Abstract
Animal growth and development depend on the precise control of gene expression at the level of transcription. A central role in the regulation of developmental transcription is attributed to transcription factors that bind DNA enhancer elements, which are often located far from gene transcription start sites. Here, we review recent studies that have uncovered significant regulatory functions in developmental transcription for the TFIID basal transcription factors and for the DNA core promoter elements that are located close to transcription start sites.
Introduction
Proper organismal growth and development depend on the timing, location and level of protein-coding and regulatory RNA gene transcription by RNA polymerase II (Pol II). Defects in these transcription parameters can result in a variety of phenotypes, including cell fate changes and lethality. Thus, much effort has gone into determining the mechanisms that regulate the transcription of developmentally important genes. These efforts have focused on mechanisms that are mediated by cis-acting DNA sequences and trans-acting protein factors.
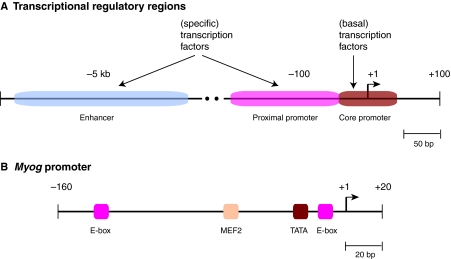
Some signals that regulate transcription are hardwired into genes via commonly occurring genomic DNA sequence elements that act as binding sites for the transcription machinery. These elements are classified as enhancer, proximal promoter, or core promoter elements based on their location within genes and their corresponding binding proteins (Fig. 1A). Enhancer and proximal promoter elements are both bound by sequence-specific DNA-binding proteins (commonly called transcription factors), but these elements are found at different locations relative to the gene transcription start site (TSS). The location of enhancer elements varies greatly between genes and can be many kilobases upstream or downstream of the TSS, whereas proximal promoter elements are typically restricted to within a couple of hundred base-pairs of the TSS. By contrast, core promoter elements are bound by basal transcription factors and are located within ~100-bp zones centered at the TSS.
Fig. 1.
Cis-acting DNA transcription-regulatory elements. (A) The generic organization of transcription-regulatory regions of a eukaryotic gene, depicting the organization and location of enhancer, proximal promoter, and core promoter elements, as well as the regulatory factors that bind these elements. (B) Promoter structure of the mouse Myog gene, which is used as an example throughout the text. Known elements in the core and proximal promoter regions are indicated.
Transcription initiation by Pol II is directed, in part, by the basal transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH that assemble on core promoters (Thomas and Chiang, 2006). Basal transcription factors are so called because they direct a low or basal level of transcription in vitro in the absence of additional transcription factors. This is in contrast to a higher or activated level of transcription in vitro that is directed by the combined activities of basal and additional transcription factors. Biochemical studies indicate that basal transcription factors assemble on core promoters in a defined manner, commencing with binding to core promoter elements. The original and quintessential example is binding of the TATA-binding protein (TBP) subunit of TFIID to the TATA box element, which nucleates assembly of TFIIA and TFIIB, followed by assembly of TFIIF and Pol II as a pre-assembled complex, and culminating in assembly of TFIIE and TFIIH.
The impetus behind this review comes from recent reports that implicate TFIID complexes and core promoter elements as being vital to the regulation of developmental transcription. This finding is likely to be surprising to those who do not closely follow the transcription literature because the textbook view is that the regulation of developmental transcription is mediated mostly by transcription factors and enhancer sequences. To convey the new advances, we will use transcription of the myogenin (Myog) gene as a central example and touch upon the transcription of other genes to illustrate particular points. Fundamental to the new advances has been the discovery that both the core promoter elements and TFIID complexes, which contain most of the proteins that bind core promoter elements, are highly diverse. This diversity appears to contribute to the capacity of regulatory signals that emanate from enhancers and transcription factors to generate the strikingly complex patterns of transcription that are necessary for proper organismal growth and development.
Introduction to Myog transcription
In contrast to housekeeping genes, such as Gapdh (glyceraldehyde-3-phosphate dehydrogenase), which are transcribed at relatively constant levels in all cell types throughout development, many genes, such as Myog, are transcribed in distinct cell types to control specific developmental events. Myog encodes a skeletal muscle-specific transcription factor involved in myogenic determination (Berkes and Tapscott, 2005). In mice, Myog is first transcribed at embryonic day (E) 8.5 in proliferating myogenic precursors in the somite myotome (Sassoon et al., 1989). Myog transcription is turned off as these cells leave the somite and invade the limb bud and is then turned on at high levels at E11.5 in the hindlimb and forelimb. In agreement with the Myog expression pattern, knockout of Myog results in the accumulation of myocytes that are arrested in their terminal differentiation program and in neonatal death due to severe muscle defects (Hasty et al., 1993; Nabeshima et al., 1993).
What mechanisms determine this complex pattern of Myog transcription? This is a difficult question to address in whole animals, so researchers have turned to cultured myoblast cells that differentiate in response to a stimulus, such as growth factor deprivation. In culture, Myog transcription is required for C2C12 mouse skeletal muscle myoblasts to terminally differentiate into multinucleated myotubes (Blau et al., 1983; Edmonson et al., 1992). An analysis of reporter gene expression in C2C12 cells has demonstrated that a 184 bp region upstream of the Myog TSS is sufficient to confer: (1) muscle cell-specific transcription; (2) Myog transcriptional activation in response to growth factor deprivation; and (3) autoregulation of Myog transcription by the myogenin protein (Edmonson et al., 1992) (Fig. 1B). Subsequent analyses, described below, have revealed that the 184 bp DNA sequence contains proximal and core promoter elements necessary to direct cell type-specific Myog transcription.
Genomic analyses of core promoters
Peaked and broad transcription initiation patterns
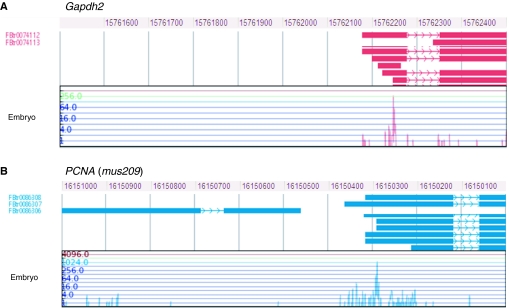
The binding of basal transcription factors to core promoter elements leads to the recruitment of Pol II to the beginning (i.e. the 5′ end) of genes and to transcription initiation. However, transcription initiation is not as clear-cut an event as is often presented in textbooks. For many genes, transcription starts at a reproducible and narrow genomic site (termed a peaked or focused initiation pattern) (Fig. 2A), but it has been known for several decades that some genes do not adhere to this pattern (Lee and Roeder, 1981; Butcher and Trifonov, 1986). Rather, the 5′ ends of some gene transcripts are broadly distributed over a range of ~100-200 bp (termed a broad or dispersed initiation pattern) (Fig. 2B).
Fig. 2.
Eukaryotic transcription initiation patterns. High-throughput genomic data on transcription start sites (TSSs) allows different transcription initiation patterns to be visualized and analyzed. Two different embryonic initiation patterns for genes with previously mapped TSSs are shown, adapted from the MachiBase database browser, which contains high-throughput sequence read data from Drosophila libraries of 5′ capped transcripts (Ahsan et al., 2009). These displays show (top) the coordinates of a selected genomic region, followed (middle) by the gene structure annotations and transcript data for this region from FlyBase: first, transcripts with FlyBase transcript (FBtr) IDs; then, expressed sequence tag (EST) evidence (for the purposes of clarity, the displays only show a selection of tags that map to the region). Finally (bottom), the distribution of embryonic 5′ reads in the selected region that align to the same genomic location are shown (read numbers are on a logarithmic scale). (A) The Gapdh2 (Glyceraldehyde-3-phosphate dehydrogenase 2) gene on chromosome 2R exhibits a peaked initiation pattern, in which most transcription events originate from a small genomic region (Tso et al., 1985). (B) The PCNA [proliferating cell nuclear antigen, also known as mutagen sensitive 209 (mus209)] gene on chromosome 2R exhibits a broad pattern, in which initiation events are spread out over a larger genomic region of typically 100-200 nucleotides (Hochheimer et al., 2002). These examples demonstrate that the major sites of transcription initiation do not necessarily correspond to the 5′ ends of transcripts annotated in genomic databases, but that they are often supported by existing transcript evidence in the form of ESTs.
Data obtained by high-throughput sequencing protocols have confirmed the existence of these different initiation patterns (Suzuki et al., 2001). In protocols such as 5′ SAGE (serial analysis of gene expression) or CAGE (capped analysis of gene expression), libraries of transcripts are constructed, and short tags, corresponding to the beginning of these presumably 5′ complete transcripts, are sequenced (Suzuki et al., 1997; Kodzius et al., 2006). Mapping of these tags back to the genome delivers high-resolution data on TSSs and has defined peaked and broad initiation patterns (Carninci et al., 2006; Kawaji et al., 2006) (Box 1). Thus far, efforts have concentrated on mammals [in particular, on mice by the FANTOM consortium (fantom.gsc.riken.jp/4/)], but recent data from Drosophila demonstrate the widespread existence of peaked and broad initiation patterns, at least in different animals (Carninci et al., 2005; Ahsan et al., 2009; FANTOM Consortium et al., 2009). Computational analyses of peaked and broad promoters have shown that these two promoter types differ in sequence features, such as GC content or the presence of CpG islands in mammals (Box 2), as well as in the preference for certain core promoter elements (Carninci et al., 2006; Sandelin et al., 2007; Rach et al., 2009). Thus, core promoter features correlate with peaked and broad initiation patterns, but the underlying mechanisms and the physiological importance of a gene having a peaked versus a broad transcription initiation pattern remain unknown.
Box 1. Sequence elements: weight matrix versus consensus sequence
Functional sequence elements in promoters are traditionally represented as consensus sequences that consist of the nucleotide(s) found most frequently at each position (see figure, top). Consensus sequences are useful for a high-level description of binding preferences; however, their use comes with certain potential pitfalls. First, they are typically reported in the initial identification of a new element, based on a handful of experimentally studied sequences, but often the number of sequences is too small to derive an unbiased (i.e. genome-wide, optimal) consensus. Second, a match to the consensus is either yes or no and does not reflect the actual binding affinity of a transcription factor to a functional element.
Promoter elements are therefore now commonly represented in the form of weight matrices (see figure, bottom), which describe the frequency of all four nucleotides at each position and which can be used to calculate scores that reflect the affinity of a transcription factor for a specific sequence (Vavouri and Elgar, 2005). Weight matrices are often visualized as sequence logos, in which all four letters are shown at their relative frequencies. The height of the nucleotides indicates the ‘information content’ at each position, measured in bit, which reflects the relative differences between the nucleotide distribution and genomic GC content (Schneider and Stephens, 1990).
Regardless of weight matrix or consensus, the number of putative functional elements can vary significantly based, for example, on the number of allowed mismatches to a consensus or on different matrix score cut-offs. These numbers can therefore be misleading if it is not assessed how often matches can be expected to occur by chance alone. As an example, a match to the consensus AC[CG]CG[TA] occurs by chance once in 1 kb, assuming that all four nucleotides are equally frequent (i.e. in 0.1% of sequences, if looking at a specific location relative to a TSS). Studies often allow a few mismatches to a consensus sequence to occur; in the above example, allowing for two mismatches to any consensus nucleotide increases the random match rate from 0.1% to ~16%. If the location of the match within the promoter is more flexible, or if the genomic region of interest has a GC content that is closer to the composition of the promoter element (and matches are therefore more probable than they are in regions with differing GC content), the random match rate is even higher. Thus, changing the search parameters can also lead to large changes in random matches.
Box 2. Chromatin-mediated transcriptional regulation
Chromatin structure can also influence transcription mechanisms. The nucleosome, which is the basic unit of chromatin, comprises ~146 bp of DNA wrapped around an octamer complex that contains two copies of each of the four core histones H2A, H2B, H3 and H4 (Luger et al., 1997). Additionally, histone H1 binds the linker DNA between nucleosomes (Happel and Doenecke, 2009). Chromatin structure is altered by four reversible post-translational histone modifications (acetylation, methylation, phosphorylation and ubiquitylation), by remodeling of histone octamer-DNA interactions by chromatin remodeling complexes, by cytosine methylation at CpG dinucleotides, and by the incorporation of variant histone proteins into nucleosomes (Henikoff et al., 2004; Bhaumik et al., 2007; Ko et al., 2008; Delcuve et al., 2009). When these events take place at core promoter DNA, they can affect the assembly of basal transcription factors (Jiang and Pugh, 2009). For example, in yeast there are three general arrangements of nucleosomes for the ~20% of gene core promoters that contain a TATA box (Ioshikhes et al., 2006). TATA boxes are located in nucleosome-free regions of DNA with an adjacent upstream nucleosome in one case or adjacent upstream and downstream nucleosomes in a second case. In the third case, the TATA box is located within a nucleosome. These distinct nucleosome organizations could permit differential transcriptional regulation through subtle changes in nucleosome positioning imparted by chromatin remodeling complexes. Such nucleosome positioning might also determine the location of the TSS in core promoters that lack canonical sequence elements by enabling TFIID to be directly recruited through the interactions of TAFs with modified histones. TFIID assembled at the core promoter can also alter chromatin structure through intrinsic activities or by recruiting proteins with chromatin-altering activities. For example, TAF1 not only post-translationally modifies histones but also interacts with the histone chaperone CIA (CCG1-interacting factor A, also known as ASF1A), which functions in the assembly and disassembly of nucleosomes (Mizzen et al., 1996; Pham and Sauer, 2000; Natsume et al., 2007) (see Fig. 4A). Thus, a give-and-take relationship probably exists between TFIID and chromatin structure that mediates transcriptional regulation at the core promoter.
Canonical core promoter elements
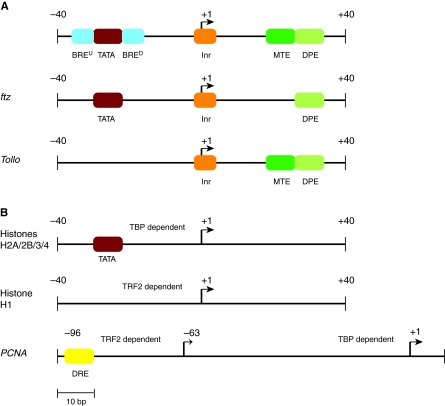
Over the years, comparisons of core promoter sequences within and between eukaryotic genomes have identified the commonly occurring elements — the TATA box, initiator (Inr) and downstream promoter element (DPE) — collectively termed canonical core promoter elements (Table 1) (Fig. 3A). However, given the overall evolutionary conservation of basal transcription factors (Aoyagi and Wassarman, 2000), canonical core promoter elements are not as widely shared among genes in a given organism or among eukaryotic organisms as one might think. For example, the TATA box [so named because the nucleotide sequence TATA occurs in the consensus sequence [CG]TATA[AT]A[AT][AG] (Table 1)] was the first eukaryotic promoter element to be identified (M. L. Goldberg, PhD thesis, Stanford University, 1979). It is one of the most extensively studied promoter elements but is present in the core promoters of only ~10-20% of eukaryotic genes (Ohler et al., 2002; Basehoar et al., 2004; Jin et al., 2006). The Inr, which is located at the TSS, serves as a good example of species-specific core promoter element differences. In Drosophila, the Inr is a well-defined 5-6 bp motif, whereas in humans and yeast, the overall consensus is much weaker and corresponds to a [CT]A dinucleotide (Ohler et al., 2002; Carninci et al., 2006; Zhang and Dietrich, 2005). In addition, even if a core promoter element occurs widely among eukaryotic organisms, its positional preference relative to the TSS may vary. In animals, the TATA box is located precisely ~30 bp upstream of the TSS, but in yeast it is more broadly located within 50-125 bp upstream of the TSS (Basehoar et al., 2004; Zhang and Dietrich, 2005; Ponjavic et al., 2006). Thus, there are no universal eukaryotic core promoter elements, suggesting that core promoter elements are uniquely adapted to the transcription initiation machinery of specific cells and organisms.
Table 1.
Eukaryotic core promoter elements
| Element | Consensus | Location | Factor | Species | References |
| Inr* | TCA[GT]T[CT] | −2 | TAF1, TAF2 | All eukaryotes | Smale and Baltimore, 1989 |
| TATA box* | [CG]TATA[AT]A[AT][AG] | −31 | TBP | All eukaryotes | M. L. Goldberg, PhD thesis, Stanford University, 1979 |
| DPE* | [GT]CGGTT[CG][GT] | +26 | TAF6, TAF9 | Animals | Burke and Kadonaga, 1996 |
| MTE* | C[CG]A[AG]C[CG][CG]A | +18 | TFIID (unknown TAF) | Animals | Lim et al., 2004 |
| BREU | [GC][GC][AG]CGCC | −39 | TFIIB | Animals (adenovirus) | Lagrange et al., 1998 |
| BRED* | [AG]T[AGT][GT][GT][GT][GT] | −23 | TFIIB | Animals (adenovirus) | Deng and Roberts, 2005 |
| DRE† | [AT]ATCGAT[AT] | Upstream (variable) | DREF-TRF2 complex | Drosophila | Hochheimer et al., 2002 |
| Motif 1† | [CT]GGTCACACT[AG] | Upstream (variable) | Unknown | Drosophila | Ohler et al., 2002 |
| Motif 6† | [CT][AG]GTAT[AT]TT[CT] | ||||
| Motif 7† | CA[GT]CNCT[AG] |
The DRE forms a complex with TRF2, but the identity of the factors that bind to the other ‘orphan’ elements remains unknown. With the exception of the BRE elements (Legrange et al., 1998; Deng and Roberts, 2005), consensus descriptions of elements have been combined from the Drosophila analyses of Ohler et al. (Ohler et al., 2002) and Fitzgerald et al. (Fitzgerald et al., 2006). The location refers to the first nucleotide of the consensus as given here. References are to papers that describe the role of an element in core promoters (not necessarily the first report of the functional element). BRE, TFIIB recognition elements; DPE, downstream promoter element; DRE, DNA replication-related element; Inr, initiator; MTE, motif ten element.
Elements associated with peaked promoters, as reflected by their precise location relative to the TSS.
Sequence motifs repeatedly identified in Drosophila genes with a broad initiation pattern.
Fig. 3.
Core promoter element configurations. (A) Promoters with canonical position-specific elements. (Top) The relative locations of known elements (see Table 1). (Middle) fushi tarazu (ftz) contains an Inr, a TATA box and DPE. ftz activation by Caudal is primarily dependent on the DPE site (Juven-Gershon et al., 2008). (Bottom) Tollo contains both of the Drosophila downstream elements, the MTE and DPE, which both contribute to transcriptional activation (Lim et al., 2004). (B) Promoters with non-position-specific elements. (Top) The Drosophila genes encoding histones H2A, H2B, H3 and H4 have canonical TATA boxes, but (middle) the H1 gene is TRF2 dependent and not bound by TBP (Isogai et al., 2007a). TRF2 is a conserved TBP-replacing factor, which does not directly associate with DNA but rather interacts with specific transcription factors, such as DREF, which binds the DRE (Hochheimer et al., 2002). (Bottom) TBP- and TRF2-dependent promoters can regulate the same gene via alternative TSSs, as is the case for PCNA, in which one TSS is TRF2 dependent via its interaction with DREF, and a second downstream promoter is TBP dependent (Hochheimer et al., 2002). Only experimentally validated elements are shown.
Newly identified core promoter elements
TSSs are to some degree defined by core promoter elements. A repertoire of elements collectively defines the TSS, with elements occurring on their own or as part of ‘modules’ that consist of two or three elements in a specific configuration (Fig. 3A). With the increasing availability of high-throughput data on the location of TSSs, computational approaches for confirming reported core promoter elements and for searching for novel core promoter elements are now available. Indeed, a first genome-scale computational analysis of Drosophila core promoters identified ten enriched elements, including the three canonical elements (Ohler et al., 2002). One of the novel sequences, the motif ten element (MTE, see Table 1), was subsequently shown to promote Pol II transcription in Drosophila, as well as in human in vitro systems; however, sequence searches have not identified an MTE-like motif as enriched in human core promoters (Lim et al., 2004; Jin et al., 2006).
The TATA box, Inr, DPE and MTE show clear position-specific bias with respect to the TSS, and the incidence of such elements is higher in peaked than in broad promoters (Ohler et al., 2002; Carninci et al., 2006; Sandelin et al., 2007; Megraw et al., 2009; Rach et al., 2009). However, not all position-specific sequences in or near core promoters are directly linked to transcription initiation but rather to ‘neighboring’ events; for example, nucleosomes are depleted upstream of many TSSs and this is related to the presence of poly(dA:dT) stretches that are disfavored from being incorporated within a nucleosome (Mavrich et al., 2008; Kaplan et al., 2009). Additionally, specific sequence elements downstream of TSSs are associated with Pol II stalling in Drosophila embryonic development (Hendrix et al., 2008; Lee et al., 2008).
Other computationally predicted Drosophila core promoter elements have so far shown less specificity in their position relative to the TSS and are enriched more broadly around the TSS or up to ~100 bp upstream. One of these elements corresponds to a previously identified sequence motif, the DNA replication-related element (DRE, Table 1) (Hochheimer et al., 2002; Ohler et al., 2002) (Fig. 3B). Other Drosophila promoter sequence motifs, including the Ohler 1, Ohler 6 and Ohler 7 elements (Table 1), have been reported by independent computational analyses (Sharan and Myers, 2005; Fitzgerald et al., 2006; Isogai et al., 2007a), but comparative computational analyses of mammalian datasets have not identified these apparently Drosophila-specific motifs (Fitzgerald et al., 2006). Functional studies are beginning to define the activities of these sequence elements. For example, the DRE is enriched in genes that are transcribed in the female germline, as well as in genes transcribed maternally and deposited in developing oocytes, and the canonical elements (TATA box, Inr or DPE) are largely absent from these genes (Fitzgerald et al., 2006; Down et al., 2007; Rach et al., 2009). In turn, canonical elements are enriched in embryonically transcribed zygotic genes, particularly those that encode developmental regulators (Engström et al., 2007; Rach et al., 2009).
Additional elements identified and studied in individual human or viral genes include two TFIIB recognition elements (BREU and BRED), the downstream core element (DCE), and the X core promoter element (XCPE) (Table 1) (Lewis et al., 2000; Tokusumi et al., 2007; Anish et al., 2009) (Fig. 3A). Although most of these canonical promoter elements have been repeatedly experimentally validated in individual promoters, they have not been found to be significantly enriched above background, indicating that their role might not be as widespread. For example, BREU and BRED, which have different consensus sequences, are positioned directly upstream and downstream, respectively, of the TATA box and affect transcription levels (Lagrange et al., 1998; Deng and Roberts, 2005). Contrary to the finding that BREU only functions upstream of a TATA box, matches to the published consensus BREU sequence actually occur at a higher rate in TATA-less promoters, and are found both upstream and downstream of the TATA box. Thus, it is not possible to distinguish from such patterns whether the BREU is a genuine core promoter element, enriched at a particular location above background, or is simply a reflection of the increased GC content around the TATA box (Sandelin et al., 2007). It does not refute the biological evidence for the function of BREs in individual promoters; as shown by studies of the MTE in humans, the functionality of a core promoter element does not automatically imply that it is a widespread and necessary feature of an organism's transcription machinery.
Lastly, analyses of human datasets have shown that sequence motifs bound by specific transcription factors, such as by CREB (cAMP response element binding protein), E2F and YY1 (yin-yang 1), are enriched close to TSSs (Fitzgerald et al., 2004; Xi et al., 2007; Tharakaraman et al., 2008). Such analyses have also identified a few additional ‘orphan’ elements that are enriched in core promoters, raising the intriguing possibility that the distinction between proximal and core promoter elements is blurred in mammals, in as much as transcription factors might also contribute to the positioning of Pol II at TSSs (Megraw et al., 2009). Thus, the availability of genome-wide data on TSSs has allowed investigators to computationally validate the presence of known elements, as well as to identify new sequence elements enriched in core promoter regions. Future work is expected to increasingly reconcile orphan elements with possible specific functions in transcription initiation.
Alternative TSSs
Analyses of available transcript data indicate that in vertebrates, 20-50% of genes contain alternative TSSs (Davuluri et al., 2008). In contrast to broad initiation patterns, which are linked to the flexibility of the initiation process at one site, alternative TSSs are typically separated by a clear spacer region and are often active under different conditions. For instance, the two TSSs of the Drosophila hunchback gene are separated by more than 3 kb, define non-overlapping first exons, and are associated with early embryonic versus adult and broader embryonic transcription (Bender et al., 1988; Schröder et al., 1988). Alternative TSSs may thus reflect an increased complexity of the transcription process, raising the problem of how enhancers communicate with specific core promoters (see discussion of enhancer-core promoter element specificity below). On the other hand, alternative TSSs might simply be indicators of ongoing cis-regulatory evolution. In a comparison of human and mouse high-throughput CAGE data, some alternative TSSs were found to show strong signs of turnover: whereas transcriptional activity was detected from two alternative TSSs for one gene in both species, different TSSs had apparently been selected for, and one TSS was found to be preferentially active in humans and the other in mouse (Frith et al., 2006). Alternative TSSs therefore increase the complexity of transcriptional regulation, both by increasing the flexibility with which promoters can interact with distal regulatory regions and by being able to accommodate evolutionary changes due to the presence of multiple sites.
Proximal and core promoter elements in Myog
The mouse Myog gene has a peaked TSS (designated +1), and within the −184 to +1 region, which is sufficient for muscle-specific transcription, the only identified core promoter element is a TATA box-like sequence (TAAAT) located at −29 to −25 (Edmonson et al., 1992) (Fig. 1B). This region contains proximal promoter sequences that are bound by muscle-specific transcription factors: E-box binding sites for MyoD (myogenic differentiation 1, or MYOD1) located upstream and downstream of the TATA box at −141 to −136 and −15 to −10, respectively, and a single binding site for MEF2 (myocyte-specific enhancer factor 2) at −67 to −59. So, Myog is an example of a gene in which the distinction between proximal and core promoter elements is blurred. Analyses of reporter genes in cultured cells have revealed that E-boxes are required for high-level Myog transcription in myotubes and for transcriptional activation by MyoD in non-myogenic cells and that the Myog TATA box is essential for transcription in myotubes. Similar transcriptional requirements for the proximal promoter sequences were observed in subsets of myogenic precursors in mouse embryos (Cheng et al., 1993). Chromatin structure is also important in Myog transcription (Box 2). Methylation of histone H3 by the PRMT5 arginine methyltransferase is involved in recruiting the SWI/SNF (switch/sucrose non-fermentable) chromatin remodeling complex to Myog, and these events are crucial for the stable binding of MyoD to the proximal promoter and for Myog transcription (Dacwag et al., 2007; Dacwag et al., 2009). Thus, in accordance with the textbook model, proximal promoter elements bound by transcription factors dictate Myog transcription parameters.
Core promoter recognition
Enhancer-core promoter element specificity
Most enhancer elements can activate transcription irrespective of the types of associated core promoter elements, but some enhancer elements exhibit core promoter element specificity. The latter finding is unexpected based on three lines of evidence. First, in transgenic animals, artificial transcription-regulatory units that contain enhancer sequences from one gene and core promoter sequences from another can drive transcription in a pattern similar to that driven by the endogenous enhancer sequences (e.g. Phelps and Brand, 1998). Recently, this methodology was used to identify enhancers that direct gene expression in subsets of cells in the adult Drosophila brain, the findings of which indicated that the Drosophila genome contains over 50,000 enhancers (Pfeiffer et al., 2008). Second, in enhancer-trap studies, enhancers that direct transcription in particular patterns during development have been identified by the random genomic insertion of a plasmid that contains a core promoter upstream of a reporter gene (e.g. Brand and Perrimon, 1993). Finally, enhancer-bound transcription factors can activate the transcription of natural target genes that have different core promoter elements and depend on different basal transcription factors. For example, the mouse hematopoietic-specific transcription factor EKLF (erythroid Krupple-like factor, or KLF1) activates the transcription of both the adult β-globin gene and the Ahsp (α-hemoglobin-stabilizing protein) gene, but β-globin transcription depends on its core promoter DCE and on a particular basal transcription factor (the TFIID subunit TAF9, as discussed below), whereas the Ahsp core promoter lacks a DCE and its transcription is TAF9 independent (Sengupta et al., 2009). Thus, enhancer elements and transcription factors are often compatible with multiple core promoter elements and basal transcription factors.
Conversely, during mouse development evidence for enhancer-core promoter specificity has emerged. For instance, the Eif1a gene has alternative TATA-containing and TATA-less core promoters that are differentially utilized during early development (Davis and Schultz, 2000). Transcripts derived from the TATA-containing core promoter predominate in the fully grown oocyte, whereas TATA-less transcripts predominate at the two-cell and blastocyst stages. More direct evidence for enhancer-core promoter specificity has come from competition experiments in Drosophila in which enhancers showed a preference for a TATA-containing over a TATA-less promoter (Ohtsuki et al., 1998). Finally, an enhancer-trap experiment revealed that in 4 of 18 cases examined, endogenous Drosophila enhancers showed a preference for a DPE-containing core promoter over a TATA-containing core promoter, or vice versa (Butler and Kadonaga, 2001). Thus, core promoter elements can affect the activity of enhancers.
Based on these findings, it was anticipated that individual transcription factors would display core promoter specificity. In fact, the Drosophila Caudal transcription factor, a regulator of genes involved in establishing the embryonic body plan, such as fushi tarazu, preferentially activates transcription from DPE-containing, as opposed to TATA-containing, core promoters (Juven-Gershon et al., 2008). Furthermore, many Caudal target genes in the Drosophila genome contain a conserved DPE, which provides evolutionary support for the functional link between Caudal and the DPE. Thus, core promoters can influence the specificity of transcription factor function and might do so in vivo to ensure that transcription factors function specifically with their cognate promoters rather than with the plethora of other promoters with which they are faced.
TFIID binds core promoter elements
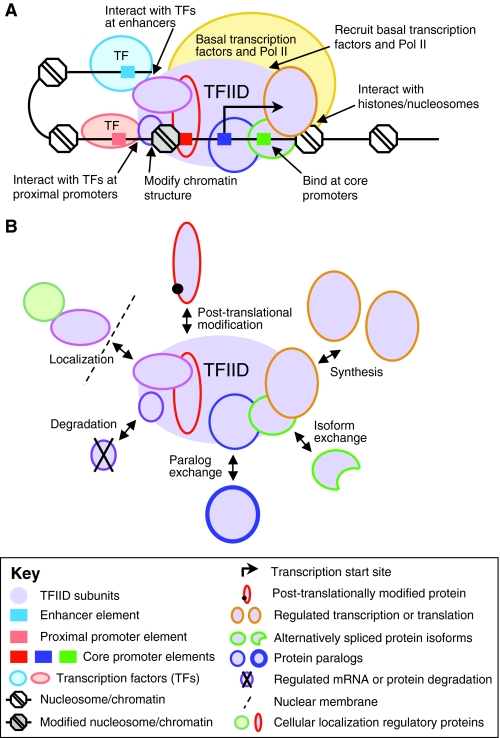
Since only a fraction of core promoters contains a TATA box element for binding by TBP, it was postulated early on that other basal transcription factors are required for binding TATA-less core promoters. Indeed, several examples have been documented, the majority of which involve TFIID components. The TFIID complex was initially biochemically purified from Drosophila embryos and human cultured cells and shown to contain TBP and ~15 TBP-associated factors (TAFs 1-15), most of which are conserved in all eukaryotes (Dynlacht et al., 1991; Takada, 1992; Tora, 2002). In a human cell line, TAF1 associates with the core promoters of 60% of the 7282 transcriptionally active genes but with only 9% of the 7143 inactive genes, indicating that core promoter binding by TFIID correlates with the transcriptional activation of many genes (Kim et al., 2005) (Fig. 4A). Despite this finding, direct binding of TAFs has been implicated for only a few core promoter elements: TAF1 and TAF2 bind the Drosophila Inr element, TAF1 binds the human DCE, and TAF6 and TAF9 bind the Drosophila DPE (Chen et al., 1994; Chalkley and Verrijzer, 1999; Burke and Kadonaga, 1997; Wu et al., 2001; Lee et al., 2005).
Fig. 4.
General functions of TFIID and mechanisms that regulate the diversity of TFIID subunits. (A) The general functions of TFIID that facilitate transcription initiation by Pol II. (B) The general mechanisms by which the structure and function of TFIID are modified. Specific examples are described in the text.
A major DNA-binding activity within TFIID is probably imparted by the histone-fold domain (HFD)-containing TAFs (TAFs 3, 4, 6, 8-13) (Gangloff et al., 2001). HFDs in histone proteins are involved in the heterodimerization of core histones H3 with H4 and H2A with H2B and their assembly into a histone octamer, the basic protein unit of chromatin (Box 2) (Luger et al., 1997). The HFD-mediated interaction enhances binding of the TAF6-TAF9 pair to the DPE (Shao et al., 2005). Other HFD-containing TAFs also bind DNA, and in the case of TAF4 the DNA-binding activity maps to a region within the HFD. These findings suggest that TAF HFDs specify binding to core promoter elements; however, the DNA-binding sequence specificity of four pairs of HFD-containing TAFs (TAF4-TAF12, TAF3-TAF10, TAF8-TAF10 and TAF11-TAF13) remains to be determined.
Interactions of TAFs with modified histones might aid core promoter recognition by TFIID (Box 2) (Fig. 4A). For example, TAF1 contains two bromodomains that bind acetylated lysine (K) and have high affinity for histone H4 doubly acetylated at K5/K12 or K8/K16, and TAF3 contains a PHD (plant homeodomain) domain that has high affinity for histone H3 trimethylated at K4 (Jacobson et al., 2000; Vermeulen et al., 2007), a histone modification known to mark the 5′ end of genes. Additionally, protein-protein interactions between transcription factors and TAFs may recruit TFIID to core promoters. For example, the glutamine-rich activation domains of the transcription factors Sp1 and CREB associate with Drosophila TAF4, but strong evidence that TAF-transcription factor interactions take place in vivo in the context of TFIID has been elusive (Wang et al., 2007). Thus, TAF interactions with modified histones and transcription factors might stabilize TFIID-core promoter interactions that are specified by the binding of TAFs to core promoter elements.
Finally, core promoter recognition by TFIID is regulated by post-translational modification (Fig. 4B). During mitosis, when Pol II transcription is inhibited, TBP and several TAFs are phosphorylated (Segil et al., 1996). TFIID purified from mitotic cells is unable to direct activated transcription in vitro; however, this activity is restored by dephosphorylation, suggesting that TFIID phosphorylation inhibits Pol II transcription. Other modifications of TAFs have also been described, including methylation of TAF10 by the SET9 (SETD7) methyltransferase, which increases the transcription of some, but not all, TAF10-dependent genes, and sumoylation of several human TAFs, which interferes with the binding of TFIID to promoter DNA (Kouskouti et al., 2004; Boyer-Guittaut et al., 2005). Similarly, human TAF1 is phosphorylated by casein kinase 2, which in vitro experiments suggest changes the binding specificity of TFIID for core promoter elements (Lewis et al., 2005). Phosphorylation appears to block TAF1 binding to the DCE and enables TAF6-TAF9 binding to the DPE. Metazoan TAF1 also undergoes various auto-modifications, but their functional consequences remain unknown (Dikstein et al., 1996; Mizzen et al., 1996; Pham and Sauer, 2000; Auty et al., 2004). Lastly, acetylation of TAFs may also be involved in core promoter recognition (Galasinski et al., 2000). Thus, post-translational modification of TFIID subunits is a crucial, but poorly understood, component of Pol II transcription-regulatory mechanisms that occur at core promoters.
Abundant TFIID does not support Myog transcription
The textbook model would predict that once MyoD is stably bound at the proximal promoter, it interacts with a TAF subunit of TFIID and, together with the binding of TBP to the TATA box, productively engages TFIID at the core promoter, resulting in activated Myog transcription. To test this model, Deato et al. used an in vitro transcription system comprising purified protein factors and a reporter gene that contained the 184 bp Myog transcription-regulatory region (Deato et al., 2008). These studies revealed that TBP alone, or the whole TFIID complex, binds the Myog core promoter, and, in the absence of MyoD, TFIID induces a basal level of transcription. Surprisingly, the addition of MyoD and its typical heterodimer partner E47 (TCF3) to the reaction did not increase transcription over the basal level. These data suggest that MyoD requires a core promoter recognition complex other than abundant TFIID to activate Myog transcription.
Diverse TFIID complexes
The existence of orphan core promoter elements with unknown binding proteins suggests that other basal transcription factors possess core promoter element binding activity. Based on previous findings, TFIID subunits are the most likely candidates. To date, the only method used for de novo identification of TFIID subunits has been the biochemical purification of TFIID from cultured mammalian cells or from Drosophila embryos by co-purification with TBP (Dynlacht et al., 1991; Takada et al., 1992). Owing to the limitations of this approach, only highly abundant, widely expressed TAFs have been identified. However, as described below, the searching of predicted metazoan proteomes for proteins that share sequence similarity with abundant TFIID subunits has identified additional TBP-like and TAF-like proteins. Those identified might represent the complete collection of TBP-like and TAF-like proteins, but it remains possible that low-abundance, cell type-specific, tissue-specific or developmental stage-specific TBP-like and TAF-like proteins with novel sequences remain to be identified, possibly by traditional biochemical purification schemes.
TBP paralogs
Three TBP paralogs have been described. The Drosophila genome encodes two TBP paralogs, termed TBP-related factor 1 (TRF1) and TRF2, other metazoans encode TRF2, and vertebrates also encode a third TBP paralog, TRF3 (TRF1-3 are also known as TBPL1-3) (Reina and Hernandez, 2007; Torres-Padilla and Tora, 2007) (Fig. 4B). Since TRF1 predominantly regulates the transcription of Pol III genes, we will focus on TRF2 and TRF3 (Isogai et al., 2007b). Multiple lines of evidence suggest that TBP, TRF2 and TRF3 play different roles in regulating transcription during development. In Xenopus, each of the TBP-like proteins is essential for embryonic development (Veenstra et al., 2000; Jallow et al., 2004; Jacobi et al., 2007). TRF2 and TRF3 are required for the transcription of many more genes than TBP in the early gastrula embryo, with TRF2 preferentially required for genes linked to carbohydrate metabolism, TRF3 for genes with ventral-specific expression, and TBP for the transcription of maternal-effect genes and genes that are more abundantly expressed in adult stages (Jacobi et al., 2007). TRF2 is also essential for early embryonic transcription and for development in C. elegans, zebrafish and Drosophila, and for early stages of metamorphosis in Drosophila (Dantonel et al., 2000; Kaltenbach et al., 2000; Veenstra et al., 2000; Müller et al., 2001; Bashirullah et al., 2007).
Interestingly, TRF2 is not essential for viability in mice but is essential for spermatogenesis, indicating that TRF2 performs different functions in different species or that the developmental function of TRF2 can be compensated by TBP or TRF3 in mice (Zhang et al., 2001). Assuming that the functions of TRF2 in mice and Drosophila are mechanistically similar, compensation by TBP is unlikely as a comparison of TBP and TRF2 DNA-binding sites in Drosophila S2 cells has revealed that the TATA box element is prevalent in TBP-bound core promoters but is scarce in TRF2-bound core promoters (Isogai et al., 2007a). Instead, the DRE is most commonly enriched in TRF2-bound core promoters, which is consistent with the identification of the DRE-binding protein DREF as a component of a TRF2-containing complex (Hochheimer et al., 2002). Compensation by TRF3 is also unlikely as TRF3 has a DNA-binding domain that is almost identical in sequence to that of TBP and, as might be expected, binds the TATA box element (Bártfai et al., 2004; Jallow et al., 2004). These data indicate that TRF2 is essential for transcription of some, but not all, Pol II genes and regulates specific developmental events that are distinct in different species (Fig. 3B).
Studies in multiple organisms have established roles for TRF3 in regulating transcription during oogenesis and early embryogenesis (Bártfai et al., 2004; Jallow et al., 2004; Yang et al., 2006; Xiao et al., 2006; Gazdag et al., 2007; Jacobi et al., 2007). Accordingly, TRF3 expression (as detected by analysis of mRNA) is limited to testes, ovaries and early-stage embryos in Xenopus and zebrafish, and to ovaries and early-stage embryos in mice (Bártfai et al., 2004; Xiao et al., 2006; Gazdag et al., 2007). However, TRF3 expression (as detected by analysis of protein) in humans and mice is not limited to ovaries and testes but instead occurs in all tissues and cells examined, including mouse skeletal muscle tissue, C2C12 cells, and primary myoblasts and myofibers (Persengiev et al., 2003; Deato and Tjian, 2007). These conflicting reports of TRF3 expression in mouse skeletal muscle cells impact the interpretation of functional studies of TRF3 during Myog transcription in skeletal muscle cells (as discussed below). Nevertheless, what is clear is that TBP, TRF2 and TRF3 function as gene-specific transcriptional regulators during development.
TAF paralogs
TAF paralogs are expressed predominantly in gonads and germ cells and are required for fertility in many organisms (Kolthur-Seetharam et al., 2008; Freiman et al., 2009). In mice, the TAF4 paralog TAF4b is expressed in somatic granulosa cells in ovaries and gonocytes in post-natal testes (Freiman et al., 2001; Falender et al., 2005). In TAF4b-deficient mice, granulosa cells have impaired proliferative capacity and undergo apoptosis, and maintenance of spermatogenesis is impaired. Gene expression array analyses of TAF4b-deficient mouse ovaries or granulosa cells indicate that TAF4b is necessary for the transcription of genes central to oogenesis (Geles et al., 2006). In Drosophila, TAF4, TAF5, TAF6, TAF8 and TAF12 paralogs are predominantly expressed in primary spermatocytes in testes and are required for the transcription of genes necessary for the entry of primary spermatocytes into meiosis (Hiller et al., 2001; Hiller et al., 2004). Thus, TAF paralogs are crucial transcriptional regulators of gametogenesis in both vertebrates and invertebrates.
Regulated TFIID subunit expression and Myog transcription
In 2001, Perletti et al. found that retinoic acid-induced terminal differentiation of F9 embryonal carcinoma cells into primitive or visceral mesoderm was accompanied by proteosome-dependent degradation of TAF4 and TBP but not of other TAFs (TAF5, TAF7, TAF12 or TAF13) (Perletti et al., 2001). The pathways that signal particular TFIID subunits for degradation were not investigated but possibly involve post-translational modifications of these subunits (Fig. 4B). Perletti et al. further found that sustained expression of TAF4 impairs differentiation, suggesting that the regulated degradation of particular TFIID subunits is required for differentiation. Finally, they found that similar reductions in TFIID subunit expression accompany serum deprivation-induced C2C12 myoblast-to-myotube differentiation. Collectively, these data suggest that the regulated degradation of abundant TFIID subunits is generally required for terminal differentiation.
Deato and Tjian added to this story by examining the expression of other proteins in response to serum deprivation-induced C2C12 myoblast-to-myotube differentiation (Deato and Tjian, 2007). They found reduced levels of TAF1, but not of TRF3, nor of the basal transcription factors TFIIA, TFIIB, TFIIE, TFIIF, TFIIH or Pol II. Additionally, they found that myoblast-to-myotube differentiation requires constant expression of TAF3 and TRF3, as knockdown of either protein reduced Myog transcription and blocked differentiation. TAF3 and TRF3 appear to directly regulate Myog transcription because a TAF3-TRF3 complex associates with the Myog core promoter in differentiated myotubes but not in proliferating myoblasts (Fig. 1B). Thus, altering the relative amounts of TFIID complexes comprises part of the Myog transcription-regulatory mechanism.
Developmental transcription
Master transcriptional control by TAFs
Studies in mammalian systems have not only echoed the importance of distinct TFIID complexes in transcriptional regulation but have also refined our knowledge of the underlying mechanisms involved. In human cells, as in Drosophila cells, distinct TFIID complexes regulate specific developmental programs. For example, in response to apoptotic cell death signals, human HeLa cells express an alternatively spliced isoform of TAF6, TAF6δ, which is a component of a TFIID complex that lacks TAF9 (the HFD partner of TAF6) (Bell et al., 2001). Particularly illustrative of the regulatory power of alternative TAFs is that overexpression of TAF6δ, or induction of endogenous TAF6δ expression, in HeLa cells induces apoptotic cell death and the transcription of pro-apoptotic genes in the absence of an apoptotic signal. Surprisingly, the pro-apoptotic transcription factor p53 (TP53) is not required for TAF6δ-dependent transcription or apoptosis (Wilhelm et al., 2008). Similarly, overexpression of TAF4b in non-granulosa cells, such as mouse NIH/3T3 fibroblasts, results in the expression of genes that are TAF4b dependent in ovaries and granulosa cells and that are not normally expressed in non-granulosa cells (Geles et al., 2006). Thus, some TAFs are master regulators of developmental transcription programs. The TAF4b and TAF6δ overexpression results are unexpected because transcriptional activation of some genes can occur in the absence of specific transcription factors: ovary-specific transcription factors, in the case of TAF4b in non-ovary cells, and pro-apoptotic transcription factors, in the case of TAF6δ in cells not stimulated to undergo apoptosis. Thus, TFIID complexes may regulate gene-specific transcription both dependently and independently of transcription factors.
Transcriptional control by regulated TFIID assembly
The TAF4b and TAF6δ overexpression results also suggest that alternative TAFs can compete with paralogous abundant TAFs for integration into TFIID and that the resultant equilibrium controls the transcription profile of the cell (Fig. 4B). Support for this model comes from studies of TAF4-deficient mouse fibroblasts and in vitro studies of purified TAF4-TFIID and TAF4b-TFIID complexes (Mengus et al., 2005; Liu et al., 2008). Furthermore, if integral subunits of the TFIID complex, such as TAF4, TAF4b, TAF6 and TAF6δ, are able to exchange, then the TFIID complex as a whole is probably highly dynamic, providing additional avenues for regulation. In this context, analysis of the p21 (CDKN1A) gene in human cells suggests that transcriptional activation involves the sequential assembly of TFIID at the core promoter (Li et al., 2007). Initially, TAF4, TAF5 and TBP localize to the p21 core promoter and TAF1 associates with the enhancer-bound transcription factor p53 through binding of TAF1 bromodomains to diacetylated p53. Subsequently, DNA looping juxtaposes the enhancer and core promoter to facilitate TAF1 assembly with the TFIID subunits at the core promoter. Thus, developmental transcription is likely to be under the control of mechanisms that regulate the assembly of TFIID, including exchange of paralogous TAFs. These mechanisms might involve unspecified chaperone proteins or ATP-dependent remodeling complexes comparable to those involved in the exchange of histones in nucleosomes (Jin et al., 2005).
Transcriptional control by regulated TAF localization
TFIID activity is regulated by the availability of TAFs (Fig. 4B). In C. elegans, the cytoplasmic sequestration of TAF-4 regulates the transition from maternal to zygotic gene transcription (Guven-Ozkan et al., 2008). Phosphorylation of the cytoplasmic OMA-1 protein (a regulator of oocyte maturation) by the MBK-2 kinase at the maternal-to-zygotic transition facilitates the binding of a TAF12-like HFD in the OMA-1 protein to the TAF4 HFD, leading to retention of TAF4 in the cytoplasm and Pol II transcription repression in early germline blastomeres. Although details of how cytoplasmic OMA-1 comes into contact with nuclear TAF4 remain to be resolved, this study highlights the importance of TFIID subunit localization for the regulation of developmental transcription. Indeed, in mammalian cells, an interaction with an HFD partner is necessary for the nuclear localization of TAF10, which lacks a nuclear localization signal, and regulated localization of TAF10 may play a role in male germ cell differentiation (Soutoglou et al., 2005). Lastly, cytoplasmic sequestration of TRF3 may regulate transcription after mitosis. In human cells, it has been observed that the re-importation of TRF3 into the nucleus after mitosis is delayed relative to that of TBP and other basal transcription factors (Persengiev et al., 2003). Therefore, subcellular localization of TAFs might be a general mechanism by which transcription is regulated at critical stages of cell proliferation or development.
A TAF3-TRF3 complex supports Myog transcription
The work of Deato et al. (Deato et al., 2008) caps off our current understanding of the role of core promoter elements and TFIID complexes in Myog transcriptional regulation during terminal differentiation. Using the reconstituted transcription system described above, they found that, unlike the abundant TFIID complex, an alternative TFIID complex that contains TAF3 and TRF3 could support the MyoD-dependent transcription of Myog (Fig. 1B). The mechanism involves binding of the TATA box by TRF3 and a direct interaction between TAF3 and MyoD. Intriguingly, the region of TAF3 that interacts with MyoD includes the HFD, suggesting that the binding of MyoD to TAF3 might alter the binding of TAF3 to TAF10 or to an unknown HFD protein or might alter the binding of TAF3 to DNA. In summary, the transcriptional activation of Myog during the terminal differentiation of myoblasts to myotubes requires multiple events: (1) the degradation of particular abundant TFIID subunits; (2) the binding of a TAF3-TRF3 complex to the TATA box core promoter element; (3) the binding of MyoD to E-box elements in the proximal promoter; and (4) the binding of MyoD to TAF3. Confirmation of this model will require the generation of Trf3-knockout mice, which would be predicted to accumulate myocytes that are arrested in their terminal differentiation program.
Conclusions
Here, we have described many lines of evidence that support roles for core promoter elements and TFIID complexes in the regulation of developmental transcription. Fundamental to this is the diversity of core promoter elements and TFIID complexes. Now that the importance and extent of this diversity have been recognized, the challenge is to understand the molecular mechanisms involved in their differing functions. For instance, how are the expression and function of TFIID complexes developmentally regulated? How do TFIID complexes recognize the core promoters of specific genes? How is transcription-regulatory information conveyed between enhancer elements and core promoter elements? Answers to these questions and many others will be necessary to fully understand how the timing, location and level of protein-coding gene transcription is achieved for proper organismal growth and development.
Acknowledgements
We thank E. Rach, L. Pile, A. Keels and the anonymous reviewers for comments on the manuscript and D. Corcoran for assistance with data analysis. Work on transcriptional regulation in the Ohler laboratory is supported by the NIH and in the Wassarman laboratory by the NSF. Deposited in PMC for release after 12 months.
References
- Ahsan B., Saito T. L., Hashimoto S., Muramatsu K., Tsdua M., Sasaki A., Matsushima K., Aigaki T., Morishita S. (2009). MachiBase: a Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res. 37, D49-D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anish R., Hossain M. B., Jacobson R. H., Takada S. (2009). Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS One 4, e5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi N., Wassarman D. A. (2000). Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: Diversity in TFIIA and TFIID components contributes to gene specific transcription regulation. J. Cell Biol. 150, F45-F49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auty R., Steen H., Myers L. C., Persinger J., Bartholomew B., Gygi S. P., Buratowski S. (2004). Purification of active TFIID from Saccharomyces cerevisiae: Extensive promoter contact and co-activator function. J. Biol. Chem. 279, 49973-49981 [DOI] [PubMed] [Google Scholar]
- Bártfai R., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orbán L., Müller F. (2004). TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 14, 593-598 [DOI] [PubMed] [Google Scholar]
- Basehoar A. D., Zanton S. J., Pugh B. F. (2004). Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699-709 [DOI] [PubMed] [Google Scholar]
- Bashirullah A., Lam G., Yin V. P., Thummel C. S. (2007). dTrf2 is required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. Dev. Dyn. 236, 3173-3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D., Scheer E., Tora L. (2001). Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8, 591-600 [DOI] [PubMed] [Google Scholar]
- Bender M., Horikami S., Cribbs D., Kaufman T. C. (1988). Identification and expression of the gap segmentation gene hunchback in Drosophila melanogaster. Dev. Genet. 9, 715-732 [DOI] [PubMed] [Google Scholar]
- Berkes C. A., Tapscott S. J. (2005). MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16, 585-595 [DOI] [PubMed] [Google Scholar]
- Bhaumik S. R., Smith E., Shilatifard A. (2007). Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008-1016 [DOI] [PubMed] [Google Scholar]
- Blau H. M., Chiu C. P., Webster C. (1983). Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32, 1171-1180 [DOI] [PubMed] [Google Scholar]
- Boyer-Guittaut M., Birsoy K., Potel C., Elliot G., Jaffray E., Desterro J. M., Hay R. T., Oelgeschläger T. (2005). SUMO-1 modification of human transcription factor (TF) IID complex subunits: inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J. Biol. Chem. 280, 9937-9945 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 [DOI] [PubMed] [Google Scholar]
- Burke T. W., Kadonaga J. T. (1996). Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10, 711-724 [DOI] [PubMed] [Google Scholar]
- Burke T. W., Kadonaga J. T. (1997). The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020-3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher P., Trifonov E. N. (1986). Compilation and analysis of eukaryotic POL II promoter sequences. Nucleic Acids Res. 14, 10009-10026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. E. F., Kadonaga J. T. (2001). Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. (2005). The transcription landscape of the mammalian genome. Science 309, 1559-1563 [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C. A., Taylor M. S., Engström P. G., Frith M. C., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626-635 [DOI] [PubMed] [Google Scholar]
- Chalkley G. E., Verrijzer C. P. (1999). DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 18, 4835-4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. L., Attardi L. D., Verrijzer C. P., Yokomori K., Tjian R. (1994). Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcription activators. Cell 79, 93-105 [DOI] [PubMed] [Google Scholar]
- Cheng T. C., Wallace M. C., Olson E. N. (1993). Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261, 215-218 [DOI] [PubMed] [Google Scholar]
- Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. (2007). The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol. 27, 384-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacwag C. S., Bedford M. T., Sif S., Imbalzano A. N. (2009). Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell. Biol. 29, 1909-1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel J. C., Quintin S., Lakatos L., Labouesse M., Tora L. (2000). TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol. Cell 6, 715-722 [DOI] [PubMed] [Google Scholar]
- Davis W., Schultz R. M. (2000). Developmental changes in TATA-box utilization during preimplantation mouse development. Dev. Biol. 218, 275-283 [DOI] [PubMed] [Google Scholar]
- Davuluri R. V., Suzuki Y., Sugano S., Plass C., Huang T. H. (2008). The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24, 167-177 [DOI] [PubMed] [Google Scholar]
- Deato M. D. E., Tjian R. (2007). Switching of the core transcription machinery during myogenesis. Genes Dev. 21, 2137-2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M. D. E., Marr M. T., Sottero T., Inouye C., Hu P., Tjian R. (2008). MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol. Cell 32, 96-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve G. P., Rastegar M., Davie J. R. (2009). Epigenetic control. J. Cell. Physiol. 219, 243-250 [DOI] [PubMed] [Google Scholar]
- Deng W., Roberts S. G. (2005). A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 19, 2418-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R., Ruppert S., Tjian R. (1996). TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell 84, 781-790 [DOI] [PubMed] [Google Scholar]
- Down T. A., Bergman C. M., Su J., Hubbard T. J. (2007). Large-scale discovery of promoter motifs in Drosophila melanogaster. PLoS Comput. Biol. 3, e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. (1991). Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66, 563-576 [DOI] [PubMed] [Google Scholar]
- Edmonson D. G., Cheng T.-C., Cserjesi P., Chakraborty T., Olson E. N. (1992). Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12, 3665-3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P. G., Ho Sui S. J., Drivenes O., Becker T. S., Lenhard B. (2007). Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 17, 1898-1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falender A. E., Freiman R. N., Geles K. G., Lo K. C., Hwang K., Lamb D. J., Morris P. L., Tjian R., Richards J. S. (2005). Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 19, 794-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium. Suzuki H., Forrest A. R., van Nimwegen E., Daub C. O., Balwierz P. J., Irvine K. M., Lassmann T., Ravasi T., Hasegawa Y., et al. (2009). The transcription network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 41, 553-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P. C., Shlyakhtenko A., Mir A. A., Vinson C. (2004). Clustering of DNA sequences in human promoters. Genome Res. 14, 1562-1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P. C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C. (2006). Comparative genomics of Drosophila and human core promoters. Genome Biol. 7, R53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R. N. (2009). Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim. Biophys. Acta 1789, 161-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R. N., Albright S. R., Zheng S., Sha W. C., Hammer R. E., Tjian R. (2001). Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293, 2084-2087 [DOI] [PubMed] [Google Scholar]
- Frith M. C., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A. (2006). Evolutionary turnover of mammalian transcription start sites. Genome Res. 16, 713-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galasinski S. K., Lively T. N., Grebe de Baron A., Goodrich J. A. (2000). Acetyl coenzyme A stimulates RNA polymerase II transcription and promoter binding by transcription factor IID in the absence of histones. Mol. Cell. Biol. 20, 1923-1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y. G., Romier C., Thuault S., Werten S., Davidson I. (2001). The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26, 250-257 [DOI] [PubMed] [Google Scholar]
- Gazdag E., Rajkovic A., Torres-Padilla M. E., Tora L. (2007). Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggest different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction 134, 51-62 [DOI] [PubMed] [Google Scholar]
- Geles K. G., Freiman R. N., Liu W.-L., Zheng S., Voronina E., Tjian R. (2006). Cell type-specific induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc. Natl. Acad. Sci. USA 103, 2594-2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R. (2008). Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135, 149-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Doenecke D. (2009). Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431, 1-12 [DOI] [PubMed] [Google Scholar]
- Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., Klein W. H. (1993). Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501-506 [DOI] [PubMed] [Google Scholar]
- Hendrix D. a., Hong J. W., Zeitlinger J., Rokhsar D. S., Levine M. S. (2008). Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 105, 7762-7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., Ahmed K. (2004). Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 20, 320-326 [DOI] [PubMed] [Google Scholar]
- Hiller M. A., Lin T.-Y., Wood C., Fuller M. T. (2001). Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15, 1021-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.-Y., Marino S., Fuller M. T. (2004). Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131, 5297-5308 [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Zhou S., Zheng S., Holmes M. C., Tjian R. (2002). TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420, 439-445 [DOI] [PubMed] [Google Scholar]
- Ioshikhes I. P., Albert I., Zanton S. J., Pugh B. F. (2006). Nucleosome positions predicted through comparative genomics. Nat. Genet. 38, 1210-1215 [DOI] [PubMed] [Google Scholar]
- Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R. (2007a). Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 21, 2936-2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Takada S., Tjian R., Keles S. (2007b). Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J. 26, 76-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi U. G., Akkers R. C., Pierson E. S., Weeks D. L., Dagle J. M., Veenstra G. J. C. (2007). TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. EMBO J. 26, 3900-3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson R. H., Ladurner A. G., King D. S., Tjian R. (2000). Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422-1425 [DOI] [PubMed] [Google Scholar]
- Jallow Z., Jacobi U. G., Weeks D. L., Dawid I. B., Veenstra G. J. C. (2004). Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc. Natl. Acad. Sci. USA 101, 13525-13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Pugh B. F. (2009). Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Cai Y., Conaway R. C., Workman J. L., Conaway J. W., Kusch T. (2005). In and out: histone variant exchange in chromatin. Trends Biochem. Sci. 30, 680-687 [DOI] [PubMed] [Google Scholar]
- Jin V. X., Singer G. A., Agosto-Pérez F. J., Liyanarachchi S., Davuluri R. V. (2006). Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics 7, 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Hsu J.-Y., Kadonaga J. T. (2008). Caudal, a key developmental regulator, is a DPE-specific transcription factor. Genes Dev. 22, 2823-2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L., Horner M. A., Rothman J. H., Mango S. E. (2000). The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6, 705-712 [DOI] [PubMed] [Google Scholar]
- Kaplan N., Moore I. K., Fondufe-Mittendorf Y., Gossett A. J., Tillo D., Field Y., LeProust E. M., Hughes T. R., Lieb J. D., Widom J., et al. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 363-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H., Frith M. C., Katayama S., Sandelin A., Kai C., Kawai J., Carnici P., Hayashizaki Y. (2006). Dynamic usage of transcription start sites within core promoters. Genome Biol. 7, R118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876-880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Sohn D. H., Chung H., Seong R. H. (2008). Chromatin remodeling, development and disease. Mutat. Res. 647, 59-67 [DOI] [PubMed] [Google Scholar]
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., et al. (2006). CAGE: cap analysis of gene expression. Nat. Methods 3, 211-222 [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam U., Martianov I., Davidson I. (2008). Specialization of the general transcription machinery in male germ cells. Cell Cycle 7, 3493-3498 [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. (2004). Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 14, 175-182 [DOI] [PubMed] [Google Scholar]
- Lagrange T., Kapanidis A. N., Tang H., Reinberg D., Ebright R. H. (1998). New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12, 34-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Li X., Hechmer A., Eisen M., Biggin M. D., Venters B. J., Jiang C., Li J., Pugh B. F., Gilmour D. S. (2008). NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 28, 3290-3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. C., Roeder R. G. (1981). Transcription of adenovirus type 2 genes in a cell-free system: apparent heterogeneity of initiation at some promoters. Mol. Cell. Biol. 1, 635-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Gershenzon N., Gupta M., Ioshikhes I. P., Reinberg D., Lewis B. A. (2005). Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 25, 9674-9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. A., Kim T. K., Orkin S. H. (2000). A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc. Natl. Acad. Sci. USA 97, 7172-7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. A., Simm R. J., Lane W. S., Reinberg D. (2005). Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell 18, 471-481 [DOI] [PubMed] [Google Scholar]
- Li A. G., Piluso L. G., Cai X., Gadd B. J., Ladurner A. G., Liu X. (2007). An acetylation switch in p53 mediates holo-TFIID recruitment. Mol. Cell 28, 408-421 [DOI] [PubMed] [Google Scholar]
- Lim C. Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J. T. (2004). The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18, 1606-1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.-L., Coleman R. A., Grob P., King D. S., Florens L., Washburn M. P., Geles K. G., Yang J. L., Ramey V., Nogales E., et al. (2008). Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol. Cell 29, 81-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mõder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251-260 [DOI] [PubMed] [Google Scholar]
- Mavrich T. N., Ioshikhes I. P., Venters B. J., Jiang C., Tomsho L. P., Qi J., Schuster S. C., Albert I., Pugh B. F. (2008). A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18, 1073-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Pereira F., Jensen S. T., Ohler U., Hatzigergiou A. G. (2009). A transcription factor affinity-based code for mammalian transcription initiation. Genome Res. 19, 644-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. (2005). TAF4 inactivation in embryonic fibroblasts activates TGF beta signaling and autocrine growth. EMBO J. 24, 2753-2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C. A., Yang X. J., Kokubo T., Brownell J. E., Bannister A. J., Owen-Hughes T., Workman J., Wang L., Berger S. L., Kouzarides T., et al. (1996). The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87, 1261-1270 [DOI] [PubMed] [Google Scholar]
- Müller F., Lakatos L., Dantonel J.-C., Strähle U., Tora L. (2001). TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 11, 282-287 [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993). Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364, 532-535 [DOI] [PubMed] [Google Scholar]
- Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007). Structure and function of the histone chaperone CIA/ASF1 complexed with histone H3 and H4. Nature 446, 338-341 [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G. C., Niemann H., Rubin G. M. (2002). Computational analyses of core promoters in the Drosophila genome. Genome Biol. 3, RESEARCH0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., Cai H. N. (1998). Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12, 547-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perletti L., Kopf E., Carré L., Davidson I. (2001). Coordinate regulation of RARgamma2, TBP, and TAFII135 by targeted proteolysis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. BMC Mol. Biol. 2, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev S. P., Zhu X., Dixit B. L., Mastoon G. A., Kittler E. L. W., Green M. R. (2003). TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc. Natl. Acad. Sci. USA 100, 14887-14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H., Laverty T. R., et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A. D., Sauer F. (2000). Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289, 2357-2360 [DOI] [PubMed] [Google Scholar]
- Phelps C. B., Brand A. H. (1998). Ectopic gene expression in Drosophila using GAL4 systems. Methods 14, 367-379 [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Lenhard B., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A. (2006). Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 7, R78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach E. A., Yuan H.-Y., Majoros W. H., Tomancak P., Ohler U. (2009). Motif composition, conservation, and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10, R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina J. H., Hernandez N. (2007). On a roll for new TRF targets. Genes Dev. 21, 2855-2860 [DOI] [PubMed] [Google Scholar]
- Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D. A. (2007). Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 8, 424-436 [DOI] [PubMed] [Google Scholar]
- Sassoon D., Lyons G., Wright W. E., Lin V., Lassar A., Weintraub H., Buckingham M. (1989). Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341, 303-307 [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stephens R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097-6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder C., Tautz D., Seifert E., Jäckle H. (1988). Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 7, 2881-2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N., Guermah M., Hoffmann A., Roeder R. G., Heintz N. (1996). Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10, 2389-2400 [DOI] [PubMed] [Google Scholar]
- Sengupta T., Cohet N., Morlé F., Bieker J. J. (2009). Distinct modes of gene regulation by a cell-specific transcription activator. Proc. Natl. Acad. Sci. USA 106, 4213-4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Revach M., Moshonov S., Tzuman Y., Gazit K., Albeck S., Unger T., Dikstein R. (2005). Core promoter binding by histone-like TAF complexes. Mol. Cell. Biol. 25, 206-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R., Myers E. W. (2005). A motif-based framework for recognizing sequence families. Bioinformatics 21, i387-i393 [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. (1989). The “initiator” as a transcription control element. Cell 57, 103-113 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Demény M. A., Scheer E., Fienga G., Sassone-Corsi P., Tora L. (2005). The nuclear inport of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol. Cell. Biol. 25, 4092-4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. (1997). Construction and characterization of a full length-enriched and 5′-end-enriched cDNA library. Gene 200, 149-156 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Taira H., Tsunoda T., Mizushima-Sugano J., Sese J., Hata H., Ota T., Isogai T., Tanaka T., Morishita S., et al. (2001). Diverse transcription initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2, 388-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada R., Nakatani Y., Hoffmann A., Kokubo T., Hasegawa S., Roeder R. G., Horikoshi M. (1992). Identification of human TFIID components and direct interaction between a 250-kDa polypeptide and the TATA box-binding protein (TFIID tau). Proc. Natl. Acad. Sci. USA 89, 11809-11813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K., Bodenreider O., Landsman D., Spouge J. L., Mariño-Ramírez L. (2008). The biological function of some human transcription factor binding motifs varies with position relative to the transcription start site. Nucleic Acids Res. 36, 2777-2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. C., Chiang C. M. (2006). The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105-178 [DOI] [PubMed] [Google Scholar]
- Tokusumi Y., Ma Y., Song X., Jacobson R. H., Takada S. (2007). The new core promoter element XCPE1 (X core promoter element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol. Cell. Biol. 27, 1844-1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L. (2002). A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16, 673-675 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M. E., Tora L. (2007). TBP homologues in embryo transcription: what does what? EMBO Rep. 8, 1016-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Wu R. (1985). Structure of two unlinked Drosophila melanogaster glyceraldehyde-3-phosphate dehydrogenase genes. J. Biol. Chem. 260, 8220-8228 [PubMed] [Google Scholar]
- Vavouri T., Elgar G. (2005). Prediction of cis-regulatory elements using binding site matrices-the successes, the failures and the reasons for both. Curr. Opin. Genet. Dev. 15, 395-402 [DOI] [PubMed] [Google Scholar]
- Veenstra G. J., Weeks D. L., Wolffe A. P. (2000). Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290, 2312-2315 [DOI] [PubMed] [Google Scholar]
- Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58-69 [DOI] [PubMed] [Google Scholar]
- Wang X., Truckses D. M., Takada S., Matsumura T., Tanese N., Jacobson R. H. (2007). Conserved region 1 of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc. Natl. Acad. Sci. USA 104, 7839-7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm E., Pellay F.-X., Benecke A., Bell B. (2008). TAF6δ controls apoptosis and gene expression in the absence of p53. PLoS ONE 3, e2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H., Madabusi L., Nishioka H., Emanuel P., Sypes M., Arkhipova I., Gilmour D. S. (2001). Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 21, 1593-1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Yu Y., Fu Y., Foley J., Halees A., Weng Z. (2007). Analysis of overrepresented motifs in human core promoter reveals dual regulatory roles of YY1. Genome Res. 17, 798-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Kim M., DeJong J. (2006). Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr. Patterns 6, 409-419 [DOI] [PubMed] [Google Scholar]
- Yang Y., Cao J., Huang L., Fang H. Y., Sheng H. Z. (2006). Regulated expression of TATA-binding protein-related factor 3 (TRF3) during early embryogenesis. Cell Res. 16, 610-621 [DOI] [PubMed] [Google Scholar]
- Zhang D., Penttila T.-L., Morris P. L., Teichmann M., Roeder R. G. (2001). Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292, 1153-1155 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S. (2005). Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 33, 2838-2851 [DOI] [PMC free article] [PubMed] [Google Scholar]