Abstract
In healthy cells, mitochondria continually divide and fuse to form a dynamic interconnecting network. The molecular machinery that mediates this organelle fission and fusion is necessary to maintain mitochondrial integrity, perhaps by facilitating DNA or protein quality control. This network disintegrates during apoptosis at the time of cytochrome c release and prior to caspase activation, yielding more numerous and smaller mitochondria. Recent work shows that proteins involved in mitochondrial fission and fusion also actively participate in apoptosis induction. This review will cover the recent advances and presents competing models on how the mitochondrial fission and fusion machinery may intersect apoptosis pathways.
Keywords: Bax, Bcl-2, Drp1, Mfn2, OPA1, fission
Apoptosis mediates the catabolism of eukaryotic cells that is crucial for metazoan development, adult tissue turnover, host defense pathways, and protection from cancer. All pathways of apoptosis converge upon the activation of caspases, proteases that orchestrate the efficient and noninflammatory demolition of cells. Two main pathways leading to caspase activation have been well characterized: the extrinsic route initiated by cell surface receptors leading directly to caspase 8 activation, and the intrinsic path that is regulated by mitochondria. The mitochondrial stage of apoptosis control is upstream of caspase activation and is mediated by the Bcl-2 family of proteins. One well understood role of mitochondria in caspase activation is to regulate the release of proteins from the space between the inner and outer mitochondrial membranes to the cytosol. When cytochrome c is released from mitochondria, it binds to APAF1 in the cytosol, activating the assembly of the apoptosome that activates caspase 9 (Bao and Shi 2007). Other proteins released from mitochondria, such as SMAC/Diablo, have less crucial accessory roles in caspase activation, perhaps most important in long-lived cells (Potts et al. 2005).
Bcl-2 family proteins regulate the release of cytochrome c and other proteins through the outer mitochondrial membrane (OMM) (Adams and Cory 2007; Chipuk and Green 2008). Some members of the Bcl-2 family inhibit apoptosis, such as Bcl-2, Bcl-xL, and Mcl-1, whereas others, such as Bax and Bak, activate apoptosis. Bax and Bak actively induce cytochrome c release from mitochondria within cells and in cell-free systems, both of which are inhibited by anti-apoptotic Bcl-2 family members. As the anti-apoptotic Bcl-2 family members closely resemble the proapoptotic members in structure, they may function as dominant negative inhibitors by binding and inhibiting Bax and Bak. Another class of disparate proteins including Puma and Bim, called BH3-only proteins, shares a short motif with Bcl-2 family proteins and regulates their activity. One model posits that upon apoptosis initiation, BH3-only proteins are induced and then bind and inhibit anti-apoptotic Bcl-2 family proteins, allowing pro-apoptotic Bax and Bak to permeabilize the mitochondrial outer membrane releasing cytochrome c and other proteins to activate caspases and induce cell demolition (for review, see Youle and Strasser 2008).
Prior to, or simultaneous with, cytochrome c release and upstream of caspase activation, mitochondria fragment into multiple small units. Blocking of this mitochondrial fission inhibits cytochrome c release and delays cell death, linking the morphogenesis machinery of this organelle to cell death induction. This review will focus on how this fragmentation occurs and particularly on how mitochondrial fission and fusion machinery may participate in outer mitochondrial membrane permeabilization and apoptosis activation.
Mitochondrial fission and fusion machinery
Mitochondrial morphology is much more dynamic than initially thought. In many adherent cell types, mitochondria form elongated tubules that continually divide and fuse to form a dynamic interconnecting network. The fusion process is critical for maintenance of mitochondrial function, as interruption of mitochondrial fusion results in a loss of inner mitochondrial membrane potential (Chen et al. 2003). Many of the gene products mediating the fission and fusion processes have been identified in yeast screens, and most are conserved in mammals, including the fission mediators Drp1 (Dnm1 in yeast) and Fis1, as well as the fusion mediators Mfn1 and 2 (Fzo1 in yeast) and OPA1 (Mgm1 in yeast) (Westermann 2008). However, mammalian homologs of the yeast fission mediator Mdv1, the Mdv1 homolog Caf4, and the yeast fusion mediator Ugo1 have not been found in mammals. Additional proteins have been found in mammals that appear absent in yeast, such as Bax and the Bax-binding protein, Endophilin B1, which affect mitochondrial morphogenesis and will be considered here in the context of apoptosis regulation.
Fission machinery in mammalian cells
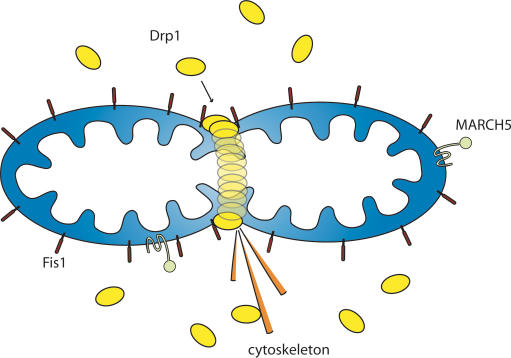
Dynamin-related protein 1 (Drp1), a large GTPase, mediates mitochondrial fission in mammalian cells (for reviews, see Okamoto and Shaw 2005; Hoppins et al. 2007). It appears to share common mechanisms with its homolog, dynamin, a protein involved in endosome scission from the plasma membrane. Dynamin assembles from the cytosol into spirals around the neck of endosomes. These dynamin helices constrict lipid tubules (Danino et al. 2004) and twist upon GTP cleavage to mediate lipid tubule scission in vitro if both ends of the tubule are anchored (Roux et al. 2006). Most Drp1 is found soluble in the cytosol of cells from where it shuttles onto and off mitochondria (Smirnova et al. 2001; Wasiak et al. 2007). Assuming analogy to studies of yeast Dnm1 (Ingerman et al. 2005), Drp1 assembles into spirals at division sites around the outer mitochondrial membrane to drive the fission process (Fig. 1). Dnm1 spirals are 109 nm in internal diameter, about five times larger than dynamin spirals (20 nm i.d.), and matching the diameter of mitochondrial constriction sites (111 nm) (Ingerman et al. 2005). Mutations that block Drp1 GTPase activity yield dominant negative mutants that inhibit mitochondrial fission, resulting in excessively tubular mitochondrial networks due to ongoing fusion (Otsuga et al. 1998; Smirnova et al. 2001). Thus, Drp1 appears to constrict mitochondria either due to the assembly of the spiral or to subsequent changes in helical diameter.
Figure 1.
Mitochondrial fission machinery. Schematic of the localization of three proteins (Drp1, Fis1, and MARCH5) involved in mitochondrial fission in mammalian cells.
The mechanism for recruitment of Drp1 to the mitochondria for spiral formation and mitochondrial fission remains unclear. In yeast, this assembly requires Fis1, a tetratricopeptide domain protein anchored into and evenly coating the entire OMM (Fig. 1; Suzuki et al. 2003; Dohm et al. 2004). In yeast, Fis1 recruits Dnm1 to the mitochondria through one of two adaptor proteins, Mdv1 or Caf4 (Mozdy et al. 2000; Tieu and Nunnari 2000; Griffin et al. 2005). However, orthologs of these adaptor proteins have not been identified in mammals, and mammalian Drp1 appears to assemble on mitochondria in the absence of Fis1 (Lee et al. 2004). It remains to be determined if Drp1 directly binds Fis1, although cross-linking studies and fluorescence resonance energy transfer (FRET) techniques are consistent with this hypothesis (Yoon et al. 2003). In mammalian cells, actin filaments and microtubules also appear to function in recruitment of Drp1 to mitochondria (Varadi et al. 2004; De Vos et al. 2005).
It remains unclear how the inner mitochondrial membrane is divided. Mitochondria in certain algae such as Cyanidioschyzon merolae (Kuroiwa et al. 2006) and the slime mold Dictyostelium (Gilson et al. 2003) retain homologs of the prokaryotic GTPase that mediates bacterial division, FtsZ, which assembles in rings within the mitochondrial matrix compartment (Kuroiwa et al. 2006) to mediate organelle division (Gilson et al. 2003). Cyanidioschyzon merola also has Drp1 and Mdv1 homologs (Nishida et al. 2007) that likely mediate OMM fission, but FtsZ is not found in yeast or mammals. In organisms lacking FtsZ, based on suborganelle location in the inner mitochondrial membrane, the dynamin homolog, OPA1, would be a candidate to mediate inner mitochondrial membrane fission if its dominant activity in inner mitochondrial membrane fusion can work in reverse.
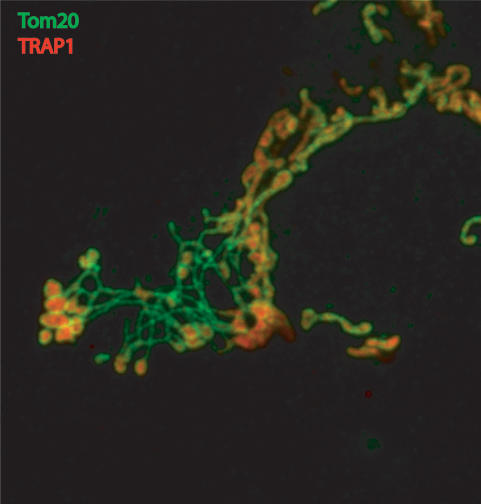
Another protein involved in mitochondrial morphogenesis in mammals is Endophilin B1 (also called Bif-1 and SH3GLB1), a BAR domain-containing protein that localizes to the cytosol of cells and interacts with Bax (Cuddeback et al. 2001; Pierrat et al. 2001). Endophilin B1, like Endophilin A1 that binds dynamin and participates in endosome scission, appears to regulate membrane curvature (Peter et al. 2004) and deforms lipid bilayers into tubules in vitro (Farsad et al. 2001). Down-regulation of Endophilin B1 in cells results in OMM dissociation from the inner mitochondrial membrane, forming long tubules of OMM (Karbowski et al. 2004b). As shown in Figure 2, in certain cells lacking Endophilin B1, nets of interconnected OMM link separated inner mitochondrial membrane-bound matrix compartments. This may occur from inner mitochondrial membrane scission continuing in the absence of OMM scission. Drp1 appears to activate the inner mitochondrial membrane scission in the Endophilin B1 knockdown cells despite the lack of OMM scission, as the combined Drp1/Endophilin B1 knockdown yields elongated mitochondria similar to that seen with Drp1 knockdown alone (Karbowski et al. 2004b).
Figure 2.
Outer mitochondrial morphology effects of Endophilin B1 knockdown. Confocal image of Endophilin B1/Bif1/SH3GLB1 knockdown by RNA interference in HeLa cells shows webs of OMM tubules (green) linking fragmented matrix compartments (red). Immunofluorescence of OMM is in green (α-Tom20) and the mitochondrial matrix is shown in red (α-TRAP1) (image by Chunxin Wang).
MTP18 (mitochondria protein, 18 kDa) and ganglioside-induced differentiation associated protein 1 (GDAP1) are two additional proteins that influence mitochondrial fission. MTP18, a downstream effector of phosphatidylinositol 3 kinase (PI3K) signaling, has been proposed to regulate mitochondrial fission through recruitment of Drp1 to the OMM (Tondera et al. 2004, 2005). Knockdown of MTP18, which localizes to the intermembrane space, leads to cytochrome c release from mitochondria, although whether this effect is direct has yet to be elucidated (Tondera et al. 2004, 2005). GDAP1 is a mitochondrial outer membrane protein (Pedrola et al. 2005) that is mutated in Charcot-Marie-Tooth disease type 4A (CMT-4A). Overexpression of GDAP1 causes fragmentation of mitochondria with no effect on cell death (Niemann et al. 2005), while down-regulation of GDAP1 or the expression of certain patient mutant forms, which are truncated and no longer localize to mitochondria, tend to elongate mitochondria, indicating that GDAP1 plays a normal function in mitochondrial fission.
Fusion machinery in mammalian cells
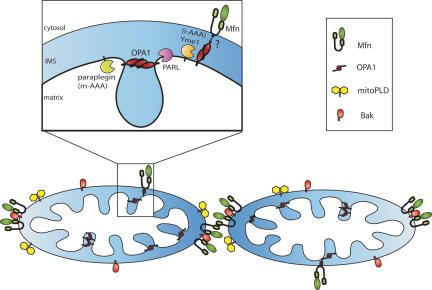
Mitochondrial fission is normally balanced by fusion. In yeast, this process has been extensively studied and reviewed (Cerveny et al. 2007; Hoppins et al. 2007; Merz et al. 2007), and here we will consider the fusion machinery in mammalian cells, omitting Ugo1 identified thus far only in yeast. Conserved between yeast and mammals, three large GTPases in the dynamin family, Mfn1, Mfn2, and OPA1, mediate mitochondrial fusion (Fig. 3). Cell-free fusion assays indicate that Mfn1 and 2 mediate OMM fusion whereas OPA1 mediates inner mitochondrial membrane fusion (Meeusen et al. 2004, 2006). OPA1 is localized on the inner mitochondrial membrane, facing the intermembrane space (Olichon et al. 2002). OPA1 in mammals is encoded by a single gene with eight transcript variants resulting from alternative splicing (Delettre et al. 2001). The various splice variants of OPA1 are differentially proteolyzed into long and short forms, yielding an array of different OPA1 isoforms in the mitochondria. Cleaved forms of OPA1 are more loosely attached to the inner mitochondrial membrane than long forms that retain a hydrophobic domain (Ishihara et al. 2006). Studies of the yeast homolog, Mgm1, show that the shorter, proteolytically processed form of Mgm1 is required in combination with the long form to mediate mitochondrial fusion (Herlan et al. 2003). A variety of proteases including members of the rhomboid family (Herlan et al. 2003; McQuibban et al. 2003; Sesaki et al. 2003; Cipolat et al. 2006), parapalegin (Ishihara et al. 2006), and Yme1 (Griparic et al. 2007; Song et al. 2007) have been implicated in OPA1 processing. OPA1 cleavage is accelerated when the electrochemical potential across the inner mitochondrial membrane is reduced, suggesting that OPA1 processing and thus its activity could be regulated to coordinate the energetic state of mitochondria with organelle morphology (Griparic et al. 2004; Duvezin-Caubet et al. 2006; Ishihara et al. 2006; Meeusen et al. 2006).
Figure 3.
Mitochondrial fusion machinery. Schematic of the submitochondrial localization of mammalian proteins involved in mitochondrial fusion in healthy cells, including the mitofusins, OPA1, mitoPLD, and Bak (see key). Insert shows the localization of proteases proposed to function in OPA1 cleavage. Presenilin-associated rhomboid-like (PARL) is one member of the rhomboid family.
Mitofusins 1 and 2 (Mfn1, Mfn2) integrate into the OMM with two transmembrane regions separated by only two to three amino acids facing the intermembrane space (Santel and Fuller 2001) orienting their N-terminal GTPase domain and C-terminal coiled-coil regions toward the cytosol (Rojo et al. 2002). The GTPase activity of mitofusins is essential for mitochondrial fusion (Hales and Fuller 1997; Hermann et al. 1998; Santel and Fuller 2001). The C-terminal coiled-coil region of Mfn1 and Mfn2 mediates tethering between mitochondria through homo- or heterotypic complexes formed between adjacent mitochondria (Chen et al. 2003; Ishihara et al. 2004; Koshiba et al. 2004; Meeusen et al. 2004).
Several Mfn2-binding proteins have been identified to date, including certain Bcl-2 family members, MIB, and Stoml2. Bax and Bak, two pro-apoptotic Bcl-2 family members, have recently been shown to interact with Mfn2 and Mfn1 by coimmunoprecipitation and fluorescence resonance energy transfer (FRET) studies (Brooks et al. 2007). Bcl-2, Bcl-xL, and Caenorhabditis elegans CED-9 have been shown to specifically interact by coimmunoprecipitation with Mfn2 but not Mfn1 (Delivani et al. 2006). The interactions between Bcl-2 family members and the mitofusins will be discussed in detail in later sections. Mitofusin-binding protein (MIB), a member of the medium-chain dehydrogenase/reductase superfamily, is mainly localized in the cytosol. Knockdown of MIB leads to elongation of the mitochondrial network, indicating that MIB regulates mitochondrial fission (Eura et al. 2006). Stomatin-like protein 2 (Stoml2) was identified in a complex with Mfn2 in healthy cells (Hajek et al. 2007) and is associated with the inner mitochondrial membrane facing the intermembrane space. Knockdown of Stoml2 reduces membrane potential without affecting mitochondrial morphology.
Mitochondrial phospholipase D (mitoPLD) is another protein implicated in mitochondrial fusion (Choi et al. 2006). Loss of mitoPLD expression induced by RNA interference (RNAi) leads to fragmentation of the mitochondrial network. Overexpression of mitoPLD with a mutation in the catalytic domain fails to reconstitute mitochondrial fusion in mitoPLD knockdown cells and leads to mitochondrial fragmentation in wild-type cells, indicating that enzyme activity is necessary for mitochondrial fusion. MitoPLD is a dimeric enzyme bound to the OMM through a C-terminal transmembrane anchor with an N-terminal catalytic domain facing the cytosol. Based on the topology of mitoPLD, the catalytic domain may insert into an adjacent OMM and modify lipids following mitofusin-mediated mitochondrial tethering. MitoPLD has been shown to hydrolyze cardiolipin, generating phosphatidic acid. While the role of phosphatidic acid in mitochondrial fusion remains unknown, it may function to induce membrane curvature or recruit other proteins to mitochondria through altering the membrane lipid environment, either of which could play a role in membrane fusion events.
Post-translational modification of Drp1 and Mfn2
Phosphorylation has recently been identified as an important post-translational modification to regulate Drp1-mediated mitochondrial fission. During mitosis, Drp1 is activated by CDK1/cyclinB phosphorylation of Ser618 (human Drp1; 585 in rat Drp1), resulting in mitochondrial fragmentation (Taguchi et al. 2007). Conversely, Drp1 GTPase activity is inactivated by phosphorylation at Ser637 (human Drp1; Ser656 in rat Drp1) within the GTPase effector domain (GED) mediated by cAMP-dependent protein kinase A (PKA), resulting in mitochondrial fusion (Chang and Blackstone 2007; Cribbs and Strack 2007). Thus, Drp1 activity can be positively and negatively regulated by kinases.
Drp1 also has been shown to be a substrate for sumoylation and to interact with SUMO1 by yeast two-hybrid (Harder et al. 2004). Overexpression of SUMO1 leads to excessive Drp1 sumoylation and results in mitochondrial fragmentation (Harder et al. 2004). SENP5, a SUMO protease, can desumoylate Drp1, and overexpression of cytosolic SENP5 can reverse SUMO1-induced mitochondrial fragmentation (Zunino et al. 2007). Thus, sumoylation of Drp1 appears to positively regulate its activity in mitochondrial fission.
MARCH5 is a newly identified membrane-spanning ubiquitin E3 ligase that localizes to mitochondria and plays a role in regulation of mitochondrial morphology (Nakamura et al. 2006; Yonashiro et al. 2006; Karbowski et al. 2007). It contains an N-terminal RING finger domain and four transmembrane domains localizing MARCH5 to the OMM (Fig. 1). The Yonashiro and Nakamura groups report that inhibition of MARCH5 activity by expression of a dominant negative mutant causes mitochondrial fragmentation (Nakamura et al. 2006; Yonashiro et al. 2006). However, Karbowski and colleagues subsequently reported that inhibition of MARCH5 increases elongation of and interconnection between mitochondria and causes excessive recruitment of Drp1 to mitochondria (Karbowski et al. 2007). In contrast to wild-type MARCH5 that localizes evenly around mitochondria, RING mutants of MARCH5 localize in foci with Drp1 on the mitochondrial surface. Thus, MARCH5 seems to be involved in mitochondrial fission, perhaps through disassembly or turnover of Drp1 spirals at fission sites.
The yeast mitofusin, Fzo1, and likely Mfn1 and 2, is regulated by ubiquitination. During mating, yeast mitochondria fragment and Fzo1 levels decrease. Interestingly, this loss of Fzo1 is attenuated by chemical inhibition (Neutzner and Youle 2005) or genetic inactivation (Escobar-Henriques et al. 2006) of the proteasome. Thus, Fzo1 appears to be removed from the OMM and degraded as a specific regulatory mechanism. During vegetative growth, Fzo1 has been shown to be Lys48 ubiquitinated (Neutzner et al. 2007), suggesting the model of a proteasome-mediated degradative process on the outer mitochondrial membrane, resembling that of the ER-associated degradation (ERAD) pathway of membrane-spanning protein removal and degradation. Expression levels of mammalian Mfn1 and 2 can be increased by treating cells with proteasome inhibitors (Karbowski et al. 2007), suggesting that the ubiquitin proteasome pathway also regulates mitofusins in mammals.
Role of mitochondrial fission machinery in apoptosis
Fission machinery plays a critical role in programmed cell death
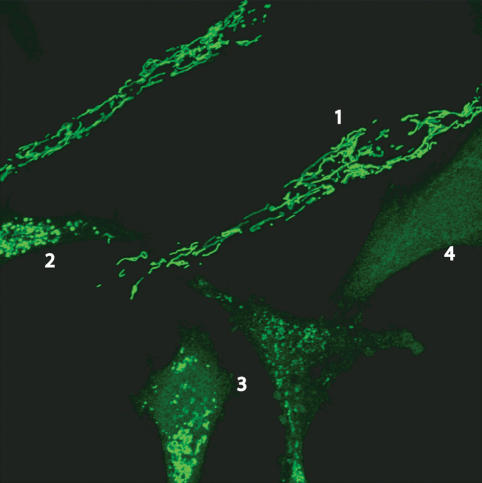
Mitochondrial fission is an early event during apoptosis, occurring before caspase activation and membrane blebbing. The process occurs very close in time to cytochrome c release, either just prior to or simultaneously, as fragmented mitochondria can readily be seen retaining cytochrome c (Fig. 4). Although this fragmentation appears to be universally associated with apoptosis, excessive mitochondrial fission can occur in the absence of apoptosis, such as that which occurs upon exposure to uncoupling agents that disrupt inner mitochondrial membrane electrochemical potential. FCCP-induced mitochondrial fragmentation, for example, is reversible upon drug removal, leaving normal mitochondria and healthy cells. Other stimuli, such as viral infection, also can induce mitochondrial fragmentation that is not linked to apoptosis. For instance, expression of a cytomegaloviral protein that interacts with Bcl-2 family members to block apoptosis at the mitochondria (Arnoult et al. 2004; Poncet et al. 2004; Karbowski et al. 2006), vMIA (viral mitochondria-localized inhibitor of apoptosis), leads to fragmentation of the mitochondrial network while functioning to block cell death (McCormick et al. 2003). Thus, while apoptosis is only one of a variety of conditions that induce mitochondrial fission, it appears to be invariably associated with mitochondrial fragmentation. Moreover, the excessive mitochondrial fission appears to be a requisite step in intrinsic apoptosis pathways, at least for the normal rate of cytochrome c release and caspase activation, as several components of the mitochondrial fission machinery, Drp1, Fis1, and Endophilin B1, have been implicated in programmed cell death progression.
Figure 4.
Cytochrome c release during apoptosis. Confocal image of multiple HeLa cells treated with actinomycin D and z-VAD-FMK (caspase inhibitor). Green represents cytochrome c immunofluorescence. (1) Healthy cells with long mitochondria and cytochrome c present in mitochondria. (2) During early stages of apoptosis, mitochondria fragment while some cytochrome c is retained in mitochondria. (3) Progressively later stages of apoptosis also have fragmented mitochondria, but cytochrome c is beginning to become visible in the cytosol. (4) Following complete release, cytochrome c is no longer within the mitochondria (image by Mariusz Karbowski). The time course for cytochrome c has been shown to be relatively fast and complete, indicating that mitochondrial fragmentation occurs immediately prior to or during the cytochrome c release process.
Drp1 mediates cytochrome c release during apoptosis
Upon apoptotic stimulation, Drp1 is recruited to the mitochondrial outer membrane (Frank et al. 2001; Breckenridge et al. 2003; Arnoult et al. 2005b; Wasiak et al. 2007), where it colocalizes with Bax and Mfn2 at fission sites (Karbowski et al. 2002). Drp1 function is required for apoptotic mitochondrial fission, as expression of a dominant negative mutant (Drp1K38A) or down-regulation of Drp1 by RNAi delays mitochondrial fragmentation, cytochrome c release, caspase activation, and cell death (Frank et al. 2001; Breckenridge et al. 2003; Lee et al. 2004; Germain et al. 2005; Barsoum et al. 2006). Similarly, a cell line with an endogenous loss of activity mutation in Drp1 displays resistance to hydrogen peroxide-induced cell death, an apoptotic process (Tanaka et al. 2006). Inhibiting Drp1 activity by RNAi (Lee et al. 2004; Germain et al. 2005) inhibits apoptosis at a unique step, downstream from Bax translocation and prior to cytochrome c release, placing Drp1 very close to the site of action of Bax on mitochondria.
Post-translational modification of Drp1 regulates its role in apoptosis
Expression of a phosphomimetic mutant Drp1 S656D in cells leads to mitochondrial elongation and increased resistance to staurosporine- and etoposide-mediated apoptosis (Cribbs and Strack 2007). On the other hand, elimination of the conserved PKA phosphorylation site with an S656A mutation in Drp1 results in increased mitochondrial fragmentation and a higher sensitivity to apoptotic stimuli (Cribbs and Strack 2007).
Drp1 sumoylation is also linked to apoptosis. During apoptosis, Drp1 sumoylation increases (Wasiak et al. 2007), along with increased Drp1 association with the OMM and increased mitochondrial fission. This study demonstrates Drp1 association with mitochondria is stabilized in a Bax/Bak-dependent manner after mitochondrial fragmentation but prior to cytochrome c release. Thus, post-translational modifications of Drp1 impact cell sensitivity to apoptosis and further link the mitochondrial fission machinery with programmed cell death.
One important proviso is that, in contrast to some nonmammalian examples below, inhibition of Drp1 in mammalian systems is typically incomplete and represents a delay rather than a block. This may reflect that inhibition of Drp1 activity is itself lethal or damaging to mammalian cells and/or that Drp1 is only indirectly connected to the activity of Bax and Bak that more centrally mediates programmed cell death. Drp1 impact on Mfn1/2, OPA1, or even Miro-1, an OMM GTPase recently found to mediate caspase activation in Drosophila (Yi et al. 2007), may be the key. If death stimuli are so great that Bcl-xL only partially protects cells, Drp1 inhibition may have little effect on cell viability even when an inhibition of cytochrome c release is apparent (Parone et al. 2006).
Beyond mammals, Drp1 is essential for programmed cell death in a number of other eukaryotes, including fungi, round worms, and flies. In the filamentous ascomycete Podospora anserina, an experimental model of aging, a link has been made between mitochondrial dynamics, apoptosis, and life-span control (Scheckhuber et al. 2007). Aging of P. anserina correlates with a disruption of the normally filamentous mitochondrial network into punctate units. Through subtractive hybridization analysis, Scheckhuber and colleagues identified an increased expression of a gene encoding a Drp1 ortholog, PaDnm1p, during the senescent phase of aging that correlates with the increase in mitochondrial fragmentation (Scheckhuber et al. 2007). Notably, strains lacking PaDnm1 display an 11-fold increase in mean life span. Reintroduction of PaDnm1 reverts this phenotype. Also, reorganization of mitochondrial DNA, a molecular hallmark of aging in P. anserina, is not observed in the PaDnm1 mutants. Similarly, generation of reactive oxygen species (ROS) is decreased in the PaDnm1 deletion strain, possibly due to a higher efficiency of oxidative phosphorylation in the longer, filamentous mitochondria. Also, PaDnm1 deletion strains are resistant to the exogenous DNA damaging agent, etoposide. The impact of mitochondrial dynamics on aging has been also shown in Saccharomyces cerevisiae through deletion of the Dnm1 or Fis1 genes (Scheckhuber et al. 2007). These deletion strains displayed an increased life span and improved fitness of old mother cells.
While the roles in unicellular organisms are less clear than in metazoans, yeast are considered to undergo a type of programmed cell death (Hardwick and Cheng 2004). For example, yeast may require cell death pathways to respond to environmental stress or infection by pathogens. Dnm1, the yeast ortholog of Drp1, has been shown to promote mitochondrial fission and cell death following exposure to environmental stress (Fannjiang et al. 2004). Also, most yeast strains are persistently infected by associated satellite dsRNA M viruses (“M killer viruses”) that are transmitted to new hosts via cell–cell fusion during mating. It has been shown that yeast cells are susceptible to M1 and M2 killer strains, while mutants lacking Dnm1 are significantly resistant to cell death induced by these viruses (Ivanovska and Hardwick 2005). These studies show the existence of cell death pathways in yeast that involve the mitochondrial fission machinery.
C. elegans displays well-defined developmental programmed cell deaths that utilize an established genetic pathway involving Bcl-2 family members and caspases (Metzstein et al. 1998). In this model it has been found that reduction of Drp1 activity using the dominant-negative Drp1 mutant (Drp1K40A) causes the inappropriate survival of ∼15% of the cells normally destined to die, equivalent to the number of cells that survive with weak loss-of-function mutations in known pro-apoptotic genes ced-3, ced-4, and egl-1. On the other hand, overexpression of drp1(wt) during embryogenesis results in increased mitochondrial fragmentation and apoptosis in ∼20% of cells normally destined to survive (Jagasia et al. 2005).
Two recent studies have shown that Drp1-mediated fragmentation of the mitochondrial network during apoptosis is also conserved in Drosophila melanogaster (Abdelwahid et al. 2007; Goyal et al. 2007). Disruption of the mitochondrial network has been shown to occur in vivo during physiological developmental programmed cell death and when primary fly cells cultured in vitro are exposed to a variety of apoptotic stimuli, including etoposide, actinomycin D, cyclohexamide, and C6-ceramide (Goyal et al. 2007). Interestingly, inhibition of Drp1 using RNAi-mediated knockdown or genetic mutants inhibited both the induction of mitochondrial fragmentation and cell death (Goyal et al. 2007). The central nervous system is enlarged in Drp1 mutant flies, also indicating that developmental cell death requires the mitochondrial fission machinery (Goyal et al. 2007). Expression of Hid and Reaper in fly cells has been shown to result in alterations in mitochondrial morphology and cytochrome c release (Abdelwahid et al. 2007). It has been demonstrated that these changes are related to caspase activation, although not as a direct consequence since caspase activation alone does not result in these mitochondrial disruptions. Furthermore, decreasing Drp1 activity by mutation decreases embryo sensitivity to apoptosis (Abdelwahid et al. 2007).
Most recently, an inhibitor of Dnm1, called mdivi-1, identified using yeast screens of chemical libraries, has been found to block mitochondrial division in yeast and mammalian cells (Cassidy-Stone et al. 2008). Upon apoptosis induction, mdivi-1 inhibits Drp1 translocation to mitochondria, cytochrome c release, and the appearance of phosphatidylserine on the outer cell membrane, a classic marker of dying cells. Importantly, this compound also blocks Bax- and Bid-mediated permeabilization of isolated mitochondria, showing very direct evidence for a role of Drp1 in Bax-mediated permeabilization of mitochondria.
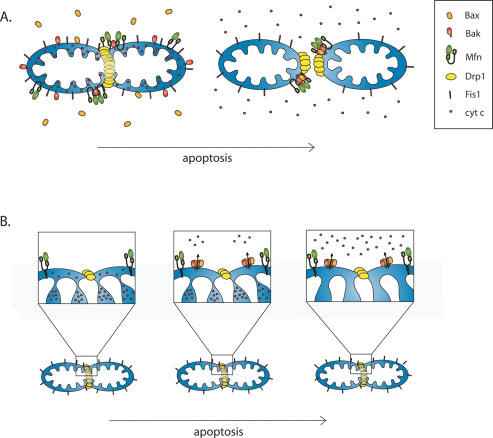
Together, these results suggest that Drp1, perhaps in concert with other components of the fission or fusion machinery, contributes to the process that mediates mitochondrial membrane permeabilization. Membrane fusion events typically utilize pore-forming domains to initiate the melding of lipid bilayers, and such pore formers that are presumably involved in mitochondrial fusion may be hijacked by the apoptosis machinery to permeabilize mitochondria. This hijacking may occur at the point when Bax and Bak meet Drp1 in fusion/fission foci on mitochondria (Fig. 5A).
Figure 5.
Competing models of mitochondrial morphogenesis machinery and apoptosis progression. (A) Mitochondrial foci model. From left to right, Bax (orange) translocates to mitochondria and coalesces into foci with Drp1, mitofusins, and Bak (see key), leading to mitochondrial division and release of cytochrome c (small red circles) from mitochondrial stores. (B) Cristae model of apoptosis. Drp1 (yellow) forms constriction sites on the OMM (left), then small amounts of cytochrome c from the intermembrane space are released through Bax/Bak pores (middle). (Right) Finally, cristae undergo remodeling, opening the junctions to release the larger amount of cytochrome c stored within the matrix.
An alternative model is suggested by the finding that Drp1 variably affects mitochondrial release of different proteins. For example, cytochrome c release may be blocked more efficiently than Smac/DIABLO in Drp1-depleted cells (Parone et al. 2006; Estaquier and Arnoult 2007). A model originally suggested by Scorrano and colleagues (Scorrano et al. 2002) states that mitochondrial fission and fusion components regulate mitochondrial cristae to effect release of intermembrane space proteins that may be selectively retained by tight membrane junctions within cristae where they approach the OMM (Fig. 5B). Consistent with this model, Drp1 has been shown to regulate cristae remodeling initiated by the pro-apoptotic BH3-only protein Bik that may control cytochrome c release from within inner mitochondrial membrane cristae folds (Germain et al. 2005).
Fis1 regulates cell death and senescence
Down-regulation of either Fis1 or Drp1 inhibits mitochondrial fission and reduces apoptosis, but they appear to act at different steps in the pathway, as only down-regulation of Fis1 inhibits Bax translocation and conformational change (Lee et al. 2004). Reduction of Fis1 expression levels using shRNA also results in senescence-associated phenotypic changes, including a reduction in cell proliferation, increased cellular granularity, and flattening of cells, in addition to the mitochondrial phenotype of sustained elongation (Lee et al. 2007). Cell senescence can occur following extended cell proliferation and division (replicative senescence) or due to sublethal stresses (stress-induced premature senescence). Interestingly, reduction of Fis1 expression acts as a sublethal stress, as prolonged mitochondrial elongation is associated with increased ROS levels and a loss of mitochondrial membrane potential. Simultaneous or sequential depletion of Fis1 and the fusion protein OPA1 reverses the elongation phenotype and the induction of senescence, providing strong evidence that excessive mitochondrial fusion is the cause of senescence-associated cellular changes. However, apoptosis resistance is not reversed by simultaneous knockdown of Fis1 and OPA1, showing that the apoptosis phenotype is not an indirect consequence of the senescent state of the cells or directly due to the excessive fission of the mitochondria (Lee et al. 2004).
Fis1 overexpression induces mitochondrial fission and apoptosis (James et al. 2003), further linking Fis1 and cell death. However, it is important to note that overexpression of proteins can lead to a number of nonphysiological stresses, and overexpression of a variety of OMM proteins, such as Tom20, can induce mitochondrial fragmentation. Fis1K148R, which has a point mutation in the region protruding into the intermembrane space, retains the ability to fragment mitochondria but fails to induce cell death (Alirol et al. 2006), suggesting that the function of Fis1 in mitochondrial fission and apoptosis can be separated.
Role of Endophilin B1/Bif-1/SH3GLB1 in Bax activity and apoptosis
Bax translocates to mitochondria during apoptosis and concentrates into foci on the mitochondrial surface when cells become committed to die (Youle and Strasser 2008). Sites on mitochondria where Bax forms foci typically colocalize with foci of Drp1 (and Mfn2) and frequently become sites of mitochondrial division (Karbowski et al. 2002). Discovered independently by two groups using yeast two-hybrid screens for Bax-binding proteins (Cuddeback et al. 2001; Pierrat et al. 2001), Endophilin B1 (also known as Bif1 and SH3GLB1) is a protein that directly links mitochondrial morphogenesis processes with apoptosis induction. Endophilin B1 binds Bax in yeast as well as in mammalian cells, although the latter interaction seems to be only transiently maintained following apoptosis induction (Takahashi et al. 2005). Endophilin B1−/− mice display an increased rate of spontaneous tumor development (Takahashi et al. 2007), and either Endophilin B1 RNAi or knockout suppresses both Bax translocation and cytochrome c release induced by apoptotic stimuli in cultured cells (Takahashi et al. 2005). Furthermore, Endophilin B1 is required in healthy cells for the maintenance of OMM morphology (Karbowski et al. 2004b), the same membrane that is breached during apoptosis. Interestingly, Endophilin B1 is able to alter membrane curvature (Farsad et al. 2001) and therefore may change the mitochondrial membrane shape to induce Bax insertion and oligomerization during apoptosis. Bax insertion into membrane bilayers is promoted by lipids that change monolayer curvature (Basanez et al. 2002), suggesting that translocation of Endophilin B1 to the OMM during apoptosis may promote Bax insertion into specific regions of the mitochondrial membrane through altering the OMM curvature. Additionally, Endophilin B1 contains an SH3 domain that, in analogy with Endophilin A1 interacting with dynamin, might bind the PXXP motifs in mitofusins or Drp1. Most recently, cells from Endophilin B1−/− mice were found to be deficient in autophagy, and Endophilin B1 was found localized partially in foci on autophagosomes (Takahashi et al. 2007) suggesting connections between mitochondrial fission and autophagy, as recently made with Drp1 inhibition (Twig et al. 2008).
Mitochondrial fusion machinery and apoptosis
Mitofusins and apoptosis
Mitochondrial fragmentation during apoptosis may result from excessive fission and/or insufficient fusion. The accumulation of Drp1 on mitochondria during apoptosis suggests that a stimulation of the fission rate may mediate the mitochondrial fragmentation process. However, mitochondrial fusion directly measured using a photoactivatable GFP method was found to be blocked during apoptosis at the point of Bax foci formation on the mitochondrial outer membrane (Karbowski et al. 2004a).
More direct evidence also indicates that inhibiting mitochondrial fusion promotes apoptosis. Silencing of Mfn1 or Mfn2 results in mitochondrial fragmentation and an increase in sensitivity to apoptotic stimuli (Sugioka et al. 2004). Furthermore, overexpression of Mfn1 or 2, in addition to increasing mitochondrial connectivity, results in delayed Bax activation, cytochrome c release, and apoptotic death, suggesting a role for mitofusins in cell death.
Overexpression of a dominant active Mfn2, with the residues within the GTPase P-loop of Mfn2 replaced by those of activated Ras (Mfn2G12V), was found to inhibit apoptosis better than wild-type Mfn2 (Neuspiel et al. 2005). Mfn2G12V displays increased nucleotide exchange rates and reduced levels of hydrolysis compared with wild-type Mfn2, indicating a state of constitutive binding to GTP, which impacts apoptosis but does not further increase mitochondrial fusion activity (Neuspiel et al. 2005; Jahani-Asl et al. 2007). Another Mfn2 mutant, which has a mutation in one of the three conserved residues exposed to the mitochondrial intermembrane space (Mfn2W631P), is unable to protect cells from apoptosis induced by staurosporine and UV treatment (Neuspiel et al. 2005). However, the protection is recovered with a double mutant containing both the Mfn2G12V and Mfn2W631P substitutions.
Mfn2 has also been shown to inhibit the ERK/MAPK signaling pathway (Chen et al. 2004). An Mfn2 mutant lacking the C-terminal coiled-coil domain, which is required for mitochondrial fusion activity, still inhibits ERK1/2 activation (Chen et al. 2004), indicating that Mfn2 regulation of ERK1/2 signaling is independent of its role in mitochondrial fusion. Since ERK activation has been shown to mediate induction of apoptosis (Cheung and Slack 2004), Mfn2 may protect cells by inhibiting ERK signaling independent of Mfn2 mitochondrial fusion activity. Two recent studies have shown that overexpression of Mfn2 in heart muscle cells (Shen et al. 2007) or vascular smooth muscle cells (Guo et al. 2007) induces apoptosis through inhibition of Ras–PI3K–Akt signaling. Here, Mfn2 is suggested to act in a pro-apoptotic manner.
OPA1 is required to protect cells from apoptosis
Loss of OPA1 induces spontaneous apoptosis of cells, providing another important link between apoptosis and mitochondrial morphogenesis (Olichon et al. 2003). This pioneering study found that Bcl-2 overexpression inhibits the cell death caused by OPA1 RNAi, placing the role of mitochondrial fusion in cell death upstream of mitochondrial outer membrane permeabilization. Overexpression of OPA1 prevents mitochondrial fission and protects cells from apoptotic death occurring through the mitochondrial pathway, but not apoptosis induced through the extrinsic pathway circumventing the mitochondria (Frezza et al. 2006). OPA1 mutants lacking functional GTPase and C-terminal GTPase effector domains (GED) are unable to protect cells from cell death (Frezza et al. 2006). These domains are required for the OPA1 mitochondrial fusion activity (Cipolat et al. 2004), suggesting these two functions, protection against apoptosis and mitochondrial fusion, could be linked. However, ectopic OPA1 expression also protected Mfn1, Mfn2 double-knockout cells without reversing their fusion deficit, indicating that OPA1 can block apoptosis independently of mediating mitochondrial fusion. The mammalian opa1 gene has eight RNA transcript variants from alternative splicing of exons 4, 4b, and 5b (Delettre et al. 2001). Knockdown of OPA1 transcripts using 4, 4b, and 5b siRNAs suggested that OPA1 transcripts containing exon 4 are important for mitochondrial fusion, while those variants containing exons 4b or 5b function to regulate cytochrome c release (Olichon et al. 2007).
OPA1, cristae remodeling, and apoptosis
Down-regulation of OPA1 not only induces spontaneous apoptosis and mitochondrial fragmentation, it also disrupts normal cristae structure (Olichon et al. 2003; Arnoult et al. 2005a; Frezza et al. 2006). Therefore, since the bulk of the cytochrome c seems to be localized within cristae in healthy cells, it has been suggested that complete cytochrome c release from the mitochondria cannot occur without cristae remodeling (Scorrano et al. 2002). Cristae junctions appear to be held together by OPA1 oligomers comprised of integral inner membrane long forms and more soluble protease-processed short forms. Incubating isolated mitochondria with tBid induces a disruption of OPA1 oligomers (Frezza et al. 2006) and alters cristae morphology (Arnoult et al. 2005a; Frezza et al. 2006). Furthermore, the tBid-induced increase of cytochrome c release into the intermembrane space from within the cristae is blocked by overexpression of OPA1, whereas expression of an Opa1 gene mutated in the GTPase domain increases the width of cristae junctions (Frezza et al. 2006). Taken together, these data suggest that GTPase activity is important for the oligomerization of OPA1 isoforms and in retaining cytochrome c in the mitochondrial matrix.
However, cristae junctions in hypercondensed mitochondria are wide enough to allow a small protein, like cytochrome c, to pass into the intermembrane space (Reed and Green 2002). Other retention mechanisms may play roles in inhibiting cytochrome c release from permeabilized mitochondria such as binding of cytochrome c to cardiolipin (Ott et al. 2002). Using the higher resolution technique of transmission electron microscopy to study changes in the ultrastructure of cristae upon apoptotic stimulation, it has been concluded that, although remodeling of the inner mitochondrial membrane occurs during apoptosis, it occurs after cytochrome c release (Sun et al. 2007). The cristae remodeling was also found to require active caspases, as it was blocked by a general caspase inhibitor, placing the process downstream from cytochrome c release.
OPA1 is released from mitochondria following the release of cytochrome c (Arnoult et al. 2005a). Arnoult and colleagues have proposed the model that initial leakage of soluble OPA1 results in alteration in cristae structure due to disruption of OPA1 oligomers at cristae junctions, thereby promoting the release of additional pools of OPA1 and cytochrome c into the cytosol (Arnoult et al. 2005a). However, Drp1K38A blocks OPA1 release from mitochondria (Estaquier and Arnoult 2007), suggesting that the release is downstream from Drp1 and mitochondrial fragmentation during apoptosis. Thus, there is evidence for and against each of the models in Figure 5, suggesting that additional hypotheses for how mitochondrial morphogenesis proteins modulate apoptosis should be considered.
Bcl-2 family members regulate mitochondrial morphology in healthy cells
While Bax and Bak have an established proapoptotic capacity, a recent study demonstrates a role for these proteins in healthy cells in the regulation of mitochondrial fusion (Karbowski et al. 2006). In Bax/Bak double-knockout (DKO) cells, mitochondrial fusion is reduced and inhibition of Drp1 activity using the dominant negative inhibitor, Drp1K38A, or RNAi does not promote elongation of mitochondria, indicating that Bax and Bak activate mitochondrial fusion without interfering with the normal fission process. Consistent with this model, Mfn2 complex assembly, mobility, and distribution along mitochondria in healthy cells is altered by the presence of Bax and Bak (Karbowski et al. 2006).
Consistent with the evidence that Bax and Bak can activate Mfn2 to promote mitochondrial fusion, Bax and Bak were shown to bind Mfn1 and Mfn2 in healthy cells, indicating that they may regulate mitofusin function through direct interaction (Brooks et al. 2007). However, this likely amounts to a minor fraction of Bax in healthy cells, as most Bax appears monomeric in the cytosol. Interestingly, Brooks et al. demonstrated that the affinity of Bak for Mfn2 decreases while the affinity for Mfn1 increases during apoptosis (Brooks et al. 2007). Upon apoptotic stimulation, Bax translocates to mitochondria. Thus, it is curious that no change in Bax–Mfn2 interaction is seen during apoptosis (Brooks et al. 2007). It is unclear how Bax and Bak may function to promote mitochondrial fusion in healthy cells and also participate in mitochondrial fission during apoptosis. Bax and Bak both change submitochondrial localization and conformation during apoptosis, which likely alters their interaction with proteins such as Mfn2 and Endophilin B1 and, consequently, their effects on the mitochondrial morphogenesis machinery.
Other Bcl-2 family members have been shown to interact with Mfn2. CED-9, the Bcl-2 homolog in C. elegans, suppresses apoptosis by binding CED-4, the APAF1 homolog. During apoptosis, EGL-1, the C. elegans BH3-only protein, binds to CED-9, releasing CED-4 to promote cell death. Delivani and colleagues show that expression of CED-9, as well as Bcl-xL, results in mitochondrial fusion through direct interactions with Mfn2 (Delivani et al. 2006). CED-9 overexpressed in mammalian cells causes mitochondrial fusion without inhibiting apoptosis, decoupling these two processes (Delivani et al. 2006). This study also shows that mitochondrial morphology is abnormal in CED-9 mutant worms, supporting a role for anti-apoptotic Bcl-2 proteins in maintenance of the mitochondrial network in healthy cells.
A cytomegalovirus-encoded protein, vMIA, inhibits apoptosis through interaction with the pro-apoptotic Bax and Bak proteins (Arnoult et al. 2004; Poncet et al. 2004; Karbowski et al. 2006). Based on its primary sequence, vMIA has very little similarity to Bcl-xL, although both proteins protect cells from a variety of apoptotic stimuli (Goldmacher et al. 1999). vMIA expression also leads to a dramatic disruption of the mitochondrial network (McCormick et al. 2003). The anti-apoptotic and mitochondrial phenotypes possibly occur through inactivation of Bax and Bak, which are required for mitofusin-dependent fusion. Supporting this, ectopic Bax is able to reverse the fragmented phenotype resulting from vMIA expression, whereas expression of Bax lacking the BH3 domain, which renders Bax inactive in apoptosis induction, is unable to elongate mitochondria when vMIA is expressed (Karbowski et al. 2006). Interestingly, a mutation in the BH3 domain of Bak also alters its ability to change conformation and dissociate from Mfn2 during apoptosis (Brooks et al. 2007). Taken together, these studies suggest a possible connection between the function of the Bax and Bak BH3 domains in both promoting apoptosis as well as regulating mitochondrial morphology in healthy cells.
Summary
The identity and mechanisms of the molecular machines mediating mitochondrial fission and fusion are rapidly becoming understood. This affords tools to explore the proposed physiological roles of mitochondrial morphogenesis in, for example, protein and DNA quality control, programmed cell death, aging, autophagy, and ROS production. Evidence from a variety of experiments indicates that the morphology of mitochondria per se is not directly involved in apoptosis, but members of the fission and fusion machinery are. However, the molecular basis for this role remains unknown. A unifying hypothesis for the function of Bcl-2 family proteins is currently lacking, as they are thought to regulate apoptosis in C. elegans and mammals by different mechanisms. Understanding how Bcl-2 family members are linked to the mitochondrial fission and fusion machinery may reveal a common underlying function.
Acknowledgments
We thank Chunxin Wang and Mariusz Karbowski for images (Figures 2 and 4, respectively) and Atsushi Tanaka and Megan Cleland for critical comments and reading of the manuscript.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1658508.
References
- Abdelwahid E., Yokokura T., Krieser R.J., Balasundaram S., Fowle W.H., White K. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Adams J.M., Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alirol E., James D., Huber D., Marchetto A., Vergani L., Martinou J.C., Scorrano L. The mitochondrial fission protein hFis1 requires the endoplasmic reticulum gateway to induce apoptosis. Mol. Biol. Cell. 2006;17:4593–4605. doi: 10.1091/mbc.E06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D., Bartle L.M., Skaletskaya A., Poncet D., Zamzami N., Park P.U., Sharpe J., Youle R.J., Goldmacher V.S. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. 2004;101:7988–7993. doi: 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D., Grodet A., Lee Y.J., Estaquier J., Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005a;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- Arnoult D., Rismanchi N., Grodet A., Roberts R.G., Seeburg D.P., Estaquier J., Sheng M., Blackstone C. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr. Biol. 2005b;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Bao Q., Shi Y. Apoptosome: A platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Barsoum M.J., Yuan H., Gerencser A.A., Liot G., Kushnareva Y., Graber S., Kovacs I., Lee W.D., Waggoner J., Cui J., et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basanez G., Sharpe J.C., Galanis J., Brandt T.B., Hardwick J.M., Zimmerberg J. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 2002;277:49360–49365. doi: 10.1074/jbc.M206069200. [DOI] [PubMed] [Google Scholar]
- Breckenridge D.G., Stojanovic M., Marcellus R.C., Shore G.C. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C., Wei Q., Feng L., Dong G., Tao Y., Mei L., Xie Z.J., Dong Z. Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins. Proc. Natl. Acad. Sci. 2007;104:11649–11654. doi: 10.1073/pnas.0703976104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Tamura Y., Zhang Z., Jensen R.E., Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Chang C.R., Blackstone C. Cyclic-AMP-dependent protein kinase phosphorylation of Drp1 regulates its gtpase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.H., Guo X., Ma D., Guo Y., Li Q., Yang D., Li P., Qiu X., Wen S., Xiao R.P., et al. Dysregulation of HSG triggers vascular proliferative disorders. Nat. Cell Biol. 2004;6:872–883. doi: 10.1038/ncb1161. [DOI] [PubMed] [Google Scholar]
- Cheung E.C., Slack R.S. Emerging role for ERK as a key regulator of neuronal apoptosis. Sci. STKE. 2004;2004:PE45. doi: 10.1126/stke.2512004pe45. [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., Green D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Cipolat S., de Martins Brito O., Dal Zilio B., Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Cribbs J.T., Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddeback S.M., Yamaguchi H., Komatsu K., Miyashita T., Yamada M., Wu C., Singh S., Wang H.G. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J. Biol. Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- Danino D., Moon K.H., Hinshaw J.E. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Delettre C., Griffoin J.M., Kaplan J., Dollfus H., Lorenz B., Faivre L., Lenaers G., Belenguer P., Hamel C.P. Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 2001;109:584–591. doi: 10.1007/s00439-001-0633-y. [DOI] [PubMed] [Google Scholar]
- Delivani P., Adrain C., Taylor R.C., Duriez P.J., Martin S.J. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol. Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- De Vos K.J., Allan V.J., Grierson A.J., Sheetz M.P. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr. Biol. 2005;15:678–683. doi: 10.1016/j.cub.2005.02.064. [DOI] [PubMed] [Google Scholar]
- Dohm J.A., Lee S.J., Hardwick J.M., Hill R.B., Gittis A.G. Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N.G., et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 2006;281:37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estaquier J., Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Oka T., Mihara K. Identification of a novel protein that regulates mitochondrial fusion by modulating mitofusion (Mfn) protein function. J. Cell Sci. 2006;119:4913–4925. doi: 10.1242/jcs.03253. [DOI] [PubMed] [Google Scholar]
- Fannjiang Y., Cheng W.C., Lee S.J., Qi B., Pevsner J., McCaffery J.M., Hill R.B., Basanez G., Hardwick J.M. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes & Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K., Ringstad N., Takei K., Floyd S.R., Rose K., De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., de Martins Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Germain M., Mathai J.P., McBride H.M., Shore G.C. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson P.R., Yu X.C., Hereld D., Barth C., Savage A., Kiefel B.R., Lay S., Fisher P.R., Margolin W., Beech P.L. Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot. Cell. 2003;2:1315–1326. doi: 10.1128/EC.2.6.1315-1326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmacher V.S., Bartle L.M., Skaletskaya A., Dionne C.A., Kedersha N.L., Vater C.A., Han J.W., Lutz R.J., Watanabe S., Cahir-McFarland E.D., et al. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal G., Fell B., Sarin A., Youle R.J., Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev. Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E.E., Graumann J., Chan D.C. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., van der Wel N.N., Orozco I.J., Peters P.J., van der Bliek A.M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A.M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Chen K.H., Guo Y., Liao H., Tang J., Xiao R.P. Mitofusin 2 triggers vascular smooth muscle cell apoptosis via mitochondrial death pathway. Circ. Res. 2007;101:1113–1122. doi: 10.1161/CIRCRESAHA.107.157644. [DOI] [PubMed] [Google Scholar]
- Hajek P., Chomyn A., Attardi G. Identification of a novel mitochondrial complex containing mitofusion 2 and stomatin-like protein 2. J. Biol. Chem. 2007;282:5670–5681. doi: 10.1074/jbc.M608168200. [DOI] [PubMed] [Google Scholar]
- Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Harder Z., Zunino R., McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hardwick J.M., Cheng W.C. Mitochondrial programmed cell death pathways in yeast. Dev. Cell. 2004;7:630–632. doi: 10.1016/j.devcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A.S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E., Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Hardwick J.M. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jahani-Asl A., Cheung E.C., Neuspiel M., Maclaurin J.G., Fortin A., Park D.S., McBride H., Slack R.S. Mitofusin 2 protects cerebellar granule neurons against injury-induced cell death. J. Biol. Chem. 2007;282:23788–23798. doi: 10.1074/jbc.M703812200. [DOI] [PubMed] [Google Scholar]
- James D.I., Parone P.A., Mattenberger Y., Martinou J.C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Lee Y.J., Gaume B., Jeong S.Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C.L., Youle R.J. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Arnoult D., Chen H., Chan D.C., Smith C.L., Youle R.J. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 2004a;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Jeong S.Y., Youle R.J. Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 2004b;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Norris K.L., Cleland M.M., Jeong S.Y., Youle R.J. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R.J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M., Chan D.C. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T., Nishida K., Yoshida Y., Fujiwara T., Mori T., Kuroiwa H., Misumi O. Structure, function and evolution of the mitochondrial division apparatus. Biochim. Biophys. Acta. 2006;1763:510–521. doi: 10.1016/j.bbamcr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Jeong S.Y., Lim W.C., Kim S., Park Y.Y., Sun X., Youle R.J., Cho H. Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J. Biol. Chem. 2007;282:22977–22983. doi: 10.1074/jbc.M700679200. [DOI] [PubMed] [Google Scholar]
- McCormick A.L., Smith V.L., Chow D., Mocarski E.S. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 2003;77:631–641. doi: 10.1128/JVI.77.1.631-641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban G.A., Saurya S., Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Merz S., Hammermeister M., Altmann K., Durr M., Westermann B. Molecular machinery of mitochondrial dynamics in yeast. Biol. Chem. 2007;388:917–926. doi: 10.1515/BC.2007.110. [DOI] [PubMed] [Google Scholar]
- Metzstein M.M., Stanfield G.M., Horvitz H.R. Genetics of programmed cell death in C. elegans: Past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M., Zunino R., Gangaraju S., Rippstein P., McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical-induced depolarization. J. Biol. Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- Neutzner A., Youle R.J. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Neutzner A., Youle R.J., Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found. Symp. 2007;287:4–14,. 14–20. discussion . [PubMed] [Google Scholar]
- Niemann A., Ruegg M., La Padula V., Schenone A., Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: New implications for Charcot-Marie-Tooth disease. J. Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K., Yagisawa F., Kuroiwa H., Yoshida Y., Kuroiwa T. WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. 2007;104:4736–4741. doi: 10.1073/pnas.0609364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J.M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Olichon A., Emorine L.J., Descoins E., Pelloquin L., Brichese L., Gas N., Guillou E., Delettre C., Valette A., Hamel C.P., et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Olichon A., Elachouri G., Baricault L., Delettre C., Belenguer P., Lenaers G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted funciton in apoptosis. Cell Death Differ. 2007;14:682–692. doi: 10.1038/sj.cdd.4402048. [DOI] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J.M. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Robertson J.D., Gogvadze V., Zhivotovsky B., Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc. Natl. Acad. Sci. 2002;99:1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parone P.A., James D.I., Da Cruz S., Mattenberger Y., Donze O., Barja F., Martinou J.C. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrola L., Espert A., Wu X., Claramunt R., Shy M.E., Palau F. GDAP1, the protein causing Charcot-Marie-Tooth disease type 4A, is expressed in neurons and is associated with mitochondria. Hum. Mol. Genet. 2005;14:1087–1094. doi: 10.1093/hmg/ddi121. [DOI] [PubMed] [Google Scholar]
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J., Evans P.R., McMahon H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pierrat B., Simonen M., Cueto M., Mestan J., Ferrigno P., Heim J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- Poncet D., Larochette N., Pauleau A.L., Boya P., Jalil A.A., Cartron P.F., Vallette F., Schnebelen C., Bartle L.M., Skaletskaya A., et al. An anti-apoptotic viral protein that recruits Bax to mitochondria. J. Biol. Chem. 2004;279:22605–22614. doi: 10.1074/jbc.M308408200. [DOI] [PubMed] [Google Scholar]
- Potts M.B., Vaughn A.E., McDonough H., Patterson C., Deshmukh M. Reduced Apaf-1 levels in cardiomyocytes engage strict regulation of apoptosis by endogenous XIAP. J. Cell Biol. 2005;171:925–930. doi: 10.1083/jcb.200504082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.C., Green D.R. Remodeling for demolition: Changes in mitochondrial ultrastructure during apoptosis. Mol. Cell. 2002;9:1–3. doi: 10.1016/s1097-2765(02)00437-9. [DOI] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Roux A., Uyhazi K., Frost A., De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Santel A., Fuller M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Scheckhuber C.Q., Erjavec N., Tinazli A., Hamann A., Nystrom T., Osiewacz H.D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- Scorrano L., Ashiya M., Buttle K., Weiler S., Oakes S.A., Mannella C.A., Korsmeyer S.J. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Hobbs A.E., Jensen R.E. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- Shen T., Zheng M., Cao C., Chen C., Tang J., Zhang W., Cheng H., Chen K.H., Xiao R.P. Mitofusin-2 is a major determinant of oxidative stress-mediated heart muscle cell apoptosis. J. Biol. Chem. 2007;282:23354–23361. doi: 10.1074/jbc.M702657200. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka R., Shimizu S., Tsujimoto Y. Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J. Biol. Chem. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
- Sun M.G., Williams J., Munoz-Pinedo C., Perkins G.A., Brown J.M., Ellisman M.H., Green D.R., Frey T.G. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat. Cell Biol. 2007;9:1057–1065. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Jeong S.Y., Karbowski M., Youle R.J., Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J. Mol. Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Karbowski M., Yamaguchi H., Kazi A., Wu J., Sebti S.M., Youle R.J., Wang H.G. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol. Cell. Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H.D., Sun M., Sato Y., Liang C., Jung J.U., Cheng J.Q., Mul J.J., et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Kobayashi S., Fujiki Y. Peroxisome division is impaired in a CHO cell mutant with an inactivating point-mutation in dynamin-like protein 1 gene. Exp. Cell Res. 2006;312:1671–1684. doi: 10.1016/j.yexcr.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D., Santel A., Schwarzer R., Dames S., Giese K., Klippel A., Kaufmann J. Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J. Biol. Chem. 2004;279:31544–31555. doi: 10.1074/jbc.M404704200. [DOI] [PubMed] [Google Scholar]
- Tondera D., Czauderna F., Paulick K., Schwarzer R., Kaufmann J., Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi A., Johnson-Cadwell L.I., Cirulli V., Yoon Y., Allan V.J., Rutter G.A. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 2004;117:4389–4400. doi: 10.1242/jcs.01299. [DOI] [PubMed] [Google Scholar]
- Wasiak S., Zunino R., McBride H.M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Molecular machinery of mitochondrial fusion and fission. J. Biol. Chem. 2008;283:13501–13505. doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- Yi C.H., Sogah D.K., Boyce M., Degterev A., Christofferson D.E., Yuan J. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 2007;179:619–626. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Krueger E.W., Oswald B.J., McNiven M.A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H.M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]