Abstract
Nuclear retinoic acid receptors (RARs) are transcriptional regulators controlling the expression of specific subsets of genes in a ligand-dependent manner. The basic mechanism for switching on transcription of cognate target genes involves RAR binding at specific response elements and a network of interactions with coregulatory protein complexes, the assembly of which is directed by the C-terminal ligand-binding domain of RARs. In addition to this scenario, new roles for the N-terminal domain and the ubiquitin-proteasome system recently emerged. Moreover, the functions of RARs are not limited to the regulation of cognate target genes, as they can transrepress other gene pathways. Finally, RARs are also involved in nongenomic biological activities such as the activation of translation and of kinase cascades. Here we will review these mechanisms, focusing on how kinase signaling and the proteasome pathway cooperate to influence the dynamics of RAR transcriptional activity.
Introduction
Nuclear retinoic acid (RA) receptors (RARs) consist of three subtypes, α (NR1B1), β (NR1B2) and γ (NR1B3) encoded by separate genes [Germain et al., 2006a; Germain et al., 2006c], which function as ligand-dependent transcriptional regulators heterodimerized with retinoid X receptors (RXRs). For each subtype, there are at least 2 isoforms, which are generated by differential promoter usage and alternative splicing and differ only in their N-terminal regions. Activation of RARs by cognate ligands triggers transcriptional events leading to the activation or repression of subsets of target genes involved in cellular differentiation, proliferation and apoptosis ([Bour et al., 2006], and references therein).
The compounds that bind RARs and modulate their activity are referred to as retinoids. This generic term covers molecules that include natural vitamin A (retinol) metabolites and active synthetic analogs. Retinoids are hydrophobic, lipid-soluble, and of small size, so that they can easily cross the lipid bi-layer of cell membranes.
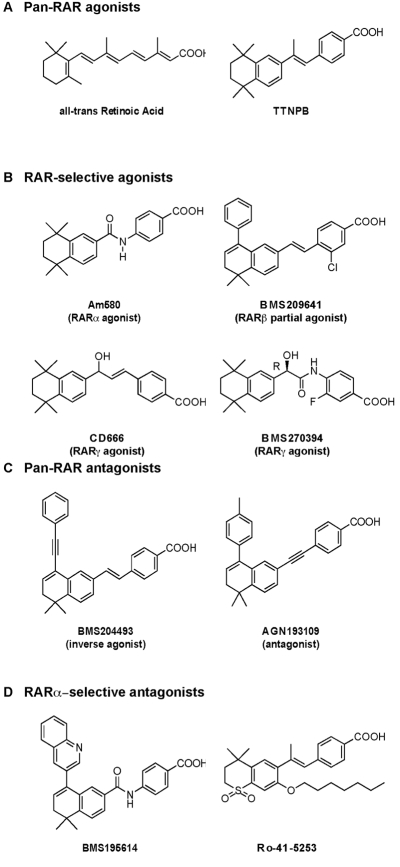
Natural retinoids, exemplified by all-trans RA, are produced in vivo from the oxidation of vitamin A [Chambon, 2005; Sporn et al., 1994] (Figure 1). An isomerization product of RA, 9-cis RA, also binds RARs with high affinity, but whether this compound is a natural bioactive retinoid remains controversial [Germain et al., 2006b].
Figure 1. Chemical structure, transcriptional activity, and selectivity of main retinoids.
See text for details.
Beyond the natural compounds, major research efforts in retinoid chemistry have been directed towards the identification of potent synthetic molecules and led to the generation of several classes of compounds with a panel of activities ranging from agonists to antagonists, selective or not to RAR subtypes [de Lera et al., 2007] (Figure 1).
Note that for (B) and (D) in Figure 1, a given ligand may be considered as selective for a certain RAR subtype when it exhibits an affinity difference greater than 100-fold between its primary target and other receptors (see the recommended usage of terms in the field of nuclear receptors [Germain et al., 2006c]).
RARs have a well-defined domain organization and structure, consisting mainly of a central DNA-binding domain (DBD) linked to a C-terminal ligand-binding domain (LBD) (Figure 2). In the past 20 years, it has been established that the basic mechanism for transcriptional regulation by RARs relies on DNA binding to specific sequence elements located in the promoters of target genes and on ligand-induced conformational changes in the LBD that direct the dissociation/association of several coregulator complexes [Chambon, 1996; Germain et al., 2003; Laudet and Gronemeyer, 2001; Lefebvre et al., 2005]. The description of the crystallographic structures of these domains and the characterization of the multiprotein complexes that specify the transcriptional activity of RARs provided a wealth of information on how these receptors regulate transcription. However, recent years have witnessed the importance of the ubiquitin-proteasome system and that of the N-terminal domain (NTD), which also interacts with specific coregulators, despite its native disordered structure [Bour et al., 2007]. Moreover, according to recent studies, RARs are involved in other nongenomic biological activities such as the activation of translation and of kinase cascades. These kinases target RARs and their coregulators, adding more complexity to the understanding of RAR-mediated transcription. In this review we will focus, in addition to the basic scenario (DNA and ligand binding, dynamics of coregulator exchanges at the LBD), on recent advances in the nongenomic effects of RA, and on how phosphorylation cascades, the NTD and the ubiquitin-proteasome system cooperate for fine-tuning RAR activity.
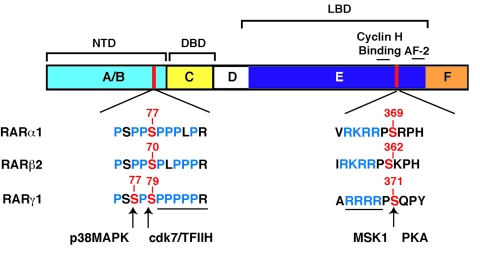
Figure 2. Schematic representation of the RAR proteins with the functional domains and the main phosphorylation sites.
RARs have a modular structure composed of six conserved regions designated A to F. The C region contains the DBD. The E region contains several domains, the LBD, the AF-2 domain, the cyclin H binding domain and the dimerisation domain. It also contains a phosphorylation site for several kinases (PKA and MSK1). The N-terminal domain (NTD) corresponds to regions A and B and contains a proline-rich motif with phosphorylation sites for Cdks and MAPKs.
Structure/function analysis
As with most nuclear receptors, RARs exhibit a modular structure composed of 6 regions of homology (designated A to F, from the N-terminal to the C-terminal end) (Figure 2) harboring specific functions [Chambon, 1996; Laudet and Gronemeyer, 2001]. Regions C and E, which encompass the DBD and the LBD, respectively, are the most conserved and important domains and govern the classical model of RAR transcriptional activity. In contrast, the A/B, D, and F regions are poorly conserved.
The DNA binding domain (DBD)
The DBD, which confers sequence-specific DNA recognition, is composed of two zinc-nucleated modules, two α-helices and a COOH-terminal extension (CTE) [Zechel et al., 1994a; Zechel et al., 1994b]. Helix 1 and helix 2 cross at right angles to form the core of the DBD folding into a single globular domain that has been determined by nuclear magnetic resonance and crystallographic studies [Lee et al., 1993]. The DBD includes several highly-conserved sequence elements, referred to as P, D, T and A boxes, that have been shown to define or contribute to the response element’s specificity, to a dimerization interface within the DBDs and to contacts with the DNA backbone and residues flanking the DNA core recognition sequence [Germain et al., 2003; Germain et al., 2006c].
RARs bind as asymmetrically-oriented heterodimers with RXRs, to specific DNA sequences or RA response elements (RAREs), composed typically of two direct repeats of a core hexameric motif, PuG (G/T) TCA [Germain et al., 2003; Leid et al., 1992; Mangelsdorf and Evans, 1995] and located in the regulatory sequences of target genes. The classical RARE is a 5bp-spaced direct repeat (referred to as DR5). However, the heterodimers also bind to direct repeats separated by 1bp (DR1) or 2bp (DR2). Note that RXR homodimers also bind to DR1.
RAREs have been identified in the promoters of a large number of RA target genes implicated in a wide variety of functions. For instance, the classical DR5 elements are found in the promoters of the RARβ2 gene itself [de The et al., 1990], of the CYP26A1 (cytochrome 450, family 26, subfamily a, polypeptide 1) gene [Loudig et al., 2000] and of several Homeobox (Hox) and hepatocyte nuclear factor (HNF) genes [Dupe et al., 1997; Qian et al., 2000]. DR2 elements were identified in the CRBPI (Cellular retinol binding protein I) and CRABPII (Cellular retinoic acid binding protein II) gene promoters [Durand et al., 1992; Smith et al., 1991]. The only natural DR1 element has been found in the rat CRBPII gene promoter [Mangelsdorf et al., 1991].
On DR2 and DR5 elements, in vitro, RXR occupies the 5’ hexameric motif, whereas the RAR partner occupies the 3’ motif (5’-RXR-RAR-3’) [Chambon, 1996; Laudet and Gronemeyer, 2001]. In contrast, on DR1 elements, the polarity is reversed, with the RAR DBD binding upstream and the RXR DBD downstream (5’-RAR-RXR-3’), switching the activity of the heterodimer from an activator to a repressor of target genes in the presence of RA.
So far, it has proved difficult to visualize the full-length RXR-RAR heterodimer in complex with DNA. However, the crystal structure of the DBDs in complex with DNA has been solved [Khorasanizadeh and Rastinejad, 2001; Rastinejad, 2001; Rastinejad et al., 2000]. Each DBD interacts with the DNA major groove at the level of a half-site through the P box of the first helix containing three exposed residues responsible for discrimination between different half-sites’ sequences. Then, they arrange head-to-tail, with cooperative contacts between the DBDs, leading to a mutual reinforcement of protein-protein and protein-DNA interactions. Depending on the DR spacing, different regions of the DBD of each receptor are used to create the dimerization interface, in order to achieve the required binding to the response elements. The heterodimeric DBD interface that is responsible for the binding of RXR-RAR heterodimers to DR5 elements involves the D box of the RXR second zinc-finger, and the tip of the RAR first zinc finger. However, when the heterodimers bind with reverse polarity to DR1 elements, they associate through the second zinc finger of RAR and the so-called T box (within the CTE- of RXR). This implies that the DBDs must be rotationally flexible with respect to the LBD dimerization interface and that the DNA curvature is different (for review, see [Renaud and Moras, 2000] and references therein). In conclusion, the DBDs of each heterodimerization partner dictate the specificity of RARE recognition and contribute through their dimerization to increase DNA binding efficiency.
The ligand binding domain (LBD)
The structures of the RAR LBDs are rather similar, as demonstrated by crystallographic studies [Moras and Gronemeyer, 1998; Renaud and Moras, 2000; Wurtz et al., 1996]. The LBD is formed by 12 conserved α helices and a β-turn (situated between H5 and H6). Helices 1-11 are folded into a three-layered, and parallel helical sandwich with H4, H5, H8, H9 and H11 sandwiched between H1, H2 and H3 on one side and H6, H7 and H10 on the other. In contrast, the C-terminal helix, H12, is more flexible and adopts conformations that may differ from one RAR subtype to the other (see below). The LBD is functionally complex, as it contains the ligand-binding pocket (LBP), the main dimerization domain and the ligand-dependent activation function-2 (AF-2).
The ligand-binding pocket (LBP). The ligand-binding pocket (LBP) comprises hydrophobic residues mainly from helices H3, H5, H11 and the β-sheet, and crystallographic studies revealed the structural basis of ligand recognition [Bourguet et al., 2000a; Li et al., 2003; Renaud et al., 1995]. The shape of the LBP matches the volume of the ligand, maximizing the hydrophobic contacts and contributing to the selectivity of ligand binding [Gehin et al., 1999; Klaholz et al., 2000; Klaholz et al., 1998].
Given that the precise contacts with ligands involve three divergent residues located in H3, H5 and H11, which are unique for each subtype receptor-cognate ligand pair, it has been possible to generate subtype-selective ligands [Germain et al., 2004] (Figure 1). For instance, the unique polar residues S232 and M272 located in the LBP of RARα and RARγ, respectively, have been exploited to develop ligands that are specific for RARα (Am580) or RARγ (BMS270394 or CD666). Via their amino group, such ligands establish hydrogen bonds with RARα S232 or RARγ M272, thereby increasing affinity and selectivity for RARα and RARγ, respectively. However, as the RARβ LBP does not contain specific polar residues, the development of RARβ-selective ligands is more challenging [Germain et al., 2004]. Nevertheless, due to differences in the volume and the shape of the LBPs of each RAR subtype, it has been possible to generate molecules with complex activities such as ligands that are RARβ agonists and RARα/RARγ antagonists, the larger size of RARβ LBP accounting for this mixed profile [Chen et al., 1995; Germain et al., 2004].
The heterodimerization surface. The heterodimerization surface involves residues from helices H7, H9, H10 and H11, as well as loops L8-9 and L9-10 [Bourguet et al., 2000b; Pogenberg et al., 2005]. Helices H9 and H10 contribute to more than 75% of the total dimerization surface and constitute the core of the dimer interface. It has been proposed that in RXR-RAR heterodimers, ligand binding affects the stability and propagation of signals across the heterodimerization surface, indicating that the LBP and the dimerization interface are in some way energetically linked [Brelivet et al., 2004].
The C-terminal helix 12, named AF-2. The C-terminal helix 12, named AF-2, controls the ability of RARs to interact with coregulators. The analysis of the crystal structures of the unliganded and ligand-bound LBDs of RXRα and RARα, respectively [Bourguet et al., 1995; Renaud et al., 1995], highlighted the crucial conformational flexibility of H12 and suggested a mechanism by which AF-2 becomes transcriptionally competent [Egea et al., 2001; Steinmetz et al., 2001]. Upon ligand binding, a series of intra-molecular interactions cause the repositioning of H11 in the continuity of H10 and the concomitant swinging of H12 which moves in a mouse trap model, sealing the “lid” of the LBP and being tightly packed against H3 and H4. Consequently, ligand binding is stabilized, and a new hydrophobic cleft is formed between H3, H4 and H12 that generates a defined interaction surface for transcriptional coactivators. In contrast, in the case of the RARβ and RARγ subtypes [Farboud et al., 2003; Farboud and Privalsky, 2004; Hauksdottir et al., 2003], biochemical studies proposed that, even in the absence of ligand, H12 would interact with H3 and adopt a constitutively closed conformation that approximates the conformation of liganded RARα. The importance of H12 in regulating coactivator and corepressor binding is detailed below.
The N-terminal AF-1 domain (NTD)
Early studies revealed the importance of the NTD of RARs, which corresponds to the A and B regions and includes the activation function AF-1, in the control of transcription of RA target genes [Nagpal et al., 1993; Nagpal et al., 1992]. However, they did not elucidate the underlying mechanism. It is interesting to note that within the NTD, the A region differs between the different subtypes and between isoforms [Chambon, 1996]. In contrast, the B region is rather conserved and depicts a proline-rich motif, which contains phosphorylation sites (Figure 2). Most importantly, proline-rich motifs can bind proteins with SH3 (Src-homology-3) or WW (tryptophan-tryptophan) domains, with phosphorylation preventing or favoring the interaction [Ball et al., 2005].
In contrast to the DBD and the LBD, there are still no high-resolution structures available for the NTD of RARs. Several biochemical and structural studies coupled to structure prediction algorithms suggested that the NTDs of RARs, as well as any member of the nuclear receptor family, are of naturally-disordered structure [Lavery and McEwan, 2005; Warnmark et al., 2003]. Most importantly, it has recently emerged that unstructured proteins or domains may be functional, undergoing transitions to more ordered states or folding into stable secondary or tertiary structures upon binding to DNA response elements or to coregulatory proteins [Dyson and Wright, 2005; Liu et al., 2006]. Moreover, disordered domains provide the flexibility that is needed for modification by enzymes such as kinases and ubiquitin-ligases [Dyson and Wright, 2005]. Such modifications may induce changes in the structural properties of the domain with profound impacts on its interactions with coregulators and/or on the dynamics of adjacent structural domains.
The D region
Poorly conserved, this region is considered to serve as a hinge between the DBD and the LBD, allowing rotation of the DBD. Therefore, it might allow the DBD and the LBD to adopt different conformations without creating steric hindrance problems. It also harbors nuclear localization signals.
The F region
This region extends C-terminal to helix 12 in RARs, but is absent in RXRs. It is highly variable in length and sequence among the different RAR subtypes and its three-dimensional structure is still unknown. Interestingly, region F is phosphorylated at multiple positions that might modify the properties of RARs [Bastien et al., 2000; Rochette-Egly et al., 1997]. Though the functions of region F are still poorly understood, it has been suggested that, in the absence of ligand, this region would stabilize H12 of the RARα subtype in an unclosed conformation, thereby enhancing corepressor binding [Farboud and Privalsky, 2004]. According to recent studies, this region would be also capable of binding to specific mRNA motifs [Poon and Chen, 2008].
The canonical RAR-mediated regulation of transcription with the LBD as the main actor
RARs are considered to be highly-regulated DNA-binding transcription factors that control transcription via several distinct mechanisms, including both repression and activation. Many years ago, it was established that after site-specific DNA binding, the final transcriptional activity of RARs depends on a set of associated proteins, the so-called corepressors and coactivators. From a molecular point of view, the discrimination between corepressors and coactivators is governed by the position of H12, which is directed by the ligand and contributes in a critical manner to the generation or removal of interaction surfaces.
The position of helix 12 governs the exposure of interaction surfaces for corepressors or coactivators
A recurring structural feature of corepressors and coactivators is the presence of highly-conserved motifs that are implicated in their recruitment at the LBD of RARs. The nuclear receptor corepressor (NCoR/NCoR1/RIP13) and silencing mediator for retinoid and thyroid hormone receptors (SMRT/NCoR2/TRAC) [Aranda and Pascual, 2001; Glass and Rosenfeld, 2000; Privalsky, 2004] contain in their C-terminal part two and three nuclear receptor interaction domains, respectively, with an LxxI/HIxxxI/L motif, which forms an extended α helix. Coactivators, which include essentially the p160 subfamily of steroid receptor coactivators (SRC), namely SRC-1 (also referred to as NCoA-1), SRC-2 (TIF-2, GRIP-1) and SRC-3 (pCIP, ACTR, AlB1, TRAM1, RAC3) [Glass and Rosenfeld, 2000; Lefebvre et al., 2005; Perissi et al., 1999], depict three copies of a highly-conserved LxxLL motif, which forms a short α helix.
In the absence of ligand, the surface of the RARα LBD presents a hydrophobic groove generated by H3, L3-4 and H4. It is worth noting that this surface is topologically related to that involved in coactivator interaction, but without H12 [Hu and Lazar, 1999] it can bind the extended LxxI/HIxxxI/L motif of corepressors. The N-terminal part of this motif extends in such a way that it masks the H12 interaction interface, thus explaining why the binding of corepressors and coactivators is mutually exclusive. Interestingly, the RARγ and RARβ subtypes interact poorly with corepressors [Farboud et al., 2003; Hauksdottir et al., 2003; Privalsky, 2004]. It has been proposed that hydrophobic interactions between H3 and H12 sequester H12 in a closed conformation, even in the absence of ligand, thus occluding the corepressor-docking site. Note that this closed conformation also prevents coactivator binding.
Upon ligand binding, RARα undergoes conformational changes and H12 becomes reoriented with a conserved glutamate residue forming a charge clamp with a lysine in H3. Such a charge clamp can specifically grip the ends of a helix of the specific length specified by the LxxLL motif of the coactivators, therefore allowing the leucine side chains to pack into the hydrophobic cavity. Because this ligand-activated charge clamp does not fit with the extended LxxI/HIxxxI/L motif of corepressors, it has been proposed that the alternative interactions of RARα with corepressors and coactivators originate from the difference in length of the interacting motifs that can be accommodated in the hydrophobic cleft in the two conformations [Germain et al., 2006c; Perissi et al., 1999].
Several crystal structures of RAR LBDs bound to synthetic retinoids revealed that ligand interactions with H11 and H12, or residues in their proximity, are primary determinants of helix 12 position, and that H12 can adopt not only the active and inactive positions, but also several intermediary positions. This implies that relatively subtle ligand modifications could significantly alter the conformation of the LBD and the H12 molecular switch, thereby generating distinct coregulator binding interfaces. Therefore, a panel of compounds comprising not only agonists, but also antagonists, inverse agonists and partial agonists, selective or not for RAR subtypes, have been generated [Altucci et al., 2007; de Lera et al., 2007; Vivat-Hannah and Zusi, 2005] (Figure 1).
From a structural point of view, agonists induce the repositioning of helix 12 and contribute in a critical manner to the surface recognized by the LxxLL boxes of coactivators. In contrast, antagonists which present a bulky side-chain that cannot be accommodated within the agonist binding cavity, prevent RARs from adopting this conformation. Antagonists include pure AF-2 antagonists, exemplified by BMS195614, a selective RARα antagonist (Figure 1). The crystal structure of the BMS195614-bound RARα LBD complex revealed that H12 packs on the groove formed by the carboxy terminal part of H3, Loop L3-4 and H4 [Bourguet et al., 2000a], which corresponds to the binding site for the coactivator LxxLL motif. Therefore, one can reason that BMS195614 may prevent interaction of coactivators. As the coactivator’s binding site overlaps with that for corepressors, BMS195614 may also impede the interaction of NCoR and SMRT corepressors.
In contrast, other antagonists, referred to as inverse agonists and exemplified by AGN193109 or BMS204493 (Figure 1), are highly effective at inducing corepressor interaction and thereby enhance silencing [Germain et al., 2009; Germain et al., 2002; Klein et al., 1996; Sanglier et al., 2004]. Although the molecular basis of inverse agonism has not been elucidated yet, H12 of RAR LBD bound to an inverse agonist should adopt an alternative position, which does not occlude the hydrophobic groove formed by H3 and H4.
Finally, AF-2 partial agonists-antagonists have been identified [Chen et al., 1995; Germain et al., 2004; Klein et al., 1996]. The interest in such compounds, which exhibit reduced efficacy compared to full agonists, is that they can act in a cell type-selective manner and/or activate only a subset of the cognate ligand-induced functions.
Transcriptional repression in the absence of ligand
When genes are silent, DNA is packaged into a highly-organized and compact nucleoprotein structure known as heterochromatin, which limits the access of promoter sequences to the transcriptional machinery and therefore impedes all the transcriptional steps [Richards and Elgin, 2002]. The basic unit of chromatin is the nucleosome, which consists of DNA wrapped around a protein core containing two copies each of four histone proteins. Protruding from the nucleosomes are the N-terminal “tails” of the core histones, whose interaction with DNA can be modulated upon covalent modifications (acetylation, phosphorylation, methylation, ubiquitination, etc.) [Narlikar et al., 2002].
According to the current model of gene regulation by RARs [Dilworth and Chambon, 2001], in a context of chromatin where the nucleosomes do not impede binding to RAREs, the RARα subtype is a strong repressor of target gene expression in the absence of ligand. This repressive activity reflects the ability of RARα to bind corepressors such as NCoR or SMRT (see above), which do not harbor intrinsic enzymatic activities, but reside in or recruit high molecular weight complexes endowed with histone deacetylase activity (HDACs). Such complexes have a well-characterized role in transcriptional repression by deacetylating lysine residues in the N-terminal tails of histones and generating a condensed chromatin structure over the target promoter. The corepressor complexes also contain other components such as TBL1 (Transducin β-like 1) and TBLR1 (TBL1-related protein 1), which serve as adaptors regulating corepressor assembly and function [Perissi et al., 2004].
It is worth noting that according to recent studies, in the absence of RA, RAR target genes have also been shown to interact with other kinds of repressors such as Topoisomerase IIβ [McNamara et al., 2008], Polycomb group proteins (PcG) [Gillespie and Gudas, 2007a; Gillespie and Gudas, 2007b] or calmodulin kinase IIγ (CaMKIIγ) [Si et al., 2007], which also dissociate in response to RA. PcG proteins act in large multimeric complexes that mediate gene silencing, while CaMKIIγ phosphorylates RARs, thereby enhancing their interaction with corepressors.
Transcriptional activation upon ligand binding: coregulator exchange at the LBD
To activate gene expression, RXR-RARα heterodimers bound at DR5 elements will have to contend with the repressive chromatin structures in order to allow the recruitment of the transcriptional machinery. Today, it is widely accepted that upon binding of a RARα agonist, bound corepressors are released and RXR-RARα heterodimers interact with coactivators of the p160 family (see above) that upon association with larger complexes with chromatin modifying and remodeling activities will decompact repressive chromatin.
Interestingly, an RXR agonist cannot activate RXR-RARα heterodimers unless the RARα partner is liganded first. Several models have been proposed to account for this phenomenon referred to as “subordination” or “silencing” (for review see [Greschik and Moras, 2003] and references therein), but most of them require more crystal structures for validation. Importantly, one model proposed that liganded RXR cannot dissociate corepressors from the RARα partner [Germain et al., 2002] and that the binding of agonists to RARα is essential for corepressor dissociation. In agreement with this idea, the RXR-RARβ heterodimer, in which RARβ only weakly recruits corepressors, responds better to a RXR agonist [Germain et al., 2002]. Finally, in the presence of both RAR and RXR agonists, there is synergy originating from the RAR agonist-induced dissociation of corepressors and the subsequent cooperative binding of coactivators to the two partners.
The p160 coactivators serve as adaptors recruiting other complexes with different enzymatic activities [Glass and Rosenfeld, 2000; Lefebvre et al., 2005; McKenna and O'Malley, 2002; Rosenfeld et al., 2006; Zhao et al., 2008]: (i) histone acetyltransferases (HATs) such as CBP/p300 (CREB binding protein) and p/CAF (p300/CBP-associated factor) (ii) histone methyl transferases (HMTs) such as CARM1 (coactivator-associated arginine methyltransferase 1) or PRMT1 (protein arginine methyl transferase 1), (iii) ubiquitinases/deubiquitinases and (iv) nucleosome remodeling complexes such as SWI/SNF (switch/sucrose non-fermenting). All these complexes alter the chromatin structure surrounding the promoter of target genes and create tags or binding sites that form a “histone code” read by particular effectors, which in turn mediate distinct outcomes [Sims and Reinberg, 2008]. In some examples, these chromatin marks function in a combinatorial manner. This code coordinates the recruitment of additional HATs or HMTs for further chromatin decompaction. It also orchestrates the recruitment of chromatin remodelers, which use the energy of ATP-hydrolysis to reposition nucleosomes through sliding them in cis or displacing them in trans, allowing the formation of nucleosome-free or nucleosome-spaced regions at the promoter [Narlikar et al., 2002]. It has also been proposed that activated RARs recruit the transcriptional machinery, including the multisubunit Mediator complex, RNA polymerase II and the general transcription factors [Bastien and Rochette-Egly, 2004; Dilworth and Chambon, 2001; Rochette-Egly, 2005; Rosenfeld et al., 2006]. This step would require the association of RARs with a specific subunit of the Mediator complex, which was identified as DRIP205/TRAP220, and which contains two LxxLL motifs [Lefebvre et al., 2005].
It must be noted that, in vivo, RARs appear to employ different programs for gene activation, depending on the target gene promoter context. Indeed, recent chromatin immunoprecipitation experiments demonstrated that in vivo, the Mediator complex and RNA PolII can already occupy with RARα the promoters of some endogenous genes, even in the absence of RA [Flajollet et al., 2006; Pavri et al., 2005; Perissi et al., 2004]. In that context, transcription initiation and the recruitment of the general transcription factors such as TFIIH at the promoter have been shown to depend on the dissociation from the Mediator complex, of the inhibitory cyclin-dependent kinase 8 (cdk8) subunit [Andrau et al., 2006; Elmlund et al., 2006]. In addition, recent studies from our laboratory revealed that in the absence of ligand, not all RA target promoters are occupied by RARs and that several events have to be initiated and coordinated to make the response elements available for RAR recruitment (see below and [Bruck et al., 2009]). Interestingly, in the context of promoters with RAREs that are distant from the transcription start site, RARs in association with coregulators and RNA PolII initiate the formation of DNA loops [Bruck et al., 2009].
Role of other unconventional coregulators
The model involving coactivators that positively modulate transcription has been challenged by the identification of three other unconventional coregulators, the receptor interacting protein of 140kDa (RIP140/NRIP1) [Augereau et al., 2006; Hu et al., 2004], the preferentially expressed antigen in melanoma (PRAME) [Epping et al., 2005] and the transcription intermediary factor-1 α (TIF1α/Trim24) [Le Douarin et al., 1995], which repress transcription despite their ligand-dependent recruitment to RARs via LxxLL motifs (one in TIF1α, seven in PRAME and nine plus a modified LxxML motif in RIP140) [Farooqui et al., 2003; Heery et al., 1997]. In fact, these coregulators would constitute a functional negative feedback mechanism limiting and/or ending RAR activity. The mechanism of TIF1α-mediated repression has not been elucidated yet [Khetchoumian et al., 2007]. However, the repressive activity of RIP140 has been attributed to the presence of four autonomous repressive domains, one of them recruiting HDACs [Wei et al., 2000], and that of PRAME to the recruitment of PcG proteins [Epping et al., 2005].
A plethora of other coregulator molecules for RA-bound RARs has been identified. Among these, are coactivators devoid of LxxLL motifs such as the thyroid receptor interacting protein-1 (TRIP1/SUG-1) [vom Baur et al., 1996], which is a subunit of the 19S regulatory subcomplex of the proteasome with an ATPase activity and which contributes to the transcription of RAR target genes [Ferry et al., 2009]. There are also coregulators that interact with domains other than the coactivators hydrophobic cleft formed between H3, H4 and H12. As such, cyclin H interacts with L8-9 and the beginning of H9 [Bour et al., 2005a], allowing the recruitment of TFIIH (see below). There is also CRABPII, which in association with cyclin D3 [Delva et al., 1999; Despouy et al., 2003], serves as a RA-channeling molecule to the receptor [Budhu and Noy, 2002; Dong et al., 1999]. Interestingly, upon deregulation of CRABPII expression, RA is shuttled to peroxisome proliferator-activated receptors (PPARβ/δ) rather than to RARs, by another intracellular lipid-binding protein, generating RA resistance [Schug et al., 2008].
New unconventional nongenomic effects of RARs: kinase activation and translation induction
Activation of the MAPK pathways
A new concept, which has recently developed is that, in addition to its well-established nuclear function to regulate gene expression, RA rapidly activates mitogen-activated protein kinases (MAPKs). Indeed, recent studies indicated that RA activates p38MAPK in several cell lines such as fibroblasts, mouse embryocarcinoma cells, mammary breast tumor cells and leukemia cells [Alsayed et al., 2001; Bruck et al., 2009; Gianni et al., 2002a; Gianni et al., 2006]. Subsequently, p38MAPK activates a downstream mitogen and stress-activated kinase, MSK1 [Bruck et al., 2009]. However, in neuronal cells (neuroblastoma cells, hippocampus neurons and P19 cells) and Sertoli cells, RA instead rapidly activates p42/44 MAPKs [Chen and Napoli, 2008; Gupta et al., 2008; Masia et al., 2007; Zanotto-Filho et al., 2008].
MAPK activation occurs very rapidly (within minutes after RA addition) subsequent to the prior activation of upstream kinases such as phosphoinositide 3-kinase (PI3K) [Bastien et al., 2006; Masia et al., 2007; Pan et al., 2005], RAC-1 [Alsayed et al., 2001] or protein kinase C δ (PKCδ) [del Rincon et al., 2004; Kambhampati et al., 2003]. This suggests an atypical, nongenomic activation event similar to that described for steroid receptors. In line with this new concept, though classically thought to reside in the nucleus, RARs have been recently reported to be present in the cytosol or in membranes, in association with PI3K or Src kinases [Dey et al., 2007; Masia et al., 2007]. Such observations strongly suggest a new paradigm by which RARs would integrate membrane/cytoplasm events that would orchestrate several post- t reductional modifications in order to fine-tune transcription (Figure 3 and Figure 4).
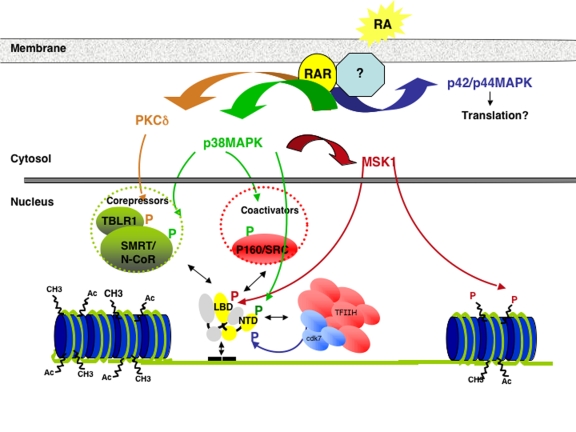
Figure 3. Crosstalk between kinase cascades and genomic pathways induced by RA.
In response to RA, PKCδ, p38MAPK and the downstream protein kinase MSK1 are activated through rapid nongenomic effects that occur in the cytosol or at the membrane. MSK1 phosphorylates RARα at S369 located in the LBD, subsequently allowing the docking of cyclin H within TFIIH and the formation of a RARα/TFIIH complex. Then the cdk7 subunit of TFIIH phosphorylates the NTD of RARα at S77. Finally, RARα phosphorylated and associated with TFIIH is recruited to response elements located in the promoter of target genes. P38MAPK, MSK1 and PKCδ also phosphorylate corepressors, coactivators and histones. All these phosphorylation processes cooperate to coordinate and fine-tune the dynamic exchanges between RARs, coactivators, corepressors and the promoters of target genes
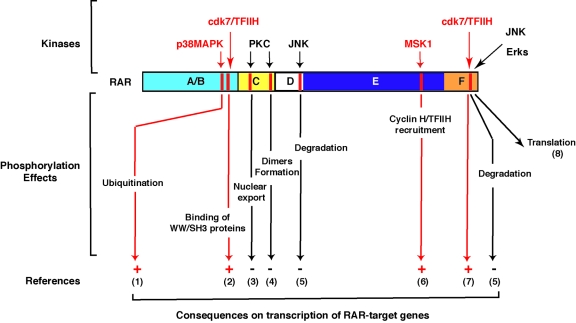
Figure 4. Recapitulation of the different signaling pathways involved in RAR phosphorylation.
The pathways that are induced by RA are in red. The consequences (positive or negative) of RAR phosphorylation on RA target genes transcription are also indicated. (1) Gianni et al., 2002a. (2) Bour et al., 2005b. (3) Sun et al., 2007. (4) Delmotte et al., 1999. (5) Srinivas et al., 2005. (6) Bruck et al., 2009. (7) Rochette-Egly et al., 1997. (8) Poon and Chen, 2008.
RNA binding and translation control
Very recent studies suggested another new nongenomic role of RARα as a RNA-binding protein. Two laboratories independently demonstrated that RA activates dendrite protein synthesis via a process that requires RARα (and not the other RAR subtypes) and is independent of transcriptional regulation [Chen and Napoli, 2008; Maghsoodi et al., 2008]. Indeed, they found that RARα is exported to neuronal dendrites, where it is associated with a subset of mRNAs such as the glutamate receptor 1 (GluR1) mRNA [Chen et al., 2008]. This binding, which is mediated by the C-terminal F region of RARα, directly represses the translation of these mRNA [Poon and Chen, 2008]. Interestingly, RA binding to RARα reduces its association with GluR1 mRNA and relieves translational repression. Such effects have been correlated to synaptic function and plasticity [Aoto et al., 2008]. As RA also activates MAPKs in neuronal cells [Chen and Napoli, 2008] and as the F region can be phosphorylated [Rochette-Egly et al., 1997], further studies are required to delineate the influence of the kinase signaling pathways on translation [Chen and Napoli, 2008]. Overall, these new nongenomic effects expand the scope of the biologic functions of RARα beyond its role as a regulator of gene transcription.
Crosstalk between kinase cascades and genomic pathways induced by RARs
The RA-activated p38MAPK/MSK1 pathway leads to a coordinated phosphorylation cascade targeting RARα, their coregulators and histones
Recent studies from our laboratory demonstrated that RARα becomes rapidly phosphorylated in response to RA at two serine residues, one being located in the LBD (serine 369) and the other one in the NTD (serine 77) [Bruck et al., 2009] (Figure 2). Interestingly, serine 369 is an exposed residue located between helices 9 and 10 within the LBD. It belongs to an arginine-lysine-rich motif that corresponds to a consensus phosphorylation motif for several kinases including the cyclic AMP-dependent protein kinase (PKA) and MSK1, suggesting that it might integrate several signaling pathways [Bruck et al., 2009; Gaillard et al., 2006]. In contrast, serine 77 located in the NTD, belongs to a proline-rich motif. The kinase responsible for phosphorylating this site has been identified as cdk7 [Rochette-Egly et al., 1997], the activity of which depends on its association with cyclin H and MAT1 to form the ternary cyclin-dependent kinase (CDK)-activating kinase (CAK) complex of TFIIH, a general transcription factor composed of 10 subunits [Giglia-Mari et al., 2004]. The correct positioning of the cdk7 kinase and thereby the efficiency of the NTD phosphorylation by cdk7 rely on the docking of cyclin H at a specific site of the LBD located in L8-9 and the N-terminal part of H9 [Bour et al., 2005a; Gaillard et al., 2006]. To our knowledge, it was the first example of cooperation between the N- and C-terminal domains of RARs through a kinase complex. The docking site of cyclin H is conserved between RARs, but not in other nuclear receptors and other cdk7 targets, and differs from the other cyclin docking sites. Therefore, it would elicit the specificity of RAR phosphorylation by cdk7.
Recently, Bruck et al. demonstrated that the RA-induced phosphorylation of RARα results from a coordinated phosphorylation cascade starting with the phosphorylation of serine 369 by MSK1 [Bruck et al., 2009]. As serine 369 is in close proximity to the docking site for cyclin H, phosphorylation of this residue induces an allosteric network, which promotes the binding of cyclin H [Gaillard et al., 2006] and thereby of cdk7, with a characteristic downstream consequence on the phosphorylation of the NTD at serine 77.
The phosphorylated serine residues located in the LBD and in the NTD are conserved between RARs. Accordingly, the RARγ subtype is also phosphorylated at the NTD (serine 79 in RARγ1 and serine 68 in RARγ2) by the cdk7 subunit of TFIIH [Bastien and Rochette-Egly, 2004]. Interestingly, RARγ (but not RARα) is also phosphorylated at an additional nearby serine residue (serine 77 in RARγ1 and serine 66 in RARγ2) by p38MAPK [Gianni et al., 2002a; Gianni et al., 2002b] (Figure 2). However, whether RARγ can be phosphorylated at the LBD site (serine 371 in RARγ1 and serine 360 in RARγ2) [Rochette-Egly et al., 1995] by MSK1 in response to RA remains to be demonstrated.
RA also activates p42/p44MAPK, but whether this kinase is able to phosphorylate RARs has not been elucidated yet. Note, however, that in neuronal cells, RA-activated p42/p44MAPK phosphorylates other activators of transcription, switching them into repressors of other gene programs [Gupta et al., 2008], while in Sertoli cells, they induce apoptosis [Zanotto-Filho et al., 2008].
Recently, the p160 SRC-3 coactivator [Gianni et al., 2006] and TBLR1, a component of the corepressor complexes [Perissi et al., 2008], have also been shown to be phosphorylated in response to RA by p38MAPK and PKCδ, respectively. Interestingly, the p160 family of coactivators [Lopez et al., 2001; Rowan et al., 2000; Wu et al., 2007; Wu et al., 2004; Yi et al., 2008], as well as p300/CBP [Vo and Goodman, 2001], RIP140 [Gupta et al., 2005; Huq et al., 2005] and SMRT [Jonas and Privalsky, 2004], are also targets for MAPKs or other kinases, but whether their phosphorylation occurs in response to RA will require further investigations. Finally, MSK1 was recently found to be recruited to RARα target promoters in the active phosphorylated form, leading to increased phosphorylation of serine 10 on histone H3 [Bruck et al., 2009].
RAR phosphorylation and the transcription of RA target genes
Early studies demonstrated the importance of TFIIH-mediated phosphorylation of RARs in the transcription of RA target genes [Bastien et al., 2000; Rochette-Egly et al., 1997]. This has been corroborated in studies using cells from patients suffering from Xeroderma pigmentosum, which harbor mutations in a TFIIH subunit [Keriel et al., 2002]. Such cells are characterized by a hypophosphorylation of the RARα subtype and a deficient RA response, but the underlying mechanism was not elucidated.
The recent results by Bruck et al. [Bruck et al., 2009] provided new information on how phosphorylation influences the transcriptional activity of RARα. Indeed, they demonstrated that the RA-induced phosphorylation cascade initiated by MSK1 and ending with the phosphorylation of the NTD by cdk7, promotes RARα recruitment to target promoters. Such results corroborate the importance of NTD phosphorylation in the transcriptional activity of RARs. However, it raises the question as to how phosphorylation of this unstructured domain can actually regulate DNA binding. In contrast, in the case of RARγ, the promoters are already occupied by the receptor in the absence of ligand [Gillespie and Gudas, 2007a; Gillespie and Gudas, 2007b] and RARγ phosphorylation occurs at the promoters (our unpublished results), suggesting that phosphorylation of the NTD might modulate transcription through controlling protein-protein interactions [Lavery and McEwan, 2005; Warnmark et al., 2003].
Phosphorylation of coactivators and corepressors that occurs in response to RA is also crucial for RARα transcriptional activity [Perissi et al., 2004; Rosenfeld et al., 2006]. Analogous to steroid nuclear receptors [Dennis and O'Malley B, 2005; Wu et al., 2005; Yi et al., 2005], it has been suggested that SRC-3 phosphorylation [Gianni et al., 2006] fine-tunes the dynamics of the exchanges of the coactivator with RARα and/or other coregulators. In contrast, phosphorylation of TBLR1 mediates ubiquitination and degradation of NCoR/SMRT [Perissi et al., 2008] and thereby overcomes SMRT/NCoR-dependent repression.
Finally, MSK1-mediated phosphorylation of H3 contributes to RARα target gene induction, probably as a chromatin mark accounting, in cooperation with other histone modifications, for the dissociation of repressive complexes and/or the recruitment of chromatin-remodeling complexes [Bruck et al., 2009; Vicent et al., 2006].
In conclusion, these recent findings challenged the conventional view of stable RAR-based, template-bound complexes and suggested a dynamic model with rapid, and sequential series of exchanges between RARs and coregulators at the promoter [Bour et al., 2007; Rochette-Egly, 2005] (Figure 3). Such exchanges, carefully coordinated by phosphorylations, would act as a “transcriptional time clock”, so that at the end, the correct proteins are present with the right activity, at the right place and at the right time.
RAR phosphorylation and other posttranslational modifications
According to recent studies, RARs are also targets for other modifications such as ubiquitination and methylation. Today, it is accepted that interplay between different posttranslational modifications is an important mechanism to achieve an integrated regulation of RAR activity, suggesting that a protein code similar to that proposed for histones could be applied to RARs [Sims and Reinberg, 2008]. The best example of crosstalk between modifications is the phosphorylation-dependent ubiquitination and subsequent proteasomal degradation of the RARγ subtype [Gianni et al., 2002a; Kopf et al., 2000]. Central to this phosphorylation function, was the observation that RARγ with the serines located in the NTD substituted with alanine residues exhibit reduced ubiquitination and degradation by the 26S proteasome upon cognate ligand binding. Whether phosphorylation controls the recruitment of the ubiquitin-proteasome machinery directly or indirectly through conformational changes would require further investigations.
RARs can also be methylated. Accordingly, the group of Li-Na Wei reported that RARα can be tri-methylated at a lysine residue located in the LBD [Huq et al., 2007] and mono-methylated at another lysine located in the DBD [Huq et al., 2008]. In both cases, methylation plays a positive role in RARα transcriptional activity, but the basal molecular mechanisms, as well as the methylases involved, remain to be characterized. Whether methylation functions in a combinatorial manner with phosphorylation also remains to be demonstrated. Finally, whether RARs, as most nuclear receptors, can also be targeted by other modifications such as acetylation or SUMOylation would require further investigations.
RAR phosphorylation in response to other cellular signaling pathways: abrogation of the RA response
Today, new roles for phosphorylation in regulation of RAR-mediated transcription are being discovered at an accelerated pace and it is increasingly clear that RAR phosphorylation can also occur in the absence of ligand. Indeed, several exogenous signals such as growth factors, insulin, stress or cytokines activate cytosolic kinase cascade pathways, ending at Akt, PKC or c-Jun N-terminal kinases (JNKs), which can enter the nucleus and phosphorylate RARs at residues different from those that are normally phosphorylated in response to RA (Figure 4). While such phosphorylation processes usually activate the transcriptional activity of steroid receptors [Faus and Haendler, 2006; Moggs and Orphanides, 2001], they instead inactivate RARα activity. As an example, PKC can phosphorylate the DBD of RARs, resulting in nuclear export [Sun et al., 2007] or in abrogation of RARs heterodimerization and binding to DNA [Delmotte et al., 1999]. Moreover, in cancers characterized by amplified or deregulated JNK or Akt activities, due to amplification of aberrant activity of receptor or cytoplasm tyrosine kinases [Blume-Jensen and Hunter, 2001], several residues located at the C-terminal end of RARα are phosphorylated, contributing to RARα degradation [Srinivas et al., 2005; Srinivas et al., 2006]. SMRT is also phosphorylated by Akt [Lefebvre et al., 2006], with a subsequent stabilization of the RAR-SMRT interaction. The nongenomic effects of RARs on the p38MAPK/MSK1 pathway are also abrogated [Bruck et al., 2009]. Finally, such cancers are RA-resistant [Neri et al., 2003; Tari et al., 2002].
In conclusion, RAR transcriptional activity, which is controlled by multiple players acting in a fine-tuned, spatially- and temporally-coordinated manner, requires the integrity of the signaling pathways.
New actors in RAR-mediated transcription: the ubiquitin-proteasome system and coregulators of the NTD
New roles for the ubiquitin-proteasome pathway, with and without proteolysis
A new aspect is that, in response to RA, RARs [Gianni et al., 2002a; Kopf et al., 2000; Zhu et al., 1999], as well as their coactivators such as SRC-3 [Gianni et al., 2006], are ubiquitinated and degraded by the 26S, which consists of the 20S proteolytic core capped by the 19S regulatory complex that recognizes the ubiquitinated proteins and prepares them for entry into the 20S core (reviewed in [Pickart and Cohen, 2004]). The degradation of RARs depends on the RA-induced recruitment at the AF-2 domain of the ubiquitin and/or degradation machineries [Kopf et al., 2000]. In the particular case of the RARγ subtype, degradation and ubiquitination of the receptor depends on the prior phosphorylation of the NTD [Gianni et al., 2002a; Kopf et al., 2000]. Degradation of SRC-3 also depends on the phosphorylation of the coactivator by p38MAPK [Gianni et al., 2006].
The exact role of the proteolytic activity of the proteasome in RAR-mediated transcription is not yet clear [Bastien and Rochette-Egly, 2004; Bour et al., 2007]. Since RAR and SRC-3 degradation occurs rather late after RA addition, when transcription has declined, a radical hypothesis would be that as for most “activators” of transcription, proteasomal degradation provides an efficient way to limit RAR function and/or to signal the end of the transcriptional process [Tansey, 2001]. However, according to the newly-arising concept that dynamic exchanges of coregulators are required for transcription to proceed, proteolysis might serve to clear out corepressors and/or coactivators so that other coregulators can subsequently bind [Collins and Tansey, 2006; Dennis and O'Malley B, 2005; Lipford and Deshaies, 2003]. In line with this theme, two components of the RAR coregulatory complexes, TBL1 and TBLR1, have been shown to serve as specific adaptors for the recruitment of the proteasome and the degradation of corepressors, in order to mediate their exchange for coactivators [Perissi et al., 2004; Perissi et al., 2008].
However, there is an increasing body of evidence that the ubiquitin-proteasome system would also play a role in the control of RA-mediated transcription without proteolysis. In that context, it is worth noting that SUG-1, one of the six ATPases of the 19S, interacts with RARs [vom Baur et al., 1996] and SRC-3 [Ferry et al., 2009], and as such, is recruited to the promoters of RA target genes, thereby contributing to their transcription [Ferry et al., 2009]. It has been proposed that the ATPase subunits of the 19S regulatory subcomplex would unfold or refold the components of the regulatory transcriptional complexes, such as the SAGA (spt-Ada-Gcn5-acetyl-transferase) complex [Lee et al., 2005], in order to facilitate their loading and/or removal at promoters. They would also reconfigure local chromatin in order to promote recruitment of appropriate histone modifiers [Ezhkova and Tansey, 2004]. Thus, the proteasome pathway would control the composition of complexes at the promoters of RAR target genes and the coordination of the different steps at the promoter, consistent with the idea that transcription is a dynamic process with continual exchange and turnover of coregulators [Collins and Tansey, 2006; Muratani and Tansey, 2003; Rochette-Egly, 2005].
A new role for the NTD of RARs via the binding of proteins with WW or SH3 domains
The NTD phosphorylation site belongs to a proline-rich motif (Figure 2). Most importantly, such motifs bind proteins with SH3 or WW domains, with phosphorylation preventing or favoring the interaction [Kay et al., 2000; Macias et al., 2002; Sudol et al., 2001; Zarrinpar and Lim, 2000]. In line with this, the phosphorylated NTD of RARα has been shown to bind the proline isomerase Pin1 (protein interacting with NIMA (never in mitosis A)) [Brondani et al., 2005; Gianni et al., 2009], a WW domain-containing protein which induces cis-trans isomerisation of the proline residues that follow the phosphorylated serines in order to create new specific recognition sites for interacting factors [Wulf et al., 2005].
A recent study performed in our laboratory has uncovered an interaction between the unphosphorylated NTD of the RARγ subtype and vinexin β [Bour et al., 2005b], which is one of a growing number of actin-binding proteins that are also present in the nucleus and modulate transcription [Bour et al., 2007]. Vinexin β is devoid of any enzymatic activity and is an adaptor characterized by the presence of three SH3 domains [Kioka et al., 2002], with the third one, proximal to the C-terminus, being involved in the interaction with the proline-rich motif of RARγ. As vinexin β is a repressor of RARγ-mediated transcription, it has been proposed that it would act as a scaffold between RARγ and nuclear proteins that interact with its two other SH3 domains [Mitsushima et al., 2006a; Mitsushima et al., 2006b], potentially forming a trimeric complex that maintains RARγ in an inactive state [Bour et al., 2007]. In support of this hypothesis, is the recent description of a direct interaction between vinexin β and SAFB2 (scaffold attachment factor B2 protein), a novel nuclear receptor corepressor [Townson et al., 2003]. Importantly, the interaction with vinexin β occurs only when the cdk7 phosphorylation site within the proline-rich motif, is not phosphorylated [Bour et al., 2005b] and vinexin β is released in response to RA (Lalevée and Rochette-Egly, unpublished results). The relative contribution of the ligand and of the cdk7-mediated phosphorylation of the NTD, in vinexin β dissociation, is a current matter of investigation in our laboratory.
Recently, other coregulators such as Acinus-S’, a nuclear protein implicated in apoptotic chromatin condensation and mRNA processing, have been shown to interact with the C-terminal part of the NTD, thereby repressing RAR transcriptional activity [Vucetic et al., 2008]. However, this interaction involves residues distinct from those of the proline-rich motif, suggesting another mechanism of action.
Transrepression by RARs (anti-AP1)
Evidence has accumulated over the past few years that the action of RARs is not restricted to the regulation (positive or negative) of cognate target gene expression, but also concerns several other gene programs by interfering with other nuclear receptors [Gupta et al., 2008] or with transcription factor complexes such as activating protein-1 (AP-1) (for review see [Germain et al., 2003; Lefebvre et al., 2005]). The AP-1 complex regulates the expression of several genes involved in oncogenic transformation and cell proliferation. Its activity is determined by the composition of the dimers, which include members of the fos (c-fos, FosB, Fra-1 and Fra-2) and jun (c-jun, junB and junD) families, and which is controlled by a complex network of phosphorylations. The inhibition of AP-1-driven transactivation by RARs is the prototype of transrepression and has been revealed in the context of several genes such as the collagenase and stromelysin genes [Lafyatis et al., 1990; Lin et al., 2000; Nicholson et al., 1990]. Generally, AP-1 inhibition is driven by liganded RARs, though AP-1 has been found to be inhibited in a ligand-independent manner by the RARβ subtype [De-Castro Arce et al., 2007; De-Castro Arce et al., 2004]. How RARs interfere with AP-1 activity and through which domains, is complex, and several models have been proposed ([Altucci and Gronemeyer, 2001; Lefebvre et al., 2005] and references therein), such as (i) a direct interaction with Jun/Fos family members, (ii) disruption of Jun-Fos dimerization (iii) competition with the recruitment of a common transcriptional coactivator such as CBP, or (iv) inhibition of JNKs, thereby preventing the phosphorylation-dependent activation of c-jun [Dedieu and Lefebvre, 2006]. Despite these models, the molecular basis of the anti-AP-1 activity of RARs has remained elusive and debated [Benkoussa et al., 2002; Suzukawa and Colburn, 2002].
In general, AP-1 inhibition correlates with the growth inhibitory effect of retinoids on tumors [Karamouzis and Papavassiliou, 2005]. Therefore, the capacity of RARs to inhibit AP-1-responsive genes seems to be the basis for the chemo preventive and chemotherapeutic effects of RA in the treatment of hyperproliferative diseases [Altucci and Gronemeyer, 2001; Dong et al., 1994; Li et al., 1996]. In this context, selective retinoids that dissociate the inhibition of AP-1 activity from the classical RARE-dependent activation of transcription have been generated [Chen et al., 1995; Fanjul et al., 1994; Resche-Rigon and Gronemeyer, 1998]. Such compounds are promising therapeutic agents and provide valuable tools to address the mechanism of the RAR/AP-1 crosstalk, the importance of which for growth control and cancer is now established.
Conclusion
The well-established function of RARs is to regulate gene expression in the nucleus. In this context, it is clear that switching on RAR transcriptional activity relies on conformational changes induced by ligand binding and on dynamic series of exchanges with coregulatory complexes. However, today there is mounting evidence that RARs have a wider spectrum of biological activities, through nonconventional, nongenomic mechanisms. Indeed, in response to RA, RARs can activate translation, as well as signaling pathways, independently of transcriptional mechanisms. Further work is needed to determine how RARs activate MAPKs and how RAR phosphorylation at the NTD controls the association/dissociation of specific partners with SH3 or WW domains to modulate the transcription of RA target genes. Up to now, only a few proteins have been shown to interact with the NTD of RARs [Bour et al., 2005b; Brondani et al., 2005; Vucetic et al., 2008] and further studies are required to investigate whether the NTDs of the different RAR subtypes can interact with other proteins involved in processes such as mRNA processing and splicing, or are endowed with chaperone or actin-binding activities. As phosphorylation of the RARγ NTD marks the receptor for ubiquitination and proteasomal degradation, ubiquitin ligases might also be interesting candidates. Ultimately, the challenge is to decipher at the molecular level how coregulator exchanges at the NTD cooperate with phosphorylations to modulate transcription.
Finally, given that aberrant retinoid signaling mechanisms have been associated with several diseases [Keriel et al., 2002] or cancers [Neri et al., 2003; Tari et al., 2002], more work is required to understand the molecular pathways that are controlled by RARs, notably those underlying the antiproliferative and anticancer activities of retinoids, with a particular effort on the role of phosphorylation processes. Moreover, in line with the improved use of retinoids in therapy, novel synthetic RAR ligands harboring increased selective properties with little toxicity compared with classical retinoids, will have to be generated. Finally, combination of retinoids with agents targeting kinases [Vitoux et al., 2007] or epigenetic modifications such as HDAC inhibitors [Cras et al., 2007; Egger et al., 2004; Feinberg and Tycko, 2004] are increasingly being sought in order to improve the retinoid response and/or to overcome retinoid resistance.
Acknowledgments
We are grateful to all the past and present members of the teams and to P. Chambon for constant support. This work was supported by funds from CNRS and INSERM. Work by CRE was also supported by the Agence Nationale pour la Recherche (ANR-05-BLAN-0390-02), the Institut National du Cancer (INCa-PL06-095, INCa-PL07-96099) and the Association pour la Recherche sur le Cancer (ARC P02/3/3169, A03/3/3103, A05/2/3139 and A07/4/3169). Work by PG was supported by funds from the University of Montpellier II and ARC.
Abbreviations
- AF-1
activation function 1
- AF-2
activation function 2
- AP-1
activating protein-1
- CAK
Cdk activating complex
- CAMK
calmodulin kinase
- CARM1
coactivator-associated arginine methyltransferase 1
- CBP
CREB (cyclic AMP response element binding protein) binding protein
- Cdk
cyclin-dependent kinase
- CRABP
cellular retinoic acid binding protein
- CRBP
cellular retinol binding protein
- CTE
C-terminal extension
- Cyp26A1
cytochrome 450, family 26, subfamily a, polypeptide 1
- DBD
DNA-binding domain
- DR
direct repeat
- DRIP
vitamin D receptor interacting protein
- GluR1
glutamate receptor 1
- HAT
histone acetyl transferase
- HDAC
histone deacetyl transferase
- HMT
histone methyl transferase
- HNF
hepatocyte nuclear factor
- Hox
homeobox gene
- JNK
c-jun N-terminal kinase
- LBD
ligand binding domain
- LBP
ligand binding pocket
- MAPK
mitogen-activated protein kinase
- MSK
mitogen and stress-activated kinase
- NCoR
nuclear receptor corepressor
- NTD
N-terminal domain
- p/CAF
p300/CBP-associated factor
- PcG
polycomb group
- PI3K
phosphoinositide 3-kinase
- PIN1
protein interacting with NIMA (never in mitosis A)
- PKC
protein kinase C
- PRAME
preferentially expressed antigen in melanoma
- PRMT1
protein arginine methyl transferase
- RA
retinoic acid
- RAR
nuclear retinoic acid receptor
- RXR
nuclear retinoid X receptor
- RARE
RA response element
- RIP140
receptor interacting protein of 140kDa
- SAFB2
scaffold attachment factor B2 protein
- SAGA
Spt-Ada-Gcn5-acetyl-transferase
- SH3
Src-homology 3
- SMRT
silencing mediator for retinoid and thyroid hormone receptors
- SRC
steroid receptor coactivator
- SWI/SNF
switch/sucrose non-fermenting
- TBL1
transducin β-like 1
- TBLR1
TBL1-related protein 1
- TIF1α
transcription intermediary factor-1 α
- TRAP
thyroid receptor associated protein
- TRIP-1
thyroid receptor interacting protein-1
- WW
trytophan-tryptophan
References
- Alsayed Y., Uddin S., Mahmud N., Lekmine F., Kalvakolanu D. V., Minucci S., Bokoch G., Platanias L. C. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J Biol Chem. 2001;276:4012–9.. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- Altucci L., Gronemeyer H. Nuclear receptors in cell life and death. Trends Endocrinol Metab. 2001;12:460–8.. doi: 10.1016/s1043-2760(01)00502-1. [DOI] [PubMed] [Google Scholar]
- Altucci L., Leibowitz M. D., Ogilvie K. M., de Lera A. R., Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- Andrau J. C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., van de Peppel J., Werner M., Holstege F. C. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–92. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Aoto J., Nam C. I., Poon M. M., Ting P., Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–20. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A., Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Augereau P., Badia E., Carascossa S., Castet A., Fritsch S., Harmand P. O., Jalaguier S., Cavailles V. The nuclear receptor transcriptional coregulator RIP140. Nucl Recept Signal. 2006;4:e024. doi: 10.1621/nrs.04024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L. J., Kuhne R., Schneider-Mergener J., Oschkinat H. Recognition of Proline-Rich Motifs by Protein-Protein-Interaction Domains. Angew Chem Int Ed Engl. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- Bastien J., Rochette-Egly C. Nuclear Retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Bastien J., Adam-Stitah S., Riedl T., Egly J. M., Chambon P., Rochette-Egly C. TFIIH interacts with the retinoic acid receptor γ and phosphorylates its AF-1-activating domain through cdk7. J Biol Chem. 2000;275:21896–904.. doi: 10.1074/jbc.M001985200. [DOI] [PubMed] [Google Scholar]
- Bastien J., Plassat J. L., Payrastre B., Rochette-Egly C. The phosphoinositide 3-kinase/Akt pathway is essential for the retinoic acid-induced differentiation of F9 cells. Oncogene. 2006;25:2040–2047. doi: 10.1038/sj.onc.1209241. [DOI] [PubMed] [Google Scholar]
- Benkoussa M., Brand C., Delmotte M. H., Formstecher P., Lefebvre P. Retinoic acid receptors inhibit AP1 activation by regulating extracellular signal-regulated kinase and CBP recruitment to an AP1-responsive promoter. Mol Cell Biol. 2002;22:4522–34. doi: 10.1128/MCB.22.13.4522-4534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P., Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–65.. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bour G., Gaillard E., Bruck N., Lalevee S., Plassat J. L., Busso D., Samama J. P., Rochette-Egly C. Cyclin H binding to the RAR{α} activation function (AF)-2 domain directs phosphorylation of the AF-1 domain by cyclin-dependent kinase 7. Proc Natl Acad Sci U S A. 2005a;102:16608–16613. doi: 10.1073/pnas.0505556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour G., Taneja R., Rochette-Egly C. Nuclear Receptors in development. Elsevier Press Inc; 2006. Mouse Embryocarcinoma F9 cells and Retinoic Acid. A model to study the molecular mechanisms of endodermal differentiation. pp. 211–253. [Google Scholar]
- Bourguet W., Vivat V., Wurtz J. M., Chambon P., Gronemeyer H., Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol.Cell. 2000b;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α [see comments] Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Germain P., Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000a;21:381–8.. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- Bour G., Lalevee S., Rochette-Egly C. Protein kinases and the proteasome join in the combinatorial control of transcription by nuclear retinoic acid receptors. Trends Cell Biol. 2007;17:302–309. doi: 10.1016/j.tcb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Bour G., Plassat J. L., Bauer A., Lalevee S., Rochette-Egly C. Vinexin {β} Interacts with the Non-phosphorylated AF-1 Domain of Retinoid Receptor {γ} (RAR{γ}) and Represses RAR{γ}-mediated Transcription. J Biol Chem. 2005b;280:17027–37. doi: 10.1074/jbc.M501344200. [DOI] [PubMed] [Google Scholar]
- Brelivet Y., Kammerer S., Rochel N., Poch O., Moras D. Signature of the oligomeric behaviour of nuclear receptors at the sequence and structural level. EMBO Rep. 2004;5:423–9. doi: 10.1038/sj.embor.7400119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondani V., Schefer Q., Hamy F., Klimkait T. The peptidyl-prolyl isomerase Pin1 regulates phospho-Ser77 retinoic acid receptor α stability. Biochem Biophys Res Commun. 2005;328:6–13. doi: 10.1016/j.bbrc.2004.12.130. [DOI] [PubMed] [Google Scholar]
- Bruck N., Vitoux D., Ferry C., Duong V., Bauer A., de The H., Rochette-Egly C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. Embo J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A. S., Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22:2632–41. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology ot retinoic acid receptors. FASEB.J. 1996;10:940–954. [PubMed] [Google Scholar]
- Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol. 2005;19:1418–28. doi: 10.1210/me.2005-0125. [DOI] [PubMed] [Google Scholar]
- Chen N., Napoli J. L. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. Faseb J. 2008a;22:236–45. doi: 10.1096/fj.07-8739com. [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Penco S., Ostrowski J., Balaguer P., Pons M., Starrett J. E., Reczek P., Chambon P., Gronemeyer H. RAR-specific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J. 1995;14:1187–1197. doi: 10.1002/j.1460-2075.1995.tb07102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Onisko B., Napoli J. L. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J Biol Chem. 2008b;283:20841–7. doi: 10.1074/jbc.M802314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins G. A., Tansey W. P. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Cras A., Darsin-Bettinger D., Balitrand N., Cassinat B., Soulie A., Toubert M. E., Delva L., Chomienne C. Epigenetic patterns of the retinoic acid receptor beta2 promoter in retinoic acid-resistant thyroid cancer cells. Oncogene. 2007;26:4018–24. doi: 10.1038/sj.onc.1210178. [DOI] [PubMed] [Google Scholar]
- de Lera A. R., Bourguet W., Altucci L., Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nat Rev Drug Discov. 2007;6:811–20. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- de The H., Vivanco-Ruiz M. M., Tiollais P., Stunnenberg H., Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- De-Castro Arce J., Soto U., van Riggelen J., Schwarz E., Hausen H. Z., Rosl F. Ectopic expression of nonliganded retinoic acid receptor β abrogates AP-1 activity by selective degradation of c-Jun in cervical carcinoma cells. J Biol Chem. 2004;279:45408–16. doi: 10.1074/jbc.M401818200. [DOI] [PubMed] [Google Scholar]
- De-Castro Arce J., Gockel-Krzikalla E., Rosl F. Retinoic acid receptor β silences human papillomavirus-18 oncogene expression by induction of de novo methylation and heterochromatinization of the viral control region. J Biol Chem. 2007;282:28520–9. doi: 10.1074/jbc.M702870200. [DOI] [PubMed] [Google Scholar]
- Dedieu S., Lefebvre P. Retinoids interfere with the AP1 signalling pathway in human breast cancer cells. Cell Signal. 2006;18:889–98. doi: 10.1016/j.cellsig.2005.08.001. [DOI] [PubMed] [Google Scholar]
- del Rincon S. V., Guo Q., Morelli C., Shiu H. Y., Surmacz E., Miller W. H. Retinoic acid mediates degradation of IRS-1 by the ubiquitin-proteasome pathway, via a PKC-dependant mechanism. Oncogene. 2004;23:9269–79. doi: 10.1038/sj.onc.1208104. [DOI] [PubMed] [Google Scholar]
- Delmotte M. H., Tahayato A., Formstecher P., Lefebvre P. Serine 157, a retinoic acid receptor α residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J Biol Chem. 1999;274:38225–31.. doi: 10.1074/jbc.274.53.38225. [DOI] [PubMed] [Google Scholar]
- Delva L., Bastie J. N., Rochette-Egly C., Kraiba R., Balitrand N., Despouy G., Chambon P., Chomienne C. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol Cell Biol. 1999;19:7158–67. doi: 10.1128/mcb.19.10.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis A. P., O'Malley B W. Rush hour at the promoter: How the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–51. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Despouy G., Bastie J. N., Deshaies S., Balitrand N., Mazharian A., Rochette-Egly C., Chomienne C., Delva L. Cyclin D3 is a cofactor of retinoic acid receptors, modulating their activity in the presence of cellular retinoic acid-binding protein II. J Biol Chem. 2003;278:6355–62. doi: 10.1074/jbc.M210697200. [DOI] [PubMed] [Google Scholar]
- Dey N., De P. K., Wang M., Zhang H., Dobrota E. A., Robertson K. A., Durden D. L. CSK controls retinoic acid receptor (RAR) signaling: a RAR-c-SRC signaling axis is required for neuritogenic differentiation. Mol Cell Biol. 2007;27:4179–97. doi: 10.1128/MCB.01352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth F. J., Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–54.. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Dong Z., Birrer M. J., Watts R. G., Matrisian L. M., Colburn N. H. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc Natl Acad Sci U S A. 1994;91:609–13. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D., Ruuska S. E., Levinthal D. J., Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–8. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- Dupe V., Davenne M., Brocard J., Dolle P., Mark M., Dierich A., Chambon P., Rijli F. M. In vivo functional analysis of the Hoxa-1 3' retinoic acid response element (3'RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Durand B., Saunders M., Leroy P., Leid M., Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Egea P. F., Rochel N., Birck C., Vachette P., Timmins P. A., Moras D. Effects of ligand binding on the association properties and conformation in solution of retinoic acid receptors RXR and RAR. J Mol Biol. 2001;307:557–76. doi: 10.1006/jmbi.2000.4409. [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Elmlund H., Baraznenok V., Lindahl M., Samuelsen C. O., Koeck P. J., Holmberg S., Hebert H., Gustafsson C. M. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:15788–93. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping M. T., Wang L., Edel M. J., Carlee L., Hernandez M., Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–47. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Ezhkova E., Tansey W. P. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–42. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Fanjul A., Dawson M. I., Hobbs P. D., Jong L., Cameron J. F., Harlev E., Graupner G., Lu X. P., Pfahl M. A new class of retinoids with selective inhibition of AP-1 inhibits proliferation. Nature. 1994;372:107–11. doi: 10.1038/372107a0. [DOI] [PubMed] [Google Scholar]
- Farboud B., Hauksdottir H., Wu Y., Privalsky M. L. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors β and γ interferes with corepressor recruitment and prevents transcriptional repression. Mol Cell Biol. 2003;23:2844–58. doi: 10.1128/MCB.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Privalsky M. L. Retinoic acid receptor-{α} is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–2853. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- Farooqui M., Franco P. J., Thompson J., Kagechika H., Chandraratna R. A., Banaszak L., Wei L. N. Effects of retinoid ligands on RIP140: molecular interaction with retinoid receptors and biological activity. Biochemistry. 2003;42:971–9. doi: 10.1021/bi020497k. [DOI] [PubMed] [Google Scholar]
- Faus H., Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–8. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Ferry C., Gianni M., Lalevee S., Bruck N., Plassat J. L., Raska I., Jr., Garattini E., Rochette-Egly C. Sug-1 plays proteolytic and non-proteolytic roles in the control of RA-target genes via its interaction with SRC-3. J Biol Chem. 2009;284:8127–8135. doi: 10.1074/jbc.M808815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajollet S., Lefebvre B., Rachez C., Lefebvre P. Distinct roles of the steroid receptor coactivator 1 and of MED1 in retinoid-induced transcription and cellular differentiation. J Biol Chem. 2006;281:20338–48. doi: 10.1074/jbc.M603023200. [DOI] [PubMed] [Google Scholar]
- Gaillard E., Bruck N., Brelivet Y., Bour G., Lalevee S., Bauer A., Poch O., Moras D., Rochette-Egly C. Phosphorylation by Protein Kinase A potentiates retinoic acid repeptor a activity by means of increasing interaction with and phosphorylation by cyclin H/cdk7. Proc Natl Acad Sci U S A. 2006;103:9548–9553. doi: 10.1073/pnas.0509717103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin M., Vivat V., Wurtz J. M., Losson R., Chambon P., Moras D., Gronemeyer H. Structural basis for engineering of retinoic acid receptor isotype-selective agonists and antagonists. Chem Biol. 1999;6:519–29. doi: 10.1016/S1074-5521(99)80084-2. [DOI] [PubMed] [Google Scholar]
- Germain P., Iyer J., Zechel C., Gronemeyer H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature. 2002;415:187–92.. doi: 10.1038/415187a. [DOI] [PubMed] [Google Scholar]
- Germain P., Gaudon C., Pogenberg V., Sanglier S., Van Dorsselaer A., Royer C. A., Lazar M. A., Bourguet W., Gronemeyer H. Differential action of coregulator interaction defines inverse retinoid agonists and neutral antagonists. Chem Biol. 2009:in press. doi: 10.1016/j.chembiol.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006a;58:712–25. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev. 2006c;58:760–72. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- Germain P., Altucci L., Bourguet W., Rochette-Egly C., Gronemeyer H. Pure Appl. Chem., 2003. Nuclear receptor superfamily: Principles of signaling; pp. 1619–1664. [Google Scholar]
- Germain P., Staels B., Dacquet C., Spedding M., Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006b;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- Germain P., Kammerer S., Perez E., Peluso-Iltis C., Tortolani D., Zusi F. C., Starrett J., Lapointe P., Daris J. P., Marinier A., de Lera A. R., Rochel N., Gronemeyer H. Rational design of RAR-selective ligands revealed by RARbeta crystal stucture. EMBO Rep. 2004;5:877–82. doi: 10.1038/sj.embor.7400235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni M., Kopf E., Bastien J., Oulad-Abdelghani M., Garattini E., Chambon P., Rochette-Egly C. Down-regulation of the Phosphatidylinositol 3-Kinase/Akt Pathway Is Involved in Retinoic Acid-induced Phosphorylation, Degradation, and Transcriptional Activity of Retinoic Acid Receptor γ 2. J Biol Chem. 2002b;277:24859–24862.. doi: 10.1074/jbc.C200230200. [DOI] [PubMed] [Google Scholar]
- Gianni M., Boldetti A., Guarnaccia V., Rambaldi A., Parrella E., Raska I., Jr., Rochette-Egly C., Del Sal G., Rustighi A., Terao M., Garattini E. Inhibition of the peptidyl-prolyl-isomerase Pin1 enhances the responses of acute myeloid leukemia cells to retinoic acid via stabilization of RARalpha and PML-RARalpha. Cancer Res. 2009;69:1016–26. doi: 10.1158/0008-5472.CAN-08-2603. [DOI] [PubMed] [Google Scholar]
- Gianni M., Parrella E., Raska I., Gaillard E., Nigro E. A., Gaudon C., Garattini E., Rochette-Egly C. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–51. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni M., Bauer A., Garattini E., Chambon P., Rochette-Egly C. Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-induced RAR γ degradation and transactivation. Embo J. 2002a;21:3760–9. doi: 10.1093/emboj/cdf374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G., Coin F., Ranish J. A., Hoogstraten D., Theil A., Wijgers N., Jaspers N. G., Raams A., Argentini M., van der Spek P. J., Botta E., Stefanini M., Egly J. M., Aebersold R., Hoeijmakers J. H., Vermeulen W. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–9. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- Gillespie R. F., Gudas L. J. Retinoic acid receptor isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic response elements. J Biol Chem. 2007a;282:33421–34. doi: 10.1074/jbc.M704845200. [DOI] [PubMed] [Google Scholar]
- Gillespie R. F., Gudas L. J. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007b;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Greschik H., Moras D. Structure-activity relationship of nuclear receptor-ligand interactions. Curr Top Med Chem. 2003;3:1573–99. doi: 10.2174/1568026033451736. [DOI] [PubMed] [Google Scholar]
- Gupta P., Huq M. D., Khan S. A., Tsai N. P., Wei L. N. Regulation of co-repressive activity of and HDAC recruitment to RIP140 by site-specific phosphorylation. Mol Cell Proteomics. 2005;4:1776–84. doi: 10.1074/mcp.M500236-MCP200. [DOI] [PubMed] [Google Scholar]
- Gupta P., Ho P. C., Huq M. M., Ha S. G., Park S. W., Khan A. A., Tsai N. P., Wei L. N. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc Natl Acad Sci U S A. 2008;105:11424–9. doi: 10.1073/pnas.0710561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauksdottir H., Farboud B., Privalsky M. L. Retinoic acid receptors β and γ do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol Endocrinol. 2003;17:373–85. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Huq M. D., Tsai N. P., Khan S. A., Wei L. N. Lysine trimethylation of retinoic acid receptor-α: a novel means to regulate receptor function. Mol Cell Proteomics. 2007;6:677–88. doi: 10.1074/mcp.M600223-MCP200. [DOI] [PubMed] [Google Scholar]
- Huq M. D., Khan S. A., Park S. W., Wei L. N. Mapping of phosphorylation sites of nuclear corepressor receptor interacting protein 140 by liquid chromatography-tandem mass spectroscopy. Proteomics. 2005;5:2157–66. doi: 10.1002/pmic.200401090. [DOI] [PubMed] [Google Scholar]
- Huq M. D., Ha S. G., Wei L. N. Modulation of retinoic acid receptor α activity by lysine methylation in the DNA binding domain. J Proteome Res. 2008;7:4538–45. doi: 10.1021/pr800375z. [DOI] [PubMed] [Google Scholar]
- Hu X., Chen Y., Farooqui M., Thomas M. C., Chiang C. M., Wei L. N. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J Biol Chem. 2004;279:319–25. doi: 10.1074/jbc.M307621200. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Jonas B. A., Privalsky M. L. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J Biol Chem. 2004;279:54676–86. doi: 10.1074/jbc.M410128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati S., Li Y., Verma A., Sassano A., Majchrzak B., Deb D. K., Parmar S., Giafis N., Kalvakolanu D. V., Rahman A., Uddin S., Minucci S., Tallman M. S., Fish E. N., Platanias L. C. Activation of protein kinase C δ by all-trans-retinoic acid. J Biol Chem. 2003;278:32544–51. doi: 10.1074/jbc.M301523200. [DOI] [PubMed] [Google Scholar]
- Karamouzis M. V., Papavassiliou A. G. Retinoid receptor cross-talk in respiratory epithelium cancer chemoprevention. Trends Mol Med. 2005;11:10–6. doi: 10.1016/j.molmed.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Kay B. K., Williamson M. P., Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14:231–41. [PubMed] [Google Scholar]
- Keriel A., Stary A., Sarasin A., Rochette-Egly C., Egly J. M. XPD Mutations Prevent TFIIH-Dependent Transactivation by Nuclear Receptors and Phosphorylation of RARalpha. Cell. 2002;109:125–35.. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- Khetchoumian K., Teletin M., Tisserand J., Mark M., Herquel B., Ignat M., Zucman-Rossi J., Cammas F., Lerouge T., Thibault C., Metzger D., Chambon P., Losson R. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor α. Nat Genet. 2007;39:1500–6. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S., Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–90. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- Kioka N., Ueda K., Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27:1–7. doi: 10.1247/csf.27.1. [DOI] [PubMed] [Google Scholar]
- Klaholz B. P., Renaud J. P., Mitschler A., Zusi C., Chambon P., Gronemeyer H., Moras D. Conformational adaptation of agonists to the human nuclear receptor RAR γ. Nat Struct Biol. 1998;5:199–202. doi: 10.1038/nsb0398-199. [DOI] [PubMed] [Google Scholar]
- Klaholz B. P., Mitschler A., Moras D. Structural basis for isotype selectivity of the human retinoic acid nuclear receptor. J Mol Biol. 2000;302:155–70. doi: 10.1006/jmbi.2000.4032. [DOI] [PubMed] [Google Scholar]
- Klein E. S., Pino M. E., Johnson A. T., Davies P. J., Nagpal S., Thacher S. M., Krasinski G., Chandraratna R. A. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J Biol Chem. 1996;271:22692–6. doi: 10.1074/jbc.271.37.22692. [DOI] [PubMed] [Google Scholar]
- Kopf E., Plassat J. L., Vivat V., de The H., Chambon P., Rochette-Egly C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors α and γ through the ubiquitin-proteasome pathway. J Biol Chem. 2000;275:33280–8.. doi: 10.1074/jbc.M002840200. [DOI] [PubMed] [Google Scholar]
- Lafyatis R., Kim S. J., Angel P., Roberts A. B., Sporn M. B., Karin M., Wilder R. L. Interleukin-1 stimulates and all-trans-retinoic acid inhibits collagenase gene expression through its 5' activator protein-1-binding site. Mol Endocrinol. 1990;4:973–80. doi: 10.1210/mend-4-7-973. [DOI] [PubMed] [Google Scholar]
- Laudet V., Gronemeyer H. Nuclear Receptor Factsbook. London: Acedemic Press; 2001. [Google Scholar]
- Lavery D. N., McEwan I. J. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–64. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Zechel C., Garnier J. M., Lutz Y., Tora L., Pierrat P., Heery D., Gronemeyer H., Chambon P., Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. Embo J. 1995;14:2020–33. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Kliewer S. A., Provencal J., Wright P. E., Evans R. M. Structure of the retinoid X receptor α DNA binding domain: a helix required for homodimeric DNA binding. Science. 1993;260:1117–21. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., Workman J. L. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–36. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Lefebvre B., Brand C., Flajollet S., Lefebvre P. Down-regulation of the tumor suppressor gene retinoic acid receptor beta2 through the phosphoinositide 3-kinase/Akt signaling pathway. Mol Endocrinol. 2006;20:2109–21. doi: 10.1210/me.2005-0321. [DOI] [PubMed] [Google Scholar]
- Lefebvre P., Martin P. J., Flajollet S., Dedieu S., Billaut X., Lefebvre B. Transcriptional activities of retinoic acid receptors. Vitam Horm. 2005;70:199–264. doi: 10.1016/S0083-6729(05)70007-8. [DOI] [PubMed] [Google Scholar]
- Leid M., Kastner P., Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci. 1992;17:427–33. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- Li Y., Lambert M. H., Xu H. E. Activation of nuclear receptors: a perspective from structural genomics. Structure. 2003;11:741–6. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- Li J. J., Dong Z., Dawson M. I., Colburn N. H. Inhibition of tumor promoter-induced transformation by retinoids that transrepress AP-1 without transactivating retinoic acid response element. Cancer Res. 1996;56:483–9. [PubMed] [Google Scholar]
- Lin F., Xiao D., Kolluri S. K., Zhang X. Unique anti-activator protein-1 activity of retinoic acid receptor β. Cancer Res. 2000;60:3271–80. [PubMed] [Google Scholar]
- Lipford J. R., Deshaies R. J. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol. 2003;5:845–50. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–88. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G. N., Turck C. W., Schaufele F., Stallcup M. R., Kushner P. J. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J Biol Chem. 2001;276:22177–82.. doi: 10.1074/jbc.M010718200. [DOI] [PubMed] [Google Scholar]
- Loudig O., Babichuk C., White J., Abu-Abed S., Mueller C., Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–97. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Macias M. J., Wiesner S., Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–7. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Maghsoodi B., Poon M. M., Nam C. I., Aoto J., Ting P., Chen L. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 2008;105:16015–20. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Umesono K., Kliewer S. A., Borgmeyer U., Ong E. S., Evans R. M. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–61. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Evans R. M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Masia S., Alvarez S., de Lera A. R., Barettino D. Rapid, nongenomic actions of retinoic Acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic Acid receptor. Mol Endocrinol. 2007;21:2391–402. doi: 10.1210/me.2007-0062. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–5. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- McNamara S., Wang H., Hanna N., Miller W. H., Jr. Topoisomerase IIbeta negatively modulates retinoic acid receptor α function: a novel mechanism of retinoic acid resistance. Mol Cell Biol. 2008;28:2066–77. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsushima M., Takahashi H., Shishido T., Ueda K., Kioka N. Abl kinase interacts with and phosphorylates vinexin. FEBS Lett. 2006b;580:4288–95. doi: 10.1016/j.febslet.2006.06.072. [DOI] [PubMed] [Google Scholar]
- Mitsushima M., Sezaki T., Akahane R., Ueda K., Suetsugu S., Takenawa T., Kioka N. Protein kinase A-dependent increase in WAVE2 expression induced by the focal adhesion protein vinexin. Genes Cells. 2006a;11:281–92. doi: 10.1111/j.1365-2443.2006.00932.x. [DOI] [PubMed] [Google Scholar]
- Moggs J. G., Orphanides G. Estrogen receptors: orchestrators of pleiotropic cellular responses. EMBO Rep. 2001;2:775–81. doi: 10.1093/embo-reports/kve185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–91. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Muratani M., Tansey W. P. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Saunders M., Kastner P., Durand B., Nakshatri H., Chambon P. Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell. 1992;70:1007–19. doi: 10.1016/0092-8674(92)90250-g. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Friant S., Nakshatri H., Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. Embo J. 1993;12:2349–60. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87.. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neri L. M., Borgatti P., Tazzari P. L., Bortul R., Cappellini A., Tabellini G., Bellacosa A., Capitani S., Martelli A. M. The phosphoinositide 3-kinase/AKT1 pathway involvement in drug and all-trans-retinoic acid resistance of leukemia cells. Mol Cancer Res. 2003;1:234–46. [PubMed] [Google Scholar]
- Nicholson R. C., Mader S., Nagpal S., Leid M., Rochette-Egly C., Chambon P. Negative regulation of the rat stromelysin gene promoter by retinoic acid is mediated by an AP1 binding site. Embo J. 1990;9:4443–54. doi: 10.1002/j.1460-2075.1990.tb07895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Kao Y. L., Joshi S., Jeetendran S., Dipette D., Singh U. S. Activation of Rac1 by phosphatidylinositol 3-kinase in vivo: role in activation of mitogen-activated protein kinase (MAPK) pathways and retinoic acid-induced neuronal differentiation of SH-SY5Y cells. J Neurochem. 2005;93:571–83. doi: 10.1111/j.1471-4159.2005.03106.x. [DOI] [PubMed] [Google Scholar]
- Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Perissi V., Aggarwal A., Glass C. K., Rose D. W., Rosenfeld M. G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–26. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Scafoglio C., Zhang J., Ohgi K. A., Rose D. W., Glass C. K., Rosenfeld M. G. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29:755–66. doi: 10.1016/j.molcel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Cohen R. E. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Pogenberg V., Guichou J. F., Vivat-Hannah V., Kammerer S., Perez E., Germain P., de Lera A. R., Gronemeyer H., Royer C. A., Bourguet W. Characterization of the interaction between RAR/RXR heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280:1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- Poon M. M., Chen L. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A. 2008;105:20303–8. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky M. L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Qian A., Cai Y., Magee T. R., Wan Y. J. Identification of retinoic acid-responsive elements on the HNF1alpha and HNF4alpha genes. Biochem Biophys Res Commun. 2000;276:837–42. doi: 10.1006/bbrc.2000.3549. [DOI] [PubMed] [Google Scholar]
- Rastinejad F. Retinoid X receptor and its partners in the nuclear receptor family. Curr Opin Struct Biol. 2001;11:33–8. doi: 10.1016/s0959-440x(00)00165-2. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Wagner T., Zhao Q., Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. Embo J. 2000;19:1045–54. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J. P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–9. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Renaud J. P., Moras D. Structural studies on nuclear receptors. Cell Mol Life Sci. 2000;57:1748–69. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resche-Rigon M., Gronemeyer H. Therapeutic potential of selective modulators of nuclear receptor action. Curr Opin Chem Biol. 1998;2:501–7. doi: 10.1016/s1367-5931(98)80126-9. [DOI] [PubMed] [Google Scholar]
- Richards E. J., Elgin S. C. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C. Dynamic Combinatorial Networks in Nuclear Receptor-mediated Transcription. J Biol Chem. 2005;280:32565–8. doi: 10.1074/jbc.R500008200. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Oulad-Abdelghani M., Staub A., Pfister V., Scheuer I., Chambon P., Gaub M. P. Phosphorylation of the retinoic acid receptor-α by protein kinase A. Mol Endocrinol. 1995;9:860–71. doi: 10.1210/mend.9.7.7476969. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Adam S., Rossignol M., Egly J. M., Chambon P. Stimulation of RAR α activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Lunyak V. V., Glass C. K. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Rowan B. G., Weigel N. L., O'Malley B. W. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen- activated protein kinase pathway. J Biol Chem. 2000;275:4475–83.. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- Sanglier S., Bourguet W., Germain P., Chavant V., Moras D., Gronemeyer H., Potier N., Van Dorsselaer A. Monitoring ligand-mediated nuclear receptor-coregulator interactions by noncovalent mass spectrometry. Eur J Biochem. 2004;271:4958–67. doi: 10.1111/j.1432-1033.2004.04466.x. [DOI] [PubMed] [Google Scholar]
- Schug T. T., Berry D. C., Toshkov I. A., Cheng L., Nikitin A. Y., Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/δ to RAR. Proc Natl Acad Sci U S A. 2008;105:7546–51. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J., Mueller L., Collins S. J. CaMKII regulates retinoic acid receptor transcriptional activity and the differentiation of myeloid leukemia cells. J Clin Invest. 2007;117:1412–21. doi: 10.1172/JCI30779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R. J., 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–20. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Smith W. C., Nakshatri H., Leroy P., Rees J., Chambon P. A retinoic acid response element is present in the mouse cellular retinol binding protein I (mCRBPI) promoter. Embo J. 1991;10:2223–30. doi: 10.1002/j.1460-2075.1991.tb07758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn M. B., Roberts A. B., Goodman D. S. The Retinoids. Biology, Chemistry, and Medecine. New York: Raven Press; 1994. [Google Scholar]
- Srinivas H., Xia D., Moore N. L., Uray I. P., Kim H., Ma L., Weigel N. L., Brown P. H., Kurie J. M. Akt phosphorylates and suppresses the transactivation of retinoic acid receptor α. Biochem J. 2006;395:653–62. doi: 10.1042/BJ20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas H., Juroske D. M., Kalyankrishna S., Cody D. D., Price R. E., Xu X. C., Narayanan R., Weigel N. L., Kurie J. M. c-Jun N-terminal kinase contributes to aberrant retinoid signaling in lung cancer cells by phosphorylating and inducing proteasomal degradation of retinoic acid receptor α. Mol Cell Biol. 2005;25:1054–69. doi: 10.1128/MCB.25.3.1054-1069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz A. C., Renaud J. P., Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu Rev Biophys Biomol Struct. 2001;30:329–59. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- Sudol M., Sliwa K., Russo T. Functions of WW domains in the nucleus. FEBS Lett. 2001;490:190–5.. doi: 10.1016/s0014-5793(01)02122-6. [DOI] [PubMed] [Google Scholar]
- Sun K., Montana V., Chellappa K., Brelivet Y., Moras D., Maeda Y., Parpura V., Paschal B. M., Sladek F. M. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol. 2007;21:1297–311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- Suzukawa K., Colburn N. H. AP-1 transrepressing retinoic acid does not deplete coactivators or AP-1 monomers but may target specific Jun or Fos containing dimers. Oncogene. 2002;21:2181–90. doi: 10.1038/sj.onc.1205281. [DOI] [PubMed] [Google Scholar]
- Tansey W. P. Transcriptional activation: risky business. Genes Dev. 2001;15:1045–50.. doi: 10.1101/gad.896501. [DOI] [PubMed] [Google Scholar]
- Tari A. M., Lim S. J., Hung M. C., Esteva F. J., Lopez-Berestein G. Her2/neu induces all-trans retinoic acid (ATRA) resistance in breast cancer cells. Oncogene. 2002;21:5224–32. doi: 10.1038/sj.onc.1205660. [DOI] [PubMed] [Google Scholar]
- Townson S. M., Dobrzycka K. M., Lee A. V., Air M., Deng W., Kang K., Jiang S., Kioka N., Michaelis K., Oesterreich S. SAFB2, a new scaffold attachment factor homolog and estrogen receptor corepressor. J Biol Chem. 2003;278:20059–68. doi: 10.1074/jbc.M212988200. [DOI] [PubMed] [Google Scholar]
- Vicent G. P., Ballare C., Nacht A. S., Clausell J., Subtil-Rodriguez A., Quiles I., Jordan A., Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–81. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Vitoux D., Nasr R., de The H. Acute promyelocytic leukemia: New issues on pathogenesis and treatment response. Int J Biochem Cell Biol. 2007;39:1063–1070. doi: 10.1016/j.biocel.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Vivat-Hannah V., Zusi F. C. Retinoids as therapeutic agents: today and tomorrow. Mini Rev Med Chem. 2005;5:755–60. doi: 10.2174/1389557054553820. [DOI] [PubMed] [Google Scholar]
- Vo N., Goodman R. H. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–8.. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- vom Baur E., Zechel C., Heery D., Heine M. J., Garnier J. M., Vivat V., Le Douarin B., Gronemeyer H., Chambon P., Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. Embo J. 1996;15:110–24. [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z., Zhang Z., Zhao J., Wang F., Soprano K. J., Soprano D. R. Acinus-S' represses RAR-regulated gene expression through interaction with the B-domain of RARs. Mol Cell Biol. 2008;28:2549–2558. doi: 10.1128/MCB.01199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnmark A., Treuter E., Wright A. P., Gustafsson J. A. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–9. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- Wei L. N., Hu X., Chandra D., Seto E., Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–7. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- Wulf G., Finn G., Suizu F., Lu K. P. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435–41. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- Wurtz J. M., Bourguet W., Renaud J. P., Vivat V., Chambon P., Moras D., Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors [see comments] [published erratum appears in Nat Struct Biol 1996 Feb; 3(2):206] Nat.Struct.Biol. 1996;3:87–94. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- Wu R. C., Qin J., Yi P., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–49. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–40. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Wu R. C., Smith C. L., O'Malley B W. Transcriptional Regulation by Steroid Receptor Coactivator Phosphorylation. Endocr Rev. 2005;26:393–399. doi: 10.1210/er.2004-0018. [DOI] [PubMed] [Google Scholar]
- Yi P., Feng Q., Amazit L., Lonard D. M., Tsai S. Y., Tsai M. J., O'Malley B. W. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29:465–76. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P., Wu R. C., Sandquist J., Wong J., Tsai S. Y., Tsai M. J., Means A. R., O'Malley B. W. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1) Mol Cell Biol. 2005;25:9687–99. doi: 10.1128/MCB.25.21.9687-9699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto-Filho A., Cammarota M., Gelain D. P., Oliveira R. B., Delgado-Canedo A., Dalmolin R. J., Pasquali M. A., Moreira J. C. Retinoic acid induces apoptosis by a non-classical mechanism of ERK1/2 activation. Toxicol In Vitro. 2008;22:1205–12. doi: 10.1016/j.tiv.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A., Lim W. A. Converging on proline: the mechanism of WW domain peptide recognition. Nat Struct Biol. 2000;7:611–3.. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- Zechel C., Shen X. Q., Chambon P., Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. Embo J. 1994a;13:1414–24. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel C., Shen X. Q., Chen J. Y., Chen Z. P., Chambon P., Gronemeyer H. The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. Embo J. 1994b;13:1425–33. doi: 10.1002/j.1460-2075.1994.tb06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H. G., Krasnov A., Georgieva S. G., Schule R., Takeyama K., Kato S., Tora L., Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gianni M., Kopf E., Honore N., Chelbi-Alix M., Koken M., Quignon F., Rochette-Egly C., de The H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor α (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A. 1999;96:14807–12. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]