SUMMARY
ATM and ATR are two master checkpoint kinases activated by double-strand DNA breaks (DSBs). ATM is critical for the initial response and the subsequent ATR activation. Here, we show that ATR activation is coupled with loss of ATM activation, an unexpected ATM-to-ATR switch during the biphasic DSB response. ATM is activated by DSBs with blunts ends or short single-strand overhangs (SSOs). Surprisingly, the activation of ATM in the presence of SSOs, like that of ATR, relies on single- and double-stranded DNA junctions. In a length-dependent manner, SSOs attenuate ATM activation and potentiate ATR activation through a swap of DNA damage sensors. Progressive resection of DSBs directly promotes the ATM-to-ATR switch in vitro. In cells, the ATM-to-ATR switch is driven by both ATM and the nucleases participating in DSB resection. Thus, single-stranded DNA orchestrates ATM and ATR to function orderly and reciprocally in two distinct phases of DSB response.
INTRODUCTION
Double-strand DNA breaks (DSBs) are among the most deleterious DNA lesions that threaten genomic integrity. DSBs are generated not only by exogenous DNA-damaging agents, but also by normal cellular processes such as V(D)J recombination, meiosis, and DNA replication. Furthermore, increased amounts of DSBs are induced by oncogenic stresses during the early stage of tumorigenesis (Bartkova et al., 2005). In response to DSBs, the ATM kinase phosphorylates and regulates a large number of substrates involved in DNA repair, DNA replication, and other cellular processes important for genomic stability (Matsuoka et al., 2007). In addition to DSBs, ATM also responds to other cellular stresses such as hypoxia and chromatin alterations (Bakkenist and Kastan, 2003; Bencokova et al., 2008; Gibson et al., 2005). Mutations of ATM in humans result in ataxia-telangiectasia (AT), a genetic disorder associated with radiation sensitivity, neuron degeneration, immune deficiencies, premature aging, and predisposition to cancers (Shiloh and Kastan, 2001). ATM is also one of the most frequently mutated kinases in human cancers (Greenman et al., 2007). All evidence points to that ATM is a crucial guardian of genomic integrity.
The mechanisms by which ATM is activated have been under intensive investigation (Harper and Elledge, 2007). The activation of ATM coincides with the autophosphorylation of ATM at Ser1981 and the conversion of ATM oligomers to monomers (Bakkenist and Kastan, 2003). The Mre11-Rad50-Nbs1 (MRN) complex is a sensor of DSBs and a direct activator of the ATM kinase (Lee and Paull, 2005). While ATM is not solely regulated by MRN in vivo (Kanu and Behrens, 2007), its activation at DSBs is primarily mediated by MRN (Berkovich et al., 2007; Falck et al., 2005; Kitagawa et al., 2004; You et al., 2005). After the initial ATM activation by DSBs, ATM executes specific functions around the breaks through a chromatin-mediated mechanism involving H2AX, Mdc1, and other proteins (Lou et al., 2006; Stewart et al., 2003; Stucki et al., 2005). Direct tethering of a large number of ATM molecules or its regulators to an array of binding sites activates ATM even in the absence of DSBs (Soutoglou and Misteli, 2008), indicating that a critical function of DSBs in ATM activation is to nucleate ATM and its regulators at sites of DNA damage. The activation of ATM at and around actual DSBs is a step-wise process initiated by the breaks. Despite the clear involvement of DSBs in ATM activation, the exact DNA structural determinants for ATM activation have not been clearly defined. Furthermore, how the structures of DNA at DSBs contribute to ATM activation is not well understood.
In addition to ATM, DSBs also activate ATR, another master checkpoint kinase that has overlapping substrate specificity with ATM. Like ATM, ATR is critical for the full checkpoint response to DSBs (Brown and Baltimore, 2003; Cortez et al., 2001), indicating that ATM and ATR have non-redundant functions in this process. Unlike ATM, however, ATR also responds to a broad spectrum of DNA damage besides DSBs, especially the damage interfering with DNA replication. The recruitment of ATR to DSBs requires RPA-coated single-stranded DNA (RPA-ssDNA), a structure generated by the nuclease-mediated resection of DSBs (Zou and Elledge, 2003). The junctions between single- and double-stranded DNA, another structure associated with resected DSBs, are also important for ATR activation (MacDougall et al., 2007; Zou, 2007). Several nucleases and helicases, including MRN, CtIP, Exo1, and BLM have been implicated in the resection of DSBs (Gravel et al., 2008; Lengsfeld et al., 2007; Limbo et al., 2007; Mimitou and Symington, 2008; Sartori et al., 2007; Schaetzlein et al., 2007; Zhu et al., 2008). Interestingly, ATM is required for the efficient resection of DSBs and the activation of ATR by DSBs (Jazayeri et al., 2006; Myers and Cortez, 2006; Yoo et al., 2007).
The sequential activation of ATM and ATR by DSBs suggests that the checkpoint response to DSBs is biphasic. Although ATM is clearly critical for the initial response to DSBs, how ATM and ATR orchestrate the second phase of checkpoint response is unclear. A particularly interesting question is how ATM and ATR are coordinated at the DSBs undergoing resection, a dynamic structure that integrates checkpoint signaling with DNA repair. While the respective activation of ATM and ATR by DSBs has been extensively studied, these kinases and the DNA structures regulating them have rarely been characterized as a whole. Furthermore, the fundamental question of how exactly ATM and ATR distinguish DNA damage structures remains to be addressed.
In this study, using a newly developed ATM/ATR activation assay, we show that the activation of ATM is regulated by multiple DNA structural elements of DSBs. More importantly, we reveal that ATM and ATR are activated by similar yet distinct DNA structures at resected DSBs. While both ATM and ATR depend on the junctions of single- and double-stranded DNA for activation, they are oppositely regulated by the lengthening of single-stranded overhangs (SSOs). SSOs simultaneously attenuate ATM activation and potentiate ATR activation, thereby promoting an ATM-to-ATR switch during the process of DSB resection. These findings provide mechanistic insights into how the DNA-damage specificities of ATM and ATR are distinct from each other and, furthermore, how ATM and ATR function in concert to bring about the biphasic DSB response.
RESULTS
Double-stranded DNA- and Nbs1-dependent ATM activation in vitro
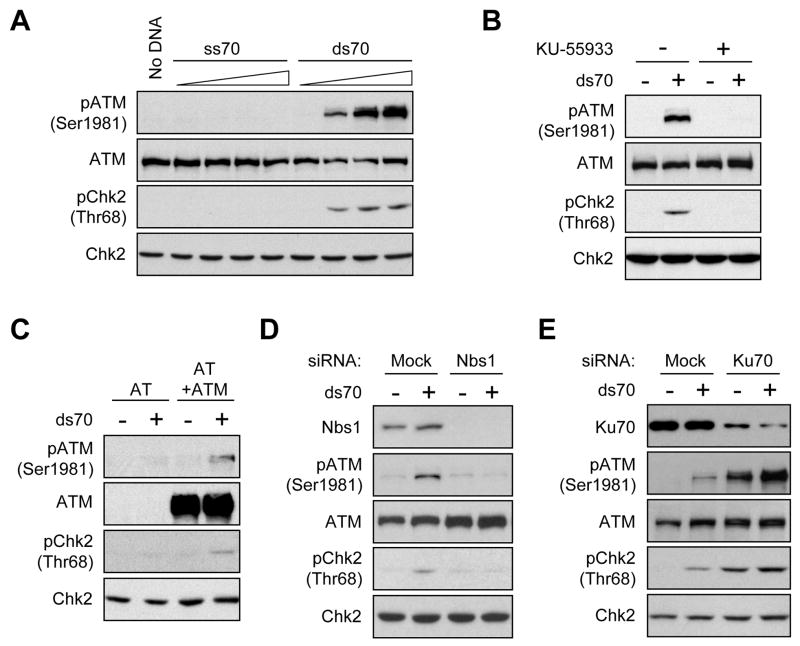
Biochemical studies using purified proteins or Xenopus extracts have shown that ATM can be activated by DNA fragments in vitro (Dupre et al., 2006; Lee and Paull, 2005; Yoo et al., 2004; You et al., 2007). To reveal the DNA structural determinants for ATM activation, we devised an in vitro ATM activation assay using human cell extracts and defined DNA structures. A 70-bp dsDNA fragment with blunt ends was generated using two complementary ssDNA oligomers. In HeLa cell nuclear extracts, dsDNA but not ssDNA induced the phosphorylation of ATM at Ser1981 in a concentration-dependent manner (Fig. 1A). The phosphorylation of Chk2 at Thr68, a known ATM substrate site in cells, was also induced by dsDNA (Fig. 1A). The dsDNA-induced phosphorylation of ATM and Chk2 was inhibited by KU-55933, a specific ATM inhibitor, suggesting that these phosphorylation events are ATM-dependent (Fig. 1B). The dsDNA-induced phosphorylation of ATM and Chk2 was not detected in AT cell extracts, but was detected in the extracts of the AT cells complemented with ATM (Fig. 1C), confirming that the phosphorylation of Chk2 is ATM-dependent.
Figure 1. Double-stranded DNA-induced ATM Activation in Human Cell Extracts.
(A) dsDNA but not ssDNA induces the phosphorylation of ATM and Chk2 in a concentration-dependent manner. Increasing concentrations (1.25, 12.5, 125, and 1250 nM) of ssDNA (ss70) or dsDNA (ds70) was incubated in HeLa cell nuclear extracts. In all the panels in this figure, the levels of phospho-ATM (Ser1981), ATM, phospho-Chk2 (Thr68), and Chk2 were analyzed by Western blotting. (B) dsDNA-induced phosphorylation of ATM and Chk2 is inhibited by ATM inhibitor. ds70 (125 nM) was incubated in extracts in the presence or absence of 10 μM KU-55933. (C) dsDNA-induced Chk2 phosphorylation depends on ATM in extracts. ds70 (125 nM) was incubated in the nuclear extracts derived from AT cells or the AT cells complemented with ATM. (D) dsDNA-induced phosphorylation of ATM and Chk2 is Nbs1-dependent. ds70 (125 nM) was incubated in the extracts derived from cells treated with Nbs1 siRNA or cells mock treated. (E) dsDNA-induced phosphorylation of ATM and Chk2 is Ku70-independent. ds70 (125 nM) was incubated in the extracts derived from cells treated with Ku70 siRNA or cells mock treated.
To further assess if the dsDNA-induced phosphorylation of ATM and Chk2 indeed reflects the activation of ATM in extracts, we asked if it is dependent on Nbs1 or Ku70. In cells, Nbs1 is critical for the activation of ATM at DSBs, whereas Ku70 is required for the activation of DNA-PKcs, another kinase responsive to DSBs. We generated extracts from the HeLa cells in which Nbs1 or Ku70 was depleted by siRNA. The induction of ATM and Chk2 phosphorylation by dsDNA was significantly diminished in the Nbs1-depleted extracts compared to the controls (Fig. 1D). In marked contrast, in the extracts with reduced levels of Ku70, ATM and Chk2 were substantially phosphorylated even when no dsDNA was added (Fig. 1E). This phosphorylation of ATM and Chk2 may be due to the genomic instability in Ku70-depleted cells, or the binding of MRN to the residual genomic DNA in extracts when Ku70 was removed. Despite this basal phosphorylation, ATM and Chk2 were further phosphorylated when dsDNA was added to the Ku70-depleted extracts. These results suggest that the DSB-induced phosphorylation of ATM and Chk2 in extracts, like that in cells, is dependent on Nbs1 but not DNA-PKcs.
To directly determine if ATM is activated by dsDNA in extracts, we measured the kinase activity of ATM. As revealed by in vitro kinase assays with immunoprecipitated ATM, dsDNA stimulated the kinase activity of ATM by approximately 2-fold in extracts (Fig. S1). Similar elevations of ATM kinase activity were observed in cells treated with ionizing radiation (IR) (Pandita et al., 2000). Collectively, these results suggest that the activation of ATM by dsDNA in extracts closely resembles the activation of ATM by DSBs in cells.
dsDNA regulates ATM activation through length- and end-dependent mechanisms
Using the in vitro assay above, we sought to systematically characterize the DNA structural determinants for ATM activation. Studies using purified proteins or Xenopus extracts have shown that ATM is activated by dsDNA in a length-dependent manner (Lee and Paull, 2005; You et al., 2007). In these studies, only the DNA fragments longer than 200 bp efficiently activated ATM (Lee and Paull, 2005; You et al., 2007). In HeLa extracts, however, even 12.5 nM of 70-bp dsDNA (1.5×1010 DNA ends μl−1) induced substantial ATM phosphorylation (Fig. 2A). The high sensitivity of this assay allowed us to analyze short dsDNA fragments with defined structural features.
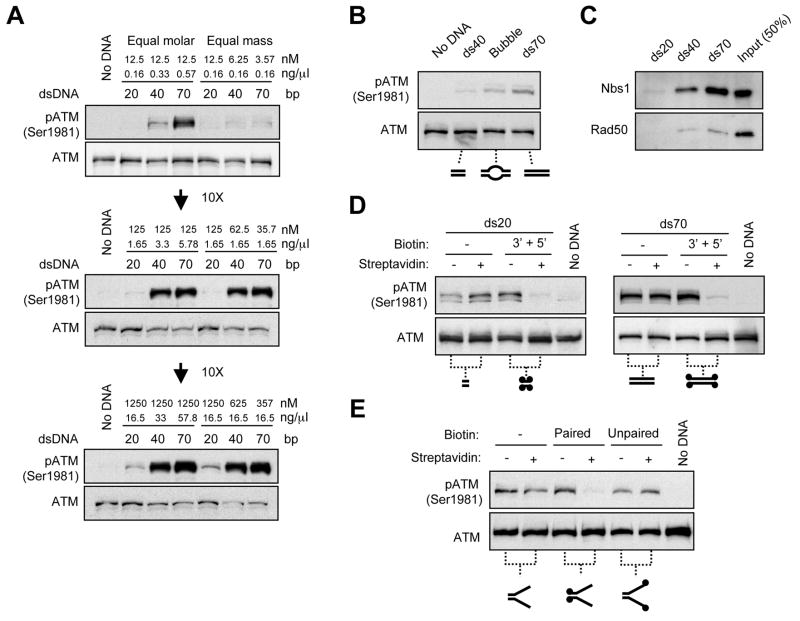
Figure 2. The Activation of ATM by dsDNA is Length- and End-Dependent.
(A) dsDNA activates ATM in a length-dependent manner. ds20, ds40, and ds70 were incubated in HeLa cell nuclear extracts at the indicated concentrations. The three DNA fragments were used at either equal molar concentrations (the left half) or equal DNA mass (the right half). The three panels represent the experiments done with three different sets of DNA concentrations, which increase by 10-fold from top to middle, and from middle to bottom panel. ATM and phospho-ATM were analyzed by Western blotting. (B) The dsDNA proximal to breaks contributes to ATM activation. ds40, bubble (30-nt ssDNA flanked by 20-bp dsDNA on each side), and ds70 (all at 12.5 nM) were incubated in extracts and analyzed as above. (C) The binding of MRN to dsDNA is length-dependent. Biotinylated ds20, ds40, and ds70 (all at 6.25 pmole) were attached to streptavidin-coated beads and incubated with purified MRN. The DNA-bound Nbs1 and Rad50 were detected by Western blotting. (D) ATM activation depends on DNA ends. ds20 (left) and ds70 (right) were unmodified or biotinylated at all 4 DNA ends. ds20 (1.25 μM) and ds70 (12.5 nM) were incubated with or without streptavidin and added to extracts. (E) Paired DNA ends are required for ATM activation. The paired or unpaired ends of a fork structure (20-bp dsDNA and 50-nt ssDNA; 1.25 μM) were blocked with streptavidin as indicated, and were analyzed in extracts as above. The DNA structures used in each experiment are depicted below the corresponding panel, with the dots representing biotin.
We first asked whether and how ATM is activated by dsDNA in a length-dependent manner in human cell extracts. When present at the same molar concentrations or the same DNA mass, dsDNA of 70 bp, 40 bp, or 20 bp induced ATM phosphorylation in a length-dependent manner (Fig. 2A). Since these dsDNA fragments are much shorter than the DNA of a single nucleosome, a length-dependent mechanism for ATM activation may operate on the nucleosome-free dsDNA immediately flanking the breaks. To investigate how the length of dsDNA contributes to ATM activation, we generated a “bubble” DNA structure by converting an internal 30-bp region of the 70-bp dsDNA into a single-stranded region (Fig. 2B). The ability of the bubble structure to induce ATM phosphorylation was between those of the 70-bp and the 40-bp dsDNA (Fig. 2B), showing that the internal region of the 70-bp dsDNA contributes to the length-dependent activation of ATM. Using purified MRN complexes, we found that greater amounts of Nbs1 and Rad50 associated with 70-bp dsDNA than 40- and 20-bp dsDNA (Fig. 2C). These results suggest that the MRN complex associates with nucleosome-free dsDNA in a length-dependent manner, providing a possible mechanism for ATM activation along dsDNA.
To assess if the ends of dsDNA are critical for ATM activation, we biotinylated all four DNA ends of the 20- and 70-bp fragments (5′ and 3′ ends of both strands). The biotinylated dsDNA efficiently induced ATM phosphorylation in the absence of streptavidin, but lost this activity when the ends were blocked by streptavidin (Fig. 2D). When only the 5′ or 3′ ends of 70-bp dsDNA were blocked, the ability of the fragment to activate ATM was substantially reduced (Fig. S2). Since the streptavidin on one DNA strand may block access to both strands, it was not possible to resolve how 5′ or 3′ ends contribute to ATM activation. Nonetheless, blockage of DNA ends inhibited ATM activation regardless of the length of dsDNA, suggesting that the length-dependent mechanism for ATM activation needs to be initiated from DNA ends, or act through the ends.
The ends of dsDNA could potentially be processed by helicases and/or nucleases in extracts. To assess how unwinding of dsDNA affects ATM activation in extracts, we generated a fork-like DNA structure that possesses both paired and unpaired DNA ends (Fig. 2E). The ability of the fork structure to activate ATM was lost when the paired ends were blocked, but was unaffected when the unpaired ends were blocked (Fig. 2E). Therefore, paired DNA ends are required for initiating ATM activation in extracts. These results suggest that ATM cannot be directly activated by unwound DNA ends or by the fork-like DNA structures associated with DNA replication or DNA repair.
Single-strand overhangs interfere with ATM activation by attenuating MRN binding
DSBs are not always blunt-ended in cells. The DSBs generated by the HO or I-SceI endonuclease initially have 4-nt 3′ single-strand overhangs (SSOs) (Colleaux et al., 1988; Kostriken et al., 1983). V(D)J recombination and meiosis produce DSBs with 3′ and 5′ SSOs, respectively (Schlissel, 1998; Xu and Kleckner, 1995). The DSBs resulting from collapsed replication forks or broken ssDNA gaps may possess either 3′ or 5′ SSOs (Lopes et al., 2006). The “uncapped” telomeres resemble DSBs with 3′ SSOs (Celli and de Lange, 2005). When exposed in cells, DSBs can be resected by exo- or endonucleases in the 5′-to-3′ direction (Lee et al., 1998). ATM has been implicated in the response to the various types of DSBs above, indicating that it can be activated by DSBs with SSOs. In extracts, while the bulk of dsDNA appeared unaltered (Fig. S3), a small fraction of it might be processed by nucleases. The in vivo functions of ATM in the response to SSO-bearing DSBs prompted us to investigate the role of SSOs in ATM activation.
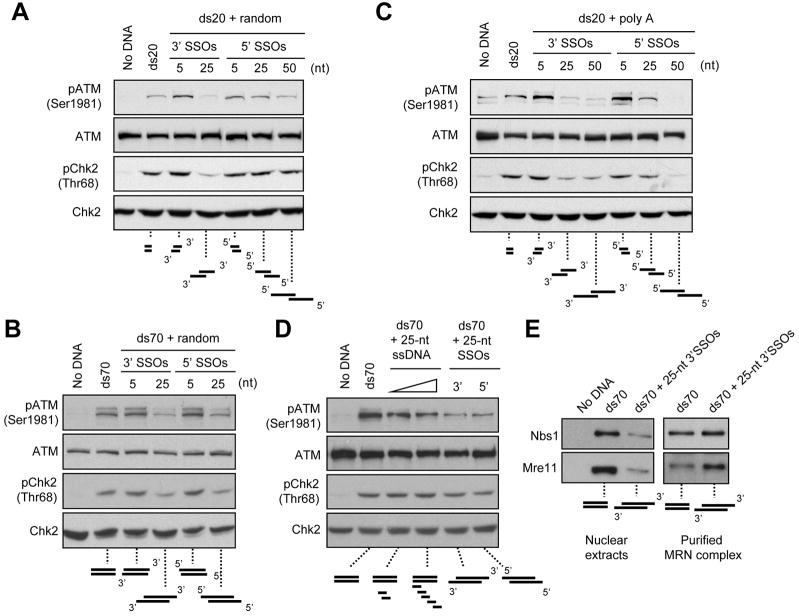
To directly assess the effects of SSOs on ATM activation, we analyzed the 20- and 70-bp dsDNA bearing either 5′ or 3′ SSOs of random sequences (Figs. 3A and 3B). Both 5′ and 3′ SSOs of 5 nt slightly enhanced ATM phosphorylation. Interestingly, both 5′ and 3′ SSOs of 25 or 50 nt attenuated ATM and Chk2 phosphorylation (Figs. 3A and 3B), suggesting that SSOs interfere with ATM activation in a length-dependent manner. SSOs of poly A also hindered ATM activation in a length-dependent manner (Fig. 3C). SSOs not only attenuated the activation of ATM by 20- and 70-bp dsDNA, but also that by linear plasmids (see Fig. 5). Together, these results suggest that SSOs may interfere with a DNA end-dependent event in ATM activation, which is independent of the length of dsDNA.
Figure 3. Regulation of ATM Activation by SSOs.
(A–B) SSOs of random sequences interfere with ATM activation in a manner dependent on the length of SSO, but independent of the length of dsDNA. (A) ds20 (1.25 μM) and (B) ds70 (12.5 nM) with 3′ or 5′ SSOs at the indicated length were incubated in extracts. ATM, phospho-ATM, Chk2, and phospho-Chk2 were analyzed by Western blotting. (C) Poly A SSOs interfere with ATM activation in a length-dependent manner. ds20 (1.25 μM) with 3′ or 5′ poly A SSOs at the indicated length was analyzed as above. (D) ssDNA interferes with ATM activation more efficiently in cis than in trans. ds70 (12.5 nM), either alone or in combination with free 25-nt random ssDNA (25 or 50 nM), and ds70 with 3′ or 5′ 25-nt SSOs of random sequences (12.5 nM) were incubated in extracts and analyzed as above. (E) SSOs attenuate the binding of MRN to dsDNA in extracts, but not in an assay using purified proteins. ds70 and ds70 with 25-nt 3′ SSOs were biotinylated at the 3′ end of one of the two strands. The DNA structures (1.25 pmole) were incubated with extracts or purified MRN complex and retrieved with streptavidin-coated beads. The Nbs1 and Mre11 associated with DNA were analyzed by Western blotting.
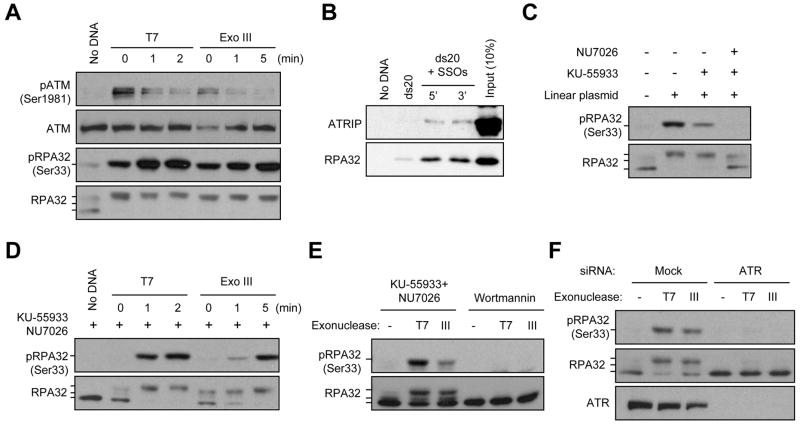
Figure 5. Resection of DNA Ends Promotes an ATM-to-ATR Switch in vitro.
(A) Inhibition of ATM activation by resection of DNA ends. A plasmid linearized with HpaI was treated with T7 exonuclease (T7) or Exonuclease III (III) to generate SSOs. The resulting DNA (12 ng/μl) was incubated in extracts. (B) Recruitment of RPA and ATRIP to SSOs. ds20 and ds20 with 5′ or 3′ 50-nt poly A SSOs (1.25 pmole) were attached to beads and incubated in extracts. The RPA and ATRIP associated with DNA were detected by Western blotting. (C) The phosphorylation of RPA32 induced by unprocessed linear plasmid is dependent on ATM and DNA-PKcs. Linear plasmid (4 ng/μl) was incubated in extracts in the presence of KU-55933 and NU7026 (10 μM). RPA and phospho-RPA (Ser33) were analyzed by Western blotting. (D) Induction of RPA phosphorylation (Ser33) by resection of DNA ends. Linear plasmid was resected as in (A) and incubated in extracts in the presence of KU-55933 and NU7026 (10 μM). (E) The SSO-induced RPA32 phosphorylation is independent of ATM and DNA-PKcs. Linear plasmid was mock treated or processed by T7 exonuclease (2 min) and Exonuclease III (5 min). The resulting DNA (4 ng/μl) was incubated in extracts with KU-55933 and NU7026 (10 μM), or Wortmannin (20 μM). (F) The SSO-induced RPA32 phosphorylation is ATR-dependent. Nuclear extracts were prepared from cells treated with ATR siRNA or cells mock treated. Linear plasmid was resected as in (E) and incubated in both extracts.
The ssDNA generated by resection may interfere with ATM activation in cis or in trans. When blunt-ended 70-bp dsDNA was added to extracts with 25-nt ssDNA at 1:2 or 1:5 molar ratios, a modest reduction of ATM activation was observed (Fig. 3D). When the 25-nt ssDNA was linked to 70-bp dsDNA as overhangs, it interfered with ATM activation more effectively. Thus, while ssDNA can interfere with ATM activation both in cis and in trans, SSOs are more potent than free ssDNA for this function.
To reveal the mechanism by which SSOs interfere with ATM activation, we asked if SSOs affect the binding of MRN to dsDNA. Indeed, 3′ SSOs of 25 nt substantially reduced the amounts of Nbs1 and Mre11 associated with 70-bp dsDNA in extracts (Fig. 3E). However, purified MRN bound to dsDNA efficiently regardless of the presence or absence of SSOs (Fig. 3E). Together, these results suggest that SSOs do not directly interfere with the binding of MRN to dsDNA, but they reduce MRN binding in the presence of other proteins.
ATM activation requires junctions of single- and double-stranded DNA
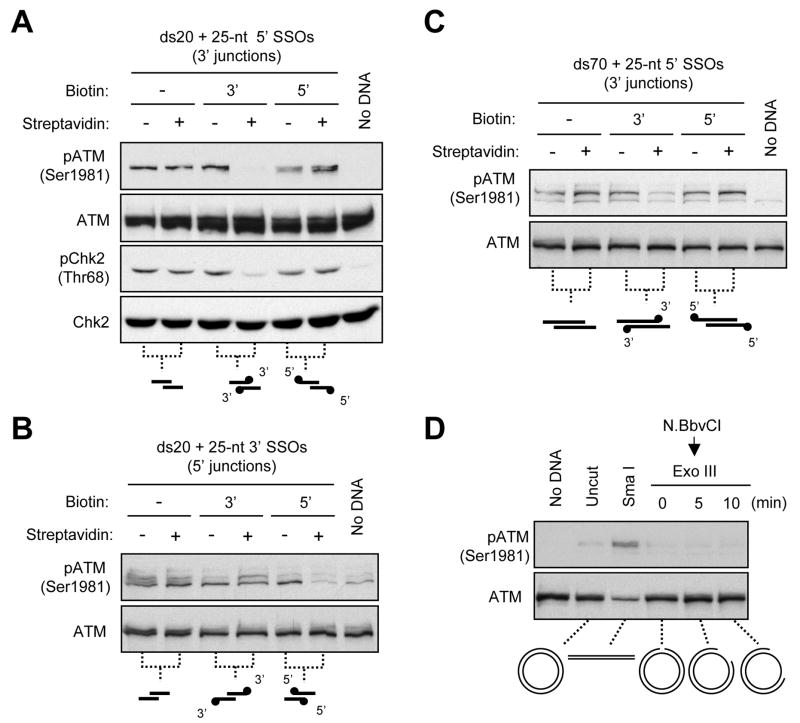
Although less potent than blunt-ended dsDNA, dsDNA bearing short SSOs retain some ability to associate with MRN and to active ATM in extracts. Our analysis of blunt-ended dsDNA suggest that ATM activation is dependent on DNA ends (Fig. 2). Two types of DNA ends are present in the DNA fragments with SSOs: the ends of the dsDNA region (the junctions of dsDNA/ssDNA) and the ends of SSOs (Fig. 4A). To assess how these DNA ends contribute to ATM activation, we tested three sets of DNA structures (20-bp dsDNA with 5′ or 3′ 25-nt SSOs and 70-bp dsDNA with 3′ 25-nt SSOs) in which either the junctions or the SSO ends were biotinylated (Figs. 4A–C). In the absence of streptavidin, all of the DNA structures with SSOs induced ATM phosphorylation at reduced levels compared to blunt-ended dsDNA (Figs. 3A–D). When the ends of the 5′ or 3′ SSOs were blocked by streptavidin, the ability of the DNA fragments to activate ATM and Chk2 was not affected (Figs. 4A–C). In striking contrast, when the 5′ or 3′ junctions of dsDNA/ssDNA were blocked by streptavidin, the DNA fragments failed to activate ATM and Chk2 (Figs. 4A–C). These results suggest that the junctions of dsDNA/ssDNA, but not the ends of SSOs, are critical for ATM activation. Furthermore, the junctions of dsDNA/ssDNA are required for ATM activation regardless of the length of dsDNA (Figs. 4A–C), suggesting that these ends are involved in an initiating event for ATM activation, possibly the DNA recognition by MRN.
Figure 4. ATM Activation Requires the Junctions of ssDNA and dsDNA.
(A–C) ATM activation is dependent on ssDNA/dsDNA junctions but not SSO ends. ds20 with 5′ (A) or 3′ (B) 25-nt SSOs, and ds70 with 5′ 25-nt SSOs (C) were unmodified or biotinylated at the junctions or the SSO ends. These ds20- and ds70-derived DNA structures (1.25 μM and 12.5 nM, respectively) were incubated with or without streptavidin and added to extracts. ATM, phospho-ATM, Chk2, and phospho-Chk2 were analyzed by Western blotting. (D) ATM is not activated by DNA nicks and gaps. A plasmid was nicked by N. BbvCI or linearized by SmaI (Fig. S4A). The resulting nicked plasmid was further treated with Exonuclease III for the indicated periods to generate ssDNA gaps. Uncut, linear, nicked, and gapped plasmids (4 ng/μl) were incubated in extracts and analyzed as above.
The junctions of dsDNA/ssDNA are present not only at DSBs, but also at single-strand DNA breaks, gaps, and DNA replication forks. To assess if dsDNA/ssDNA junctions are sufficient to activate ATM, we generated a plasmid carrying a single cleavage site of the nicking enzyme N. BbvCI (Fig. S4A). Using the nicking enzyme or a restriction enzyme that cuts the plasmid in both DNA strands, we generated nicked plasmids and linear plasmids bearing blunt ends (Fig. S4B). Like the short dsDNA fragments, linear plasmids induced ATM phosphorylation (Fig. 4D). In contrast, nicked plasmids were unable to induce any ATM phosphorylation (Fig. 4D). Moreover, when the DNA nicks were extended into ssDNA gaps by Exonuclease III (Fig. S4C), the gap-carrying plasmids were still unable to activate ATM (Fig. 4D). Thus, while the junctions of dsDNA/ssDNA are required for ATM activation at DSBs, they are not sufficient to elicit ATM response when present internally on DNA. These internal junctions may be recognized by proteins that inhibit ATM activation. Alternatively, additional structural features of DSBs, such as the topological state of DNA (Fig. S4B), may be involved in ATM activation.
Resection of DNA ends promotes an ATM-to-ATR switch
The involvement of dsDNA/ssDNA junctions in ATM activation is surprising because these structures have been implicated in the activation of ATR (MacDougall et al., 2007; Zou, 2007). These results raise the question as to how the DNA-damage specificities of ATM and ATR are distinct from each other at DSBs, and how ATM and ATR are coordinated at the DSBs undergoing resection. In human cells, ATM is required for DSB resection and ATR activation (Jazayeri et al., 2006). In this study, we show that SSOs interfere with ATM activation in a length-dependent manner (Fig. 3). In addition, we have previously shown that ssDNA coated by RPA binds to ATRIP in a length-dependent manner, allowing the ATR-ATRIP kinase complex to recognize DSBs (Zou and Elledge, 2003). These findings led us to hypothesize that following the activation of ATM and the initiation of resection, SSOs might promote an ATM-to-ATR switch at DSBs.
To directly investigate if the process of SSO generation can restrict ATM activation and induce ATR activation, we used exonucleases to resect the DNA ends of a linear plasmid (Fig. S5A). In a time-dependent manner, T7 exonuclease progressively resects DNA ends in the 5′-to-3′ direction, whereas Exonuclease III cleaves in the 3′-to-5′ direction (Fig. S5B). When the same amounts of processed or unprocessed plasmids were added to extracts, the processed plasmids exhibited a reduced ability to activate ATM compared to the unprocessed plasmids (Fig. 5A). These results confirm that the generation of SSOs progressively interferes with ATM activation (Figs. 3A-C).
When generated at DSBs in cells, SSOs are recognized by RPA, leading to a DNA-protein structure recruiting ATR-ATRIP (Zou and Elledge, 2003). In extracts, SSOs of 50 nt associated with both RPA and ATRIP (Fig. 5B). To assess if SSOs induce ATR activation, we monitored the phosphorylation of RPA32 at Ser33 (Olson et al., 2006). The phosphorylation of RPA32 was induced by linear plasmids even in the absence of exonuclease (Fig. 5C). This phosphorylation of RPA32 was partially inhibited by KU-55933 alone, and virtually abolished by the combination of KU-55933 and NU7026, a specific inhibitor of DNA-PKcs, suggesting that the RPA32 phosphorylation induced by linear plasmids involves both ATM and DNA-PKcs. Interestingly, the phosphorylation of RPA32 was progressively enhanced by the exonuclease-mediated resection of DNA ends (Figs. 5A and 5D). This is in marked contrast to the decline of ATM phosphorylation when SSOs were generated (Fig. 5A). Furthermore, the phosphorylation of RPA32 induced by resected ends was not inhibited by the combination of KU-55933 and NU7026, but was inhibited by Wortmannin, a pan-inhibitor of ATR, ATM, and DNA-PKcs (Fig. 5E). These results suggest that the SSO-induced RPA32 phosphorylation is independent of ATM and DNA-PKcs, but may be dependent on ATR.
To directly address if the increased phosphorylation of RPA32 reflects the activation of ATR, we used siRNA to knockdown ATR in HeLa cells and generated nuclear extracts from these cells. KU-55933 and NU7026 were added to the extracts to eliminate the potential contributions of DNA-PKcs and ATM to RPA32 phosphorylation. In the absence of exonucleases, linear plasmids did not induce RPA32 phosphorylation in the ATR-depleted or the control extracts (Fig. 5F). Importantly, in the presence of exonucleases, the phosphorylation of RPA32 was efficiently induced in the control extracts, but not in the extracts lacking ATR (Fig. 5F). Together, these results demonstrate that the generation of SSOs not only interferes with ATM activation, but also promotes ATR activation.
Consecutive activation of ATM and ATR in cells
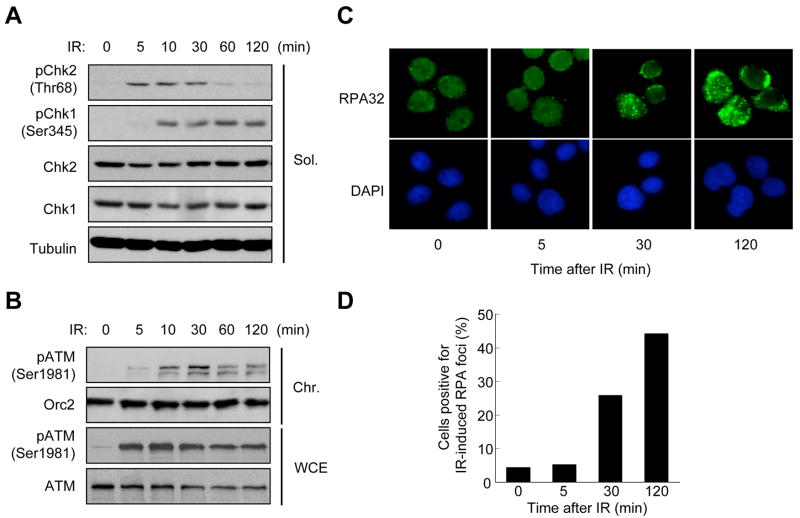
The opposite effects of SSOs on ATM activation and ATR activation prompted us to investigate if ATM and ATR are reciprocally regulated in cells. We followed the IR-induced phosphorylation of Chk2 and Chk1, two specific substrates of ATM and ATR, respectively. Within 5 min after IR, Chk2 but not Chk1 was strongly phosphorylated, showing that Chk2 is phosphorylated more rapidly than Chk1 (Fig. 6A). Furthermore, Chk2 phosphorylation started to decline 30 min after IR, whereas Chk1 phosphorylation remained at high levels until 2 hr (Fig. 6A). These results show that Chk2 is transiently phosphorylated during a window that precedes the window of Chk1 phosphorylation. Nevertheless, there was a short period (10 to 30 min) in which both Chk2 and Chk1 were strongly phosphorylated. This may be due to asynchronous resection of DSBs in individual cells (Barlow et al., 2008; Zierhut and Diffley, 2008), or asynchronous resection in different cell sub-populations (Jazayeri et al., 2006). It should be noted that IR not only induces DSBs but also other types of DNA damage (Ward, 2000), some of which may interfere with DNA replication and lead to delayed ATR/ATM response. Overall, the decline of Chk2 phosphorylation coincided with strong Chk1 phosphorylation, which is consistent with an ATM-to-ATR switch in these cells.
Figure 6. Consecutive Activation of ATM and ATR in Cells.
(A) Distinct kinetics of Chk2 and Chk1 phosphorylation after IR. HeLa cells were treated with 10 Gy of IR and were analyzed at the indicated time points. The levels of phospho-Chk2 (Thr68), Chk2, phospho-Chk1 (Ser345), Chk1, and Tubulin (a loading control) in the soluble extracts (Sol., see Procedures) were analyzed by Western blotting. (B) Transient accumulation of phosphorylated ATM on chromatin after IR. HeLa cells were treated with 10 Gy of IR and the chromatin fractions (Chr., see Procedures) and whole cell extracts (WCE) were prepared at the indicated time points. The levels of phospho-ATM (Ser1981), ATM and Orc2 (a loading control) were analyzed by Western blotting. (C) Immunostaining of RPA in IR-treated cells. HeLa cells were treated with 10 Gy of IR, and RPA32 was stained at the indicated time points. (D) Quantifications of the cells positive for IR-induced RPA foci in the experiment shown in (C). More than 200 cells were counted at each time point.
Chk2 is phosphorylated by ATM locally at DSBs and then moves throughout the nucleus (Lukas et al., 2003), prompting us to assess more directly if ATM is transiently activated at DSBs. The accumulation of phosphorylated ATM at DSBs marks one of the events during the process of ATM activation (Berkovich et al., 2007; You et al., 2005). Phosphorylated ATM appeared on chromatin within 5 min after IR and started to decline after 30 min (Fig. 6B). Since the overall levels of phosphorylated ATM in cells remain high for many hours after IR (Fig. 6B; Bakkenist and Kastan, 2003), our data indicates that Chk2 is primarily targeted by the phosphorylated ATM associated with DSBs. These results suggest that ATM is transiently activated at DSBs.
To monitor the resection of DSBs in cells, we analyzed the IR-induced RPA foci at different times after irradiation (Figs. 6C and 6D). Few RPA foci were detected 5 min after IR. At 30 min, RPA foci appeared in a significant fraction of cells. At 120 min, approximately half of the cells exhibited intense RPA foci. Thus, RPA gradually accumulated at DSBs as ATM activation was attenuated. The numerous RPA foci at 120 min suggest that the attenuation of ATM activation was not due to the completion of DNA repair. These results provide further evidence that the activation of ATM is gradually attenuated by DSB resection in cells. Furthermore, because RPA-ssDNA is a key structure involved in ATR activation, these results also provide in vivo evidence linking the attenuation of ATM activation to ATR activation.
Regulation of the ATM-to-ATR switch by ATM and nucleases
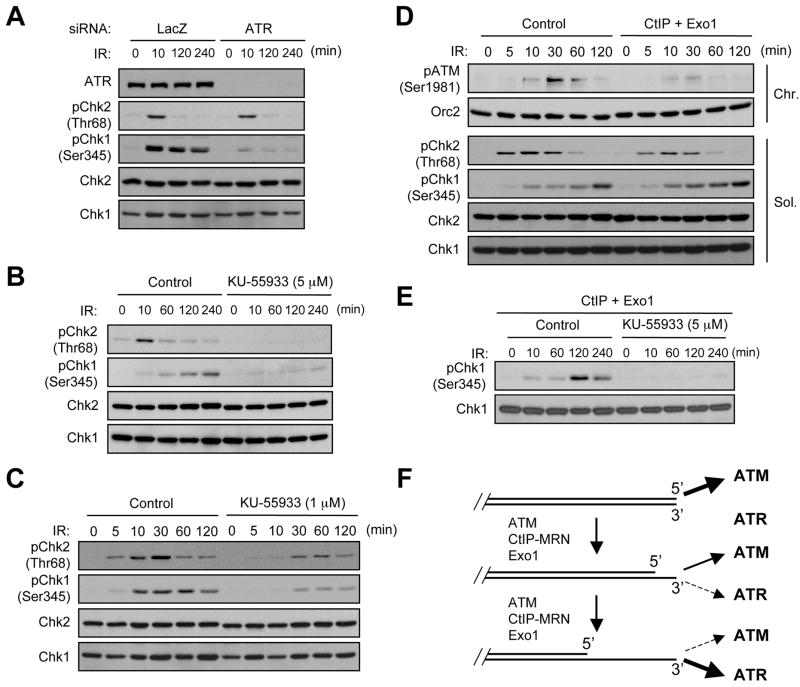
The attenuation of ATM activation at resected DSBs could be attributed to the generation of SSO or the consequent ATR activation. To distinguish these possibilities, we analyzed the phosphorylation of Chk1 and Chk2 in cells treated with ATR siRNA. While Chk1 phosphorylation was abolished in cells lacking ATR, the kinetics of Chk2 phosphorylation was not altered in these cells (Fig. 7A). These results strongly suggest that the process of DSB resection, rather than the activation of ATR, is responsible of the loss of ATM activation.
Figure 7. Regulation of the ATM-to-ATR switch by ATM, CtIP, and Exo1.
(A) ATR is not required for the attenuation of ATM activation by DSB resection. HeLa cells transfected with control siRNA or ATR siRNA were irradiated with IR (10 Gy) and analyzed at the indicated time points. The levels of ATR, phospho-Chk2 (Thr68), Chk2, phospho-Chk1 (Ser345), and Chk1 were monitored by Western blotting. (B) ATM is required for DSB-induced ATR activation. HeLa cells were treated with KU-55933 (5 μM) or mock treated with DMSO for 30 min prior to the IR treatment, and analyzed as above at the indicated time points. (C) Partial inhibition of ATM leads to delayed and reduced activation of Chk2 and Chk1. HeLa cells were treated with KU-55933 (1 μM) or DMSO for 30 min prior to IR and analyzed at the indicated time points. (D) Expression of CtIP and Exo1 promotes the ATM-to-ATR switch in cells. HeLa cells were transfected with the plasmids encoding CtIP and Exo1, and were treated with IR. Chromatin and soluble fractions were prepared from cells collected at the indicated time points. The levels of phospho-ATM (Ser1981), Orc2, phospho-Chk2, (Thr68), Chk2, phospho-Chk1 (Ser345), and Chk1 in the indicated fractions were analyzed as above. (E) Expression of CtIP and Exo1 cannot bypass the requirement of ATM for IR-induced ATR activation. Cells overexpressing CtIP and Exo1 were treated with KU-55933 (5 μM) or mock treated with DMSO for 30 min prior to IR. The levels of phospho-Chk1 (Ser345), and Chk1 were analyzed at the indicated time points. (F) A model for an SSO-orchestrated ATM-to-ATR switch at DSBs.
If the ATM-to-ATR switch is indeed driven by DSB resection in cells, one would expect that this transition is controlled by the regulators of DSB resection. The resection of DSBs in cells is regulated by ATM and exonucleases including MRN-CtIP and Exo1. To test if ATM regulates the ATM-to-ATR switch, we treated cells with KU-55933 prior to IR irradiation. In the presence of 5 μM of KU-55933, the phosphorylation of both Chk2 and Chk1 was virtually abolished (Fig. 7B). When ATM was partially inhibited by 1 μM of KU-55933, both Chk2 phosphorylation and Chk1 phosphorylation were reduced and delayed, and the two events became increasingly overlapped (Fig. 7C). Thus, a timely and synchronous transition from ATM to ATR relies on ATM activity.
To more vigorously test if the ATM-to-ATR switch is driven by DSB resection in cells, we asked if it is possible to promote the switch by enhancing the functions of nucleases. CtIP, an activator of MRN (Sartori et al., 2007), and Exo1 were co-expressed in cells (Fig. S6). In the absence of IR, expression of CtIP and Exo1 did not induce significant ATM and Chk1 phosphorylation (Fig. 7D). In response to IR, CtIP and Exo1 diminished Chk2 phosphorylation but enhanced Chk1 phosphorylation (Fig. 7D), consistent with a more efficient ATM-to-ATR switch. Although ATM phosphorylation was not compromised in cells expressing CtIP and Exo1 (Fig. S6), its retention to chromatin was reduced (Fig. 7D). Interestingly, the IR-induced Chk1 phosphorylation was abolished by KU-55933 in cells expressing CtIP and Exo1 (Fig. 7E), suggesting that ATM may act before CtIP and Exo1 during DSB resection. These results lend strong support to the conclusion that the ATM-to-ATR switch is driven by DSB resection in cells.
DISCUSSION
Both ATM and ATR are key regulators of the cellular response to DSBs, yet how exactly they function in concert is not well understood. Recent studies revealed that ATM is required for the resection of DSBs (Jazayeri et al., 2006; Myers and Cortez, 2006), a process necessary for ATR activation as well as homology-directed DNA repair. While these studies established a critical function of ATM in initiating DSB response, they have not resolved how ATM and ATR function during the dynamic process of DSB resection, a crucial period for both damage signaling and DNA repair. A major obstacle to understanding the coordination of ATM and ATR is that the DNA structural elements regulating ATM activation have not been clearly defined. In this study, we developed an extract-based in vitro assay in which both ATM and ATR can be activated by dsDNA in a DNA structure-regulated manner. Using this assay, we systematically characterized the DNA structural determinants for ATM activation, as well as the orchestration of ATM and ATR at DSBs.
The results of this study addressed two important issues with regard to the activation of ATM at resected DSBs; Can ATM be activated by resected DSBs? If so, is ATM activated by the ends of SSOs or the junctions of single/double-stranded DNA? Our results clearly demonstrated that ATM can be activated in the presence of short SSOs and, furthermore, that ATM activation relies on the junctions of single/double-stranded DNA but not the ends of SSOs (Fig. 4). Given that the activation of ATM by dsDNA requires Nbs1, it is plausible that the MRN complex directly recognizes the junctions of single/double-stranded DNA when SSOs are present. We also show that paired DNA ends are important for this recognition step (Fig. 2E). The MRN complex bound to DNA ends may directly activate ATM and/or initiate the nucleation of ATM at DSBs that leads to its activation. The recognition of DNA ends by MRN may be a prerequisite for its DNA unwinding, tethering, or nuclease activities implicated in ATM activation (Costanzo et al., 2004; Jazayeri et al., 2008; Lee and Paull, 2005; Uziel et al., 2003).
Our results also reveal that the activation of ATM is coordinately regulated by three distinct DNA structural elements of DSBs: (1) DNA ends, (2) dsDNA, and (3) ssDNA. Notably, ATM activation is oppositely regulated by the two DNA structures accompanying DNA ends: it is enhanced by flanking dsDNA, but hindered by SSOs. Both of these regulatory mechanisms operate in a length-dependent manner and may function in concert to quantitatively regulate ATM activation at DNA ends. Interestingly, ATM is involved in the resection of DNA breaks (Jazayeri et al., 2006), suggesting that the activation of ATM elicits an inhibitory feedback loop through SSO formation. While these results present a clear picture of how ATM activation is regulated by the structures of DNA at DSBs, how the DNA- and chromatin-mediated regulatory mechanisms are integrated during ATM activation remains to be determined (Bakkenist and Kastan, 2003; Lou et al., 2006; You et al., 2007). The in vitro ATM activation assay described here may provide a new basis for future biochemical studies to dissect the concerted action of DNA and chromatin in ATM activation.
The involvement of dsDNA/ssDNA junctions in ATM activation reveals an unexpected similarity between the DNA structural specificities of ATM and ATR, suggesting that the choice between activating ATM or ATR at a resected DSB is made by another DNA structure. We have previously shown that ssDNA coated by RPA is the key structure that enables the ATR-ATRIP kinase complex to recognize DSBs (Zou and Elledge, 2003). Our finding that SSOs interfere with ATM activation immediately raised the possibility that ATR activation is coupled with loss of ATM activation through ssDNA. Consistent with this model, RPA gradually accumulates at DSBs (Fig. 6C), whereas Nbs1 associates with DSBs rapidly and transiently when it is unable to retain on the flanking chromatin (Celeste et al., 2003). The activation of yeast Tel1(ATM) is attenuated as DSBs are progressively resected (Mantiero et al., 2007). We find that Chk2, a specific substrate of ATM, is rapidly and transiently phosphorylated after IR treatment. Furthermore, the ATR-dependent Chk1 phosphorylation lags behind Chk2 phosphorylation, and Chk1 phosphorylation increases as Chk2 phosphorylation declines (Fig. 6). Collectively, these results provide compelling evidence that an ATM-to-ATR switch indeed occurs in human cells in response to DSBs.
How does the ATM-to-ATR switch occur at DSBs? The progressive attenuation of ATM activation could be attributed to the loss of DNA structures that activate ATM, or to the generation of DNA structures that interfere with ATM activation. Our finding that SSOs do not directly affect the binding of purified MRN to dsDNA, and that ssDNA interferes with ATM activation in extracts both in cis and in trans support the latter possibility. Both the recruitment of ATR-ATRIP and the interference with ATM activation are dependent on the length of SSOs. As DSBs are progressively resected by nucleases, SSOs are gradually lengthened, simultaneously enhancing the abilities of SSOs to interfere with ATM activation and to promote ATR activation (Fig. 7F). We show that SSOs attenuate the binding of MRN to dsDNA in extracts (Fig. 3E), but facilitate the recruitment of RPA and ATRIP (Fig. 5B). These results suggest that SSOs promote a swap of DNA damage sensors at DSBs, revealing the underlying mechanism for the ATM-to-ATR switch. Interestingly, recent studies suggested that the resection of DSBs by nucleases is also a biphasic process: it is initiated by the MRN-CtIP complex and then extended by the Exo1- or BLM-dependent mechanisms (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). It is plausible that the release of MRN from resected DNA ends may link the nuclease switch with the ATM-to-ATR switch during DSB resection.
We propose that the ATM-to-ATR switch driven by DSB resection is the key mechanism through which the functions of ATM and ATR are coordinated and integrated. The ATM-to-ATR switch is distinct from, but complementary with the sequential activation of ATM and ATR reported previously (Jazayeri et al., 2006). Together, these two regulatory mechanisms ensure that ATM launches DSB response, whereas ATR plays a primary role in the second phase of this dynamic process. The associations of ATM and ATR with DSBs are not only temporally but also spatially distinct (Bekker-Jensen et al., 2006), and are differentially influenced by the cell cycle (Huertas et al., 2008; Ira et al., 2004; Jazayeri et al., 2006; Zierhut and Diffley, 2008). Moreover, the levels of ATM and ATR activation can be fine tuned by DSB resection (Mantiero et al., 2007; Vaze et al., 2002; Zierhut and Diffley, 2008). The ATM-to-ATR switch may enable the two kinases to target distinct sets of downstream effectors in a regulated and coordinated manner. For instance, this switch may couple ATM and ATR with distinct events during DSB repair. The ATM-to-ATR switch driven by DSB resection may provide a unifying mechanism that brings together the temporal, spatial, and quantitative regulations of ATM and ATR, thus orchestrating the collective checkpoint response in human cells. This study has set the stage for future investigations to reveal how the ATM-to-ATR switch conducts the specific functions of ATM and ATR during the biphasic DSB response.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells were cultured in DMEM supplemented with 10% FBS. ATM-deficient (AT) fibroblasts (FT169) and a derivative line complemented with the wild-type ATM (AT+ATM) were cultured in DMEM with 15 % FBS. HeLa cells were transfected twice with 100 nM siRNA using the X-treamGENE transfection reagent (Roche) and were analyzed 4 days after the first transfection. The sequences of the siRNAs used in this study are listed in the Supplemental Data. Plasmid transfections of HeLa cells were performed with Lipofectamin 2000 (Invitrogen) and the transfected cells were analyzed after 48 hr.
Preparation of Cellular Extracts
The nuclear extracts used in the ATM/ATR activation assay were prepared following the Dignam’s protocol (Dignam et al., 1983), except that 600 mM KCl was used in Buffer C. Chromatin fractionation was performed as described previously with modifications (Yang and Zou, 2006). Cells were washed with phosphate buffered saline (PBS) and resuspended in Solution A [10 mM Hepes (pH 7.9) 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mM Na2VO3, protease inhibitors], and Triton X-100 was subsequently added to a final concentration of 0.1%. After a 5-min incubation on ice, the samples were spun at low speed (1300g for 4 min) to separate soluble proteins (Sol.) and permeablized nuclei. The resulting nuclei were lysed with Solution B [3 mM EDTA, 0.2 mM EGTA, 1 mM DTT] and a chromatin-enriched fraction was isolated by centrifugation (1700g for 4 min). These pellets were subsequently extracted with Solution C [50 mM Tris (pH 8.0), 600 mM NaCl, 0.5% TritonX-100, 0.5% sodium deoxycholate, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, protease inhibitors] to release the chromatin-bound proteins (Chr.).
DNA Oligonucleotides
To generate DNA structures with dsDNA regions, equal moles of the complimentary DNA oligonucleotides were annealed. Streptavidin was mixed with DNA at a 3:1 ratio to block the biotinylated ends. The sequences of the DNA oligonucleotides used in this study are listed in the Supplemental Data.
Extract-Based ATM Activation Assay
Nuclear extracts were supplemented with the Reaction Buffer (Buffer R), which brings the final buffer compositions to 10 mM Hepes (pH 7.6), 50 mM KCl, 0.1 mM MgCl2, 1 mM PMSF, 0.5 mM DTT, 1 mM ATP, 10 μg/ml creatine kinase, and 5 mM phosphocreatine. Various DNA structures were incubated in the supplemented extracts for 30 min at 37 °C. The extracts were then subjected to Western blotting, immunoprecipitation, or the DNA binding assay (see below).
DNA Binding Assay
The biotinylated DNA structures were attached to streptavidin-coated magnetic beads according to the manufacture’s instruction (Dynal). The beads coated with DNA were incubated with purified MRN as described in (Lee and Paull, 2005), or with nuclear extracts in the binding buffer [10 mM Tris-HCl, (pH 7.5), 100 mM NaCl, 10% glycerol, 0.01 % NP-40, and 10 μg/ml bovine serum albumin]. After a 30-min incubation at room temperature, beads were retrieved and washed 3 times with the binding buffer. The proteins bound to beads were denatured in the SDS sample buffer, separated on SDS-PAGE, and analyzed by Western blotting. The MRN complex was purified from insect cells infected with the baculoviruses expressing Mre11, Rad50, and Nbs1. The baculoviruses were kindly provided by Drs. Stephen Elledge (Harvard) and Tanya Paull (U. of Texas Austin).
Supplementary Material
Acknowledgments
We thank Drs. R. Abraham, B. Chen, D. Chen, J. Chen, D. Cortez, G. Li, S. Elledge, T. Paull, and L. Rasmussen for reagents, Drs. S. Elledge and N. Dyson for comments on the manuscript, and members of the Zou lab for discussions. L. Z. is a V Scholar and an Ellison New Scholar. This work is supported by grants from NIH (GM076388), the Susan G. Komen Foundation, and the Breast Cancer Alliance. B. S. was partly supported by a fellowship from the Tosteson Foundation.
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include six figures and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. ATM activation and signalling under hypoxic conditions. Mol Cell Biol. 2008 doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Gibson SL, Bindra RS, Glazer PM. Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res. 2005;65:10734–10741. doi: 10.1158/0008-5472.CAN-05-1160. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008 doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. Embo J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kanu N, Behrens A. ATMIN defines an NBS1-independent pathway of ATM signalling. Embo J. 2007;26:2933–2941. doi: 10.1038/sj.emboj.7601733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Clerici M, Lucchini G, Longhese MP. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8:380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- Pandita TK, Lieberman HB, Lim DS, Dhar S, Zheng W, Taya Y, Kastan MB. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19:1386–1391. doi: 10.1038/sj.onc.1203444. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Kastan MB. ATM: genome stability, neuronal development, and cancer cross paths. Adv Cancer Res. 2001;83:209–254. doi: 10.1016/s0065-230x(01)83007-4. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Ward JF. Complexity of damage produced by ionizing radiation. Cold Spring Harb Symp Quant Biol. 2000;65:377–382. doi: 10.1101/sqb.2000.65.377. [DOI] [PubMed] [Google Scholar]
- Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. Embo J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Zou L. Recruitment of ATR-ATRIP, Rad17, and 9-1-1 complexes to DNA damage. Methods Enzymol. 2006;409:118–131. doi: 10.1016/S0076-6879(05)09007-5. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J. 2008 doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.