Abstract
Positive and negative feedback loops are common regulatory elements in biological signaling systems. We discuss core feedback motifs that have distinct roles in shaping signaling responses in space and time. We also discuss approaches to experimentally investigate feedback loops in signaling systems.
Feedback loops are processes that connect output signals back to their inputs. The history of biological feedback goes back at least 130 years to observations by Eduard Pflüger that organs and other living systems “satisfy their own needs” (1). Feedback became an influential concept that led to Walter Cannon's theory of physiological homeostasis (2); Alan Turing's model of pattern formation (3); as well as investigations of metabolic end-product inhibition (4), metabolic oscillations (5), and transcriptional self-repression (6). Biological feedback concepts were further influenced by chemical oscillation theories (7) and the field of cybernetics (8). It has more recently become appreciated that the concept of feedback may be useful as a framework for understanding how intracellular signaling systems elicit specific cell behavior.
Mammalian species use over 3000 signaling proteins and over 15 second messengers to build hundreds of cell-specific signaling systems. Many of the signaling components have multiple upstream regulators and downstream targets, creating a web of connectivity within and between signaling pathways (9). The presence of multiple feedback loops in these systems (10) poses a challenge to understanding how receptor inputs control cellular behavior. We discuss how recurring feedback designs, or motifs (11), mediate biological functions such as bistability, oscillation, polarization, and robustness. Our goal was to generate a comprehensive guide for feedback in signal transduction that would also be instructive for understanding transcription networks, control of metabolism, pattern formation, the cell cycle, and the behavior of circadian oscillators.
We focus on two mammalian signaling systems: the receptor-triggered Ca2+ signaling system in nonexcitable cells (12) and the phosphoinositide 3-kinase (PI3K) signaling pathway in chemotactic neutrophils (13). These were chosen because of existing knowledge of feedback mechanisms that generate both simple and complex temporal and spatial signaling responses (Fig. 1). We use graphical representations of feedback motifs, with signaling components shown as vertices and directed negative and positive regulatory steps shown as arrows with and without minus symbols, respectively (Figs. 2 to 5, gray background). An arrow may consist of multiple steps so that a single positive arrow could reflect, for example, the net effect of two serial negative regulatory steps. Feedback loops are defined as paths that begin from and return to the same vertex. We also use mathematical representations with variables corresponding to vertices and equation terms corresponding to arrows (table S1). The simulated functions for each motif (Figs. 2 and 3) can be recreated by accompanying computer programs (table S2). We conclude by suggesting experimental approaches to investigate feedback loops.
Fig. 1.
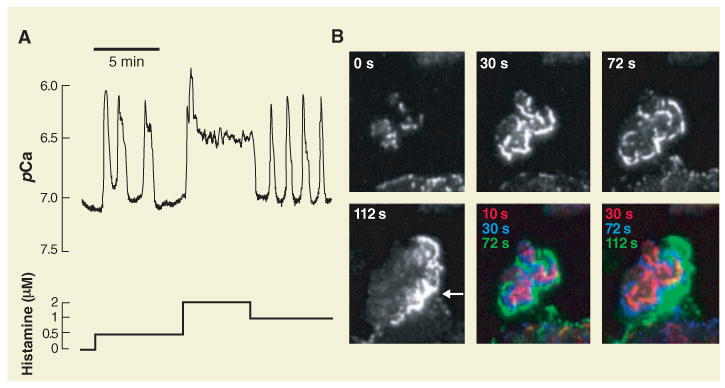
Ca2+ and chemotaxis signaling systems exhibit complex temporal and spatial dynamics. (A) Histamine-triggered cytosolic Ca2+ signals in a single epithelial cell. The response includes spikes, oscillations, and plateaus. pCa is the negative log of the Ca2+ concentration (7 and 6 correspond to 10−7 and 10−6 M, respectively). [Adapted from (28)] (B) Total internal reflection fluorescence images of a neutrophil cells stimulated by chemoattractant. Hem-1 is a regulator of actin polymerization that initially concentrates in foci. It then relocalizes as part of outwardly propagating waves of actin polarization that terminate when the leading edge is reached (denoted by arrow at 112 s). The fifth and sixth images show overlays of successive Hem-1 distributions in red, blue, and green, respectively. [Adapted from (29)]
Fig. 2.
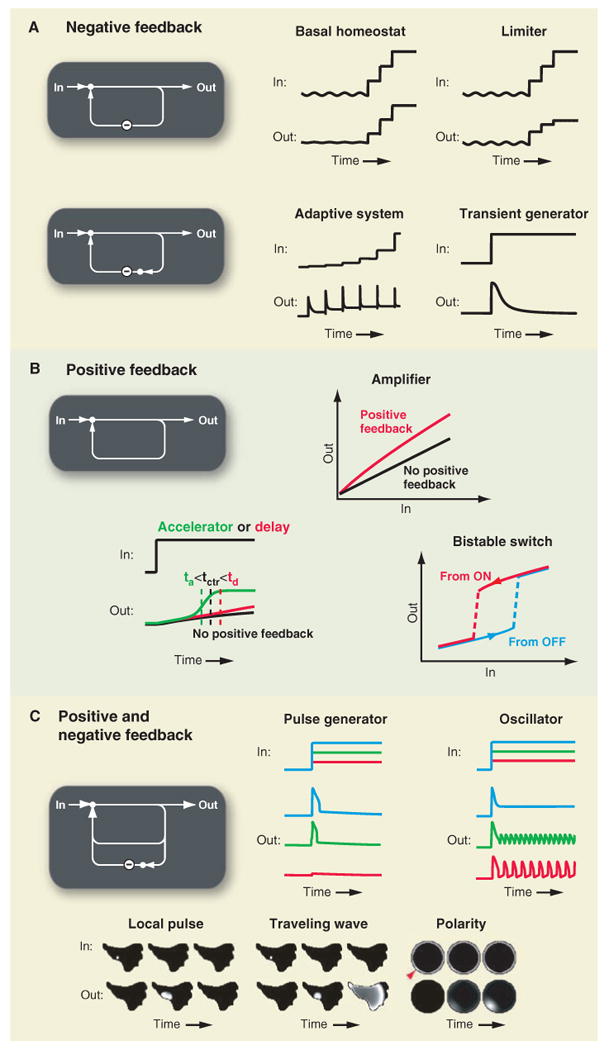
Feedback motifs have important functions in signaling systems. (A) Negative feedback can stabilize basal signaling levels, limit maximal signaling output, enable adaptive responses, or create transient signal responses. (B) Positive feedback can amplify signaling responses, alter kinetics, or create bistable switches. (C) Mixtures of positive and negative feedback can create single pulses or oscillatory signal outputs. Mixed feedbacks can also trigger local signals, self-propagating waves, or cell polarization.
Fig. 5.
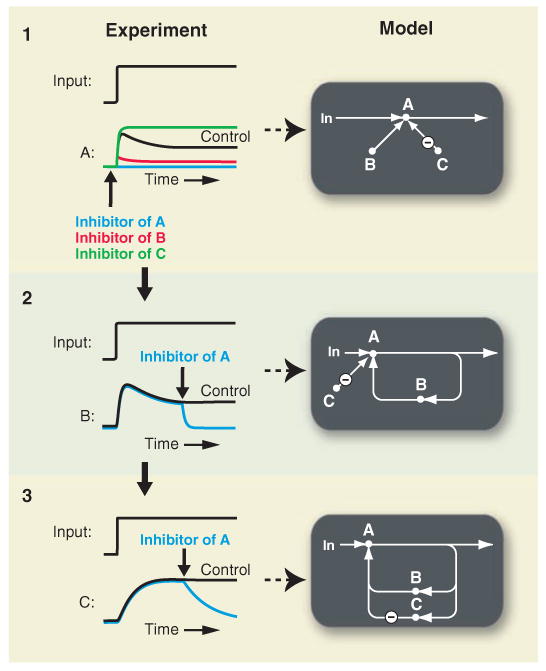
Proposed rapid perturbation and monitoring strategy to experimentally investigate feedback loops. The shown graphs with proposed experiments use computer simulations of experiments to explain how fast perturbations and fast readouts can uncover feedback. Bold arrows denote the addition of inhibitors for the activities A, B, and C. Experiments in 1 show that the two players B and C regulate A. Experiments 2 and 3 reveal that B and C are part of a positive and a negative feedback loop, respectively.
Fig. 3.
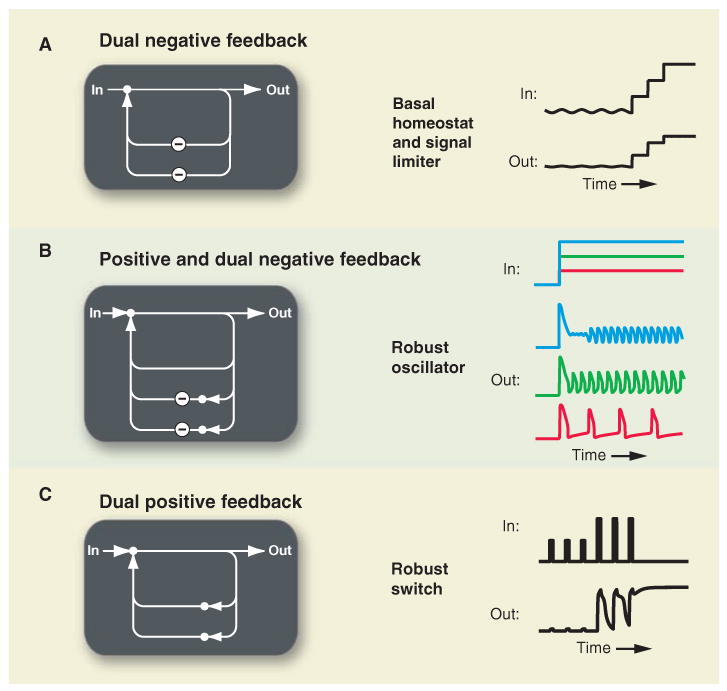
Addition of extra feedback to core functions can be used to integrate key signaling characteristics or enhance the robustness of important functions. (A) Combining two negative feedback loops can independently stabilize basal signaling and limit maximal signal output. (B) Adding a negative feedback loop to the oscillator in Fig. 2C can result in sharper spikes, an increased input range over which oscillations occur and an increased output frequency range. (C) A system made up of a dual fast and slow positive feedback loops can exhibit transient or persistent bistable states if the fast or both positive feedbacks are engaged.
Signaling with a Single Negative Feedback Loop
Negative feedback loops are found in nearly all known signaling pathways and are defined as sequential regulatory steps that feed the output signal, inverted, back to the input (Fig. 2A). Depending on its characteristics and initial conditions, a single negative feedback motif can create four distinct signaling functions: basal homeostat, output limiter, adaptation, and transient generator.
The presence of a small-amplitude negative feedback loop stabilizes the basal signaling state without preventing strong input signals from triggering maximal pathway activation (Fig. 2A, basal homeostat). In this case, small deviations of an input signal are suppressed in the output, and only large changes in the input control the output. For example, a negative feedback involving the endoplasmic reticulum (ER) Ca2+-sensing protein STIM2 (stromal interaction molecule 2) (14) keeps basal Ca2+ concentration in the cytosol and in the lumen of the ER at ∼50 nM and ∼400 μM, respectively. STIM2 triggers influx of extracellular Ca2+ at ER–plasma membrane junctions in response to a reduction in ER Ca2+ concentration, forming a negative feedback loop (14). Because many cellular processes are regulated by Ca2+, maintaining proper resting levels is crucial.
A different use of negative feedback is to limit maximum signaling output (Fig. 2A, limiter). Upon stimulation, the output signal rapidly increases but is attenuated once it passes a threshold. For example, receptor-triggered increases of Ca2+ concentration in the cytosol are clipped and stabilized by a rapid negative feedback resulting from Ca2+ uptake by mitochondria. Mitochondrial Ca2+ uptake progressively increases when cytosolic Ca2+ concentration exceeds about 0.6 μM (15) and therefore occurs only during active signaling.
Negative feedback is also necessary to generate adaptive behavior. Adaptive signaling systems are built to respond to changes in input rather than the absolute amount of input signal (Fig. 2A, adaptive). Studies of bacterial chemotaxis (16) and vertebrate visual signal transduction (17) revealed sophisticated regulatory circuits that generate optimized adaptative behavior. In neutrophil chemotaxis, cells sense relative chemoattractant gradients by a mechanism that involves partial deactivation and subsequent internalization of activated cell surface receptors. The resulting lower input level prevents saturation of downstream signaling responses and allows for subsequent signaling when chemoattractant concentrations further increase (18). Most heterotrimeric guanine nucleotide–binding protein (G-protein)–coupled receptors, which include receptors involved in chemotaxis, exhibit adaptive behavior through use-dependent receptor down-regulation (19).
Lastly, a strong negative feedback loop that is triggered after a delay converts a constant input into a transient output signals with amplitudes that increase as a function of the amplitude of the input step (Fig. 2A, transient generator). An example of this function is the delayed activation of the Ca2+/calmodulin (CaM)–regulated plasma membrane Ca2+ pump (PMCA) (20). After a 10- to 60-s delay that follows an increase in cytosolic Ca2+, enhanced PMCA activity reduces the Ca2+ signals to create a transient signaling response.
Signaling with a Single Positive Feedback Loop
Positive feedback is often associated with uncontrolled, runaway processes such as nuclear chain reactions and models of catastrophic climate change. Nevertheless, functions of positive feedback in biological systems have been described for almost 100 years (21). Positive feedback is defined as a set of regulatory steps that feeds the output signal back to the input (Fig. 2B). If signaling output activity increases, positive feedback will further increase input levels, thereby enhancing the output signal. There are three common functions of a single positive feedback motif: amplifying a signal, changing the timing of a signaling response, and creating bistable switches.
Positive feedback can provide absolute as well as relative amplification (Fig. 2B, amplifier). Both types of amplification are exemplified in the activation of the inositol 1,4,5-trisphosphate (IP3)–receptor (IP3R), a ligand-gated ER-localized Ca2+ channel. The binding of four IP3 molecules to a single IP3R is believed to partially activate the channel, inducing an initial Ca2+ release from the ER. This, in turn, triggers a positive feedback loop whereby the binding of multiple released Ca2+ molecules fully activates IP3Rs (22). Thus, as a result of binding four IP3 molecules, thousands of Ca2+ ions are released into the cytosol, providing for absolute amplification of the input signal. The ultrasensitive nature of the release process also results in relative amplification; a threefold increase in IP3 can result in a 20-fold increase in cytosolic Ca2+ levels (23). Ultrasensitive regulatory steps are characterized by a sigmoidal dose-response curve (24).
As is the case for negative feedback (limiter and transient generator), positive feedback also changes the timing of the signaling response (Fig. 2B, accelerator or delay). In the case of IP3-gated Ca2+ release, positive feedback accelerates the signaling response by opening more Ca2+ channels so that a saturating cytosolic Ca2+ concentration is more rapidly reached (green curve; half-maximal time: ta < tctr). Positive feedback in nonsaturating conditions can also prolong the time required to reach a higher steady state (red curve; td > tctr) (10).
A positive feedback loop with an ultrasensitive regulatory step can trigger a bistable switch (Fig. 2B, bistability) (25). This is arguably one of the most important regulatory motifs in cell signaling. For inputs below a critical threshold, the signaling output remains near its basal state; for inputs above the threshold, the output increases to a high, active state. Bistable systems make use of hysteresis to remain in the active state, meaning that the input stimulus required to keep the system in the active state is lower than the input required for triggering the initial transition from basal to active state. Many cell signaling processes have been proposed to use positive feedback to implement either a reversible or irreversible bistable switch, such as Ca2+ spikes (25), chemotaxis (26), and oocyte maturation (27).
Mixed Positive and Negative Feedback
Delayed negative feedback can force a bistable system back to the inactive state and create a pulse in the signaling output that is typically characterized by a fixed amplitude and duration (Fig. 2C, pulse generator). Ca2+ pulses (28) are triggered by the aforementioned IP3R fast positive feedback loop and a delayed negative feedback by which high levels of Ca2+ inhibit the IP3R (22). Similarly, in neutrophil migration, a proposed fast positive feedback loop between the scaffold protein hematopoietic 1 (Hem-1) and actin nucleation enhances actin polymerization and local extension of lamellipods (29). Recruitment of a yet-unidentified inhibitor may break apart sites of nucleation, resulting in reversible local lamellipod extension. The common design principle is that the negative feedback that terminates the positive feedback is induced only at high concentrations of the output signal or after a time delay.
After a pulse in output activity, the negative feedback inhibition often recovers so that a new positive feedback cycle can be triggered. This results in a periodic cycling between high and low output states in response to a steady input (Fig. 2C, oscillator). In cell signaling, such a role of ultrasensitive positive and negative feedback in generating Ca2+ concentration oscillations was proposed 20 years ago (25). Model calculations showed that increases in stimulus amplitude increase the frequency of oscillation while keeping the duration and the amplitude of pulses constant. This behavior has been observed experimentally (28). Although there are feedback motifs without positive feedback that can produce oscillations (30), we believe that a more common mechanism in cell signaling consists of coupled positive and negative feedback loops (31).
Spatial Aspects of Mixed Positive and Negative Feedback
Biological systems often feature positive and negative feedbacks that are active in only part of the cell rather than the whole cell. This results in distinct functions such as local pulses, waves, and cell polarization (Fig. 2C, bottom images).
Local signal activation can occur without a global response (Fig. 2C, local pulse). In the case of Ca2+ signaling under weak stimulation, local positive feedback that activates IP3R quickly inactivates, and the diffusing Ca2+ ions can be restricted to sub-micrometer distances from the source, creating local Ca2+ pulses (32). Local Ca2 + signaling pulses are useful to control cell functions such as secretion (33). In weakly stimulated neutrophils (and therefore in the absence of cell polarity), a chemoattractant triggers similar short-lived local pulses of actin polymerization (29).
If the local signal is sufficiently strong, new local pulses triggered at nearby sites can keep the signal alive and allow a self-propagating signal to move away from its source (Fig. 2C, traveling wave). Models combining positive and negative feedback with a diffusive second messenger predict the existence of such self-propagating Ca2+ waves (34). Ca2+ waves have been shown to propagate across gap junctions and synchronize astrocyte, epithelial, and other cell assemblies (35). Similarly, under stimulated conditions, neutrophils generate local waves of actin polymerization that propagate over several micrometers toward the front of a cell until they reach a cell's leading edge, where they extend local lamellopdia and help propel the cell forward (29).
Combined positive and negative feedback can also polarize cells (Fig. 2C, polarity). Polarization and migration of neutrophils has been proposed to be regulated by a positive feedback loop whereby the guanylate exchange activator of the small guanosine triphosphatase Rac, Dock, both activates lamellipod extension and is recruited to locations where lamellipods extend (13, 36). Once the positive feedback-driven local recruitment has been triggered (stochastically or in response to a local input), it diminishes the concentration of available Dock throughout the cell. This is a plausible mechanism for a negative feedback that can prevent a second front from forming. Mathematical modeling of a neuronal polarity system showed that reinforced local recruitment of a scarce activator is a robust mechanism for triggering cell polarization (37).
Enhancing Core Feedback Functions by Adding Feedback
Addition of extra feedback loops provides enhancements to the discussed core feedback functions. A design with two negative feedback loops allows cells to simultaneously stabilize the basal state and limit the activated signaling state (Fig. 3A, basal homeostat and limiter). Combined negative feedback through STIM2 and mitochondrial Ca2+ uptake is an example where two negative feedback loops function largely independent of each other. Combination of a basic oscillator (single positive and negative feedback loops) with an additional negative feedback loop can create patterns of repetitive spikes over a broader range of input stimuli and generate a wider range of output frequencies (Fig. 3B, robust oscillator). A reported calcium/calmodulin-dependent protein kinase II (CaMKII)–mediated inhibitory phosphorylation of the IP3R (38) may serve as such a second negative feedback loop that prolongs the time between Ca2+ spikes and thereby extends the output frequency range.
A key function of a second positive feedback loop is to make a bistable signaling switch more robust in the presence of cell-to-cell variations resulting from noise in external inputs or copy numbers of internal signaling components (Fig. 3C, robust switch) (39). In the example shown, the first three stimulus pulses are not sufficient to engage the positive feedbacks, the next two stimuli trigger only the fast reversible positive feedback, whereas the last stimulus also engages the slow positive feedback and triggers a persistent output increase. This same motif also increases the overall ultrasensitivity and hysteresis if the two positive feedback loops are linked by an AND gate (40). Furthermore, coupling two positive feedback loops in which one loop is composed of two negative regulatory steps can delay the exit from the active state (41). Lastly, the added slower positive feedback loop can also spatially integrate and synchronize the activity of the faster loop, converting separate local bursts of signaling into a robust global signaling response. In the case of Ca2+ signaling, Ca2+ activation of IP3R can provide rapid and localized positive feedback, whereas a second slow positive feedback from Ca2+ concentration to phospholipase C (25, 42) and the production of the rapidly diffusible second messenger IP3 yield global bistability and increased robustness to noise when triggering global Ca2+ responses.
Thus, although different types of network connectivity can generate dual positive feedback loops, these examples suggest that a common emerging property is enhanced robustness to noise. The robustness results from (i) increased hysteresis, (ii) increased spatial synchronization, (iii) delayed exit from the active state, or (iv) a combination thereof. It is plausible that addition of tailored positive or negative feedback loops to other functions described in Fig. 2 also make their functional properties more robust.
Signaling Systems Combine Multiple Feedback Functions to Orchestrate Cell Behavior
Feedback functions are combined in cells to produce a physiological output behavior. Graphic representations that reduce each of the feedback loops to a positive or negative regulatory step are useful to describe systems with multiple feedbacks. This is achieved by functionally annotating and ordering nested feedback loops according to increasing time constants. In the Ca2+ signaling system (Fig. 4A), basal homeostasis involves negative loops that include the ER Ca2+ sensor STIM2 and the Ca2+ regulation of PMCAs. In the receptor-stimulated case, the fastest positive and negative feedback loops are the Ca2+ activation and inhibition of the IP3R, which can create local or more-global spikes. Spatial synchronization of Ca2+ signals and robust bistability, oscillations, and waves are provided by a second positive feedback loop based on Ca2+ activation of phospholipase C and generation of more IP3. A second negative feedback that phosphorylates and down-regulates the IP3R may enhance the frequency range of Ca2+ oscillations. Mitochondria limit maximal Ca2+ signaling responses, whereas plasma membrane pumps ensure that cytosolic Ca2+ signals remain transient. Persistent Ca2+ signaling requires a third positive feedback in which the ER Ca2+ sensor STIM1 links the lowering of ER Ca2+ concentration to plasma membrane Ca2+ influx, which in turn triggers additional IP3R-mediated Ca2+ release and lowering of Ca2+ in the ER (43). This triple positive feedback in the Ca2+ system comprises a robust switch and is likely critical in T cells, where Ca2+ concentration has to remain elevated for many hours in order to induce T cell differentiation (44). Note that separate feedback loops in the diagram can regulate the same target. For example, IP3R is activated by Ca2+ concentrations below 0.2 μM (creating a positive feedback loop) and inhibited by higher Ca2+ levels (creating a negative feedback loop) (22).
Fig. 4.
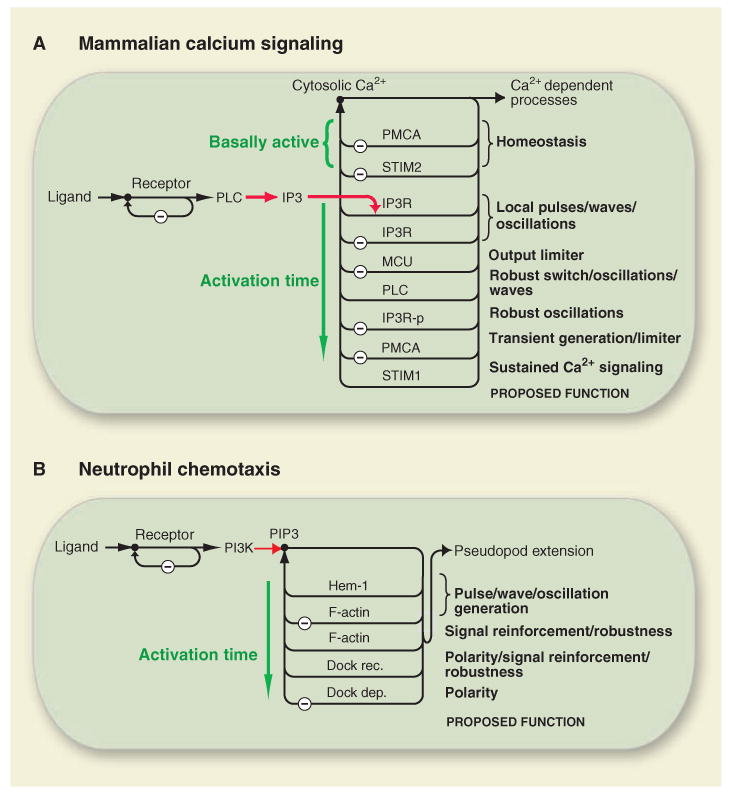
Cell signaling systems take advantage of multiple feedback loops nested in time and space to generate desired signaling responses. Schematic representations of (A) mammalian Ca2+ signaling for nonexcitable cells and (B) neutrophil chemotaxis. The input signal spreads from membrane-bound receptors to a network of interlinked feedback loops. Feedback loops are arranged according to the loop time constant. PLC, phospholipase C; MCU, mitochondrial Ca2+ uniporter; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; Dock, dedicator of cytokinesis; rec., recruitment; dep., depletion.
In neutrophil chemotaxis, the balance between positive and negative feedback loops involving Hem-l and other actin polymerization processes leads to small bursts in actin polymerization and local reversible lamellipod extensions (Fig. 4B) (29). When the proposed polarity-inducing positive feedback (e.g., Dock/Rac/PIP3) is engaged, actin polymerization and local lamellipod extension becomes restricted to the leading edge, resulting in cell migration (13, 29). Key mechanisms for polarization are likely the recruitment of Dock and other components of the global positive feedback to the front and their concomitant depletion from the back. The time constants of feedback loops and the diffusion coefficients of their components define the spatial and temporal characteristics of local lamellipod extension and global cell polarization. Chemotaxis may then result from steering the front of these polarized cells by coupling local chemoattractant sensing to local lamellipod extension and creating small turns in the direction of migration toward a chemoattractant source (45). Additional feedback loops and regulatory components may further contribute to robust chemotaxis (13, 36, 46).
Experimental Investigation of Predicted Feedback Loops
Experimental investigation of signaling feedback in cells is challenging. Genetic, RNA interference, dominant negative, or chemical attenuation of a component's activity that results in an increased signaling response might indicate that the component (i) is part of a negative feedback loop, (ii) is part of a suppression step within the signaling pathway, or (iii) plays a housekeeping role by reducing protein stability, blocking endocytosis, or dephosphorylating an activation site. How can a postulated component in a signaling pathway be identified as part of a positive or negative feedback loop?
A direct approach to demonstrate that a positive or negative feedback controls a signaling pathway is to combine rapid perturbations of signaling steps with rapid monitoring of upstream and downstream signaling events (Fig. 5). We use a thought experiment to discuss this approach. In a first set of experiments, inhibitors of components A, B, and C are used to identify B and C as positive and negative regulators, respectively, of the signaling pathway leading to A. The second experiment monitors B and uses rapid inhibition of A to demonstrate a positive feedback from A back to B. The third experiment monitors C and uses rapid inhibition of A to demonstrate a negative feedback from A to C. Additional constrictions of a feedback model are gained from the kinetics of A, B, and C activity changes. There are three experimental challenges to the described approach: to develop perturbation tools to specifically and rapidly inhibit signaling steps, to develop biosensor or biochemical assays to rapidly measure their activity, and to set up single-cell experiments or use synchronized populations to be able to infer the existence and to measure the kinetics of putative feedback loops.
This strategy has been used in Ca2+ signaling to demonstrate the existence of a positive feedback from Ca2+ to phospholipase C. A rapid increase in cytosolic Ca2+ concentration by addition of Ca2+ ionophore increased the concentration of the phospholipase C product IP3 within seconds (42). The kinetics of the other step in the feedback loop was measured by a rapid step increase in the concentration of IP3 by photorelease of an inactive IP3 precursor and measurement of the resulting subsecond Ca2+ release using a fluorescent biosensor (47). In neutrophils, the positive feedback for polarization was investigated by rapid chemical inhibition of PI3K (48) and by a chemical heterodimerization method whereby synthetic activators of endogenous PI3K and Rac proteins were forced to the plasma membrane within less than 30 s (49). These perturbations were combined with live-cell imaging of phoshoinositol 3,4,5-phosphate localization, lamellipod extension, and cell migration to infer the existence of a positive feedback (48, 49).
Conclusions
Each feedback motif has a specific function in a signaling system, such as homeostasis or adaptation for a negative feedback motif, amplification or bistability for a positive feedback motif, and polarization or oscillations for a mixed feedback motif. The function of a particular feedback motif depends on its characteristics and initial conditions. Cell signaling systems are made up of multiple feedback functions that together generate a cell's input-output behavior. We propose that working models of signaling systems can be based on graphic representations that show signaling components as part of feedback motifs with assigned functions and sorted by increasing feedback time constants. If the known feedback functions are insufficient to explain a cell's input-output behavior, this analysis can guide the search to identify control loops missing from a signaling model.
Supplementary Material
Acknowledgments
We thank M. Hammer, J. Ferrell, M. Covert, A. Salmeen, Y. Brandman, and P. Vitorino for helpful discussion and suggestions and NIH grants R01GM063702, R01GM030179, and R01MH064801 for funding the work.
Footnotes
References and Notes
- 1.Pflüger EFW. Pflügers Arch. 1877;XV:57. [Google Scholar]
- 2.Cannon WB. Physiol Rev. 1929;IX:399. [Google Scholar]
- 3.Turing AM. Philos Trans R Soc London Ser B. 1952;237:37. [Google Scholar]
- 4.Umbarger HE. Science. 1956;123:848. doi: 10.1126/science.123.3202.848. [DOI] [PubMed] [Google Scholar]
- 5.Chance B, Estabrook RW, Ghosh A. Proc Natl Acad Sci USA. 1964;51:1244. doi: 10.1073/pnas.51.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monod J, Jacob F. Cold Spring Harbor Symp Quant Biol. 1961;26:389. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 7.Prigogine I. Thermodynamics of Irreversible Processes. 2. Interscience; New York: 1961. [Google Scholar]
- 8.Wiener N. Cybernetics: Or the Control and Communication in the Animal and the Machine. MIT Press; Cambridge, MA: 1948. [Google Scholar]
- 9.Jordan JD, Landau EM, Iyengar R. Cell. 2000;103:193. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman M. Nature. 2000;408:313. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 11.Alon U. Nat Rev Genet. 2007;8:450. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 12.Clapham DE. Cell. 2007;131:1047. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Stephens L, Milne L, Hawkins P. Curr Biol. 2008;18:R485. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 14.Brandman O, Liou J, Park WS, Meyer T. Cell. 2007;131:1327. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls D, Akerman K. Biochim Biophys Acta. 1982;683:57. doi: 10.1016/0304-4173(82)90013-1. [DOI] [PubMed] [Google Scholar]
- 16.Alon U, Surette MG, Barkai N, Leibler S. Nature. 1999;397:168. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 17.Burns ME, Baylor DA. Annu Rev Neurosci. 2001;24:779. doi: 10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- 18.Zigmond SH, Sullivan SJ. J Cell Biol. 1979;82:517. doi: 10.1083/jcb.82.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Annu Rev Physiol. 2007;69:483. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 20.Bautista DM, Hoth M, Lewis RS. J Physiol. 2002;541:877. doi: 10.1113/jphysiol.2001.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lotka A. J Phys Chem. 1910;14:271. [Google Scholar]
- 22.Bezprozvanny I, Watras J, Ehrlich BE. Nature. 1991;351:751. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- 23.Mak DO, McBride S, Foskett JK. Proc Natl Acad Sci USA. 1998;95:15821. doi: 10.1073/pnas.95.26.15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshland DE, Jr, Goldbeter A, Stock JB. Science. 1982;217:220. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
- 25.Meyer T, Stryer L. Proc Natl Acad Sci USA. 1988;85:5051. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meinhardt H, Gierer A. Bioessays. 2000;22:753. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Xiong W, Ferrell JE., Jr Nature. 2003;426:460. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 28.Jacob R, Merritt JE, Hallam TJ, Rink TJ. Nature. 1988;335:40. doi: 10.1038/335040a0. [DOI] [PubMed] [Google Scholar]
- 29.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elowitz MB, Leibler S. Nature. 2000;403:335. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 31.Tsai TY, et al. Science. 2008;321:126. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker I, Yao Y. Proc Biol Sci. 1991;246:269. doi: 10.1098/rspb.1991.0154. [DOI] [PubMed] [Google Scholar]
- 33.Augustine GJ, Neher E. Curr Opin Neurobiol. 1992;2:302. doi: 10.1016/0959-4388(92)90119-6. [DOI] [PubMed] [Google Scholar]
- 34.Meyer T, Stryer L. Annu Rev Biophys Biophys Chem. 1991;20:153. doi: 10.1146/annurev.bb.20.060191.001101. [DOI] [PubMed] [Google Scholar]
- 35.Cornell-Bell AH, Finkbeiner SM, Cooper MS, Smith SJ. Science. 1990;247:470. doi: 10.1126/science.1967852. [DOI] [PubMed] [Google Scholar]
- 36.Sai J, et al. J Biol Chem. in press. [Google Scholar]
- 37.Fivaz M, et al. Curr Biol. 2008;18:44. doi: 10.1016/j.cub.2007.11.051. [DOI] [PubMed] [Google Scholar]
- 38.Foskett JK, et al. Physiol Rev. 2007;87:593. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Science. 2005;310:496. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Politi A, Gaspers LD, Thomas AP, Hofer T. Biophys J. 2006;90:3120. doi: 10.1529/biophysj.105.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrell JE., Jr Curr Biol. 2008;18:R244. doi: 10.1016/j.cub.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harootunian AT, Kao JP, Paranjape S, Tsien RY. Science. 1991;251:75. doi: 10.1126/science.1986413. [DOI] [PubMed] [Google Scholar]
- 43.Liou J, et al. Curr Biol. 2005;15:1235. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis RS. Annu Rev Immunol. 2001;19:497. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 45.Arrieumerlou C, Meyer T. Dev Cell. 2005;8:215. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Iglesias PA, Devreotes PN. Curr Opin Cell Biol. 2008;20:35. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Walker JW, Somlyo AV, Goldman YE, Somlyo AP, Trentham DR. Nature. 1987;327:249. doi: 10.1038/327249a0. [DOI] [PubMed] [Google Scholar]
- 48.Weiner OD, et al. Nat Cell Biol. 2002;4:509. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue T, Meyer T. PLoS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


