Summary
DNA recombination and repair pathways require structure-specific endonucleases to process DNA structures that include forks, flaps, and Holliday junctions. Previously, we determined that the Drosophila MEI-9-ERCC1 endonuclease interacts with the novel MUS312 protein to produce meiotic crossovers, and that MUS312 has a MEI-9-independent role in interstrand crosslink (ICL) repair. The importance of MUS312 to pathways crucial for maintaining genomic stability in Drosophila prompted us to search for orthologs in other organisms. Based on sequence, expression pattern, conserved protein-protein interactions, and ICL repair function, we determined that the mammalian ortholog of MUS312 is BTBD12. Orthology between these proteins and S. cerevisiae Slx4 helped identify a conserved interaction with a second structure-specific endonuclease, SLX1. Genetic and biochemical evidence described here and in related papers suggest that MUS312 and BTBD12 direct Holliday junction resolution by at least two distinct endonucleases in different recombination and repair contexts.
Introduction
Specialized endonucleases execute important steps in DNA repair and recombination pathways by recognizing and cleaving specific DNA structures, including 5′ and 3′ flaps, bubbles, forks, and Holliday junctions (HJs). Some such endonucleases can cut different structures in different pathways. An example is vertebrate XPF–ERCC1 and its orthologs Rad1–Rad10 in S. cerevisiae and MEI-9–ERCC1 in D. melanogaster (Ciccia et al., 2008). These enzymes were first identified for their roles in nucleotide excision repair (NER), a pathway responsible for removal of UV-damaged bases. In NER, these enzymes nick the damaged strand at the 5′ end of a bubble (Bardwell et al., 1994; Park et al., 1995). They also function in repair of double-strand breaks (DSBs) (Bergstralh and Sekelsky, 2008). It is likely that they function in multiple DSB repair pathways; one role is in single-strand annealing (SSA), where Rad1–Rad10 and XPF–ERCC1 cleave 3′-ended flaps (Al-Minawi et al., 2008; Fishman-Lobell and Haber, 1992). MEI-9–ERCC1 is important for meiotic DSB repair, where it is thought to cut double-HJ (dHJ) intermediates to generate crossovers (Radford et al., 2007). Finally, these enzymes are critical for repair of DNA interstrand crosslinks (ICLs), though their exact functions in ICL repair are not well understood (Bergstralh and Sekelsky, 2008).
The ability of enzymes like XPF–ERCC1 and orthologs to recognize different substrates in different pathways is likely dependent on specific protein-protein interactions. Such interactions may recruit the nuclease to the site of damage. For example, an interaction with the damage-binding protein XPA recruits XPF–ERCC1 for NER (Park and Sancar, 1994). Alternatively, interaction of a nuclease with another protein might directly modulate substrate specificity. A possible example is seen with Drosophila MEI-9–ERCC1, where the meiotic function, but not the DNA repair functions, requires physical interaction with the MUS312 protein (Yildiz et al., 2002).
MUS312, like MEI-9–ERCC1, functions in ICL repair. Relative sensitivities of mutants to crosslinking agents indicate that MUS312 has a more crucial function and acts independently of MEI-9–ERCC1 (Yildiz et al., 2002), raising the possibility that MUS312 partners with a different nuclease in ICL repair.
Given the central importance of MUS312 to meiotic recombination and ICL repair, we hypothesized that homologs would have similar functions in other eukaryotes. We report here the identification of BTBD12 as the vertebrate ortholog of MUS312. Expression patterns and knockdown studies suggest that BTBD12 has functions similar to MUS312. Both are orthologous to yeast Slx4 and, like Slx4, complex with at least two different endonucleases.
Results and Discussion
MUS312 is orthologous to BTBD12 and Slx4
MUS312 lacks known functional domains and is poorly conserved even within arthropods (Figure S1). We therefore conducted sequence analyses to detect conserved structural characteristics. MUS312's predicted architecture includes a short coiled-coil domain and a C-terminus with seven α-helices (Figure 1A and B). The final two predicted helices are separated by a conserved glycine. This structure is similar to the SAP domain, a DNA-binding domain found in many repair proteins (Aravind and Koonin, 2000). PSI-BLAST searches using the C-terminal sequence identified proteins with similar C-termini, including one in each vertebrate genome (Figure 1B). The mammalian protein, BTBD12, has a predicted coiled-coil domain like MUS312, and a BTB (Broad-complex, Tramtrack, Bric-a-brac) domain. BTB domains mediate hetero- and homotypic protein interactions and are commonly located N-terminal to other conserved domains (Stogios et al., 2005).
Figure 1. MUS312 is orthologous to BTBD12 and Slx4.
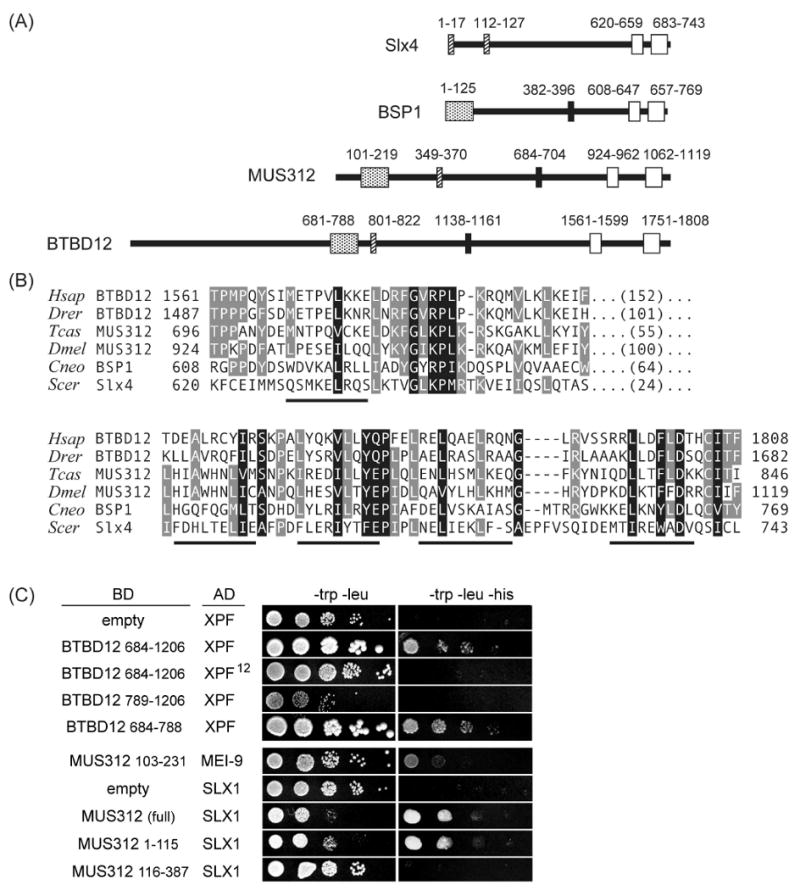
(A) Domain architecture of S. cerevisiae Slx4, C. neoformans BSP1, D. melanogaster MUS312, and H. sapiens BTBD12. Open boxes: conserved C-terminal domain; filled boxes: internal motif; hatched boxes: predicted coiled-coils; stippled boxes: BTB domain of BTBD12 and the regions on MUS312 and BSP1 that have sequence similarity (Fig. S2).
(B) Alignments of C-termini. Two divergent representatives from vertebrates, arthropods, and fungi are shown. Hsap = H. sapiens; Ggal = Gallus gallus; Tcas = Tribolium castaneum; Dm = D. melanogaster; Cgat = C. gatti; Scer = S. cerevisiae. Predicted alpha helices are underlined.
(C) Yeast two-hybrid interactions. Serial dilutions of cells expressing the indicated fusions to the Gal4 DNA binding domain (BD) or activating domain (AD) were plated on -leu -trp or -leu -trp -his dropout plates; growth on the former requires the presence of both the BD and the AD plasmid, and growth on the latter indicates a physical interaction. Top half: human proteins; bottom half: fly proteins.
MUS312 interacts physically with MEI-9, the catalytic subunit of the MEI-9–ERCC1 endonuclease (Yildiz et al., 2002). By yeast two-hybrid (Y2H) assay, we found that human BTBD12 interacts with XPF, the ortholog of MEI-9 (Figure 1C). The interacting region mapped to the BTB domain. Although MUS312 does not have a BTB domain, we detected weak sequence similarity in residues 101-219 (Figure S2). This region interacts with MEI-9 (Figure 1C). Conservation between these interacting regions indicates the biological relevance of the Y2H interactions. Additional support comes from our finding that the same single amino acid substitution in MEI-9 or XPF abolishes the interaction. The mei-912 mutation, G349E, which interaction with MUS312 (Yildiz et al., 2002). We made the equivalent substitution in XPF (G325E), and it abolishes interaction with the BTB domain of BTBD12 (Figure 1C).
Elements of the MUS312/BTBD12 architecture are also recognizable in BSP1, a protein from bipolar mating species of the Cryptococcus genus of fungal pathogens. BSP1 has sequence similarity with the C-termini of MUS312 and BTBD12 and with the N-terminal MEI-9/XPF-interaction region (Figures 1A and S2). These three proteins also share another short motif (Figures 1A and S2).
Others recently indentified MUS312 and BTBD12 when searching for orthologs of S. cerevisiae Slx4 (Fekairi et al., 2009). The previous finding that Slx4 interacts with Rad1–Rad10 supports this identification (Fricke and Brill, 2003). Slx4 also interacts with a second structure-specific endonuclease, Slx1 (Fricke and Brill, 2003). In a Y2H assay, Drosophila MUS312 and SLX1 also interact (Figure 1C); a similar interaction has been shown for human BTBD12 and SLX1 (Fekairi et al., 2009; Svendsen et al., 2009). We conclude that Drosophila MUS312, vertebrate BTBD12, and yeast Slx4 are orthologous proteins whose functions involve physical interactions with at least two different structure-specific DNA repair endonucleases.
BTBD12 expression suggests conservation of the meiotic recombination function
MUS312 is important for meiotic recombination in Drosophila; in mus312 mutants, meiotic crossovers (COs) are decreased by about 95% (Green, 1981; Yildiz et al., 2002). It has been unclear whether the MUS312 CO pathway is unique to Drosophila, but mined expression data suggest a meiotic function for Btbd12 in mice. Murine Btbd12 mRNA is most highly expressed in testes and oocytes (Figure 2A). Testis expression increases as the animal approaches sexual maturity (Schultz et al., 2003). The increase begins when spermatocytes first enter pachytene, the stage at which meiotic recombination takes place, and expression is much higher in pachytene spermatocytes than mitotically dividing pre-meiotic spermatagonia (Figure 2B and 2C) (Namekawa et al., 2006). This expression pattern suggests that mammalian BTBD12 might also have a role in generating meiotic COs.
Figure 2. Btbd12 expression is increased in cells undergoing meiotic recombination.
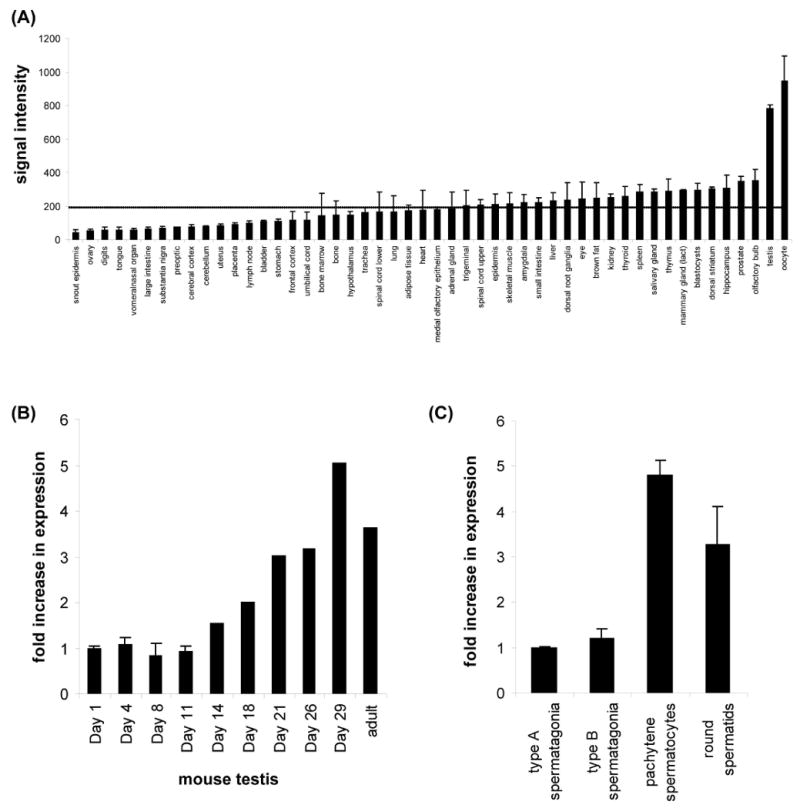
(A) Relative expression levels across multiple tissues. Btbd12 mRNA is most highly expressed in mouse testis and oocytes (biogps.gnf.org).
(B) Post-natal testis expression. Btbd12 expression increases during development, peaking at sexual maturity (Schultz et al., 2003).
(C) Stage-specific testis expression. Btbd12 expression peaks during pachytene, which is when meiotic recombination occurs (Namekawa et al., 2006).
Orthology to Cryptococcus BSP1 raises intriguing evolutionary implications for the importance of these proteins to meiosis. BSP1 is one of 26 genes at the mating type (MAT) loci of C. neoformans and C. gattii. The MAT loci of these organisms are unlike those of other fungi, but share features with animal sex chromosomes (Fraser et al., 2004). BSP1 is part of the “intermediate II” class of genes, which were incorporated into the mating locus at a period in evolution thought to be important for the genesis of bipolar mating. Three of the six genes incorporated into the MAT loci during this period have been functionally characterized, and two of these (SPO14 and RUM1) are orthologous to meiotic genes from other fungi (Honigberg et al., 1992; Quadbeck-Seeger et al., 2000). We speculate that Cryptococcus BSP1 has meiotic functions similar to those of MUS312.
MUS312 interacts with MEI-9–ERCC1 to generate meiotic COs (Yildiz et al., 2002). However, COs are decreased by >95% in mus312 mutants, but by only 85-90% in mei-9 mutants (Yildiz et al., 2004; Yildiz et al., 2002), suggesting that some COs generated by MUS312 are independent of MEI-9. It is possible that a small percentage of COs require MUS312 and another endonuclease, such as SLX1. It will be interesting to see whether, as suggested by its expression pattern, BTBD12 has a meiotic recombination function and the extent to which this requires XPF–ERCC1, SLX1, or other nucleases.
MUS312 and BTBD12 have important functions in ICL repair
mus312 mutants were first recovered in screens for hypersensitivity to DNA damaging agents (Boyd et al., 1981). mus312 mutants are mildly hypersensitive to the alkylating agent methyl methanesulfonate (MMS), but are highly hypersensitive to the nitrogen mustard mechloramine (HN2), a bifunctional agent that can cause DNA interstrand crosslinks (Boyd et al., 1981). A role for BTBD12 in responding to DNA damage is suggested by the identification of BTBD12 in proteomic screens for substrates of the DNA damage checkpoint kinases ATM and ATR (Matsuoka et al., 2007; Mu et al., 2007). S. cerevisiae Slx4 is a target of the orthologous kinases, Mec1 and Tel1 (Flott and Rouse, 2005). To determine whether BTBD12 has a role in ICL repair, we used siRNA to knock down BTBD12, XPF, and SLX1 in HeLa cells. Reduction of the tetrazolium reagent XTT was measured as an indicator of cell respiration before and after exposure to DNA damaging agents (see Experimental Procedures). Transfection with control or XPF siRNA had no effect on XTT reduction, but transfection with siRNAs for BTBD12 or SLX1 caused a 25% decrease, suggesting that BTBD12–SLX1 plays a role in cell proliferation or survival in the absence of exogenous damage (Figure 3A).
Figure 3. BTBD12 acts in ICL-repair.
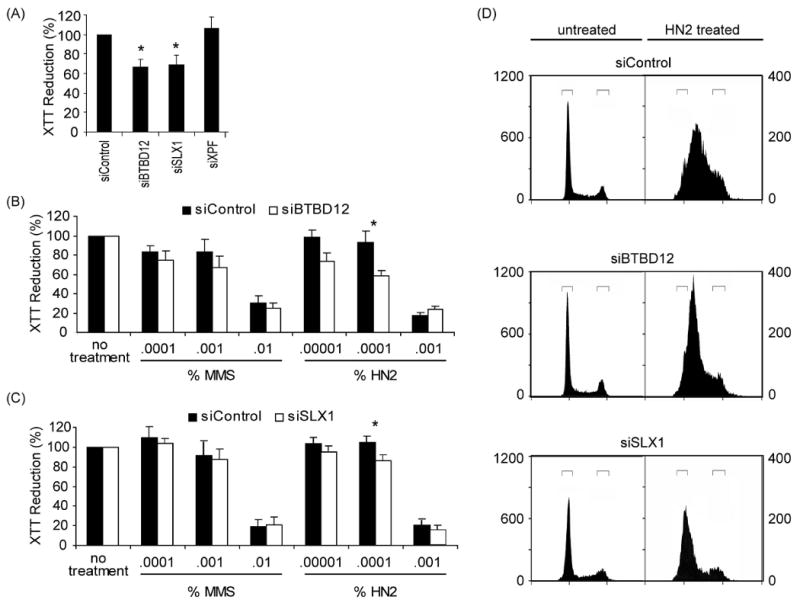
(A) Depletion of BTBD12 or SLX1 affects cell proliferation. XTT reduction was measured four days after transfection of HeLa cells with siBTBD12, siSLX1, siXPF, or siControl. siBTBD12 and siSLX1 caused identical decreases in XTT reduction, indicating slowed proliferation or cell death; siXPF and siControl had no effect. Bars indicate mean of at least five experiments, and error bars denote SEM. Asterisks indicate P<0.05 by paired t test.
(B), (C) Transfection with siRNA to deplete BTBD12 or SLX1 causes hypersensitivity to HN2. Three days after transfection with the indicated siRNA, cells were exposed to the indicated concentration of HN2 or MMS for 24 hours. Relative cell respiration was measured with the XTT assay, normalizing to decreases caused by siRNA treatment alone (panel A). Each bar represents the mean from five separate experiments, with error bars indicating S.E.M. Sensitivity was not detected at the lowest dose, and the highest dose caused extensive cell death in both control and experimental; the difference was significant at the intermediate dose, however. Asterisks indicate P <0.05 by paired t test.
(D) HN2 causes early S phase accumulation after knockdown of BTBD12 or SLX1. Cell number is plotted as a function of DNA content. Cells were transfected with siRNA, then three days later were mock treated (left) or treated with HN2 (right). Bars indicate cells with 2C (G1) and 4C (G2) DNA content.
Knockdown of BTBD12 or SLX1 caused a significant increase in sensitivity to HN2, but not to MMS (Figure 3B and 3C). Since ICL repair occurs primarily during replication (Niedernhofer et al., 2004), ICLs should slow progression through S phase, and a repair defect should arrest cells in S phase. Cell cycle profiles of cells treated with HN2 are consistent with this hypothesis: Cells transfected with control siRNA then treated with HN2 show a broader distribution through S phase than untreated cells (Figure 3D). In contrast, cells knocked down for BTBD12 or SLX1 accumulate in early S phase after treatment with HN2 (Figure 3C), suggesting that these cells arrest in S phase due to failure to repair ICLs.
Evidence that BTBD12 is required for ICL repair has also been obtained by others using different siRNAs, different sensitivity assays, and different crosslinking agents (Fekairi et al., 2009; Svendsen et al., 2009). Thus, the previously identified role of MUS312 in ICL repair appears to be broadly conserved in animals.
Synthetic lethality between mus312 and mus309 reveals an important function in cell proliferation
SLX4 was first identified in a screen for mutations that are lethal in the absence of Sgs1, the S. cerevisiae ortholog of BLM (Mullen et al., 2001). The Drosophila ortholog, DmBLM, is encoded by mus309. We found that mus312 mus309 double mutants are inviable, dying after pupariation (Figure 4A). Third-instar larvae have melanotic tumors, suggestive of elevated cell death, and lack the highly proliferative larval imaginal discs. Furthermore, larval brains of double mutants were greatly reduced in size (Figure 3B). These phenotypes indicate a severe proliferation defect and reveal an important function for MUS312 even in the absence of induced DNA damage.
Figure 4. Synthetic lethality between mus312 and mus309.
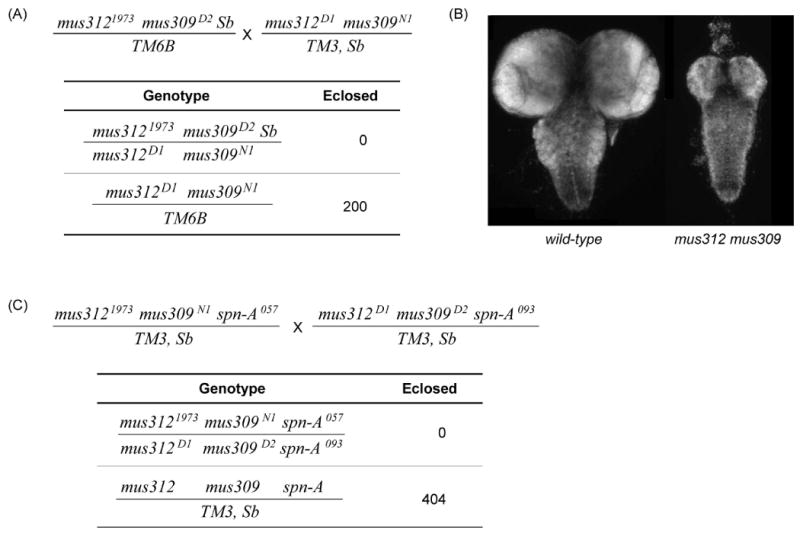
(A) Cross scheme to detect synthetic lethality. This cross generates progeny doubly mutant for mus312 and mus309. No double mutant adults (top genotype) eclosed, though there were expected to be as frequent as the lower genotype (the two genotypes not listed are inviable).
(B) mus312 mus309 double mutants have small brains. Brains were dissected from wandering L3 larvae and stained with DAPI. Brains from mus312 or mus309 single mutants are indistinguishable from wild-type brains (data not shown), but brains from double mutants are severely under-developed.
(C) mus312 mus309 synthetic lethality is not suppressed by mutation of spn-A. This cross generates progeny triply mutant for mus312, mus309, and spn-A. No triple mutant adults (top genotype) eclosed, though there were expected to be half as frequent as the lower genotype (either triple mutant chromosome over the TM3 balancer; TM3 / TM3 is inviable).
mus81; mus309 double mutants, which lack DmBLM and the structure-specific endonuclease MUS81–MMS4, also have melanotic tumors; imaginal discs are present, but apoptosis is highly elevated (Trowbridge et al., 2007). This synthetic lethality is suppressed by mutations in spn-A, which encodes the ortholog of the strand invasion protein Rad51 (Trowbridge et al., 2007). In contrast, spn-A mutations do not suppress the lethality of mus312 mus309 double mutants (Figure 4C), suggesting that mus312 mus309 lethality is not due to defects in processing a repair intermediate that requires strand invasion. This result is similar to the case in S. cerevisiae, where rad51 mutations suppress mus81 sgs1 lethality but not slx4 sgs1 lethality (Bastin-Shanower et al., 2003). It is likely that the function of MUS312 that is essential in the absence of DmBLM requires SLX1, as in S. cerevisiae (Mullen et al., 2001). Consistent with this suggestion, mei-9; mus309 double mutants are viable as adults (data not shown); unfortunately, there are currently no slx1 mutations available to directly test this hypothesis.
Roles of MUS312/BTBD12 complexes in DNA repair and recombination
MUS312 and its orthologs clearly have multiple functions in DNA repair and recombination. Since these proteins interact with at least two different structure-specific endonucleases, it is important to consider what structures are cleaved in different pathways. In vitro, S. cerevisiae and S. pombe Slx4–Slx1 are most active on 5′ flaps and structures that mimic replication forks (Coulon et al., 2004; Fricke and Brill, 2003). These activities could explain the known function for Slx4–Slx1 in maintaining rDNA stability in the absence of Sgs1 (Coulon et al., 2004; Fricke and Brill, 2003). It is thought that replication forks that stall at natural barriers in rDNA are normally processed by Sgs1, but are cut by Slx4–Slx1 if Sgs1 is not available. Slx4–Rad1–Rad10 is also thought to cut 5′ flaps in its role in SSA (Li et al., 2008).
The function of MUS312–MEI-9–ERCC1 in generating meiotic COs is not easily explained by cleavage of flaps or forks. Rather, the genetic and molecular defects in meiotic recombination seen in mei-9 mutants suggest that MEI-9–ERCC1 cuts a dHJ intermediate to generate COs (Radford et al., 2007; Yildiz et al., 2004). Although biochemical activities of MUS312–MEI-9–ERCC1 have not been reported, human BTBD12–SLX1 is reported to have HJ resolvase activity (Fekairi et al., 2009; Svendsen et al., 2009). It seems reasonable to propose that MUS312 and BTBD12 share an ability to confer HJ resolution activity on associated nucleases. S. cerevisiae Slx4-Slx1 can cut HJs in vitro, though Fricke and Brill (2003) concluded that it is not a true resolvase. This could be due to reaction conditions, a missing co-factor, or some divergence in activities of the yeast and metazoan enzymes.
HJ resolution could also account for the roles of MUS312/BTBD12–SLX1 in ICL repair. ICL repair is known to involve formation of a DSB, and HJs are predicted to be generated during processing of this DSB for replication restart (Bergstralh and Sekelsky, 2008; Li and Heyer, 2008). HJs might also be generated prior to DSB formation, by regression of the blocked fork to form a “chicken foot” structure. It is possible that MUS312/BTBD12–SLX1 cuts this structure to initiate ICL repair. However, ICL repair is poorly understood, and some models do not involve fork regression (Raschle et al., 2008).
The synthetic lethal interaction between mus312 and mus309 may also reflect an inability to process HJs. BLM helicases have HJ branch migration activity that is believed to function in several mechanisms of replication fork repair, including reversing regressed forks and dissolving dHJ intermediates (Wu, 2007). Loss of BLM might therefore leave HJs that must be processed by other mechanisms. We speculate that in some cases, perhaps depending on the type or location of blockage, HJs that cannot be acted upon by DmBLM must be cut by MUS312–SLX1 to allow replication to proceed. Our data indicate that the intermediate that requires either MUS312 or DmBLM is not generated during Rad51-mediated recombination, but HJs may be generated in other ways (e.g., replication fork regression).
Several eukaryotic proteins cut HJs in vitro have now been identified, including Mus81–Eme1 (Boddy et al., 2001), Yen1/GEN1 (Ip et al., 2008), and BTBD12–SLX4 (Fekairi et al., 2009; Svendsen et al., 2009). Although it is unknown whether these all cut HJs in vivo, their identification raises the question of whether cells need more than one resolvase enzyme. HJs are though to be formed during meiotic recombination, regression of blocked replication forks, some types of replication fork restart, and perhaps during DSB repair. These different processes may well employ different HJ cutting enzymes. It is also evident that the same function may use different enzymes in different species. For example, most meiotic COs in S. pombe require Mus81–Eme1, but the orthologous enzyme generates only a subset of COs in S. cerevisiae, and none in Drosophila (Whitby, 2005; Trowbridge et al., 2007).
In summary, MUS312, BTBD12, and Slx4 are orthologous proteins that each interact with at least two different structure-specific endonucleases. Based on previous studies, the work presented here, and recent work from other groups, we propose that MUS312 and BTBD12 are key non-catalytic subunits of Holliday junction resolvases.
Experimental Procedures
Sequence analysis
Coiled-coil domain prediction used COILS2 (www.ch.embnet.org/software/COILS_form.html). Structural analysis was performed with PHYRE (Bennett-Lovsey et al., 2008). The H. sapiens BTB domain position was determined using Pfam (pfam.sanger.ac.uk/search/).
Yeast two-hybrid analysis
Yeast two-hybrid analysis was performed as described previously (Radford et al., 2005).
Expression data
Cross-tissue mRNA expression of mouse BTBD12 was mined from BioGPS (biogps.gnf.org). Mouse testis (GDS410) and mouse sperm (GDS2390) expression patterns were mined from the NCBI GEO database (Barrett et al., 2009).
Cell lines and reagents
HeLa cells were cultured in DMEM (Gibco) with 8% FBS, 10 units/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained in 5% CO2 at 37°C. DharmaFECT 1 transfection reagent and Control, BTBD12, and XPF SMARTpool siRNA oligonucleotides were purchased from Dharmacon.
XTT assay
HeLa cells were plated into 96-well plates at ∼20% confluency. siRNA was transfected according to manufacturer's instructions. Transfection medium was replaced with 100ml of complete DMEM after two days. On day three, 100 μl treatment medium was added to each well. After 24 hours, 50 μl serum-free medium with 25 μM phenazine methosulfate and 1mg/ml XTT (2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide) was added to each well. Optical density at 450nM was measured 4 hours later.
Cell Cycle Analysis
Cell cycle analysis was performed as described previously (Bergstralh et al., 2004).
Supplementary Material
Acknowledgments
We thank Corbin Jones for help with analysis of MUS312 sequence conservation, Joe Heitman for discussions about Cryptococcus BSP1, and Monica Colaiácovo, Pierre-Henrí Gaillard for communicating results prior to publication. D.T.B. was supported by the Lineberger Comprehensive Cancer Center Postdoctoral Fellowship in Basic Sciences. This work was supported by an award from the Glenn Foundation for Medical Research and a grant from the National Institutes of Health (GM61252) to J.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program PHYRE. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bergstralh DT, Taxman DJ, Chou TC, Danishefsky SJ, Ting JP. A comparison of signaling activities induced by Taxol and desoxyepothilone B. J Chemother. 2004;16:563–576. doi: 10.1179/joc.2004.16.6.563. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Boyd JB, Golino MD, Shaw KES, Osgood CJ, Green MM. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics. 1981;97:607–623. doi: 10.1093/genetics/97.3-4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, Russell P, et al. Human SLX4 is a Holliday junction resolvase that binds multiple DNA repair/recombination endonucleases. Cell. 2009 doi: 10.1016/j.cell.2009.06.029. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Flott S, Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MM. mus(3)312D1, a mutagen sensitive mutant with profound effects on female meiosis in Drosophila melanogaster. Chromosoma. 1981;82:259–266. doi: 10.1007/BF00286110. [DOI] [PubMed] [Google Scholar]
- Honigberg SM, Conicella C, Espositio RE. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics. 1992;130:703–716. doi: 10.1093/genetics/130.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157 doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Bessho T, Matsunaga T, Sancar A. Purification and characterization of the XPF-ERCC1 complex of human DNA repair excision nuclease. J Biol Chem. 1995;230:22657–22660. doi: 10.1074/jbc.270.39.22657. [DOI] [PubMed] [Google Scholar]
- Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4 (XPF) excision repair proteins. Proc Natl Acad Sci U S A. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadbeck-Seeger C, Wanner G, Huber S, Kahmann R, Kamper J. A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol Microbiol. 2000;38:154–166. doi: 10.1046/j.1365-2958.2000.02128.x. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Goley E, Baxter K, McMahan S, Sekelsky J. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics. 2005;170:1737–1745. doi: 10.1534/genetics.104.036178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, McMahan S, Blanton HL, Sekelsky J. Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics. 2007;176:63–72. doi: 10.1534/genetics.107.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell B, Gygi SP, Elledge SJ, Harper JW. BTBD12, a mammalian ortholog of SLX4/MUS312, is a scaffold for assembly of a Holiday Junction resolvase and is required for interstrand crosslink DNA repair. Cell. 2009 doi: 10.1016/j.cell.2009.06.030. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K, McKim KS, Brill S, Sekelsky J. Synthetic lethality in the absence of the Drosophila MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 2007;176:1993–2001. doi: 10.1534/genetics.106.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936–944. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Yildiz Ö, Kearney H, Kramer BC, Sekelsky J. Mutational analysis of the Drosophila repair and recombination gene mei-9. Genetics. 2004;167:263–273. doi: 10.1534/genetics.167.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz Ö, Majumder S, Kramer BC, Sekelsky J. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


