Abstract
High-energy inositol pyrophosphates, such as IP7 (diphosphoinositol pentakisphosphate), can directly donate a β-phosphate to a prephosphorylated serine residue generating pyrophosphorylated proteins. Here, we show that the β subunit of AP-3, a clathrin-associated protein complex required for HIV-1 release, is a target of IP7-mediated pyrophosphorylation. We have identified Kif3A, a motor protein of the kinesin superfamily, as an AP3B1-binding partner and demonstrate that Kif3A, like the AP-3 complex, is involved in an intracellular process required for HIV-1 Gag release. Importantly, IP7-mediated pyrophosphorylation of AP3B1 modulates the interaction with Kif3A and, as a consequence, affects the release of HIV-1 virus-like particles. This study identifies a cellular process that is regulated by IP7-mediated pyrophosphorylation.
Keywords: phosphorylation, trafficking, kinesin
Inositol pyrophosphates such as IP7 (diphosphoinositol pentakisphosphate or PP-IP5) belong to a class of inositol polyphosphates containing highly energetic pyrophosphate moieties that undergo very rapid turnover (1, 2). Inositol pyrophosphates have been implicated in numerous important cellular events (3, 4), including apoptosis (5, 6) and insulin secretion (7, 8), and the use of radiolabeled IP7 (5β[32P]IP7) has allowed us to demonstrate that the high-energy pyrophosphate bond can participate in phospho-transfer reactions (9). One of the hallmarks of protein phosphorylation via IP7 is that the putative targets contain a serine-rich acidic region (9). IP7 substrates must initially be primed through ATP-dependent protein kinase phosphorylation (10), following which the phospho-serine becomes a substrate of IP7-mediated pyrophosphorylation (10). However, the physiological in vivo significance of this posttranslational modification remains unclear. To address the functional significance of protein pyrophosphorylation, we investigate the effect of this posttranslational modification in the functionality of the adaptor protein complex AP-3.
Results
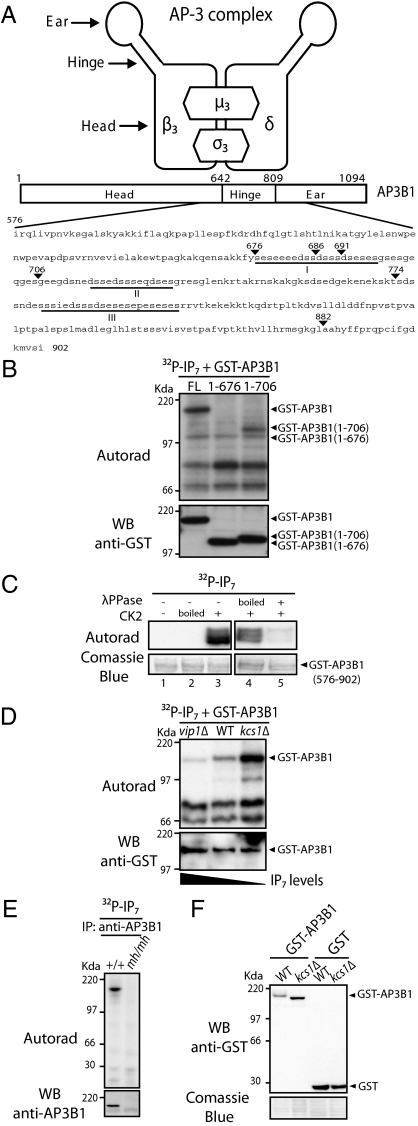
The β subunit of the adaptor protein complex AP-3 (AP3B1) (Fig. 1A) was initially identified as a potential target for IP7 phosphorylation by database searching for proteins containing serine-rich acidic regions. Adaptor protein complexes, comprising AP-1 to AP-4, mediate the sorting of transmembrane and cargo proteins to specific membrane compartments within the cell (11, 12). The AP-3 complex in particular is involved in sorting to lysosomes and related organelles (13, 14). It contains two large subunits (β and δ), a medium subunit (μ3), and a small subunit (σ3) (12). The AP3B1 subunit contains three main domains: the N-terminal head domain, the hinge, and the C-terminal ear domain (Fig. 1A and Fig. S1A). The putative target region of IP7 phosphorylation lies within the hinge domain, which contains three distinct serine-rich acidic stretches that we named regions I, II, and III (Fig. 1A and Fig. S1A). To test whether human AP3B1 is a substrate of IP7-mediated phosphorylation, we performed an IP7 phosphorylation assay on protein extracts from yeast expressing either the full-length human AP3B1 (GST-AP3B1), the N-terminus region lacking the acidic domains [GST-AP3B1 (1–676)] or the N-terminus containing region I [GST-AP3B1 (1–706)] (Fig. 1A). Both GST-AP3B1 and GST-AP3B1 (1–706) were phosphorylated by 5β[32P]IP7, whereas no phosphorylation of GST-AP3B1 (1–676) could be detected (Fig. 1B). These results demonstrate that AP3B1 is a bona fide target of IP7-mediated phosphorylation. Moreover, the lower phosphorylation levels observed in cells expressing GST-AP3B1 (1–706) indicate that the regions II and III are also likely to be targets of IP7 pyrophosphorylation (Fig. 1B). To test this hypothesis and to further map the IP7 pyrophosphorylation target regions of AP3B1, we cloned each of the serine-rich acidic domains and performed the same experiment described above (Fig. S1B). With the exception of clone GST-AP3B1 (576–686) and clone GST-AP3B1 (576–691) that contain only a small part of acidic region I, all of the clones were pyrophosphorylated by IP7. These results indicate that all three AP3B1 acidic regions are potentially pyrophosphorylated by IP7.
Fig. 1.
AP3B1 is pyrophosphorylated by IP7. (A) Schematic representation of the AP-3 complex and domain structure of AP3B1. The region amino acid 576 to 901 represents the bait used in the yeast two-hybrid screen. The acidic regions containing potential IP7-targeted serines are underlined (regions I–III). (B) In vitro phosphorylation of AP3B1. Protein extracts of kcs1Δ yeast expressing GST-AP3B1 or derivatives were incubated with 5β[32P]IP7 and resolved by NuPAGE; autoradiography was used to determine phosphorylation and immunoblotting with anti-GST antibody confirmed protein equal loading. (C) In vitro pyrophosphorylation of purified AP3B1. GST-AP3B1 (576–902) was expressed and purified from E. coli (BL21). Purified GST-AP3B1 (576–902) immobilized on glutathione beads was preincubated as follows: (i) Without, with inactive (boiled) or with active CK2 and ATP and subsequently treated with 5β[32P]IP7 as in B (lanes 1, 2, and 3, respectively); (ii) with active CK2 and ATP, treated with boiled or active λ-phosphatase, and subsequently phosphorylated with 5β[32P]IP7 as in B (lanes 4 and 5, respectively). (D) In vitro pyrophosphorylation of AP3B1 depends on the endogenous levels of IP7. Protein extracts of vip1Δ, wild-type (WT), and kcs1Δ yeast expressing GST-AP3B1 were incubated with 5β[32P]IP7 and processed as in B. (E) IP7 pyrophosphorylation of endogenous AP3B1. AP3B1 was immunoprecipitated from WT MEF (+/+) or mocha (mh/mh) cell lines, treated with 5β[32P]IP7, and processed as in B. (F) Intracellular pyrophosphorylation of AP3B1 results in a gel mobility shift. Quickly prepared cell extracts from WT and kcs1Δ yeast expressing GST-AP3B1 were boiled in sample buffer and resolved by NuPAGE. Gels are representative of at least three independent experiments.
To investigate whether the λ-phosphate of IP7 is transferred to a prephosphorylated serine (10), we used Escherichia coli purified GST-AP3B1 (576–902). When recombinant GST-AP3B1 (576–902) was incubated with casein kinase two (CK2) in the presence of γ[32P]ATP robust protein phosphorylation was observed, indicating that AP3B1 is a target for CK2 phosphorylation. To test whether CK2 phosphorylation is required for IP7-dependent pyrophosphorylation of AP3B1, recombinant GST-AP3B1 (576–902) was incubated with either native or inactivated (boiled) CK2 before performing the IP7 phosphorylation assay (Fig. 1C). IP7 pyrophosphorylation of AP3B1 was only observed following pretreatment with native CK2 (Fig. 1C, lane 3). We further confirmed these results by treating AP3B1 with λ-phosphatase after CK2 treatment and before the incubation with 5β[32P]IP7. AP3B1 pyrophosphorylation was inhibited by the λ-phosphatase pretreatment (Fig. 1C, lane 5). Taken together, these results demonstrate that CK2-dependent phosphorylation of the serine-rich acidic stretches of AP3B1 is necessary for priming IP7-mediated pyrophosphorylation.
To demonstrate that IP7-dependent protein pyrophosphorylation occurs in vivo, we tested whether GST-AP3B1 expressed in yeast strains containing different levels of endogenous IP7 show different degree of in vitro 5β[32P]IP7-driven pyrophosphorylation. We used an IP6-kinase yeast mutant that does not contain IP7 (kcs1Δ) (15, 16) (Fig. S2) and an IP7-kinase yeast mutant that accumulates large amounts of IP7 (vip1Δ) (17) (Fig. S2). We reasoned that if AP3B1 is endogenously pyrophosphorylated, it would be a weak target for in vitro 5β[32P]IP7-driven pyrophosphorylation when expressed in vip1Δ, due to the high level of endogenous IP7. Whereas AP3B1 expressed in kcs1Δ by not being naturally pyrophosphorylated due to the lack of IP7 would be a strong target for in vitro 5β[32P]IP7-driven pyrophosphorylation. Protein extracts from either WT, kcs1Δ, or vip1Δ yeast strains expressing GST-AP3B1 were tested for 5β[32P]IP7 pyrophosphorylation in vitro (Fig. 1D). As predicted, GST-AP3B1 pyrophosphorylation was lower in vip1Δ cells than in WT yeast (Fig. 1D). Importantly, exogenous pyrophosphorylation of GST-AP3B1 was substantially higher in kcs1Δ than in WT (Fig. 1D). To demonstrate that endogenous mammalian AP3B1 can be pyrophosphorylated by IP7, AP3B1 was immunoprecipitated from mouse embryonic fibroblast (MEF) and subjected to IP7 pyrophosphorylation assay (Fig. 1E). A robust phosphorylation signal was observed from WT MEF (+/+), whereas phosphorylation was undetected in MEF cells from mocha mice (mh/mh) that do not contain a functional AP-3 complex and do not possess detectable AP3B1 subunit (18, 19) (Fig. 1E).
As protein phosphorylation often results in a gel mobility shift that can be detected by Western blot analysis, we asked whether IP7-mediated protein pyrophosphorylation could also cause a mobility shift (Fig. 1F). When cell extracts of yeast expressing full-length GST-AP3B1 were rapidly processed, a dramatic gel mobility shift, which directly correlated with the endogenous IP7 levels, was detected (Fig. 1F). A similar gel mobility shift was observed for GST-NSR1 and GST-Nucleolin, two known targets of IP7-mediated pyrophosphorylation (9) (Fig. S3 A and B), whereas the gel migration of GST-Kif3A and HA-Ankyrin, proteins that are not IP7 targets, were not affected (Fig. S3C). The gel mobility shift is suggestive of high stoichiometry of phosphorylation in vivo, however the inability to visualize gel shift using 5β[32P]IP7 pyrophosphorylation (Fig. 1 B–E) is indicative of lower stoichiometry in vitro. Various scenarios can be envisaged to explain this stoichiometry difference between the in vitro and in vivo experiments: in vivo IP7-mediated pyrophosphorylation could stimulate additional CK2 phosphorylation or additional IP7 pyrophosphorylation. We can also hypothesize the existence of uncharacterized cellular components regulating protein pyrophosphorylation in vivo. Alternatively, protein pyrophosphorylation might represent a step for a more complex protein modification.
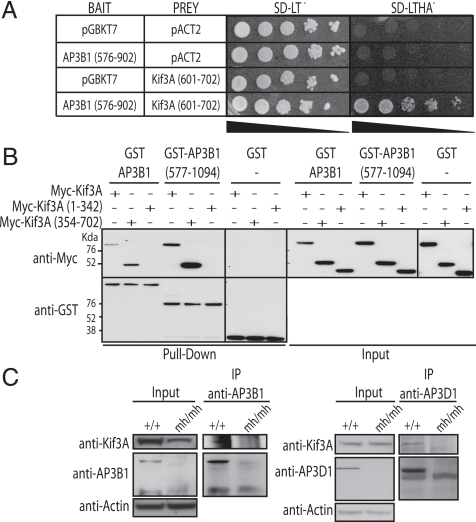
To determine the physiological consequence of IP7-mediated pyrophosphorylation of AP3B1, we investigated whether this modification could regulate protein–protein interaction. To identify interacting partners of AP3B1, we performed a yeast two-hybrid screen on a human fetal cDNA library using the target region of IP7 phosphorylation, AP3B1 (576–902), as bait (Fig. 1A and Fig. S1). The identified binding partners were screened for the ability of the interaction to be modulated by the cellular presence of IP7 (see below), and we choose to further analyze the interactor Kif3A, a motor protein that belongs to the kinesin superfamily (Fig. 2A and Fig. S4). Kif3A is a 70 Kda protein, and the region identified in the screen corresponded to the C-terminus that includes amino acids 601–702 (Fig. 2A and Fig. S4). GST pull down in mammalian cells of GST-AP3B1 or GST-AP3B1 (577–1094), with either Myc-Kif3A, Myc-Kif3A (1–342), or Myc-Kif3A (354–702), demonstrated that AP3B1 specifically interacts with the non-motor domain of Kif3A (Fig. 2B). The interaction between the AP3 complex and Kif3A was also confirmed by coimmunoprecipitation of endogenous AP3B1 and AP3D1 proteins in WT MEF, whereas no interaction was detected in the control mocha MEF cells (Fig. 2C).
Fig. 2.
AP3B1 interacts with Kif3A. (A) Yeast two-hybrid interaction between AP3B1 (amino acid 576 to 902; in pGBKT7 vector) and the C-terminal non-motor domain of Kif3A (amino acid 601 to 702; in pACT2 vector). Yeast AH109 expressing both vectors were serially diluted and spotted on yeast synthetic media lacking either leucine and tryptophane (SD-LT−, to assess growth of cotransformed yeast) or leucine, tryptophane, histidine, and adenine (SD-LTHA−, to assess interaction strength), and grown at 30 °C for 3 days. (B) Interaction between GST-AP3B1 and Myc-Kif3A in mammalian cells. HeLa cells were cotransfected with Myc-Kif3A, Myc-Kif3A (1–342), or Myc-Kif3A (354–702) together with GST-AP3B1, GST-AP3B1 (577–1094), or GST vector control. Proteins were extracted 24 h after transfection and subjected to pull-down with glutathione beads. Interactions were detected by immunoblotting with an anti-Myc antibody (Top) and an anti-GST antibody (Bottom). The inputs are shown on the Right. (C) Endogenous AP3 complex interacts with endogenous Kif3A in mammalian cells. AP3B1 and AP3D1 were immunoprecipitated (IP) from wild-type MEF and mocha (mh/mh) cell lines and immunoblotted with antibodies against Kif3A, AP3B1, and AP3Da. Actin was used as loading control.
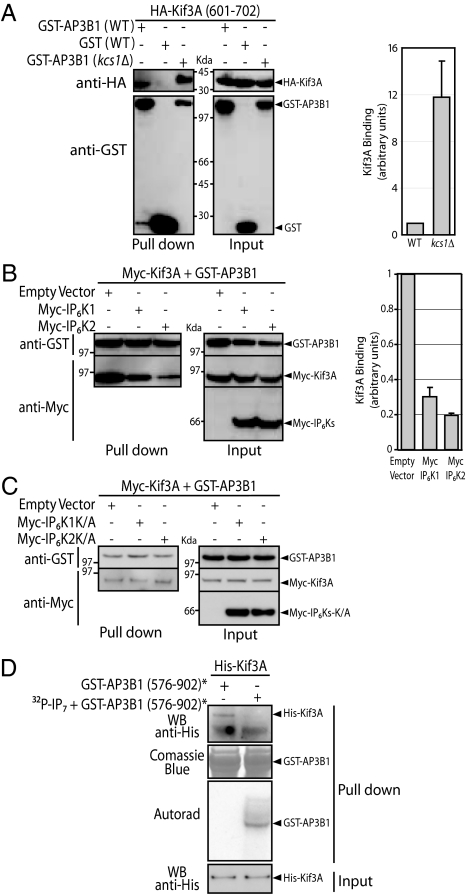
To evaluate whether IP7-dependent pyrophosphorylation of AP3B1 regulates AP3B1 and Kif3A interaction, we performed experiments in both yeast and HeLa cells. Homogenates of WT and kcs1Δ yeast expressing GST-AP3B1 were incubated for 2 h with homogenates of kcs1Δ yeast expressing HA-Kif3A (601–702) and subjected to GST pull down (Fig. 3A). In kcs1Δ yeast, AP3B1 is not pyrophosphorylated due to the lack of IP7, and its interaction with Kif3A (601–702) was on average 12 times stronger when compared to AP3B1 expressed in WT yeast (Fig. 3A). To test whether this was a general effect associated with the kcs1Δ yeast strain, we performed similar experiments with ankyrin, another AP3B1-binding protein identified in the YTH screen; however no difference in binding was observed (Fig. S5). We next investigated whether a similar regulation occurs in HeLa cells by increasing the intracellular levels of IP7 by ≈6-fold through expression of the IP6-kinases IP6K1 or IP6K2 (Fig. S6). When either IP6K1 or IP6K2 were coexpressed with GST-AP3B1 and Myc-Kif3A, we detected on average a 4-fold decrease in AP3B1-Kif3A interaction (Fig. 3B). This effect was dependent on the levels of IP7, as overexpression of a kinase-dead IP6K1K/A or IP6K2K/A had no effect on AP3B1-Kif3A interaction (Fig. 3C). We next investigated whether the reduction in the interaction between AP3B1 and Kif3A was the sole consequence of IP7 pyrophosphorylation by performing in vitro reconstitution experiments using recombinant bacterially expressed GST-AP3B1 (576–902) and His-Kif3A (Fig. 3D). GST-AP3B1 (576–902) phosphorylated by CK2 is able to interact with His-Kif3A, however the subsequent pyrophosphorylation by IP7 substantially weakens the binding ability (Fig. 3D). These results demonstrate that the interaction between AP3B1 and Kif3A is direct, and together with the previous results, that this interaction is negatively regulated by AP3B1 pyrophosphorylation.
Fig. 3.
AP3B1 pyrophosphorylation reduces interaction with Kif3A. (A) Lack of AP3B1 pyrophosphorylation increases its interaction with Kif3A in yeast cells. GST-pull downs (Left) and respective quantification (Right). Protein extracts from WT yeast expressing GST-AP3B1 or GST vector control, or from kcs1Δ yeast expressing GST-AP3B1 were incubated with protein extracts of kcs1Δ yeast expressing HA-Kif3A (601–702). Protein extracts were subjected to pull-down with glutathione beads. Interactions were detected by immunoblotting with anti-HA and anti-GST antibodies. Inputs are shown on the right. Quantification was done by taking the ratio between the bands intensities of the HA-Kif3A and the GST-AP3B1 and normalizing kcs1Δ expressed GST-AP3B1 against WT expressed protein. Data represent means ± standard deviation from three independent experiments. (B) AP3B1 pyrophosphorylation reduces its interaction with Kif3A in mammalian cells. GST-pull downs (Left) and respective quantification (Right). HeLa cells were triple transfected with Myc-Kif3A, GST-AP3B1, and either Myc-IP6K1, Myc-IP6K2, or Myc vector control. Protein extracts were subjected to pull-down with glutathione beads. Interaction between AP3B1 and Kif3A was detected by immunoblotting with anti-Myc and anti-GST antibodies. Inputs are shown on the right. Quantification was done by taking the ratio between the bands intensities of the Myc-Kif3A and the GST-AP3B1 and normalizing for IP6K1–2 overexpression compared with vector-transfected cells. Data represent means ± standard deviation of the mean from three independent experiments. (C) Interaction between AP3B1 and Kif3A is not affected by overexpression of kinase dead IP6K1 or IP6K2. HeLa cells were triple transfected with Myc-Kif3A, GST-AP3B1, and either Myc vector control, Myc-IP6K1K/A, or Myc-IP6K2K/A. Interaction between AP3B1 and Kif3A was detected as in B. (D) IP7-mediated pyrophosphorylation of AP3B1 is sufficient to reduce the interaction with Kif3A. Purified recombinant GST-AP3B1 (576–902) conjugated to glutathione beads was sequentially phosphorylated with CK2 [GST-AP3B1 (576–902)*], washed, and either subject to pyrophosphorylation by 5β[32P]IP7 or kept under the same conditions but without the addition of 5β[32P]IP7. Glutathione conjugated GST-AP3B1 (576–902)* was subsequently washed, incubated with purified recombinant His-Kif3A, and run on SDS-PAGE after being thoroughly washed. Interaction between AP3B1 and Kif3A was detected by immunoblotting with anti-His antibodies.
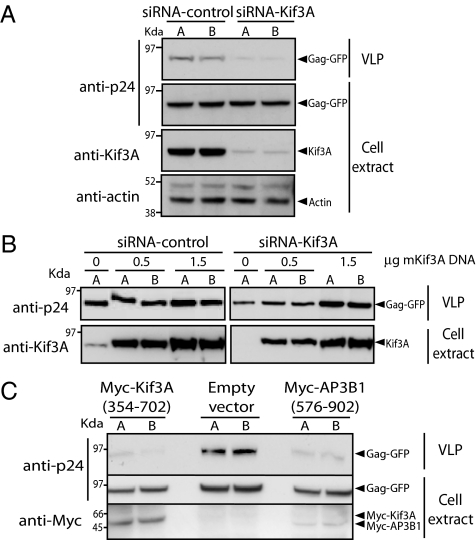
We next investigated how IP7-mediated pyrophosphorylation of AP3B1 affects AP3B1 and Kif3A function. Assembly of HIV-1 is directed by the Gag protein, the major structural protein of the virus (20). The δ subunit (AP3D1) of the AP-3 complex has been shown to bind to HIV-1 Gag and to be involved in its intracellular trafficking (21, 22). This functional role of AP3 complex led us to investigate whether IP7-mediated pyrophosphorylation could affect HIV release through modulation of AP3B1-Kif3A interaction. We first tested whether Kif3A, like the AP-3 complex (21, 22), is involved in HIV-1 Gag release. HIV-1 Gag alone contains all of the determinants required to produce noninfectious virus-like particles (VLP) in the absence of other viral proteins (23, 24). To establish whether Kif3A plays a role in HIV-1 VLP release, we used small interfering RNA (siRNA), achieving ≈75% of Kif3A depletion 96 h after transfection as evaluated by anti-Kif3A immunoblotting (Fig. 4A). Kif3A silenced cells were transfected with Gag-GFP, and a dramatic reduction of VLP release was observed when compared with control siRNA cells transfected with Gag-GFP in HeLa cells (Fig. 4A). Importantly, the intracellular levels of Gag-GFP remained unaltered, thereby excluding the possibility that Kif3A depletion affects Gag-GFP stability (Fig. 4A). We confirmed that reduction of VLP release was due to Kif3A depletion and not an off-target effect of the siRNA by complementing Kif3A-silenced HeLa cells with mouse Kif3A (Fig. 4B). We further confirmed that AP3B1 and Kif3A interaction is involved in HIV-1 Gag release, by expressing the respective AP3B1 and Kif3A binding domains. We reasoned that if these domains behaved as dominant negative forms, the VLP release should be affected. Cotransfection of Gag-GFP with vectors encoding motorless Kif3A [Myc-Kif3A (354–702)], which binds to AP3B1 (Fig. 2B) but is unable to hydrolyze ATP or to bind to microtubules, substantially reduces VLP release (Fig. 4C). Likewise, expression of Myc-AP3B1 (576–902) that contains the Kif3A-interacting domain, with Gag-GFP, consistently reduced VLP release (Fig. 4C). These results indicate that Kif3A and the AP-3 complex are involved in HIV-1 Gag VLP release from HeLa cells.
Fig. 4.
Kif3A is required for HIV-1 VLP release. (A) Kif3A depletion reduces VLP release. HeLa cells transiently expressing siRNA-Kif3A were transfected with Gag-GFP. Culture supernatant and cells lysate were collected 16–18 h later. Kif3A silencing was analyzed by immunoblotting with anti-Kif3A antibody. Gag intracellular expression and release was analyzed by immunoblotting with an anti-p24 antibody. The percentage of Kif3A silencing (see text) was analyzed by quantifying Kif3A protein from siRNA-Kif3A against siRNA-control cells both normalized against an internal endogenous control (actin). (B) Mouse Kif3A complements Kif3A in HeLa cells. HeLa cells transfected with siRNA-Kif3A were subsequently cotransfected with Gag-GFP, and increasing amounts of Myc-mKif3A (mouse Kif3A) samples were analyzed as in A. (C) AP3B1 and Kif3A dominant negative constructs reduced VLP release. HeLa cells were cotransfected with Gag-GFP and Myc-Kif3A (354–702), Myc-AP3B1 (576–902), or empty vector control. Expression of Myc-tagged proteins was analyzed by immunoblotting with an anti-Myc antibody. Gels are representative of analyses performed in duplicate (lanes A and B) in three independent experiments.
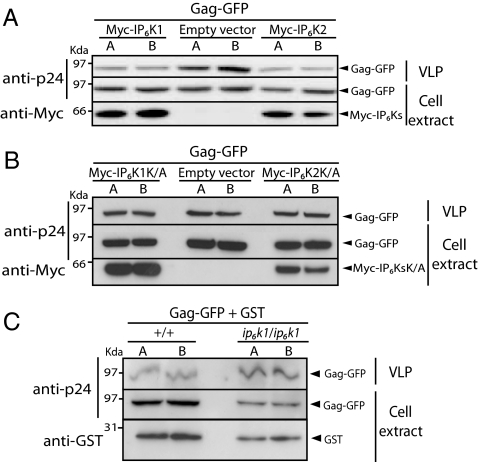
We next investigated whether changes of IP7 intracellular levels affect HIV-1 Gag release through modulation of AP3B1 and Kif3A interaction. The effect of increased IP7 intracellular content was tested in HeLa cells expressing either IP6K1 or IP6K2 (Fig. S6) together with Gag-GFP. High levels of IP7 consistently reduced VLP release by 30% (Fig. 5A). This reduction is noteworthy, considering that we are altering a dynamic enzymatic pathway and that the turnover of IP7 is already high under basal conditions (1). Importantly, no reduction of VLP release was observed upon overexpression of kinase dead versions of either IP6K1K/A or IP6K2K/A (Fig. 5B). Most mammalian cells express three IP6-kinases, and HeLa cells in particular express both IP6K1 and IP6K2. We were unable to obtain a reduction of the enzymatic activity that substantially reduced cellular IP7 through RNA interference (RNAi), and instead we used MEF cells derived from IP6K1 null mice that, although still possessing IP6K2, contain reduced levels of IP7 (7). IP6K1 null MEF were transfected with Gag-GFP and empty GST vector. The transfection efficiency of IP6K1 null MEF cells is substantially lower than WT MEF cells as seen by the lower ectopic expression of GST in the mutant cells, however the VLP release was substantially increased in these cells (Fig. 5C). Quantification of the ratio between intracellular and released VLP, revealed a ≈35% (±5%) increase of extracellular VLP from mutant cells when compared with WT MEF cells (Fig. 5C). Together, these findings demonstrate that changes in the cellular levels of IP7 alter VLP release.
Fig. 5.
IP7-mediated pyrophosphorylation of AP3B1 affects HIV-1 VLP release. Immunoblots analyzing how the modulation between AP3B1 and Kif3A interaction effects HIV-1 VLP release. (A) Increased levels of IP7 in mammalian cells decrease VLP release. HeLa cells were cotransfected with Gag-GFP and Myc-IP6K1, Myc-IP6K2, or empty vector control. Culture supernatants and cell lysates were collected 24 h posttransfection. Detection of Myc-tagged proteins was analyzed by immunoblotting with an anti-Myc antibody. Gag expression, VLP release, and data analysis were performed as in Fig. 4A. (B) Overexpression of kinase dead IP6K1 or IP6K2 has no effect on VLP release. HeLa cells were cotransfected with Gag-GFP and Myc-IP6K1K/A, Myc-IP6K2K/A, or empty vector control. The experiment was performed as described in A. (C) Decreased levels of IP7 in mammalian cells increase VLP release. MEF WT (+/+) and IP6K1 knock-out MEF (ip6k1/ip6k1) cell lines show increased HIV-1 VLP release upon Gag-GFP transfection. MEF cells IP6K1 knock-out were transfected with Gag-GFP and GST empty vector (as a control for transfection efficiency) and analyzed 48 h posttransfection. Gag expression, VLP release, and data analysis were performed as in Fig. 4A.
Discussion
In this study, we identified Kif3A as an AP3B1 interacting protein and established that this motor protein is involved in HIV-1 VLP release from HeLa cells. Evidence for the involvement of motor proteins in providing an efficient way for viruses to egress from host cell has been mounting. Kif3C, a motor protein that heterodimerizes with Kif3A, was recently identified in a whole genome RNAi screen as a late-acting protein required for viral assembly and release in HeLa cells (25). Moreover, Kif4 was shown to associate with multiple retrovirus Gag proteins and to function in Gag intracellular trafficking (26).
More importantly, we have identified AP3B1 as a target of IP7-mediated pyrophosphorylation and used its recently established function in HIV-1 release (21, 22) as a model to address the role of this mechanism of protein posttranslational modification. Through back phosphorylation experiments, we demonstrate that AP3B1 can be pyrophosphorylated by IP7 and that, in vivo, this modification affects the electrophoretic mobility of several target proteins. The observation that IP7-mediated pyrophosphorylation of proteins requires conventional ATP-dependent phosphorylation as a priming event raises the possibility that cellular functions that have been previously attributed to ATP-dependent phosphorylation may also involve pyrophosphorylation. For instance, in neurons, the AP-3 complex regulates endosomal synaptic vesicle biogenesis (27, 28), and AP3B1 phosphorylation was shown to inhibit vesicular coating and to impair synaptic vesicle formation (27). It is possible that this phenomenon depends on decreased IP7-mediated pyrophosphorylation. We have shown that IP7-mediated pyrophosphorylation of a target protein, AP3B1, modified its interaction with a specific binding partner, Kif3A. This modification has a direct effect on one of the biological functions of AP3B1, HIV-1 Gag release. However, modification of the interaction between two interacting partners is unlikely to be the only mechanism by which IP7-mediated pyrophosphorylation regulates the function of target proteins.
Methods
Unless otherwise stated, all reagents were from commercial sources. Please refer to SI Experimental Procedures for detailed reagent origins, cloning procedures, and routine methods.
Phosphorylation, Dephosphorylation, and Pyrophosphorylation Treatments.
CK2 phosphorylation and λ-phosphatase treatments were performed as described in ref. 10. 5β[32P]IP7 was produced and purified as described in ref. 9. IP7 pyrophosphorylation analysis was performed as described in ref. 9. Before the IP7 pyrophosphorylation assay of endogenous AP3B1, immunoprecipitated AP3B1 conjugated to protein G agarose beads was equilibrated in IP7 phosphorylation buffer (9). Protein extracts were run on 4–12% NuPAGE gels (Invitrogen), transferred onto PVDF membranes (Bio-Rad Laboratories), and exposed. Membranes were subsequently subject to Western blot with appropriate antibodies or directly stained with Coomassie blue.
Analysis of Protein Expression and Virus Release.
Culture supernatants of Gag-GFP transfected mammalian cells were collected 16–18 h posttransfection and clarified by centrifugation (4,000 rpm for 10 min). VLPs were pelleted through a 20% sucrose layer at 47,000 rpm for 90 min, resuspended in 1× LDS sample buffer (Invitrogen) and boiled. Cells were collected, and proteins were extracted in hypotonic buffer. VLP and protein extracts were analyzed by Western blot with the appropriate antibodies. Quantification of protein bands intensities was performed using the Quantity One program (version 4.6.5; Bio-Rad Laboratories) on scanned X-ray films (GS-800 calibrated densitomer scanner; Bio-Rad Laboratories). The percentage of HIV-Gag release represented the ratio between released VLP and the total Gag (intracellular Gag plus released VLP).
SI Experimental Procedures.
Supporting data include detailed experimental procedures and six SI figures (Figs. S1–S6).
Supplementary Material
Acknowledgments.
We thank A. C. Resnick, A. Riccio, and M. Raff for helpful comments. We thank S. H. Snyder and R. Bhandari for supplying IP6K1 knockout MEF cells, A. Peden for the mocha cells, and W. Sundquist for the Gag-GFP construct. This work was supported by the Medical Research Council (MRC) funding to the Cell Biology Unit and by a European Union (EU) Marie Curie through Institutional Research Grant (IRG; 014827). E.R.-M. was supported by Fondo de Investigación Sanitaria (FIS; CD08/00172) and Redes Temáticas de Investigación Cooperativa (RTICS; RETICS 2006 and RD06/0006/0021).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909176106/DCSupplemental.
References
- 1.Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- 2.Stephens L, et al. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- 3.Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: Metabolism and signaling. Cell Mol Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton A, Hu X, Saiardi A. Are inositol pyrophosphates signalling molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- 5.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata E, et al. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 7.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci USA. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illies C, et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 10.Bhandari R, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci USA. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchhausen T. Cell biology—Boa constrictor or rattlesnake? Nature. 1999;398:470–471. doi: 10.1038/18989. [DOI] [PubMed] [Google Scholar]
- 12.Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 13.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 14.Dell'Angelica EC, Ooi CE, Bonifacino JS. Beta3A-adaptin, a subunit of the adaptor-like complex AP-3. J Biol Chem. 1997;272:15078–15084. doi: 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- 15.Dubois E, et al. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- 16.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onnebo SM, Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signaling. Biochem J. 2009;423:109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- 18.Kantheti P, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- 19.Peden AA, Rudge RE, Lui WW, Robinson MS. Assembly and function of AP-3 complexes in cells expressing mutant subunits. J Cell Biol. 2002;156:327–336. doi: 10.1083/jcb.200107140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freed EO. HIV-1 Gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 21.Dong XH, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Garcia E, Nikolic DS, Piguet V. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic. 2008;9:200–214. doi: 10.1111/j.1600-0854.2007.00678.x. [DOI] [PubMed] [Google Scholar]
- 23.Delchambre M, et al. The Gag precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gheysen D, et al. Assembly and release of Hiv-1 precursor Pr55gag virus-like particles from recombinant baculovirus infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 25.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, et al. Cellular motor protein KIF-4 associates with retroviral Gag. J Virol. 1999;73:10508–10513. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blumstein J, et al. The neuronal form of adaptor protein-3 is required for synaptic vesicle formation from endosomes. J Neurosci. 2001;21:8034–8042. doi: 10.1523/JNEUROSCI.21-20-08034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faundez VV, Kelly RB. The AP-3 complex required for endosomal synaptic vesicle biogenesis is associated with a casein kinase I alpha-like isoform. Mol Biol Cell. 2000;11:2591–2604. doi: 10.1091/mbc.11.8.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.