability to differentiate into multiple lineages (30). The factors that influence these processes include cell-cell contacts, cell-matrix adhesion, and soluble growth factors - the so called morphogens (71). Morphogens can be secreted from the niche in close vicinity of the stem cells and act as paracrine effectors, or they can be blood-borne growth factors from other endocrine organs throughout the body (Figure 3A). The interplay of these morphogens, and possibly also autocrine factors originating from the stem cell itself, are thought to control progenitor cell preservation, lineage commitment and differentiation.
Figure 3. Proposed model of stem cell niche and morphogen gradient controlled adipocyte tissue development.
(A) The stem cell niche consists of the surrounding cells (niche cells), the extracellular matrix, and the local microvasculature. Locally secreted morphogens, such as BMPs, and the blood-borne factors affect stem cell fate differentially and depending on their concentration in the microenvironment. The extracellular matrix also represents an important regulator for stem cell function as it forms a natural barrier limiting the accessibility and diffusion of endocrine and paracrine factors. (B) Morphogen gradients are known to regulate multiple developmental processes in the maturing embryo and during adult tissue regeneration. The family of BMP proteins has been implicated in these processes. A localized group of surrounding (stromal) cells secretes instructive factors which then distribute throughout the microenvironment to form a so called “morphogen gradient. Local concentrations are higher at the proximal site and lower at distal regions. This gradient provides distinct effects to the stem cells located at proximal versus distal sites. It is hypothesized that BMPs, and other morphogens, could affect adipose tissue formation in a similar manner. This model could apply to white versus brown adipocyte formation, as well as determination of different white fat depots.
BMPs are known as one of the niche factors which provide instructive signals to the pluripotent stem cells in proximity or at a distance. During early embryonic development, BMPs form a morphogen gradient to instruct body patterning (72). For example, in the fruit fly, Drosophila melanogaster, BMPs have been implicated in embryogenesis following the formation of concentration gradients within the developing embryo (73). The effect of BMPs on formation of fat appears to be evolutionally conserved. The Drosophila BMP-7 homologue glass bottom boat (gbb-60A) plays an indispensible role in fat body formation, as larvae lacking gbb-60A display severe morphological abnormalities of the fat body (74). Based on the role of morphogens in guiding tissue/organ formation during embryonic development, we speculate that a similar morphogen gradient, presumably established by BMPs and/or other developmental regulators, may instruct the formation of different fat depots distributed in various locations of the body (Figure 3B). Whether BMPs can be directly secreted from different cell types residing in the adipose vasculature or they are secreted at a distant site, and then travel to the adipose tissue via circulation remains to be determined. In addition, the surrounding niche cells could potentially affect ligand binding to the appropriate receptors. Because fat distribution is tightly associated with metabolic phenotype, the embryonic morphogen gradient may influence the susceptibility of developing obesity and other metabolic disorders later in life.
Several lines of evidence have suggested that BMPs provide inductive signals for adipose cell fate determination in mammalian systems (55). The effects of BMPs on adipogenesis appear to depend on the stage of cell development and the dosage of different BMP ligands. In embryonic stem cell-derived embryoid bodies, BMP-4, presumably through interaction with retinoic acid (75;76), can promote adipogenesis (77). In bone marrow stromal cells, the predominant effect of BMPs, in particular BMP-2, is to promote osteogenic differentiation and inhibit adipogenesis (78–82); however, low concentrations of BMPs modestly stimulate adipocyte differentiation (79). The effects of BMPs in the pluripotent mesenchymal cell line C3H10T1/2 are more complex and tightly controlled by the dosages and types of BMPs used in the system as well as by the presence of other extracellular and intracellular factors. The C3H10T1/2 cells are mouse embryonic fibroblasts established from 14- to 17-day-old embryos of the C3H mouse strain (83). These cells functionally resemble mesenchymal stem/progenitor cells that possess the ability to differentiate into multiple lineages, including myoblast, adipocyte, chondrocyte, and osteoblast (84–86). In these cells, low concentrations of BMP-2 and BMP-7 induce adipogenic differentiation whereas high concentrations promote differentiation toward chondrocyte and osteoblast (85;87). Stable expression of cDNAs encoding different BMPs induces C3H10T1/2 cells to differentiate into osteogenic, chondrogenic and adipogenic lineages (86;88). These BMPs appear to have differential effects on adipogenesis in this system, with BMP-4 having the greatest effect on induction of lipid accumulation and expression of markers for mature adipocytes (88).
While these early studies suggest BMPs regulate adipogenesis in the multipotent progenitors, the effect of different BMP members on determination of brown versus white fat cell fate has not been established until recently (Figure 2). Treatment of C3H10T1/2 with BMP-4 has been shown to induce commitment and subsequent differentiation into white adipocytes (89;90). We have recently discovered that BMP-7 specifically triggers commitment of the multipotent mesenchymal cells into the brown fat lineage, and implantation of C3H10T1/2 cells treated with BMP-7 into nude mice results in the formation of a UCP-1 positive brown fat pad (91). In NIH-3T3 cells, a cell line with no adipogenic character, both BMP-7 and BMP-4 induce lipid accumulation and expression of adipogenic marker PPARγ, while only BMP-7 is able to induce expression of brown fat-specific markers, such as PRDM16 and UCP-1 expression. Moreover, BMP-7 in combination with a hormonal induction cocktail and rosiglitazone produces similar effects on other mouse embryonic fibroblast cell lines and primary culture of stromal-vascular fraction isolated from interscapular brown fat (91). Together, these data highlight the fact that BMP-7 can not only trigger commitment of mesenchymal cells to a brown adipocyte lineage, but also can act in concert with other differentiating agents to induce characteristics of brown fat in more primitive fibroblastic cells.
The notion that BMP-7 serves as the inductive signal for brown fat development in vivo has also been established. In 1992, Loncar et al., demonstrated that engraftment of mesoderm from E9 rat embryos into the kidney capsule (renal tissue being the main source of BMP-7 in adult animals (92)) results in the implant exclusively differentiating into brown fat (93). The direct evidence for a BMP-7 role in embryonic brown fat development comes from examination of BMP-7 knockout embryos. Both E17.5 and E18.5 embryos of BMP-7 knockout mice show a marked paucity of brown fat and near complete absence of UCP-1 protein (91), suggesting that BMP-7 is absolutely required for formation of functional brown adipose tissue during embryonic development.
4.2. Regulation of adipocyte differentiation in committed preadipocytes
BMPs can also stimulate differentiation in committed preadipocytes. The two most prominent white preadipocyte cell lines are 3T3-L1 and 3T3-F442A. BMP-2 can induce a mature white fat phenotype in both cell lines suggesting that BMPs not only regulate progenitor cell commitment as discussed above, but also promote terminal adipogenic differentiation (94;95). The transcription factor PPARγ is a key regulator of the adipogenic process. Some findings suggest a cross talk between BMP signaling and PPARγ action, since adipogenesis induced by BMP-2 treatment in committed preadipocytes can be further enhanced following the treatment with the PPARγ agonist rosiglitazone (96). The synergistic effect of BMP-2 and PPARγ ligand may be explained, at least in part, by the ability of BMP-2 to upregulate PPARγ expression (97).
BMP-7, as discussed above, triggers progenitor cell commitment towards the brown adipocyte lineage. Furthermore, BMP-7 also promotes brown adipogenesis in committed brown preadipocytes even in the absence of normally required induction cocktail, while it does not affect the differentiation of committed white preadipocytes under the same conditions (91). Taken together, these data suggest that different members of the BMP family exert differential effects on brown versus white adipocyte differentiation, with BMP-2 and BMP-4 as white adipogenic factors and BMP-7 as the unique brown fat inducer (Figure 2).
4.3. Molecular mechanisms
At the molecular level, Hata et al. have reported that both Smad1 and p38 MAPK pathways are involved in regulating the expression and activity of PPARγ during BMP-2-induced adipogenesis in C3H10T1/2 cells (97). In addition, Schnurri (Shn)-2, a zinc finger-containing protein that enters the nucleus upon BMP-2 stimulation, is found to cooperate with Smad1/4 and C/EBPα to induce PPARγ gene expression (98). Interestingly, Shn-2 knockout mice display reduced white, but not brown, fat mass, suggesting that BMP-2 utilizes the Smad/Shn-2 pathway to regulate white adipogenesis in vivo. In committed brown preadipocytes, BMP-7 activates a full program of brown adipogenesis including suppression of adipogenic inhibitors, induction of early regulators of brown fat fate PRDM16 and PGC-1α, increased expression of adipogenic transcription PPARγ and C/EBPs, and mitochondrial biogenesis (91). Interestingly, while BMP-7 is able to activate both Smad and p38 MAPK pathways in brown preadipocytes, activation of p38 MAPK is essential for BMP-7-induced thermogenic program, while this pathway appears to be dispensable for BMP-7’s effect on lipid accumulation.
5. Effects of BMPs on systemic energy metabolism
Compared to the substantial amount of data concerning the roles of BMPs in different aspects of embryonic development and morphogenesis, very little is known about the role of BMPs in adipocyte development in vivo and in systemic energy homeostasis. This is partially due to embryonic lethality in many of the knockout models of BMPs and/or because severe defects in other tissues/organs overshadow the adipose phenotype. Nevertheless, high expression levels of BMP-3 in white fat positively correlates with increased susceptibility to high fat diet-induced obesity in inbred stain of mice (99). In vivo overexpression of GFD-3, a member of the BMP subfamily, increases adiposity and hepatic steatosis in mice fed a high fat diet (100), while GDF-3 deficiency protects mice from diet-induced obesity by selectively targeting white adipose tissue (101). Deletion of myostatin, also known as GDF-8, not only confers muscle hypertrophy but also results in reduced adipose tissue mass (102;103). Lastly, increasing circulating BMP-7 levels by adenoviral-mediated gene transfer results in a significant increase in brown, but not white, fat mass and leads to an increase in energy expenditure and reduced weight gain (91), consistent with the specific role of BMP-7 in brown fat differentiation and function.
As discussed above, the cellular response of BMPs is mediated by ligand binding to the cell surface receptors. Of the different BMP receptor isoforms, BMPR1A is particularly interesting to adipocyte development since it has been shown to specialize in adipocyte differentiation in vitro (104). Notably, BMPR1A binds to BMP-2 and BMP-4 with high affinity, while it exerts low binding capacity to BMP-7 (105). Recently, an increased expression of BMPR1A was found in visceral and subcutaneous white fat depots in overweight and obese human subjects (106), consistent with the role of BMP-2 and BMP-4 on formation of white fat. Furthermore, an association of BMPR1A-SNPs with obesity-linked quantitative trait loci was identified which could potentially affect the pathophysiology of human obesity. Because mice with whole-body knockout of BMPR1A are embryonic lethal (107), we recently generated a conditional knockout model with adipocyte-specific deletion of BMPR1A. These mice display a significant reduction in body weight as well as a trend toward reduced fat pad weight on both standard and high fat diets (108). Together, these data suggest a critical, yet complex, role of the BMP superfamily in systemic energy metabolism via regulation of adipocyte development and function.
6. Conclusion
Adipocyte development is a complex process, involving a multitude of interactions between the progenitor cells and inductive signals. Here we have discussed compelling evidence that establishes a critical role of BMPs in adipogenesis and energy metabolism. BMPs are involved in many aspects of adipocyte development, including adipose cell fate determination, differentiation of committed preadipocytes, and function of mature adipocyte. Adipose tissue plays an important role in systemic energy metabolism. It not only serves as an energy reservoir in the form of white fat, but also functions in energy expenditure, which mainly occurs in brown fat. Furthermore, this tissue is also an important source of adipokines that influences appetite, glucose and lipid homeostasis. While detailed mechanisms by which BMPs regulate adipocyte differentiation and function remain to be elucidated, BMPs and their downstream signaling components provide a new avenue to develop potential therapies for the treatment of obesity.
Acknowledgments
We thank A. M. Cypess and K. L. Townsend for a critical reading of the manuscript. T.J.S. is supported by a fellowship from the German Research Foundation. This work was supported in part by an NIH R01 grant DK077097, and research grants from the Eli Lilly Research Foundation, the Harvard Stem Cell Institute, and the Harvard Catalyst/Harvard Clinical and Translational Science Center (to Y.-H. T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 2.Hsu IR, Kim SP, Kabir M, Bergman RN. Metabolic syndrome, hyperinsulinemia, and cancer. Am J Clin Nutr. 2007;86(3):s867–s871. doi: 10.1093/ajcn/86.3.867S. [DOI] [PubMed] [Google Scholar]
- 3.Craft S. Insulin resistance and Alzheimer’s disease pathogenesis: potential mechanisms and implications for treatment. Curr Alzheimer Res. 2007;4(2):147–152. doi: 10.2174/156720507780362137. [DOI] [PubMed] [Google Scholar]
- 4.Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, et al. The metabolic syndrome. Endocr Rev. 2008;29(7):777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finkelstein EA, Trogdon JG, Cohen JW, Dietz W. Annual Medical Spending Attributable To Obesity: Payer- And Service-Specific Estimates. Health Aff (Millwood) 2009 doi: 10.1377/hlthaff.28.5.w822. [DOI] [PubMed] [Google Scholar]
- 6.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131(2):242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Friedman JM. Obesity in the new millennium. Nature. 2000;404(6778):632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- 8.Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocr Rev. 2006;27(7):762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 9.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinti S. The adipose organ. Prostaglandins Leukot Essent Fatty Acids. 2005;73(1):9–15. doi: 10.1016/j.plefa.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 12.Kissebah AH, Krakower GR. Regional adiposity and morbidity. PMID. 1994;74(4):761–811. doi: 10.1152/physrev.1994.74.4.761. [DOI] [PubMed] [Google Scholar]
- 13.Ghorbani M, Claus TH, Himms-Hagen J. Hypertrophy of brown adipocytes in brown and white adipose tissues and reversal of diet-induced obesity in rats treated with a beta3-adrenoceptor agonist. Biochem Pharmacol. 1997;54(1):121–131. doi: 10.1016/s0006-2952(97)00162-7. [DOI] [PubMed] [Google Scholar]
- 14.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J Clin Invest. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowell BB, Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366(6457):740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 16.Lean ME, James WP, Jennings G, Trayhurn P. Brown adipose tissue uncoupling protein content in human infants, children and adults. Clin Sci (Lond) 1986;71(3):291–297. doi: 10.1042/cs0710291. [DOI] [PubMed] [Google Scholar]
- 17.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 19.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 20.Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58(7):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009 doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch J, Batchelor B. Adipose tissue cellularity in human obesity. Clin Endocrinol Metab. 1976;5(2):299–311. doi: 10.1016/s0300-595x(76)80023-0. [DOI] [PubMed] [Google Scholar]
- 23.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 24.Tang W, Zeve D, Suh J, Bosnakovski D, Kyba M, Hammer B, et al. White Fat Progenitor Cells Reside in the Adipose Vasculature. Science. 2008 doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135(2):240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 26.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104(11):4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103 (Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 29.Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc Natl Acad Sci U S A. 2007;104(7):2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 31.Moulin K, Truel N, Andre M, Arnauld E, Nibbelink M, Cousin B, et al. Emergence during development of the white-adipocyte cell phenotype is independent of the brown-adipocyte cell phenotype. Biochem J. 2001;356(Pt 2):659–664. doi: 10.1042/0264-6021:3560659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 34.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73(4):725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 35.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 36.Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci U S A. 2004;101(12):4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scime A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, et al. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005;2(5):283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol. 2005;7(6):601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 39.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 41.Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111(7):931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Qi C, Krones A, Woodring P, Zhu X, Reddy JK, et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 2006;3(2):111–122. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Steel JH, White R, Parker MG. Role of the RIP140 corepressor in ovulation and adipose biology. J Endocrinol. 2005;185(1):1–9. doi: 10.1677/joe.1.05896. [DOI] [PubMed] [Google Scholar]
- 44.Hansen JB, Kristiansen K. Regulatory circuits controlling white versus brown adipocyte differentiation. Biochem J. 2006;398(2):153–168. doi: 10.1042/BJ20060402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lacroix P. Recent investigations on the growth of bone. Nature. 1945;156:576. [Google Scholar]
- 46.Urist MR. Bone: formation by autoinduction. Science. 1965;150(698):893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 47.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242(4885):1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 48.Reddi AH. Role of morphogenetic proteins in skeletal tissue engineering and regeneration. Nat Biotechnol. 1998;16(3):247–252. doi: 10.1038/nbt0398-247. [DOI] [PubMed] [Google Scholar]
- 49.Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8(2):133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 50.Zhao GQ. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35(1):43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 51.Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 2004;16(3):291–299. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Xiao YT, Xiang LX, Shao JZ. Bone morphogenetic protein. Biochem Biophys Res Commun. 2007;362(3):550–553. doi: 10.1016/j.bbrc.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto Y, Oelgeschlager M. Regulation of bone morphogenetic proteins in early embryonic development. Naturwissenschaften. 2004;91(11):519–534. doi: 10.1007/s00114-004-0575-z. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22(4):233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 55.Tseng YH, He TC. Bone Morphogenetic Proteins and Adipocyte Differentiation. Cellscience Review. 2007;3(3):342–360. [Google Scholar]
- 56.Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16(3):265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Halbrooks PJ, Ding R, Wozney JM, Bain G. Role of RGM coreceptors in bone morphogenetic protein signaling. J Mol Signal. 2007;2:4. doi: 10.1186/1750-2187-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, et al. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem. 2002;277(7):5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- 59.Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, et al. Determinants of specificity in TGF-beta signal transduction. Genes Dev. 1998;12(14):2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 61.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389(6651):622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 62.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature. 1999;397(6721):710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 63.Liu X, Nagarajan RP, Vale W, Chen Y. Phosphorylation regulation of the interaction between Smad7 and activin type I receptor. FEBS Lett. 2002;519(1–3):93–98. doi: 10.1016/s0014-5793(02)02718-7. [DOI] [PubMed] [Google Scholar]
- 64.Kandror KV. A long search for Glut4 activation. Sci STKE. 2003;2003(169):E5. doi: 10.1126/stke.2003.169.pe5. [DOI] [PubMed] [Google Scholar]
- 65.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 66.Kimura N, Matsuo R, Shibuya H, Nakashima K, Taga T. BMP2-induced apoptosis is mediated by activation of the TAK1-p38 kinase pathway that is negatively regulated by Smad6. J Biol Chem. 2000;275(23):17647–17652. doi: 10.1074/jbc.M908622199. [DOI] [PubMed] [Google Scholar]
- 67.Shibuya H, Iwata H, Masuyama N, Gotoh Y, Yamaguchi K, Irie K, et al. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17(4):1019–1028. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, et al. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18(1):179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Caestecker M. The transforming growth factor-beta superfamily of receptors. Cytokine Growth Factor Rev. 2004;15(1):1–11. doi: 10.1016/j.cytogfr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1–2):7–25. [PubMed] [Google Scholar]
- 71.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graff JM. Embryonic patterning: to BMP or not to BMP, that is the question. Cell. 1997;89(2):171–174. doi: 10.1016/s0092-8674(00)80196-8. [DOI] [PubMed] [Google Scholar]
- 73.O’Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133(2):183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khalsa O, Yoon JW, Torres-Schumann S, Wharton KA. TGF-beta/BMP superfamily members, Gbb-60A and Dpp, cooperate to provide pattern information and establish cell identity in the Drosophila wing. Development. 1998;125(14):2723–2734. doi: 10.1242/dev.125.14.2723. [DOI] [PubMed] [Google Scholar]
- 75.Dani C, Smith AG, Dessolin S, Leroy P, Staccini L, Villageois P, et al. Differentiation of embryonic stem cells into adipocytes in vitro. J Cell Sci. 1997;110 (Pt 11):1279–1285. doi: 10.1242/jcs.110.11.1279. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Schwarz EM, Zuscik MJ, Rosier RN, Ionescu AM, Puzas JE, et al. Retinoic acid stimulates chondrocyte differentiation and enhances bone morphogenetic protein effects through induction of Smad1 and Smad5. Endocrinology. 2003;144(6):2514–2523. doi: 10.1210/en.2002-220969. [DOI] [PubMed] [Google Scholar]
- 77.Taha MF, Valojerdi MR, Mowla SJ. Effect of bone morphogenetic protein-4 (BMP-4) on adipocyte differentiation from mouse embryonic stem cells. Anat Histol Embryol. 2006;35(4):271–278. doi: 10.1111/j.1439-0264.2006.00680.x. [DOI] [PubMed] [Google Scholar]
- 78.Gimble JM, Morgan C, Kelly K, Wu X, Dandapani V, Wang CS, et al. Bone morphogenetic proteins inhibit adipocyte differentiation by bone marrow stromal cells. J Cell Biochem. 1995;58(3):393–402. doi: 10.1002/jcb.240580312. [DOI] [PubMed] [Google Scholar]
- 79.Chen TL, Shen WJ, Kraemer FB. Human BMP-7/OP-1 induces the growth and differentiation of adipocytes and osteoblasts in bone marrow stromal cell cultures. J Cell Biochem. 2001;82(2):187–199. doi: 10.1002/jcb.1145. [DOI] [PubMed] [Google Scholar]
- 80.Pereira RC, Delany AM, Canalis E. Effects of cortisol and bone morphogenetic protein-2 on stromal cell differentiation: correlation with CCAAT-enhancer binding protein expression. Bone. 2002;30(5):685–691. doi: 10.1016/s8756-3282(02)00687-7. [DOI] [PubMed] [Google Scholar]
- 81.Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144(12):5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- 82.Song C, Guo Z, Ma Q, Chen Z, Liu Z, Jia H, et al. Simvastatin induces osteoblastic differentiation and inhibits adipocytic differentiation in mouse bone marrow stromal cells. Biochem Biophys Res Commun. 2003;308(3):458–462. doi: 10.1016/s0006-291x(03)01408-6. [DOI] [PubMed] [Google Scholar]
- 83.Reznikoff CA, Brankow DW, Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33(12):3231–3238. [PubMed] [Google Scholar]
- 84.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 85.Wang EA, Israel DI, Kelly S, Luxenberg DP. Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors. 1993;9(1):57–71. doi: 10.3109/08977199308991582. [DOI] [PubMed] [Google Scholar]
- 86.Ahrens M, Ankenbauer T, Schroder D, Hollnagel A, Mayer H, Gross G. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol. 1993;12(10):871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- 87.Asahina I, Sampath TK, Hauschka PV. Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp Cell Res. 1996;222(1):38–47. doi: 10.1006/excr.1996.0005. [DOI] [PubMed] [Google Scholar]
- 88.Bachner D, Ahrens M, Schroder D, Hoffmann A, Lauber J, Betat N, et al. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2) Dev Dyn. 1998;213(4):398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 89.Butterwith SC, Wilkie RS, Clinton M. Treatment of pluripotential C3H 10T1/2 fibroblasts with bone morphogenetic protein-4 induces adipocyte commitment. Biochem Soc Trans. 1996;24(2):163S. doi: 10.1042/bst024163s. [DOI] [PubMed] [Google Scholar]
- 90.Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101(26):9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454(7207):1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalluri R, Zeisberg M. Exploring the connection between chronic renal fibrosis and bone morphogenic protein-7. Histol Histopathol. 2003;18(1):217–224. doi: 10.14670/HH-18.217. [DOI] [PubMed] [Google Scholar]
- 93.Loncar D. Brown adipose tissue as a derivative of mesoderm grafted below the kidney capsule. A model for differentiation of isolated rat mesoderm. Int J Dev Biol. 1992;36(2):265–274. [PubMed] [Google Scholar]
- 94.Ji X, Chen D, Xu C, Harris SE, Mundy GR, Yoneda T. Patterns of gene expression associated with BMP-2-induced osteoblast and adipocyte differentiation of mesenchymal progenitor cell 3T3-F442A. J Bone Miner Metab. 2000;18(3):132–139. doi: 10.1007/s007740050103. [DOI] [PubMed] [Google Scholar]
- 95.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol Cell Biol. 2003;23(20):7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sottile V, Seuwen K. Bone morphogenetic protein-2 stimulates adipogenic differentiation of mesenchymal precursor cells in synergy with BRL 49653 (rosiglitazone) FEBS Lett. 2000;475(3):201–204. doi: 10.1016/s0014-5793(00)01655-0. [DOI] [PubMed] [Google Scholar]
- 97.Hata K, Nishimura R, Ikeda F, Yamashita K, Matsubara T, Nokubi T, et al. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol Biol Cell. 2003;14(2):545–555. doi: 10.1091/mbc.E02-06-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, et al. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell. 2006;10(4):461–471. doi: 10.1016/j.devcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 99.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2(5):e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang W, Yang Y, Meng Y, Shi Y. GDF-3 is an adipogenic cytokine under high fat dietary condition. Biochem Biophys Res Commun. 2004;321(4):1024–1031. doi: 10.1016/j.bbrc.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 101.Shen JJ, Huang L, Li L, Jorgez C, Matzuk MM, Brown CW. Deficiency of growth differentiation factor 3 protects against diet-induced obesity by selectively acting on white adipose. Mol Endocrinol. 2009;23(1):113–123. doi: 10.1210/me.2007-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin J, Arnold HB, Della-Fera MA, Azain MJ, Hartzell DL, Baile CA. Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem Biophys Res Commun. 2002;291(3):701–706. doi: 10.1006/bbrc.2002.6500. [DOI] [PubMed] [Google Scholar]
- 103.McPherron AC, Lee SJ. Supression of body fat accumulation in myostatin-deficient mice. J Clin Invest. 2002;109(5):595–601. doi: 10.1172/JCI13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, et al. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol. 1998;142(1):295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sebald W, Nickel J, Zhang JL, Mueller TD. Molecular recognition in bone morphogenetic protein (BMP)/receptor interaction. Biol Chem. 2004;385(8):697–710. doi: 10.1515/BC.2004.086. [DOI] [PubMed] [Google Scholar]
- 106.Bottcher Y, Unbehauen H, Kloting N, Ruschke K, Korner A, Schleinitz D, et al. Adipose Tissue Expression and Genetic Variants of the Bone Morphogenetic Protein Receptor 1A Gene (BMPR1A) are Associated with Human Obesity. Diabetes. 2009 doi: 10.2337/db08-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9(24):3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 108.Schulz TJ, Huang TL, Mishina Y, Tseng YH. Adipocyte-specific Inactivation of Type 1A Bone Morphogenetic Protein Receptor Impacts Systemic Energy Metabolism and Fat Physiology. Diabetes. 2009;58(Supplement 1) [Google Scholar]