Abstract
In the past, bile acids were considered to be just detergent molecules derived from cholesterol in the liver. They were known to be important for the solubilization of cholesterol in the gallbladder and for stimulating the absorption of cholesterol, fat-soluble vitamins, and lipids from the intestines. However, during the last two decades, it has been discovered that bile acids are regulatory molecules. Bile acids have been discovered to activate specific nuclear receptors (farnesoid X receptor, preganane X receptor, and vitamin D receptor), G protein coupled receptor TGR5 (TGR5), and cell signaling pathways (c-jun N-terminal kinase 1/2, AKT, and ERK 1/2) in cells in the liver and gastrointestinal tract. Activation of nuclear receptors and cell signaling pathways alter the expression of numerous genes encoding enzyme/proteins involved in the regulation of bile acid, glucose, fatty acid, lipoprotein synthesis, metabolism, transport, and energy metabolism. They also play a role in the regulation of serum triglyceride levels in humans and rodents. Bile acids appear to function as nutrient signaling molecules primarily during the feed/fast cycle as there is a flux of these molecules returning from the intestines to the liver following a meal. In this review, we will summarize the current knowledge of how bile acids regulate hepatic lipid and glucose metabolism through the activation of specific nuclear receptors and cell signaling pathways.
Keywords: short heterodimer partner, fibroblast growth factor 15/19, G protein coupled receptor TGR5, muscarinic receptors, cholesterol 7α-hydroxylase, sterol 12α-hydroxylase, sterol 27-hydroxylase, steroidogenic acute regulatory protein, secondary bile acids, intestine
In the past, conjugated bile acids were considered to be detergent molecules produced from cholesterol in the liver. As detergents, bile acids are important for the solubilization of cholesterol in the gallbladder and intestine by forming mixed micelles with cholesterol and phospholipids. These amphipathic molecules are also recognized as being required for the activation of certain pancreatic enzymes and for the solubilization and absorption of cholesterol, lipid soluble vitamins (A, D, K, and E), and to a lesser extent, triglycerides and fatty acids from the intestines (1). Bile acid synthesis is also recognized as a major output pathway of cholesterol from the body; the other being biliary cholesterol secretion (1).
The concept of bile acids as hormones was not apparent until 1999 when three independent laboratories reported that bile acids were ligands for the farnesoid X receptor-α (FXR-α; NR1H4) (2–4). Following this discovery, numerous genes in the liver and intestine were shown to be induced by bile acids via a functional FXR element in their promoters (see Ref. 5 for recent summary). It was discovered that many of the genes induced by bile acids encode enzymes/proteins involved in regulating bile acid synthesis, transport, conjugation, lipoprotein synthesis, uptake, and metabolism (5). After FXR was shown to be activated by bile acids, it was discovered that the preganane X receptor (PXR; NR1I2) and the vitamin D receptor (VDR; NR1I1) could be activated by lithocholic acid, a hydrophobic bile acid formed by 7α-dehydroxylation of chenodeoxycholic acid by intestinal anaerobic bacteria (6–8). Activation of PXR or VDR induces genes encoding enzymes involved in the metabolism and detoxification of lithocholic acid (6–8).
In the last few years, bile acids have also been discovered to activate various cell signaling pathways in cells in the liver and gastrointestinal tract. In this regard, bile acids have been known for many years to activate different isoforms of protein kinase C (PKC) in colonic cells and hepatocytes by poorly defined mechanisms (9–12). In 2001, bile acids were also reported to activate the c-jun N-terminal kinase (JNK) 1/2 signaling pathway in primary hepatocytes, and this was linked to the downregulation of cholesterol 7α-hydroxylase (CYP7A1) and bile acid synthesis (13). Moreover, in the same year, two independent laboratories reported that deoxycholic acid activated the epidermal growth factor receptor in hepatocytes (14, 15). This was later shown to occur in other epithelial cells in the gastrointestinal system (16, 17). In 2002, a Gαs protein-coupled receptor, TGR5, was discovered to be responsive to bile acids by two research groups (18, 19). Activation of TGR5 by bile acids was shown to be possibly linked to energy metabolism (20, 21) and hepatoprotection in the liver (22). Subsequently, investigations reported evidence for Gαi protein-coupled receptor(s) in hepatocytes activated by conjugated, but not free, bile acids. These putative bile acid-responsive Gαi protein-coupled receptor(s) were linked to the activation of the AKT (insulin signaling pathway) and ERK 1/2 signaling pathways in hepatocytes in culture (23) and in vivo (24). The rapid activation of the insulin signaling pathway by bile acids strongly suggests they may help control glucose metabolism in the liver. In total, the current evidence strongly indicates that bile acids play an important role in the regulation of their own synthesis, fatty acid, lipid, and lipoprotein synthesis, as well as glucose metabolism in the liver. Moreover, bile acids may be important in resistance to intestinal bacterial growth and translocation in the small intestine (25). Bile acids appear to primarily function as metabolic regulatory molecules during the feed/fast cycle as the flux of bile acids returning from the intestine to the liver is increased following a meal. This review will focus on how bile acid-activated nuclear receptors and cell signaling pathways interact to produce an overall physiological response regulating various biosynthetic and metabolic pathways in the liver. There are already excellent reviews focused on FXR and metabolic diseases (5, 26).
BILE ACID BIOSYNTHETIC PATHWAYS
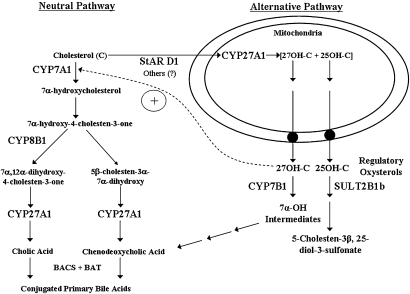
There are two major bile acid biosynthetic pathways in the liver, the neutral and alternative pathway (Fig. 1). The neutral pathway is initiated by CYP7A1, a cytochrome P450 enzyme located in the smooth endoplasmic reticulum of hepatocytes. The 7α-hydroxylation of cholesterol has been shown to be the rate-limiting step in the neutral pathway of bile acid biosynthesis (1). The gene encoding CYP7A1 is highly regulated at the transcriptional level, and its mRNA has a very short half-life (∼1.5 h). The gene encoding CYP7A1 is strongly downregulated by most bile acids, glucagon, certain cytokines [tumor necrosis factor-α (TNF-α) and interleukin 1 (IL-1)] and FGF 15/19 (27). The CYP7A1 gene has been reported to be upregulated by thyroid hormone, glucocorticoids, and oxysterols (in rodents, but not humans) (27). The neutral bile acid biosynthetic pathway consists of least 16 enzymatic steps leading to the formation of cholic acid and chenodeoxycholic acid (reviewed in Ref. 28). The ratio of cholic acid to chenodeoxycholic acid in this pathway is controlled by sterol 12α-hydroxylase (CYP8B1). The gene encoding CYP8B1 is also highly regulated at the transcriptional level by many of the same mechanisms regulating CYP7A1. However, there some differences in the regulation of the genes encoding CYP7A1 and CYP8B1 (29). The enzymes in the bile acid biosynthetic pathway are located at many different cellular locations in the hepatocyte, including smooth endoplasmic reticulum, mitochondria, peroxisomes, and cytoplasm. It is unclear how various bile acid biosynthetic intermediates in this pathway move from one cellular location to another. The neutral pathway of bile acid biosynthesis appears to be the major pathway of bile acid synthesis in humans under normal conditions. However, during various pathophysiological conditions, the alternative pathway of bile acid synthesis is the most active (30).
Fig. 1.
Pathways of bile acid biosynthesis in the liver. The neutral and alternative pathways of bile acid biosynthesis are initiated by cholesterol 7α-hydroxylase (CYP7A1) and mitochondrial sterol 27-hydroxylase (CYP27A1), respectively. Sterol 12α-hydroxylase (CYP8B1) determines the ratio of cholic acid to chenodeoxycholic acid synthesized. In humans, the neutral pathway is the major pathway of synthesis under normal physiological conditions. StAR D1, steroidogenic acute regulatory protein D1; 27OH-C, 27-hydroxycholesterol; 25OH-C, 25-hydroxycholesterol; SULT2B1b, hydroxycholesterol sulfotransferase 2B1b; CYP7B1, oxysterol 7α-hydroxylase; BAL, bile acid CoA ligase; BAT, bile acid CoA:amino acid N-acyltransferase.
The alternative pathway of bile acid biosynthesis is initiated by mitochondrial sterol 27-hydroxylase (CYP27A1). However, the rate-limiting step in this pathway appears to be transport of cholesterol to the inner mitochondrial membrane. The inner mitochondrial membrane is normally low in cholesterol concentration. It has been demonstrated that overexpression of the gene encoding StarD1, a soluble cholesterol binding protein, either in primary hepatocytes (31) or in vivo in rodent liver results in increased rates of bile acid biosynthesis (32). StarD1 is known to transport free cholesterol from intracellular membranes to the mitochondria (33). Evidence has also been presented for the presence of StarD1 mRNA and protein in primary hepatocytes (34). Whether other cholesterol transport proteins are involved in the movement of cholesterol to mitochondria is currently unknown. Unlike CYP7A1, which is expressed only in hepatocytes, mitochondrial CYP27A1 is expressed in many different tissues and cell types in the body and has been hypothesized to play a role in reverse cholesterol transport (35). CYP27A1 is a unique cytochrome P-450 enzyme in that it catalyzes the 27-hydroxylation of cholesterol as well as various bile acid biosynthetic intermediates (36). It also is capable of carrying out further oxidation of the 27-hydroxy group to yield a C-27 carboxyl group. Finally, this enzyme also has the ability to 25-hydroxylate cholesterol, yielding a potentially important regulatory oxysterol, 25-hydroxycholesterol (37). 25-Hydroxycholestrol can be further metabolized in hepatocytes to 25-hydroxycholesterol-3-sulfate. There is some evidence that this sulfated oxysterol may be a regulatory metabolite (38). 7α-Hydroxylation of 27-hydroxycholesterol and other oxysterols is carried out by oxysterol 7α-hydroxylase (CYP7B1). Therefore, in addition to bile acid biosynthesis, the alternative pathway generates 27-hydroxycholesterol and 25-hydroxycholesterol, which are regulatory oxysterols (35, 39). These oxysterols activate the liver X receptor (LXR; NR1H2/3), which is known to be important in maintaining cholesterol and fat homeostasis in the liver and other tissues (39, 40).
ENTEROHEPATIC CIRCULATION OF BILE ACIDS AND SYNTHESIS OF FIBROBLAST GROWTH FACTOR 15/19
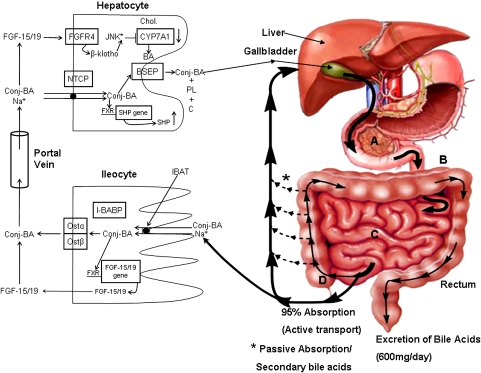
Conjugated bile acids, synthesized in the liver, are actively transported across the canalicular membrane of the hepatoycte into the bile duct system along with cholesterol and phosphatidylcholine (Fig. 2). The ratio of conjugated bile acids, cholesterol, and phospholipids in bile is tightly regulated to insure that cholesterol is solubilized as either mixed micelles of conjugated bile acids, cholesterol, and phospholipids or as cholesterol:phospholipid vesicles (1). Excess biliary cholesterol secretion relative to conjugated bile acids and phospholipids can result in cholesterol saturated bile and an increased risk for cholesterol gallstone formation. Biliary lipids are stored in the gallbladder and released into the duodenum of the small intestine in response to alimentary hormones. In the small bowel, conjugated bile acids function to activate specific pancreatic lipases and to solubilize dietary lipids, sterols, and fat-soluble vitamins by forming mixed micelles, which allows for the uptake of these nutrients into enterocytes. Bile acids move through the small bowel under the influence of peristalsis. In the ileum, bile acids are actively taken up by a sodium-dependent transporter (IBAT; SLC10A2) (41, 42). Inside the ileocyte, bile acids bind to the ileal bile acid binding protein (IBAT) and are transported to the basolateral membrane where bile acids exit the ileocyte primarily via the heterodimeric organic solute transporter OSTα/β (43–45). Bile acids absorbed from the intestines are transported back to the liver via the portal vein where they are actively transported into the hepatocyte primarily via the Na+/taurocholate cotransporting polypeptide (SLC10A1) (46).
Fig. 2.
Enterohepatic circulation of bile acids. Conjugated bile acids (Conj-BA) are actively transported from the canalicular side of the hepatocyte by the bile salt export pump (BSEP; ABC B11) along with phospholipids (PL; ABC 4) and cholesterol (C; ABC G5/G8). Conjugated bile acids are actively transported into ileocytes by the sodium-dependent intestinal bile acid transporter (IBAT). Inside the ileocyte, bile acids induce the synthesis of fibroblast growth factor 15/19 (FGF-15/19) but are also bound by the intestinal bile acid binding protein (I-BABP). They exit the ileocyte on the basolateral side via the heterodimeric organic solute transporter (Ost-α/β). Bile acids and FGF-15/19 are transported back to the liver via the portal blood. Conjugated bile acids are actively transported into the hepatocyte primarily by the Na+/taurocholate cotransporting polypeptide (NTCP). FGF-15/19 binds to and activates hepatic fibroblast growth factor receptor 4 (FGFR4), which in turn activates the JNK signaling pathway. Activation of JNK downregulates the gene encoding cholesterol 7α-hydroxylase (CYP7A1), inhibiting bile acid synthesis.
In ileocytes, bile acids stimulate the synthesis of fibroblast growth factor 15/19 (mouse FGF-15 is the ortholog of human FGF-19) via a functional FXR element in the promoter of the gene encoding FGF 15/19 (47). This hormone, when secreted from ileocytes, is believed to be transported to the liver and bind to its cognate tyrosine kinase receptor (FGFR-4) on hepatocytes, which activates the JNK 1/2 signaling pathway (48, 49). Activation of the JNK 1/2 signaling pathway in primary hepatocytes by bile acids or cytokines (i.e., TNF-α) has been shown to downregulate CYP7A1 mRNA (13). FGFR-4 is highly expressed in the liver but not in the ileum. Holt et al. (47) and Song et al. (49) have reported that bile acids can induce the synthesis of FGF19 in human primary hepatocytes in culture, resulting in the downregulation of the gene encoding CYP7A1. However, the significance of these observations in vivo is not yet clear as most evidence suggest that intestinally synthesized FGF 15/19 is probably the main mechanism of CYP7A1 repression in the liver (50, 51). It was discovered early on that FGFR-4 or β-klotho null mice had increased CYP7A1 mRNA levels and an expanded bile acid pool (52, 53). Further study showed that β-klotho is required for FGF 15/19 to interact with FGFR-4 and activate the JNK 1/2 signaling pathway (Fig. 2).
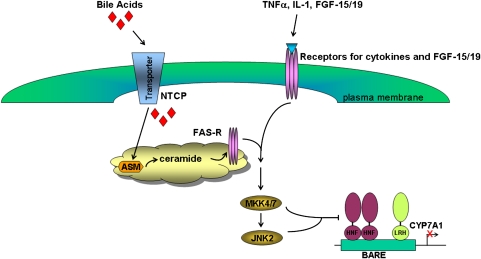
Bile acids also induce the gene encoding the short heterodimer partner (SHP; NR0B2) in the liver. SHP was originally hypothesized to play a role in regulating CYP7A1gene expression (54, 55). SHP is a nuclear receptor without a DNA binding domain that can physically interact with positive acting transcription factors, such as hepatocyte nuclear factor 4α (HNF4α; NR2A1), liver receptor homolog 1 (NR5A2), LXRα/β (NR1H2/3), and others. However, more recent studies by Mataki et al. (56) and Lee et al. (57) showed that disruption of the liver receptor homolog 1 gene in mouse hepatocytes did not alter the regulation of CYP7A1 gene expression. In contrast, sterol 12α-hydroxylase (CYP8B1) gene expression was markedly decreased in the liver of these same animals. This had the effect of decreasing cholic acid biosynthesis and altering the bile acid pool composition. The bulk of the current scientific evidence suggests that the gene encoding CYP7A1 is downregulated by activating the JNK 1/2 signaling pathway. JNK 1/2 and protein kinase A (PKA) kinases have been hypothesized to phosphorylate HNF4α and alter its ability to activate the CYP7A1 promoter (58, 59) (Fig. 3).
Fig. 3.
Downregulation of cholesterol 7α-hydroxylase (CYP7A1) by bile acids, cytokines, and FGF-15/19. Bile acids are transported into the hepatocyte via the Na+/taurocholate cotransporting polypeptide (NTCP). In vitro, bile acids activate ASM-generating ceramide, which causes the clustering and activation of the FAS receptor. This receptor then activates the JNK signaling pathway. Activation of the JNK 1/2 pathway by bile acids, TNF-α, IL-1, or fibroblast growth factor-15/19 (FGF-15/19) is hypothesized to result in the phosphorylation of HNF4α, decreasing its ability to activate the gene encoding CYP7A1. BARE, bile acid-responsive element; MKK4/7, MAP kinase kinase 4/7; RXR, retinoid X receptor.
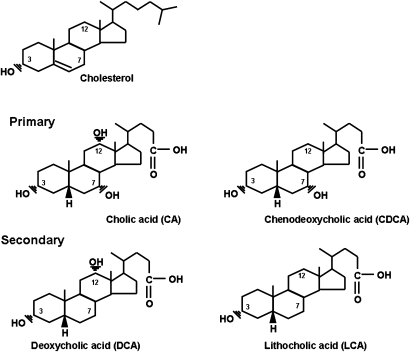
During their enterohepatic circulation, several hundred milligrams of bile acids escape this cycle each day and enter the colon where they are exposed to the indigenous intestinal microbiota. Bile acids are metabolized in a variety of ways by intestinal bacteria, including deconjugation, epimerization of hydroxy groups at C-3, C-7, and C12, and the 7α-dehydroxylation of cholic acid and chenodeoxycholic acid, yielding deoxycholic acid and lithocholic acid, respectively (60). Deoxycholic acid and lithocholic acid are referred to as secondary bile acids (Fig. 4). Secondary bile acids and metabolites are passively absorbed from the colon and are returned to the liver via the portal vein. Hence, there is a mixture of primary and secondary bile acids and metabolites returning to the liver during each enterohepatic cycle. Bile acids returning from the intestines are actively transported from the blood into hepatocytes, biotransformed, conjugated to either glycine or taurine, actively transported from the hepatocyte into bile, and become part of the circulating bile acid pool. The ratio of taurine to glycine bile acids in bile is a function of dietary habits in humans, but not rodents. High-protein diets in humans favor taurine conjugation, whereas vegetarian diets favor glycine conjugation (61). Interestingly, the percentage of deoxycholic acid in bile can vary from <1% to >50% (60). Deoxycholic acid increases the hydrophobicity of the bile acid pool, which is associated with greater toxicity and increased cholesterol secretion from the liver. The human bile acid pool contains only small amounts (1–4%) of conjugated and/or sulfated lithocholate. This is due mainly to the very hydrophobic nature and insolubility of lithocholic acid, which is formed in the colon by the 7α-dehydroxylation of chenodeoxycholic acid by anaerobic bacteria (60). Hence, only small amounts are absorbed. However, it is a very toxic bile acid and must be rapidly conjugated and/or sulfated to limit damage to hepatocytes.
Fig. 4.
Structures of unconjugated primary and secondary bile acids in humans. Cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) are synthesized from cholesterol in the liver hepatocytes. Deoxycholic acid (DCA; 3α,12α-dihydroxy-5β-cholan-24-oic acid) and lithocholic acid (LCA; 3α-hydroxy-5β0cholan-24-oic acid) are synthesized from cholic acid and chenodeoxycholic acid, respectively, by a small population of 7α-dehydroxylating intestinal anaerobic bacteria.
Bile acids appear to function as important regulatory molecules in the small intestinal tract by inducing genes, in an FXR-dependent manner, that are involved in resistance to bacterial growth and translocation into intestinal epithelial cells (25). These observations may have important pathophysiological implications for patients with cholestatic liver disease or impaired bile flow as bacterial overgrowth and uptake may enhance pro-inflammatory processes in the intestines that could affect multiple organ systems in the body.
ACTIVATION OF NUCLEAR RECEPTORS AND CELL SIGNALING PATHWAYS BY BILE ACIDS
Over the past decade, bile acids have been reported to activate several nuclear receptors (FXR, PXR, and VDR) and cell signaling pathways (AKT, ERK 1/2, and JNK 1/2) in cells in the liver and gastrointestinal tract (Fig. 5); however, the physiological significance of activating these nuclear receptors and cell signaling pathways is only now becoming apparent. Activation of the JNK 1/2 signaling cascade by bile acids can occur by direct or indirect mechanisms. The addition of either conjugated or free bile acids to primary hepatocytes in culture rapidly (∼1 h) activates the JNK 1/2 signaling pathway and downregulates CYP7A1 mRNA (13). Activation of the JNK 1/2 pathway by bile acids at the level of the hepatocyte requires the synthesis of ceramide (62). Bile acids appear to generate ceramide by activating acidic sphingomyelinase (ASM) as hepatocytes isolated from ASM null mice fail to synthesize ceramide in response to bile acids or activate the JNK 1/2 signaling pathway. However, the JNK 1/2 pathway is intact in ASM null hepatocytes as TNF-α activates this pathway to the same extent as in wild-type hepatocytes. Activation of the JNK 1/2 pathway in primary hepatocytes is hypothesized to downregulate CYP7A1 primarily by phosphorylation of HNF4α by activated JNK 1/2 kinases in this pathway (Fig. 3). In vivo, bile acids appear to activate the JNK 1/2 signaling cascade primarily by the FXR-dependent synthesis of FGF-15/19 in the ileum (48). In this scenario, bile acids activate FXR (nuclear receptor), inducing the gene encoding FGF15/19 (a hormone) that is secreted, transported to the liver, and binds to FGFR-4 (cell surface tyrosine kinase receptor) activating JNK 1/2 (cell signaling pathway), which downregulates CYP7A1 (a specific gene). Hence, this is an example of crosstalk between a bile acid-activated nuclear receptor and cell signaling pathway.
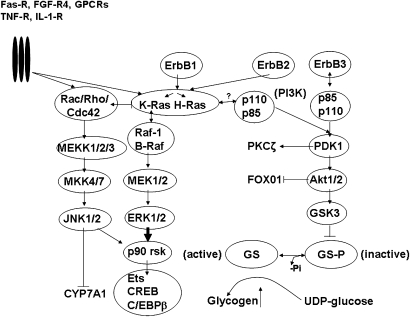
Fig. 5.
Major cell signaling pathways activated by bile acids in hepatocytes. Components of the JNK 1/2, extracellular-regulated kinases (ERK 1/2), and Ser/Thr kinase Akt/PKB (protein kinase B) signaling pathways are shown. Each signaling pathway consists of three sequentially acting kinases. GS, glycogen synthase; GSK-3, glycogen synthase kinase 3; PI3K, phosphoinositide-3-kinase; PDK-1, phosphoinositide-dependent protein kinase 1; Erb1/2/3, epidermal growth factor receptor family members; Rac/Rho;Cdc-42, small molecular weight GTP/GDP binding proteins involved in activating JNK 1/2 pathway; K-Ras and H-Ras, small molecular weight GTP/GDP binding proteins associated with activation of ERK 1/2 pathway; p90 rsk, p90 ribosomal 6 kinase; Ets, ETS domain transcription factor; CREB, c-AMP-responsive element binding protein; C/EBPβ, CCAAT enhancer binding protein β; GPCRs, G-protein-coupled receptors; FGF-R4, fibroblast growth factor receptor 4; TNF-R, tumor necrosis factor receptor; IL-1 R, interleukin 1 receptor; Fas-R, Fas receptor.
It is unclear why the intraduodenal infusion of taurocholate in the chronic bile fistula rat model does not rapidly activate JNK 1/2 in the liver (R. Cao et al., unpublished observations). However, this may be due to the rapid transport of conjugated bile acids through the hepatocyte into bile. The JNK 1/2 pathway can also be activated by pro-inflammatory cytokines, i.e., TNF-α and IL-1 (13). These cytokines are synthesized during infection and other pathophysiological conditions (i.e., excess cholesterol in macrophages). In addition, their synthesis may be stimulated by unconjugated hydrophobic bile acids returning from the intestines. In this regard, Miyake et al. (63) showed that hydrophobic bile acids induced the expression of genes encoding IL-1 and TNF-α in monocyte/macrophage cultures. The in vivo significance of these cytokines in regulating CYP7A1 expression and bile acid synthesis under normal physiological conditions is currently unclear.
Bile acids can activate AKT (insulin signaling pathway) in hepatocytes by two different mechanisms. Conjugated bile acids activate this signaling pathway primarily via Gαi protein-dependent receptor(s), while unconjugated hydrophobic bile acid appear to activate this pathway by mitochondrially generated superoxide ions (Fig. 6) (23). The activation of the AKT pathway by bile acids has been shown to occur in primary rat hepatocytes and in the liver of the chronic bile fistula rat (23, 24). Activation of the AKT pathway by bile acids allows them to function in a manner almost identical to insulin in regulating glucose metabolism in the liver (23, 24, 64). This makes physiological sense as bile acids are released from the gallbladder following a meal. This may allow bile acids returning to the liver to boost insulin signaling during the feed/fast cycle. In this regard, the addition of bile acids to primary hepatocytes activates glycogen synthase activity, an insulin target enzyme, to the same degree as insulin. The addition of both a bile acid and insulin to primary hepatocytes yields an additive effect on glycogen synthase activity (24). Taurocholate added to cultures of primary hepatocytes also increases the phosphorylation of FOX01 and downregulates the gluconeogenic genes (PEPCK and G-6-Pase) in a manner similar to insulin (R. Cao et al., unpublished observations). Moreover, it has been recently reported that activation of the AKT pathway is necessary for the optimal induction of the gene encoding SHP by TCA in primary hepatocytes. Preliminary evidence indicates that phosphorylation of FXR by PKCζ (65) or possibly other isoforms of PKC (66) may enhance the induction of SHP by TCA (R. Cao et al., unpublished observations). PKCζ is known to be activated by PDK-1, a component of the insulin signaling pathway (Fig. 6). Activation of the insulin signaling pathway and phosphorylation of FXR has been hypothesized, but not yet proven, to increase the affinity of FXR for TCA (R. Cao et al., unpublished observations). In vitro, TCA is a very poor activator of FXR (EC50 >1 mM) (2–4, 67). However, in primary hepatocytes, TCA causes induction of SHP mRNA at concentrations as low as 1–5 µM (R. Cao et al., unpublished observations). Hence, the activation of a cell signaling pathway (AKT) by bile acids may alter the functionality of a nuclear receptor (FXR) that binds bile acids. The phosphorylation of nuclear receptors by activated protein kinases in cell signaling pathways can alter their cellular location, affinity for ligand, turnover rate, DNA binding, and ability to interact with coactivators (reviewed in Refs. 68, 69).
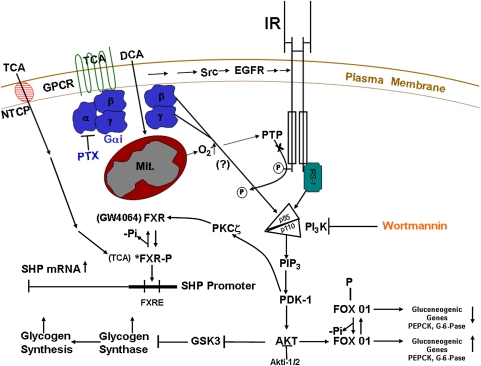
Fig. 6.
Activation of the Akt (insulin signaling pathway) and FXR by bile acids in hepatocytes. Taurocholate (TCA) activates the AKT pathway via a Gαi protein;coupled receptor(s). Phosphoinositide-dependent protein kinase 1 (PDK-1), a kinase in the insulin signaling pathway, then activates atypical protein kinase C ζ (PKCζ). Activated PKCζ is proposed to phosphorylate FXR, enhancing its ability to bind TCA and induce the gene encoding SHP. Activated Akt phosphorylates the transcription factor FOX01. The phosphorylated form of FOX01 exits the nucleus, allowing for the downregulation of gluconeogenic genes [PEP carboxykinase (PEPCK) and glucose-6-phosphatase (G-6-Pase)]. Akt also phosphorylates glycogen synthase kinase 3 (GSK-3), inactivating this kinase, which allows for the dephosphorylation and activation of glycogen synthase activity. NTCP, Na+/taurocholate cotransporting polypeptide; DCA, deoxycholic acid; Src, src family kinases; EGFR, epidermal growth factor receptor; PTP, phosphotyrosine phosphatase; PTX, pertussis toxin; IR, insulin receptor.
Bile acids appear to be able to alter glucose metabolism in hepatocytes by at least two mechanisms. First, conjugated bile acids activate the insulin signaling pathway via Gαi protein-coupled receptors or superoxide ions and function much like insulin to activate glycogen synthase and repress gluconeogenic genes. This is probably a very rapid (minutes) response to bile acids. In addition, bile acids activate (hours) the gene encoding SHP via a functional FXR site in its promoter. SHP can bind to FOX01, CEBPα, and HNF4α, transcription factors known to activate gluconeogenic genes (Fig. 7) (70, 71). Although the Gαi protein-coupled receptors in hepatocytes activating the insulin signaling pathway have not yet been identified, they could be a target for drug development for treating patients with type II diabetes. High-affinity agonists activating the putative conjugated bile acid G-protein-coupled receptor may bypass insulin resistance by activating the PI3K/PDK-1/AKT pathway via the β/γ-subunits of G-protein-coupled receptors (72).
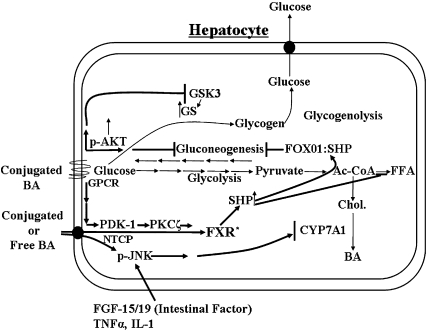
Fig. 7.
Regulation of hepatic glucose metabolism and cholesterol and fatty acid biosynthetic pathways by bile acids. Bile acids regulate glucose metabolism in hepatocytes by activating the insulin signaling pathway and by induction of the gene encoding SHP. Conjugated bile acids activate the insulin signaling pathway (pAKT) via surface G-protein-coupled receptors (GPCRs), which activates glycogen synthase activity and inhibits gluconeogenesis by phosphorylation of FOX01. SHP is known to interact with FOX01, a transcription factor known to upregulate gluconeogenic genes. Phosphoinositide-dependent protein kinase 1 (PDK-1), a kinase in the insulin signaling pathway, is known to phosphorylate and activate protein kinase Cζ (PCKζ). PKCζ is proposed to phosphorylate FXR, enhancing the ability of conjugated bile acids to induce the gene encoding SHP. SHP can also downregulate the gene encoding SREBP1-c, which controls the rate of FA synthesis. Bile acids can activate JNK 1/2 in hepatocyte cultures by stimulating ceramide synthesis. In vivo, bile acids can activate the JNK 1/2 pathway by stimulating the synthesis of fibroblast growth factor 15/19 (FGF-15/19) in the intestines. Finally, TNF-α and IL-1 can also activate the JNK 1/2 pathway downregulating CYP7A1. Bold lines indicate pathways activated by bile acids.
There is abundant literature indicating that bile acid-activated FXR and cell signaling pathways play an important role in hepatic glucose metabolism (reviewed in Ref. 73). However, the cellular mechanisms of regulating glucose metabolism by bile acids are not yet clear. It was reported in 2003 by De Fabiani et al. (74) that there was coordinate regulation between the regulation of the genes encoding CYP7A1 and PEPCK by an FXR-independent mechanism. In other studies, FXR-null mice were shown to develop fatty liver and elevated serum free fatty acids, which are associated with elevated serum glucose levels and insulin resistance (75). FXR appears to be crucial for normal glucose homeostasis during the feed-fast cycle and may also play a role in the inhibition of glycolysis via repression of liver pyruvate kinase as well as inhibiting gluconeogenic genes via induction of SHP (76). Treatment of diabetic animals with GW4064, an FXR synthetic ligand, improves insulin resistance and hyperglycemia (77). Overall, the effects of bile acids on hepatic glucose metabolism appears to involve both a bile acid activated cell signaling pathway (AKT) and the nuclear receptors FXR and SHP (78). Generally, cell signaling pathways respond much faster (seconds to minutes) than nuclear receptors (minutes to hours) to regulatory ligands. Therefore, the combination of these two regulatory mechanisms may be required for an overall physiological response.
It has been known for many years that bile acids can regulate lipid metabolism in humans. Bile acid binding resins decrease the transhepatic bile acid flux and increase serum levels of VLDL-triglycerides and HDL-cholesterol but reduce LDL-cholesterol. In contrast, treatment of cholesterol gallstone patients with chenodeoxycholic acid, which increases the transhepatic flux of bile acids, has the opposite effect on lipid metabolism (79–81). How do bile acids regulate lipid homeostasis in the liver? Current evidence suggests that bile acids regulate the rates of fatty acid, triglyceride, and VLDL synthesis primarily through the FXR, SHP, LXR, and SREBP-1c pathway (82–84). In this model, bile acids activate FXR, inducing the gene encoding SHP, which then interacts with LXR to decrease the transcriptional activity of gene encoding SREBP-1c. Sterol-regulatory element binding proteins (SREBPs) are transcription factors that are activated by a highly regulated, sterol-dependent, proteolytic cleavage process. It has been shown that expression of the gene encoding active SREBP1c specifically induces genes encoding enzymes involved in fatty acid, triglyceride, and VLDL biosynthesis (84). In contrast, SREBP2 has been shown to primarily regulate genes encoding enzymes involved in cholesterol biosynthesis (84). Watanabe et al. (82) have reported convincing evidence that SHP downregulates the gene encoding SREBP1c by interacting with LXR. LXRα is known to enhance the transcriptional activity of the gene encoding SREBP1c. In this regard, LXRα agonists have been shown to raise serum triglyceride levels (85, 86). Bile acids may also lower serum triglyceride levels by enhancing the clearance and turnover of VLDL. In this regard, bile acids induce the gene encoding apolipoprotein C-II, a coactivator of lipoprotein lipase, which is involved in the metabolism of VLDL (87). Finally, bile acids induce a number of other genes encoding enzymes/proteins involved in the uptake and metabolism of VLDL (83).
The ERK 1/2 signaling pathway is rapidly activated by either conjugated or free bile acids in primary hepatocytes and in vivo (23, 24). The activation of the ERK 1/2 signaling pathway by bile acids can also occur by two different mechanisms. Conjugated bile acids activate this signaling pathway via Gαi protein-dependent receptor(s). Alternatively, free bile acids stimulate the synthesis of mitochondrial superoxide ions, which has been reported to inactivate phosphoprotein phosphatase(s), resulting in the activation of the epidermal growth factor receptor and activation of ERK 1/2 (23). There is also evidence that bile acids activate this pathway in some epithelial cells via stimulation of matrix metalloproteinase that generates TGFα, an epidermal growth factor receptor ligand, in an autocrine/paracrine manner (17). Although the physiological significance of activation of the ERK 1/2 pathway in hepatocytes and other epithelial cells is not yet clear, its activation does help to protect the hepatocyte against bile acid-induced apopotosis (14, 88).
ACTIVATION OF TGR5 AND MUSCARINIC RECEPTORS BY BILE ACIDS
TGR5 was discovered in 2002 (18), fully characterized in 2003 (19), and shown to be a Gαs protein-coupled receptor responsive to both conjugated and free bile acids. Gαs protein-coupled receptors, once activated, stimulate the synthesis of c-AMP, which activates PKA. Activated PKA, among other things, can phosphorylate c-AMP response element binding protein. This activated transcription factor can then induce the expression of many genes that have a functional c-AMP response element in their promoter. The secondary bile acids, lithocholic acid and deoxycholic acid, were found to be the best activators of TGR5 using in vitro cell culture techniques (19). By amino acid sequence analysis, TGR5 is a member of the Rhodopsin-like subfamily of G protein-coupled receptors (89). The gene encoding TGR5 is widely expressed in different tissues in the body, with the highest expression in gallbladder, spleen, liver, intestine, adipose tissue, and immune cells (19). However, there appears to be very low expression in hepatocytes. In mice, it has been shown that bile acids induce c-AMP-dependent thyroid hormone activating enzyme type 2 iodothyronine deiodinase (D2) and other enzymes/proteins involved in energy expenditure in brown adipose tissue and muscle (20). D2 converts metabolically inactive thyroxine (T4) into T3, which is known to play a vital role in energy homeostasis in brown adipose tissue and muscle. Mice fed a high-fat diet supplemented with cholic acid gained less body weight than mice fed a high fat diet alone although both groups consumed the same amount of food. In D2 gene null mice, there was no difference in weight gain on these two diets. However, further studies of D2 null mice failed to confirm greater weight gain in male D2 mice on a high-fat diet compared with control mice (90). Moreover, in TGR5 null animals, male mice had greater expression of CYP7A1 than females. It has been hypothesized that in humans, bile acids might help regulate energy metabolism in skeletal muscle as TGR5 can regulate D2 expression in human muscle myoblasts (20). However, the physiological importance of muscle TGR5 in humans is unclear as the serum levels of bile acids are usually low (5–15 µM) and mostly bound to serum albumin and lipoproteins. Finally, hydrophobic secondary bile acids are the most powerful activators of TGR5, and their concentrations are quite variable in human bile (60). TGR5 may be important in immune cells as bile acids have been shown to have immunoregulatory properties in these cells (19). Moreover, TGR5 may also play a protective role in the liver as it is expressed in sinusoidal endothelial cells in the liver. Bile acids have been shown to induce nitric oxide synthase in these cells in a c-AMP-dependent manner (22). The physiological importance of this TGR5 is only beginning to be elucidated and will probably be tissue/cell type dependent.
In 1998, taurolithocholate was reported to activate cholinergic receptors in chief cells isolated from guinea pig stomach, resulting in the increased secretion of pepsinogen (91). No other bile acid tested (conjugated or free) altered pepsinogen secretion in these cells. This was the first evidence that specific conjugated bile acids might activate muscarinic receptors. There are five muscarinic receptors (M1 to M5), and they are coupled to different G protein families (M1,3,5 are coupled to Gαq/G11 and M2,4 to Gαi/Go-type proteins) (92). Muscarinic receptor subtypes are variably expressed in different tissues in the body, including stomach, small intestine, colon, gallbladder, intestinal smooth muscle, central nervous system, and pancreatic β-islet cells. Taurolithocholate preferentially activates the M3 receptor subtype presumably because of its structural similarity to acetylcholine (92–94). The physiological importance of activation of muscarinic receptors by taurolithocholic acid is uncertain as high concentrations are required for activation. Lithocholic acid is formed only by intestinal anaerobic bacteria, and conjugated and sulfated lithocholic acid is quite low (1–4%) in human bile.
PERSPECTIVE
During the last decade, the perception of bile acids in health and disease has dramatically changed. In the past, bile acids were considered to be just detergent molecules involved in digestive processes in the intestines and important for cholesterol solubilization in the gallbladder. However, it is now clear that bile acids are hormones that can activate specific nuclear receptors and cell signaling pathways in cells in the liver and gastrointestinal tract in a physiologically relevant manner. In general, they appear to primarily regulate biosynthetic and metabolic pathways in the liver and intestines during the feed/fast cycle. However, these molecules also have the potential to control cell proliferation and inflammatory processes in the liver and gastrointestinal tract. Although a large amount of literature has been published in the last decade related to bile acids as regulatory molecules, there are a number of important questions yet to be answered. 1) Do changes in the human bile acid pool composition affect the activation of nuclear receptors and cell signaling pathways in the liver and intestines in a physiologically significant way? In this regard, humans are different from rodents and a number of other species, in that the human liver cannot 7α-hydroxylate deoxycholic acid, which allows for this hydrophobic secondary bile acid to accumulate to high levels in the bile acid pool in some individuals (60). The percentage of deoxycholic acid in bile, relative to other bile acids, appears to be a function of levels of 7α-dehydroxylating intestinal bacteria, colonic pH, and transit time. High levels of secondary bile acids in blood, bile, and feces have been correlated with an increased incidence of colon cancer (reviewed in Ref. 95). Finally, in humans, but not rodents, dietary habits can alter the ratio of taurine to glycine conjugation of bile acids in bile (61). Hence, both diet and gut microbiota may alter the human bile acid pool composition and possibly hepatic and intestinal physiology. The possible pathophysiological significance of bile acid pool changes is not yet clear. However, current thinking suggests that inhibiting the formation of secondary bile acids in the colon will yield a more hydrophilic bile acid pool with less toxicity and pathophysiology to cells in the liver and gastrointestinal tract. 2) There are as yet unidentified Gαi protein-coupled receptors in hepatocytes and other epithelial cells in the gastrointestinal tract that are activated by conjugated bile acids. These putative G protein-coupled receptor(s) activate the AKT (insulin signaling pathway) and ERK 1/2 pathway by poorly defined mechanisms. In the liver, preliminary data indicate that activation of the AKT pathway by bile acids may be physiologically important in helping to control hepatic glucose metabolism. Identification of this putative Gαi protein-coupled receptor(s) may be important as it could be a target for drug development for controlling hepatic glucose metabolism. It is not yet clear if the same Gαi protein-coupled receptor activating the AKT pathway activates the ERK 1/2 signaling pathway in the liver. Bile acid-activated G protein-coupled receptors may also be relevant to certain pathophysiological processes in the gastrointestinal tract. For example, conjugated bile acids have been reported to activate the AKT and ERK 1/2 signaling pathways in most epithelial cells of the gastrointestinal tract, including those in the esophagus. This may have relevance to Barrett's esophagus and the development of esophageal cancer as the inappropriate activation of these signaling pathways may enhance cell proliferation following bile reflux. Conjugated bile acids may also play a role in the regulation of cholangiocyte proliferation in cholestatic liver disease through activation of these cell signaling pathways. Hence, it will be important to identify and characterize the Gαi protein-coupled receptors activated by conjugated bile acids to gain a full understanding of their role in health and disease.
Acknowledgments
The authors thank Elaine Studer for careful reading and editing of this manuscript.
Footnotes
Abbreviations:
- ASM
- acidic sphingomyelinase
- CYP7A1
- cholesterol 7α-hydroxylase
- D2
- type 2 iodothyronine deiodinase
- FXR
- farnesoid X receptor
- HNF4α
- hepatocyte nuclear factor 4α
- IL-1
- interleukin 1
- JNK
- c-jun N-terminal kinase
- LXR
- liver X receptor
- PKA
- protein kinase A
- PKC
- protein kinase C
- PXR
- preganane X receptor
- SHP
- short heterodimer partner
- SREBP
- sterol-regulatory element binding protein
- TNF-α
- tumor necrosis factor-α
- VDR
- vitamin D receptor
This work was supported by National Institutes of Health Grant DK-057543 and R01 DK52825.
REFERENCES
- 1.Vlahcevic Z. R., Heuman D. M., Hylemon P. B. 1996. Physiology and pathophysiology of enterohepatic circulation of bike acids. Zakim D., Boyer T. D., editors W. B. Sanders Co., Philadelphia, PA: 376–417 [Google Scholar]
- 2.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365 [DOI] [PubMed] [Google Scholar]
- 3.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Wilson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368 [DOI] [PubMed] [Google Scholar]
- 4.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553 [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191 [DOI] [PubMed] [Google Scholar]
- 6.Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., Mackenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. 2001. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 98: 3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie W., Radominska-Pandya A., Shi Y., Simon C. M., Nelson M. C., Ong E. S., Waxman D. J., Evans R. M. 2001. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. USA. 98: 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science. 296: 1313–1316 [DOI] [PubMed] [Google Scholar]
- 9.Craven P. A., Pfanstiel J., DeRubertis F. R. 1987. Role of activation of protein kinase C in the stimulation of colonic epithelial proliferation and reactive oxygen formation by bile acids. J. Clin. Invest. 79: 532–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pongracz J., Clark P., Neoptolemos J. B., Lord J. M. 1995. Expression of protein kinase C isoenzymes in colorectal cancer tissue and their differential activation by different bile acids. Int. J. Cancer. 61: 35–39 [DOI] [PubMed] [Google Scholar]
- 11.Stravitz R. T., Rao Y. P., Vlahcevic Z. R., Gurley E. C., Jarvis W. D., Hylemon P. B. 1996. Hepatocellular protein kinase C activation by bile acids: implication for regulation of cholesterol 7-alpha-hydroxylase. Am. J. Physiol. 271: G293–G303 [DOI] [PubMed] [Google Scholar]
- 12.Rao Y. P., Stravitz R. T., Vlahcevic Z. R., Gurley E. C., Sando J. J., Hylemon P. B. 1997. Activation of protein kinase C alpha and delta by bile acids: correlation with bile acid structure and diacylglycerol formation. J. Lipid Res. 38: 2446–2454 [PubMed] [Google Scholar]
- 13.Gupta S., Stravitz R. T., Dent P., Hylemon P. B. 2001. Down-regulation of cholesterol 7-alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276: 15816–15822 [DOI] [PubMed] [Google Scholar]
- 14.Qiao L., Studer E., Leach K., McKinstry R., Gupta S., Decker R., Kukreja R., Valerie K., Nagarkatti P., El Deiry W., et al. 2001. Deoxycholic acid (DCA) causes ligand-independ activation of epidermal growth factor receptor (EGFR) and FAS receptor in primary hepatocytes: inhibition of EGFR/mitogen activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol. Biol. Cell 12: 2629–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao Y. P., Studer E. J., Stravitz R. T., Gupta S., Qiao L., Dent P., Hylemon P. B. 2002. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 35: 307–314 [DOI] [PubMed] [Google Scholar]
- 16.Yoon J. H., Higuchi H., Werneburg N. W., Kaufmann S. H., Gores G. J. 2002. Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology. 122: 985–993 [DOI] [PubMed] [Google Scholar]
- 17.Werneburg N. W., Yoon J-H., Higuchi H., Gores G. J. 2003. Bile acids activate EGF receptor via a TGF-α-dependent mechanism in human cholangiocyte cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 285: G31–G36 [DOI] [PubMed] [Google Scholar]
- 18.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. 2002. Identification of a membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298: 714–719 [DOI] [PubMed] [Google Scholar]
- 19.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278: 9435–9440 [DOI] [PubMed] [Google Scholar]
- 20.Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. 2006. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 439: 484–489 [DOI] [PubMed] [Google Scholar]
- 21.Thomas C., Auwerx J., Schoonjans K. 2008. Thyroid economy-regulation, cell biology, thyroid hormone metabolism and action: the special edition: metabolic effects of thyroid hormones. Thyroid. 18: 167–17418279017 [Google Scholar]
- 22.Keitel V., Reinehr R., Gatsios P., Rupprecht C., Görg B., Selbach O., Häussinger D., Kubitz R. 2007. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 45: 695–704 [DOI] [PubMed] [Google Scholar]
- 23.Dent P., Fang Y., Gupta S., Studer E., Mitchell C., Spiegel S., Hylemon P. B. 2005. Conjugated bile acids promote ERK 1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 42: 1291–1299 [DOI] [PubMed] [Google Scholar]
- 24.Fang Y., Studer E., Mitchell C., Grant S., Pandak W. M., Hylemon P. B., Dent P. 2007. Conjugated bile acids regulate hepatocyte glycogen synthase activity in vitro and in vivo via G-alpha-I signaling. Mol. Pharmacol. 71: 1122–1128 [DOI] [PubMed] [Google Scholar]
- 25.Inagaki T., Moschetta A., Lee Y-K., Peng L., Zhao G., Downes M., Yu R. T., Shelton J. M., Richardson J. A., Repa J. J., et al. 2006. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA. 103: 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers F., Stroeve J. H. M., Caron S., Staels B. 2007. Bile acids, farnesoid X receptor, atherosclerosis and metabolic control. Curr. Opin. Lipidol. 18: 289–297 [DOI] [PubMed] [Google Scholar]
- 27.Gilardi F., Mitro N., Godio C., Scotti E., Caruso D., Crestani M., De Fabiani E. 2007. The pharmacological exploitation of cholesterol 7α-hydroxylase, the key enzyme in bile acid synthesis: from binding resins to chromatin remodeling to reduce plasma cholesterol. Pharmacol. Ther. 116: 449–472 [DOI] [PubMed] [Google Scholar]
- 28.Russell D. W.2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174 [DOI] [PubMed] [Google Scholar]
- 29.Lundell K., Wikvall K. 2008. Species-specific and age-dependent bile acid composition: aspects of CYP8B and CYP4A subfamilies in bile acid biosynthesis. Curr. Drug Metab. 9: 323–331 [DOI] [PubMed] [Google Scholar]
- 30.Axelson M., Sjövall J. 1990. Potential bile acid precursors in plasma-possible indicators of biosynthetic pathways to cholic acid and chenodeoxycholic acid in man. J. Steroid Biochem. 36: 631–640 [DOI] [PubMed] [Google Scholar]
- 31.Pandak W. M., Ren S., Marques D., Hall E., Redford K., Mallonee D., Bodhan P., Heuman D., Gil G., Hylemon P. 2002. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J. Biol. Chem. 277: 48158–48164 [DOI] [PubMed] [Google Scholar]
- 32.Ren S., Hylemon P. B., Marques D., Gurley E., Bodhan P., Hall E., Redford K., Gil G., Pandak W. M. 2004. Overexpression of cholesterol transporter StAR increases in vivo rate of bile acid synthesis in the rat and mouse. Hepatology. 40: 910–917 [DOI] [PubMed] [Google Scholar]
- 33.Alpy F., Tomasetto C. 2005. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118: 2791–2801 [DOI] [PubMed] [Google Scholar]
- 34.Hall E. A., Ren S., Hylemon P. B., Rodriguez-Agudo D., Redford K., Marques D., Kang D., Gil G., Pandak W. M. 2005. Detection of the steroidogenic acute regulatory protein, StAR, in human liver cells. Biochim. Biophys. Acta. 1733: 111–119 [DOI] [PubMed] [Google Scholar]
- 35.Luoma P. V.2008. Cytochrome P450 and gene activation—from pharmacology to cholesterol elimination and regression of atherosclerosis. Eur. J. Clin. Pharmacol. 64: 841–850 [DOI] [PubMed] [Google Scholar]
- 36.Norlin M., von Bahr S., Björkhem I., Wikvall K. 2003. On the substrate specificity of human CYP27A1: implications for bile acid and cholestanol formation. J. Lipid Res. 44: 1515–1522 [DOI] [PubMed] [Google Scholar]
- 37.Li X., Pandak W. M., Erickson S. K., Ma Y., Yin L., Hylemon P., Ren S. 2007. Biosynthesis of the regulatory oxysterol, 5-cholesten-3-beta,25-diol 3-sulfate, in hepatocytes. J. Lipid Res. 48: 2587–2596 [DOI] [PubMed] [Google Scholar]
- 38.Ma Y., Xu L., Rodiguez-Agudo D., Li X., Heuman D. M., Hylemon P. B., Pandak W. M., Ren S. 2008. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1- signaling pathway. Am. J. Physiol. Endocrinol. Metab. 295: E1369–E379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiévet C., Staels B. 2009. Liver X receptor modulators: Effects on lipid metabolism and potential use in the treatment of atherosclerosis. Biochem. Pharmacol. 77: 1316–1327 [DOI] [PubMed] [Google Scholar]
- 40.Chen W., Chen G., Head D. L., Mangelsdorf D. J., Russell D. W. 2007. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 5: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong M. H., Oelkers P., Craddock A. L., Dawson P. A. 1994. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J. Biol. Chem. 269: 1340–1347 [PubMed] [Google Scholar]
- 42.Craddock A. L., Love M. W., Daniel R. W., Kirby L. C., Walters H. C., Wong M. H., Dawson P. A. 1998. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am. J. Physiol. 274: G157–G169 [DOI] [PubMed] [Google Scholar]
- 43.Dawson P. A., Hubbert M., Haywood J., Craddock A. L., Zerangue N., Christian W. V., Ballatori N. 2005. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J. Biol. Chem. 280: 6960–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballatori N., Christian W. V., Lee J. Y., Dawson P. A., Soroka C. J., Boyer J. L., Madejczyk M. S., Li N. 2005. OST alpha-OST beta a major basolateral bile acid and steroid transporter in human intestinal, renal and biliary epithelia. Hepatology. 42: 1270–1279 [DOI] [PubMed] [Google Scholar]
- 45.Wang W., Seward D. J., Li L., Boyer J. L., Ballatori N. 2001. Expression cloning of two genes that together mediate organic solute and steroid transport in the liver of a marine vertebrate. Proc. Natl. Acad. Sci. USA. 98: 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kullak-Ublick G. A., Stieger B., Meier P. J. 2004. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 126: 322–342 [DOI] [PubMed] [Google Scholar]
- 47.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225 [DOI] [PubMed] [Google Scholar]
- 49.Song K-H., Li T., Owsley E., Strom S., Chiang J. Y. L. 2009. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7α-hydroxylase gene expression. Hepatology. 49: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672 [DOI] [PubMed] [Google Scholar]
- 51.Rao A., Haywood J., Craddock A. L., Belinsky M. G., Kruh G. D., Dawson P. A. 2008. The organic solute transporter alpha-beta, Ost-alpha-Ost-beta, is essential for intestinal bile acid transport and homeostasis. Proc. Natl. Acad. Sci. USA. 105: 3891–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu C., Wang F., Kan M., Jin C., Jones R. B., Weinstein M., Deng C. X., McKeehan W. L. 2000. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275: 15482–15489 [DOI] [PubMed] [Google Scholar]
- 53.Ito S., Fujimori T., Furuya A., Satoh J., Nabeshima Y., Nabeshima Y. 2005. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 115: 2202–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1 and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526 [DOI] [PubMed] [Google Scholar]
- 55.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis of feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515 [DOI] [PubMed] [Google Scholar]
- 56.Mataki C., Magnier B. C., Houten S. M., Annicotte J-S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., et al. 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27: 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y-K., Schmidt D. R., Cummins C. L., Choi M., Peng L., Zhang Y., Goodwin B., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. 2008. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol. Endocrinol. 22: 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song K. H., Chiang J. Y. 2006. Glucagon and cAMP inhibit cholesterol 7-alpha-hydroxylase (CYP7A1) gene expression in human hepatocytes: discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 43: 117–125 [DOI] [PubMed] [Google Scholar]
- 59.Li T., Jahan A., Chiang J. Y. 2006. Bile acids and cytokines inhibit the human cholesterol 7-alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridlon J. M., Kang D. J., Hylemon P. B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259 [DOI] [PubMed] [Google Scholar]
- 61.Hardison W. G.1978. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 75: 71–75 [PubMed] [Google Scholar]
- 62.Gupta S., Natarajan R., Payne S. G., Studer E. J., Spiegel S., Dent P., Hylemon P. B. 2004. Deoxycholic acid activates the c-Jun N-terminal kinase pathway via FAS receptor activation in primary hepatocytes. J. Biol. Chem. 279: 5821–5828 [DOI] [PubMed] [Google Scholar]
- 63.Miyake J. H., Wang S. L., Davis R. A. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7-alpha-hydroxylase. J. Biol. Chem. 275: 21805–21808 [DOI] [PubMed] [Google Scholar]
- 64.Han S. I., Studer E., Gupta S., Fang Y., Qiao L., Li W., Grant S., Hylemon P. B., Dent P. 2004. Bile acids enhance the activity of the insulin receptor and glycogen synthase in primary rodent hepatocytes. Hepatology. 39: 456–463 [DOI] [PubMed] [Google Scholar]
- 65.Frankenberg T., Miloh T., Chen F. Y., Ananthanarayanan M., Sun A. Q., Balasubramaniyan N., Arias I., Setchell K. D., Suchy F. J., Shneider B. L. 2008. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology. 48: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gineste R., Sirvent A., Paumelle R., Helleboid S., Aquilina A., Darteil R., Hum D. W., Fruchart J. C., Staels B. 2008. Phosphorylation of the farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol. Endocrinol. 22: 2433–2447 [DOI] [PubMed] [Google Scholar]
- 67.Maloney P. R., Parks D. J., Haffner C. D., Fivush A. M., Chandra G., Plunket K. D., Creech K. L., Moore L. B., Wilson J. G., Lewis M. C., et al. 2000. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43: 2971–2974 [DOI] [PubMed] [Google Scholar]
- 68.Weigel N. L., Moore N. L. 2007. Steroid receptor phosphorylation: a key modulator of multiple receptor functions. Mol. Endocrinol. 21: 2311–2319 [DOI] [PubMed] [Google Scholar]
- 69.Staudinger J. L., Liehti K. 2008. Cell signaling and nuclear receptors: new opportunities for molecular pharmaceuticals in liver disease. Mol. Pharm. 5: 17–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamagata K., Daitoku H., Shimamoto Y., Matsuzaki H., Hirata K., Ishida J., Fukamizu A. 2004. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J. Biol. Chem. 279: 23158–23165 [DOI] [PubMed] [Google Scholar]
- 71.Park M. J., Kong H. J., Kim H. Y., Kim H. H., Kim J. H., Cheong J. H. 2007. Transcriptional repression of the gluconeogenic gene PEPCK by the orphan nuclear receptor SHP through inhibitory interaction with C/EBPα. Biochem. J. 402: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smrcka A. V.2008. G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell. Mol. Life Sci. 65: 2191–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen A., Bouscarel B. 2008. Bile acids and signal transduction: role in glucose homeostasis. Cell. Signal. 20: 2180–2197 [DOI] [PubMed] [Google Scholar]
- 74.De Fabiani E., Mitro N., Gilardi F., Caruso D., Galli G., Crestani M. 2003. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J. Biol. Chem. 278: 39124–39132 [DOI] [PubMed] [Google Scholar]
- 75.Ma K., Saha P. K., Chan L., Moore D. D. 2006. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 116: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duran-Sandoval D., Cariou B., Percevault F., Hennuyer N., Grefhorst A., van Dijk T. H., Gonzalez F. J., Fruchart J-C., Kuipers F., Staels B. 2005. The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J. Biol. Chem. 280: 29971–29979 [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y., Lee F. Y., Barrera G., Lee H., Vales C., Gonzalez F. J., Willson T. M., Edwards P. A. 2006. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA. 103: 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L., Huang J., Saha P., Kulkarni R. N., Hu M., Kim Y., Park K., Chan L., Rajan A. S., Lee I., et al. 2006. Orphan receptor small heterodimer partner is an important mediator of glucose homeostasis. Mol. Endocrinol. 20: 2671–2681 [DOI] [PubMed] [Google Scholar]
- 79.Grundy S. M., Ahrens E. H., Jr., Salen G. 1971. Interruption of the enterohepatic circulation of bile acids in man: comparative effects of cholestyramine and ileal exclusion on cholesterol metabolism. J. Lab. Clin. Med. 78: 94–121 [PubMed] [Google Scholar]
- 80.Nestel P. J., Grundy S. M. 1976. Changes in plasma triglyceride metabolism during withdrawal of bile. Metabolism. 25: 1259–1268 [DOI] [PubMed] [Google Scholar]
- 81.Angelin B., Einarsson K., Hellstrom K., Leijd B. 1978. Effects of cholestyramine and chendeoxycholic acid on the metabolism of endogenous triglyceride in hyperlipoproteinemia. J. Lipid Res. 19: 1017–1024 [PubMed] [Google Scholar]
- 82.Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. 2004. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREPB-1c. J. Clin. Invest. 113: 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Castellani L. W., Sinal C. J., Gonzalez F. J., Edwards P. A. 2004. Peroxisome proliferators-activated receptor gamma coactivator 1alpha (PGC-alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 18: 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cha J. Y., Repa J. J. 2007. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 282: 743–751 [DOI] [PubMed] [Google Scholar]
- 87.Kast H. R., Nguyen C. M., Sinal C. J., Jones S. A., Laffitte B. A., Reue K., Gonzalez F. J., Willson T. M., Edwards P. A. 2001. Farnesoid X-activated receptor induces apolipoprotein C–II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Mol. Endocrinol. 15: 1720–1728 [DOI] [PubMed] [Google Scholar]
- 88.Qiao L., Yacoub A., Studer E., Gupta S., Pei X. Y., Grant S., Hylemon P. B., Dent P. 2002. Inhibition of the MAPK and PI3K pathways enhances UDCA-induced apoptosis in primary rodent hepatocytes. Hepatology. 35: 779–789 [DOI] [PubMed] [Google Scholar]
- 89.Foord S. M., Bonner T. I., Neubig R. R., Rosser E. M., Pin J. P., Davenport A. P., Spedding M., Marmar A. J. 2005. International union of pharmacology. XLVI. G protein-coupled receptor list. Pharmacol. Rev. 57: 279–288 [DOI] [PubMed] [Google Scholar]
- 90.Maruyama T., Tanaka K., Suzuki J., Miyoshi H., Harada N., Nakamura T., Miyamoto Y., Kanatani A., Tamai Y. 2006. Targeted disruption of the G protein coupled bile acid receptor 1 (Gpbar1/M-bar) in mice. J. Endocrinol. 191: 197–205 [DOI] [PubMed] [Google Scholar]
- 91.Raufman J. P., Zimniak P., Bartoszko-Malik A. 1998. Lithocholyltaurine interacts with cholinergic receptors on dispersed chief cells from guinea pig stomach. Am. J. Physiol. 274: G997–G1004 [DOI] [PubMed] [Google Scholar]
- 92.Wess J., Eglan R. M., Gautam D. 2007. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat. Rev. Drug Discov. 6: 721–733 [DOI] [PubMed] [Google Scholar]
- 93.Raufman J-P., Chen Y., Cheng K., Compadre C., Compadre L., Zimniak P. 2002. Selective interaction of bile acids with muscarinic receptors: a case of molecular mimicry. Eur. J. Pharmacol. 457: 77–84 [DOI] [PubMed] [Google Scholar]
- 94.Raufman J-P., Cheng K., Zimniak P. 2003. Activation of muscarinic receptor signaling by bile acids. Dig. Dis. Sci. 48: 1431–1444 [DOI] [PubMed] [Google Scholar]
- 95.McGarr S. E., Ridlon J. M., Hylemon P. B. 2005. Diet, anaerobic bacterial metabolism and colon cancer risk: a review of the literature. J. Clin. Gastroenterol. 39: 98–109 [PubMed] [Google Scholar]