Abstract
Cloning of large viral genomes into bacterial artificial chromosomes (BACs) facilitates analyses of viral functions and molecular mutagenesis. Previous derivations of viral BACs involved laborious recombinations within infected cells. We describe a single-step production of viral BACs by direct cloning of unit length genomes, derived from circular or head-to-tail concatemeric DNA replication intermediates. The BAC cloning is independent of intracellular recombinations and DNA packaging constraints. We introduced the 160-kb human herpes virus 6A (HHV-6A) genome into BACs by digesting the viral DNA replicative intermediates with the Sfil enzyme that cleaves the viral genome in a single site. The recombinant BACs contained also the puromycin selection gene, GFP, and LoxP sites flanking the BAC sequences. The HHV-6A-BAC vectors were retained stably in puromycin selected 293T cells. In the presence of irradiated helper virus, supplying most likely proteins enhancing gene expression they expressed early and late genes in SupT1 T cells. The method is especially attractive for viruses that replicate inefficiently and for viruses propagated in suspension cells. We have used the fact that the BAC cloning “freezes” the viral DNA replication intermediates to analyze their structure. The results revealed that HHV-6A-BACs contained a single direct repeat (DR) rather than a DR-DR sequence, predicted to arise by circularization of parental genomes with a DR at each terminus. HHV-6A DNA molecules prepared from the infected cells also contained DNA molecules with a single DR. Such forms were not previously described for HHV-6 DNA.
Keywords: BAC, Direct cloning, HHV-6A
Human herpes virus 6 (HHV-6) is a member of the betaherpesvirus subfamily (1). In culture, the virus replicates in T cells. HHV-6 isolates represent two distinct variants, HHV-6A and HHV-6B, which differ in their restriction enzyme patterns, antigenicity and disease association (1–3). Disease associations have been reviewed (1, 4): HHV-6B is the causative agent of roseola infantum, a prevalent children's disease with high fever and skin rash. The virus exhibits neurotropism and was found in children experiencing convulsions up to lethal encephalitis. HHV-6B reactivations in bone marrow and other transplantations were found associated with febrile illness, delayed transplant engraftment, and disease up to lethal encephalitis. HHV-6A has thus far no clear disease association, although evidence has suggested CNS tropism and aggravation of symptoms of multiple sclerosis (MS).
Viral DNA Structure and Replication.
HHV-6A and HHV-6B have linear genomes with a 143-kb unique (U) segment, flanked by 8- to 10-kb terminal direct repeats (DRs) (1, 5, 6). The DRs contain the pac-1 and pac-2 cleavage and packaging signals conserved in herpesviruses (4, 7, 8), in addition to telomeric (GGGTTA) reiterations (1, 4, 5, 8). We have previously suggested a model whereby HHV-6 DNA circularizes, prior rolling circle replication, producing large head-to-tail repeated genomes. The concatemeric genomes are cleaved at the pac-2-pac-1 signals when packaged into structural virions (8). Circularization of linear parental DNA by end to end covalent linkage was first reported for herpes simplex virus (HSV) (9). The resultant circle replicated by rolling circle yielded head-to-tail concatemers (10). This was coincidental with the finding that defective genomes and engineered amplicons replicated in the presence of helper virus by rolling circle replication (7, 11–14) producing head-to-tail concatemers. We also identified the pac-2-pac-1 signals operating to cleave the concatemers during packaging (7, 11, 12). Concatemeric junctions cleaved at the pac-2 and pac-1 signals were reported in many herpesviruses, including amplicon vectors from the lymphotropic viruses HHV-6 and HHV-7 (4, 15, 16). Alternative mechanisms were suggested whereby the endless, head-to-tail genomes were produced by intermolecular recombinations of linear or circular genomes (17, 18).
Cloning of Herpesvirus Genomes into Bacterial Artificial Chromosomes (BACs).
It has been of great interest to produce cloned BACs carrying intact viral genomes. They can serve as useful tools for DNA structural analyses, mutagenesis and derivation of packaging systems to produce helper-less amplicon vectors. Several human herpesviruses were cloned in BACs, including HSV-1 and HSV-2 (19, 20), varicella-zoster virus (VZV) (21), Epstein-Barr virus (EBV) (22), human cytomegalovirus (HCMV) (23), and Kaposi's sarcoma herpesvirus (KSHV) (24). Examplary BACs of animal herpesviruses included pseudorabies virus (PRV) (25), and the murine gamma herpesvirus 68 (MHV-68) (26). Cloning of all these BAC-viral vectors was accomplished by inserting the BAC sequences into a chosen viral DNA region by homologous recombinations in infected cells and selecting the BAC containing viruses. The recombinant herpesvirus BACs had to be purified from the wild-type (w.t.) virus, a task difficult to achieve for viruses replicating inefficiently in lymphocytes devoid of plaque selection capability. Here we describe a single-step procedure to generate herpesvirus BACs, by direct cloning of circular or unit length DNA produced from head-to-tail replication intermediates. We then used the BAC clones and infected cell DNA preparations to analyze the structure of replicative intermediates.
Results
Strategy for Direct Viral DNA Cloning into BACs.
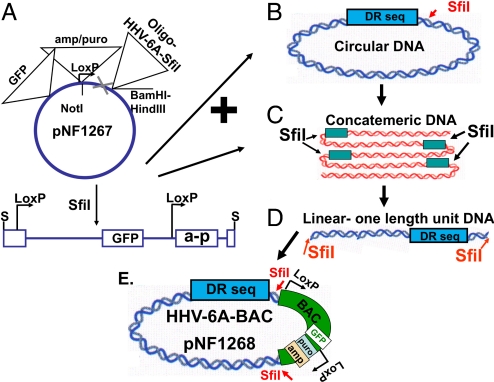
Based on the HHV-6 and 7 rolling circle DNA replication model (8), we sought an enzyme that cleaves once in the HHV-6A (U1102) genome. This would digest the circular and head-to-tail concatemers into unit length viral genomes, cloneable directly into the BAC vector. The SfiI enzyme cleaves the HHV-6A (U1102) genome once at position 9,520 with the sequence GGCC ACCT/C GGCC (PubMed accession NC_001664), whereas the pIndigoBAC-5 contains a single SfiI site GGCC GCCC/G GGCC. To allow cohesive ligation, the SfiI site in the pIndigoBAC-5 plasmid was replaced with a SfiI site matching the site in the HHV-6A genome (Fig. S1F). Replicating viral DNA obtained by the Hirt procedure (27) was cleaved by SfiI, generating unit length genomes, for direct cloning into the modified BAC vector.
Generation of the BAC pNF1267 for HHV-6A DNA Cloning.
The preparation of the BAC vector involved the following: (i) PCR introduction of point mutations in the pIndigoBAC-5 abolishing one NotI site and the SfiI site (positions 631 and 639, respectively (Fig. S1 A and B). The resultant pNF1254 had a single NotI site and no SfiI sites. (ii) An ampicillin and puromycin (amp/puro) cassette was prepared from the pMSCV plasmid by PCR of the amp and the puro segments, employing in both cases a primer with NotI at the 5′ terminus and a MluI site at the 3′ terminus. The products were digested with MluI, ligated to form the amp/puro cassette (Fig. S1C) and cloned into the NotI site of pNF1254 forming the pNF1255 (Fig. S1D). (iii) The GFP gene, prepared by PCR from pEGFP-C3 (Clontech) was inserted into the HpaI site of pNF1255, generating pNF1266 (Fig. S1E). (iv) A 75-bp oligonucleotide was prepared with the LoxP sequence, the SfiI site GGCC ACCT/C GGCC matching the site in the viral DNA. BamHI and HindIII sites were added at the termini for directional cloning. The oligomer was ligated with the pNF1266 plasmid, cleaved with BamHI and HindIII, yielding the BAC clone pNF1267 (Fig. S1F).
The Derivation of HHV-6A-BAC Vector (pNF1268).
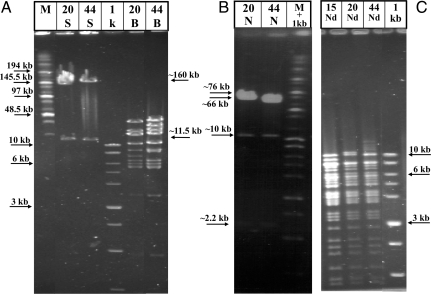
DNA prepared from HHV-6A infected cells by the Hirt procedure (27) was digested with SfiI and cloned into the pNF1267-modified BAC vector, cleaved with SfiI (Fig. 1), employing the E. coli DH10B bacteria. Colonies were screened by PCR, employing primers for BAC, HHV-6A DNA sequences, GFP, and the amp/puro cassette. Several clones with the expected PCR products were further analyzed by cleavage with the SfiI, BamHI, NotI, and NdeI enzymes. The results for clones 15, 20, and 44 (Fig. 2) can be summarized as follows. (i) Cleavage with SfiI yielded an approximate 160-kb intact HHV-6A DNA and 11.5-kb SfiI-cleaved BAC. (ii) Cleavage with the NotI enzyme generated fragments of sizes approximately 76 and 66 kb and approximately 10- and 2.2-kb fragments. (iii) BamHI and NdeI generated multiple bands, as expected from the PubMed sequences. These patterns and digests with five additional enzymes, cleaving at multiple sites were grossly similar in the three BAC clones obtained in independent BAC cloning experiments. (iv) Blot hybridization analyses with a pac-2-pac-1 probe were done as later discussed in the text.
Fig. 1.
The derivation of the HHV-6A-BAC (pNF1268) clone. (A) The pNF1267 clone, containing the amp/puro cassette, the GFP gene, and the oligonucleotide insert was cleaved with the SfiI enzyme. (B) Putative circular replication intermediate, of the HHV-6A genome. The DR sequences of the circularized genome are delineated. (C) Putative head-to-tail concatemers, produced by rolling circle replication. The SfiI site appears once per unit-length viral DNA. (D) SfiI cleavage of the replication intermediates yields unit length genomes with SfiI termini. (E) Ligation of the SfiI-cleaved HHV-6A and the SfiI-cleaved pNF1267, produced the HHV-6A-BAC (pNF1268).
Fig. 2.
Restriction enzyme digests of HHV-6A-BAC clones 15, 20, and 44 (pNF1268). The clones were digested with (A) SfiI (S) and BamHI (B), (B) NotI (N), and (C) NdeI (Nd). The digested BACs were analyzed in pulse field gels along with the high molecular weight (M) and 1-kb markers. Some of the marker sizes are noted. Arrows in between A and B point to sizes of fragments of the HHV-6A-BACs. The SfiI fragments of approximately 160 kb, corresponded to the intact HHV-6A genome and the approximate 11.5 kb corresponded to the re-engineered BAC plasmid. Arrows point to the approximate 2.2-kb NotI fragments, containing the amp/puro cassette and the fragment between positions 6,139 bp to 8,332 bp; the approximate 66-kb and 76-kb fragments represent the fragments as expected from the sequence.
Stable Retention of HHV-6A-BAC pNF1268 in 293T Cells.
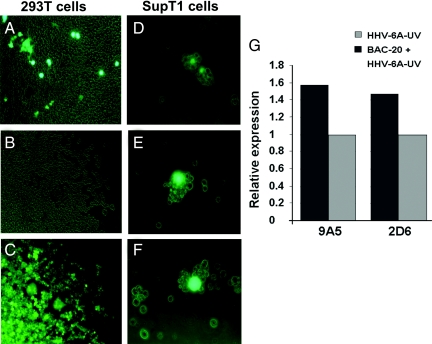
The 293T cells were transfected with pNF1268, containing the GFP marker and the puro-selection gene. Three days later, the cells were treated with puromycin and continuously passaged in the presence of the drug. At 72 h post-transfection (p.t.), an estimated 10% of the cultured cells expressed GFP. The fraction of puro-resistant GFP cells increased significantly upon passaging. At 30 days p.t., the majority of the cells expressed GFP (Fig. 3 B and C), and retained the viral genome as judged by PCR.
Fig. 3.
GFP and viral gene expression in 293T and in SupT1-transfected cells. (A) Green fluorescence of 293T transfected cells 72 h p.t. (B) Light photograph of the transfected cells 14 days after puromycin selection. (C) The same as B, but with green fluorescence filter. (D) SupT1 cells electroporated with the pNF1268 and photographed 24 h p.t. (E and F) Cells transfected with pNF1268 and then superinfected with the U.V-inactivated HHV-6A (U1102) virus. The cells were photographed 4 days (E) and 8 days (F) post-infection. (G) Viral gene expression in SupT1 T cells transfected with pNF1268 vector and superinfected with UV light inactivated HHV-6A (U1102). The p41 and gp82–105 proteins reactive with the 9A5 and 2D6 mAb, respectively, were quantified by densitometric analyses. The amounts recovered corrected for loading of the sample relative to the eIF2α “housekeeping” gene. Shown are the average ratios of proteins recovered from cells transfected with pNF1268 and superinfected with HHV-6A-UV compared to the proteins in cells infected solely with the UV-irradiated HHV-6A.
Complementation of HHV-6A-BAC Viral Gene Expression by UV Light-Inactivated Virus.
Viral gene expression was not detected in Western blots of SupT1 cells transfected with the HHV-6A-BAC, most likely due to the low transfection efficiency and the absence of virion protein(s) brought into the cells and serving to induce immediate early gene expression. For the closely related HCMV, it was reported that the addition of the pp71 tegument protein, encoded by the UL82 gene, to transfecting BAC-CMV plasmid resulted in a 30-fold increase in plaque formation (23). In the absence of a known protein serving as enhancer of HHV-6A immediate early gene expression, we tested whether UV-inactivated helper virus could supply the enhancing protein function(s). The level of UV irradiation was first calibrated to obtain loss of viral infectivity. Triplicate cultures of SupT1 cells were transfected with the pNF1268 vector. A second set of parallel cultures were exposed to the UV-irradiated virus. A third set of parallel cultures were transfected with the pNF1268 vector, and 48 h later, they received equal amounts of UV-inactivated virus. The cultures were monitored daily by light and fluorescence microscopy, and the cytopathic effect (CPE) and viral gene expression were followed. The results can be summarized as follows. (i) Approximately 1% of the transfected cells showed limited fluorescence by 24 h p.t. (Fig. 3D), reflecting inefficiency of transfection of large DNA molecules (such as the HHV-6A BACs) into lymphocytes. (ii) The relative fraction of fluorescing BAC-containing cells was further decreased upon cell division. (iii) The cultures receiving the UV virus showed very limited infection. (iv) In contrast, the third set of cultures receiving both transfected BAC and the superinfecting UV-inactivated virus showed some CPE in the GFP-containing cells, already at 4 days (Fig. 3E) and 8 days (Fig. 3F) p.i. (v) GFP expression prevailed longer in the BAC transfected-superinfected cells compared to the cells receiving BAC alone. (vi) Viral gene expression was monitored 8 days p.t. by Western blot analyses. Expression of the early p41 DNA polymerase accessory protein and the late envelope glycoprotein complex gp82-gp105 was followed employing the 9A5 and 2D6 mAbs of Bala Chandran and colleagues (28, 29). The proteins were quantified by densitometric analyses and resultant values were normalized relative to the eIF2α“housekeeping” gene expression. Fig. 3G quantifies the expression of the viral genes in cells receiving both the HHV-6A-BAC and UV-inactivated helper virus relative to the cells receiving only the UV-inactivated virus. The results revealed 1.6-fold higher recoveries of the p41 protein and 1.5-fold higher recoveries of gp82-gp105 in the cells receiving both the HHV-6A-BAC and UV-inactivated helper virus, compared to the culture receiving solely the irradiated virus. These results were significant, considering the fact that the initial BAC electroporation efficiency was low.
HHV-6A-BACs and the Viral DNA Replication Intermediates Contain a Single DR.
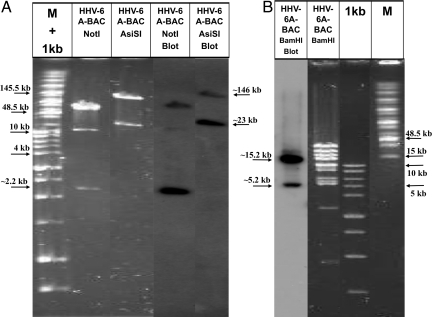
The direct BAC cloning allowed “freezing” of individual circular or concatemeric DNA replication intermediates. Restriction enzymes, blot hybridizations, and PCR analyses revealed two aspects of the replicating viral DNA: (i) The HHV-6A (U1102) currently propagated in the laboratory in SupT1 cells had short DRs of approximately 2.7 kb in size. By comparison, viruses from stocks produced earlier in cord blood mononuclear cells (CBMC) contained DRs of approximately 8 kb as reported (5, 6), and as summarized in PubMed for HHV-6A (accession NC_001664). (ii) Restriction enzyme, blot hybridizations with the pac-2-pac-1 probe, and PCR analysis revealed that the HHV-6A within the viral-BAC clones contained a single DR rather than duplicated DRs, expected if DNA circularization produced stable DR-DR junctions. Specifically, cleavage with restriction enzymes possessing a single site within the DR, including the NotI, AsiSI, and BamHI did not yield an approximate 2.7-kb band hybridizable to the pac-2-pac-1 probe. Furthermore, the NotI sites are located in DRL at 6,139 bp and in DRR at 157,372 bp. If genome circularization would yield a stable double DRs we would expect an approximate 2.7-kb fragment between these sites. No such fragment was detected. In contrast, two NotI fragments hybridized with the pac-2-pac-1 probe (Fig. 4), as expected with a single DR: (i) A NotI fragment of size approximately 2.2 kb, from the NotI site in the DR to the NotI site in the unique region. This fragment was verified by sequencing (ii) the approximate 76-kb fragment, going leftward from the NotI site in the DR into the unique region (Fig. 4A, lane 4). The NotI fragments of approximately 10 and 66 kb did not contain the pac-2-pac-1 sequences and although visible in the ethidium bromide (EtBr)-stained sample (Fig. 4A, lane 2), they did not significantly hybridize with the probe. AsiSI has sites at positions 7,333 bp in DRL at 158,566 bp in DRR, and a third site within the unique region at position 18,973 bp. No 2.7-kb fragment was detected in the blot. The approximately 24-kb and 134-kb fragments hybridized as expected (Figs. 4A, lane 5, and 5D). Finally, BamHI produced hybridizable fragments of approximately 5.2 and 15.2 kb, but not a fragment of 2.7 kb (Figs. 4B and 5D). Based on these results, the BAC clones and the majority of replicating DNA contained only a single DR. Later stages of viral DNA maturation were expected to duplicate the DRs and produce the pac-2-pac-1 signal before cleavage and packaging.
Fig. 4.
Restriction enzyme and Southern blot analyses of HHV-6A-BAC with Pac-2-Pac-1 probe. (A and B) Shown are mixtures of the large size marker (M) and the 1 kb marker (lane 1) next to HHV-6A-BAC DNA digested with the restriction enzymes NotI, AsiSI, and BamHI. The Southern blot of the gel was probed with the pac-2-pac-1 probe (A: lane 4 and 5 and B: lane 1). The hybridized bands of approximately 2.2- and 76-kb bands are visible. The band at approximately 10 kb in the blot is most likely non-specific [as apparent from its low intensity (lane 4)].
Fig. 5.
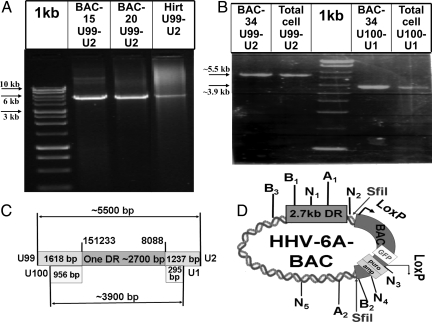
PCR analyses of BAC clones, Hirt extracts, total infected cell DNA. (A) PCR products of the BAC-HHV-6A clones 15, 20, and the Hirt preparation of HHV-6A-infected cells DNA employing the denoted primers. (B) PCR of BAC clone 34 and total HHV-6A-infected cells DNA, with the two sets of primers as denoted. (C) A scheme of the PCR results, revealing the presence of a single approximately 2,700-bp DR. The two sets of primers: first set, from U99 up to U2 (positions 149,615 bp sense to 9,325 bp anti-sense yields fragment of ≈5,500 bp). The second set, from U100 up to U1 (positions 150,277 bp sense to 8,383 bp anti-sense yields fragment of ≈3,900 bp). (D) A scheme of the HHV-6A-BAC clone with selected sites of SfiI, NotI (N), AsiSI (A), and the BamHI (B) sites hybridizable to the pac-2-pac-1 probes. The restriction sites are marked with numbers according to their location in the map.
The presence of a single DR in HHV-6A-BACs and replicating DNA was confirmed by PCR of the BAC clones 15, 20, and 34 with primers from the U99 to the U2 genes (149,615 to 9,325 bp). The three BACs yielded a fragment of approximately 5.5 kb, corresponding to a single DR and the flanking unique sequences (Fig. 5 A and B). If there were two DRs, flanked by the unique sequences the PCR would yield an approximate 8.2-kb fragment. The same size (≈5.5 kb) fragment was obtained in PCR of DNA prepared by Hirt extraction or from total DNA of HHV-6A (U1102) infected cells (Fig. 5 A and B), indicating that the single DR was a property of the replicating DNA intermediates which were cloned into the BAC. To verify the results, an additional PCR from positions 150,277–8,383 bp, was done and yielded a fragment of approximately 3.9 kb, compatible with a single DR (Fig. 5B). A fragment with double DR (≈6.6 kb) was not observed.
Finally, the PCR products from the HHV-6A-BAC and from the replicating intermediates prepared by Hirt extraction from the HHV-6A-infected cells were cloned into pGEM vectors and were sequenced starting from the flanking unique regions. The results showed that the HHV-6A-BAC clones as well as the replicating viral DNA contained grossly similar single DR sequences.
Discussion
Direct Cloning of the HHV-6A Genome into BACs.
We have described the BAC cloning of circular or concatemeric DNA replication intermediates prepared by Hirt extraction, employing SfiI which cleaves once per genome. The procedure was rapid and reproducible. Several BAC clones produced in three independent cloning experiments were found to contain intact HHV-6A DNAs with similar restriction enzyme patterns. Because the various BAC clones represented the population of replicating viral DNA molecules, minor variations were observed with enzymes possessing numerous sites in the genome. The direct BAC cloning is advantageous for viruses with low infectious virus yields and replicating in lymphocytes without plaque isolation capability. It does not require intra-cellular plasmid-viral DNA recombination. It can be potentially used in additional human and animal virus families that replicate by double stranded circular DNA intermediates. Moreover, there are herpesviruses possessing two or three restriction enzyme sites in the genome rather than a single site. The direct BAC cloning can be performed by “partially” cleaving the Hirt-extracted concatemers and screening the colonies by PCR primers from the partially cleaved genomes. Finally, the method can be potentially used with additional large cloning vectors, including yeast artificial chromosome (YAC), and phage 1 artificial chromosome (PAC).
Although the currently designed HHV-6A-BAC has the entire viral genome it is as yet not independently infectious and requires helper functions. Current studies are ongoing to modify the vector by using the Cre-lox signals to shorten the genome so it can be packaged.
Additionally, because the SfiI cleavage disrupted the U2 gene during the generation of the BAC vector, attempts will be made to insert an intact U2 gene into the vector.
The HHV-6A-BAC Cloning Supports the Model of Viral DNA Replication by the Rolling Circle Mechanism.
The production of large concatemeric genomes was found in several herpesviruses and was suggested to entail the rolling circle replication mechanism, intermolecular recombinations or both, as reviewed for HSV (17, 30). We have previously suggested that the naturally occurring defective genomes and the engineered HSV amplicons replicated by the rolling circle mechanism, as reviewed (11). The direct HHV-6A BAC cloning presented here was based on the anticipation that HHV-6 DNA replication occurred by circular or large head-to-tail concatemeric intermediates. The BAC clones produced in three independent cloning attempts appeared to be very similar providing support for the proposed model that viral replication was by the rolling circle mechanism.
The HHV-6A-BACs, as well as Parental Replicating Viral DNA Intermediates, Contain a Single DR.
The BAC cloning allowed the “freezing” of replicating DNA molecules, prior their cleavage and packaging into virions. Restriction enzyme-blot hybridizations and PCR analyses revealed that the BAC clones contained a single DR flanked by unique sequences. Moreover, analyses of infected cell DNA and Hirt DNA preparations also revealed the presence of molecules with a single DR.
The presence of single DR vs. DR-DR junctions is a cardinal question in genome replication and maturation. Although the DR region contains both pac-1 and pac-2 signals only in the DR-DR junction the signals are placed adequately for cleavage to occur (4). The findings of single DRs in replicating DNA raises the question of how these molecules are packaged. In the present paper the BAC clones from the Hirt preparations reflected the DNA molecules in the midst of replication. It is possible that the replicating concatemeric DNAs are further matured during packaging producing the juxtaposed DRs which are then cleaved at the pac-2-pac-1 signals. Indeed several groups have found HHV-6 DR-DR junctions in their analyses (6, 31–34). Furthermore, in additional experiments to be discussed elsewhere we also found packaged DNA containing double DRs in packaging compartments. The lack of PCR products with double DRs in the present study could have reflected early stages of viral DNA replication.
The question of single non reiterated terminal repeat sequences within replicating DNA pertains also to other herpesviruses. In HSV significant proportions of S-L junctions in replicating viral DNAs contained “bac” junctions with a single “a” sequence (35), as compared to lower proportions of “baac” and higher repeats of “a” sequences. We proposed that the “a” duplication occurred by the double-stranded break-gap repair model, similar to bacteriphage lambda DNA recombination (7). Duplication of the “a” sequence was found in experiments with amplicon vectors in which “bac” junctions gave rise to cleavable “baac” junctions (7, 11, 12). Reduction of reiterated elements in the concatemeric junctions was also found for guinea pig CMV genomes (36), and it was reported by McVoy and colleagues that these genomes almost exclusively contained single copies of the terminal repeat. They suggested that terminal repeat duplication occurred in conjunction with cleavage.
Based on the cumulative data we propose that recombinational events lead to reduction of duplicated DR terminal repeat sequences. Their regeneration during packaging might be a generalized pattern of herpesvirus DNA replication, perhaps by the double-stranded break-gap repair model (7). Current experiments in our laboratory are ongoing to determine: (i) at which stage one of the HHV-6A duplicated DRs is removed, most likely by intermolecular homologous recombinations and (ii) when are the DRs duplicated, producing the DR-DR junction, as necessary for cleavage and packaging to occur.
Methods
Cells and Viruses.
The SupT1 CD4+ human T cells (37) were obtained from the National Institutes of Health AIDS Research and Reference reagent program. The viruses were propagated in SupT1 cells as described before (38). The virus with the short DR is infectious both by cell-to-cell and cell-free propagation.
The Preparation of HHV-6A DNA by the Hirt Method.
The HHV-6A DNA replication intermediates were prepared by the Hirt method (27). Infected cells were rinsed with PBS and lysed in 0.6% SDS and 10 mM EDTA. Following 30-min lysis, 5 M NaCl was added to reach 1 M NaCl final concentration. After incubation at 4 °C for 24 h, cell debris was cleared by centrifugation, and DNA in the supernatant was purified by phenol-chloroform and ethanol precipitation.
Transfection of 293T and SupT1 Cells with HHV-6A-BAC.
Detailed methods can be found in the SI Text.
Pulse-Field Gel Electrophoresis and Southern Blot Analyses of HHV-6A-BAC with the pac-2-pac-1 Junction Probe.
Detailed methods including probe sequences can be found in the SI Text.
Direct Cloning of the Intact HHV-6A DNA into BAC and Screening of the Clones.
Detailed methods including primers and oligonucleotides sequences can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Ester Michael for technical assistance; Dr. N. Bala Chandran from the Rosalind Franklin University of Medicine and Science, North Chicago, Illinois, for his generous gift of the 9A5 and 2D6 antibodies. This work was supported by the Israel Science Foundation, the S. Daniel Abraham Institute for Molecular Virology, and the S. Daniel Abraham Chair for Molecular Virology and Gene Therapy, Tel Aviv University, Israel.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908504106/DCSupplemental.
References
- 1.Yamanishi K, Mori Y, Pellett PE. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Lippincott Raven publishers; 2007. pp. 2819–2845. [Google Scholar]
- 2.Ablashi D, et al. Human herpesvirus-6 strain groups: A nomenclature. Arch Virol. 1993;129:363–366. doi: 10.1007/BF01316913. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer EC, Wyatt LS, Yamanishi K, Rodriguez WJ, Frenkel N. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc Natl Acad Sci USA. 1991;88:5922–5926. doi: 10.1073/pnas.88.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenkel N, Borenstein R. Characterization of the lymphotropic Amplicons-6 and Tamplicon-7 vectors derived from HHV-6 and HHV-7. Curr Gene Ther. 2006;6:399–420. doi: 10.2174/156652306777592036. [DOI] [PubMed] [Google Scholar]
- 5.Gompels UA, Macaulay HA. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J Gen Virol. 1995;76:451–458. doi: 10.1099/0022-1317-76-2-451. [DOI] [PubMed] [Google Scholar]
- 6.Lindquester GJ, Pellett PE. Properties of the human herpesvirus 6 strain Z29 genome: G + C content, length, and presence of variable-length directly repeated terminal sequence elements. Virology. 1991;182:102–110. doi: 10.1016/0042-6822(91)90653-s. [DOI] [PubMed] [Google Scholar]
- 7.Deiss LP, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frenkel N, Roffman E. In: Fields Virology. 3rd Ed. Fields BN, Knipe DM, Howley PM, editors. Lippincott Raven publishers; 1995. pp. 2609–2622. [Google Scholar]
- 9.Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: Presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frenkel N. The history of the HSV amplicon: From naturally occurring defective genomes to engineered amplicon vectors. Curr Gene Ther. 2006;6:277–301. doi: 10.2174/156652306777591992. [DOI] [PubMed] [Google Scholar]
- 12.Deiss LP, Frenkel N. Herpes simplex virus amplicon: Cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57:933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locker H, Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin) J Virol. 1979;29:1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spaete RR, Frenkel N. The herpes simplex virus amplicon: A new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 15.Borenstein R, Singer O, Moseri A, Frenkel N. Use of amplicon-6 vectors derived from human herpesvirus 6 for efficient expression of membrane-associated and -secreted proteins in T cells. J Virol. 2004;78:4730–4743. doi: 10.1128/JVI.78.9.4730-4743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romi H, Singer O, Rapaport D, Frenkel N. Tamplicon-7, a novel T-lymphotropic vector derived from human herpesvirus 7. J Virol. 1999;73:7001–7007. doi: 10.1128/jvi.73.8.7001-7007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weller SK. In: Alpha Herpesviruses: Mol Cell Biol. Sandri-Goldin RM, editor. Caister Academic; 2006. pp. 85–104. [Google Scholar]
- 18.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 19.Stavropoulos TA, Strathdee CA. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meseda CA, Schmeisser F, Pedersen R, Woerner A, Weir JP. DNA immunization with a herpes simplex virus 2 bacterial artificial chromosome. Virology. 2004;318:420–428. doi: 10.1016/j.virol.2003.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaike K, et al. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine. 2004;22:4069–4074. doi: 10.1016/j.vaccine.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 22.Kanda T, Yajima M, Ahsan N, Tanaka M, Takada K. Production of high-titer Epstein-Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J Virol. 2004;78:7004–7015. doi: 10.1128/JVI.78.13.7004-7015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borst EM, Hahn G, Koszinowski UH, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: A new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou FC, et al. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: Application for genetic analysis. J Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith GA, Enquist LW. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97:4873–4878. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler H, Messerle M, Wagner M, Koszinowski UH. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 28.Balachandran N, Tirawatnapong S, Pfeiffer B, Ablashi DV, Salahuddin SZ. Electrophoretic analysis of human herpesvirus 6 polypeptides immunoprecipitated from infected cells with human sera. J Infect Dis. 1991;163:29–34. doi: 10.1093/infdis/163.1.29. [DOI] [PubMed] [Google Scholar]
- 29.Chang CK, Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J Virol. 1991;65:2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roizman B, Knipe DM, Whitley RJ. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Lippincott Raven publishers; 2007. pp. 2501–2601. [Google Scholar]
- 31.Severini A, Sevenhuysen C, Garbutt M, Tipples GA. Structure of replicating intermediates of human herpesvirus type 6. Virology. 2003;314:443–450. doi: 10.1016/s0042-6822(03)00451-3. [DOI] [PubMed] [Google Scholar]
- 32.Martin ME, et al. The genome of human herpesvirus 6: Maps of unit-length and concatemeric genomes for nine restriction endonucleases. J Gen Virol. 1991;72:157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- 33.Thomson BJ, Dewhurst S, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68:3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. Human herpesvirus 6B genome sequence: Coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locker H, Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: Occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979;32:429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nixon DE, McVoy MA. Terminally repeated sequences on a herpesvirus genome are deleted following circularization but are reconstituted by duplication during cleavage and packaging of concatemeric DNA. J Virol. 2002;76:2009–2013. doi: 10.1128/JVI.76.4.2009-2013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith SD, et al. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res. 1984;44:5657–5660. [PubMed] [Google Scholar]
- 38.Mlechkovich G, Frenkel N. Human Herpesviruses 6A and 6B (HHV-6A and HHV-6B) cause alterations in E2F1/Rb pathways, E2F1 localization, as well as cell cycle arrest in infected T cells. J Virol. 2007;81:13499–13508. doi: 10.1128/JVI.01496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.