Abstract
Excessive accumulation of β-amyloid peptides in the brain is a major cause for the pathogenesis of Alzheimer disease. β-Amyloid is derived from β-amyloid precursor protein (APP) through sequential cleavages by β- and γ-secretases, whose enzymatic activities are tightly controlled by subcellular localization. Delineation of how intracellular trafficking of these secretases and APP is regulated is important for understanding Alzheimer disease pathogenesis. Although APP trafficking is regulated by multiple factors including presenilin 1 (PS1), a major component of the γ-secretase complex, and phospholipase D1 (PLD1), a phospholipid-modifying enzyme, regulation of intracellular trafficking of PS1/γ-secretase and β-secretase is less clear. Here we demonstrate that APP can reciprocally regulate PS1 trafficking; APP deficiency results in faster transport of PS1 from the trans-Golgi network to the cell surface and increased steady state levels of PS1 at the cell surface, which can be reversed by restoring APP levels. Restoration of APP in APP-deficient cells also reduces steady state levels of other γ-secretase components (nicastrin, APH-1, and PEN-2) and the cleavage of Notch by PS1/γ-secretase that is more highly correlated with cell surface levels of PS1 than with APP overexpression levels, supporting the notion that Notch is mainly cleaved at the cell surface. In contrast, intracellular trafficking of β-secretase (BACE1) is not regulated by APP. Moreover, we find that PLD1 also regulates PS1 trafficking and that PLD1 overexpression promotes cell surface accumulation of PS1 in an APP-independent manner. Our results clearly elucidate a physiological function of APP in regulating protein trafficking and suggest that intracellular trafficking of PS1/γ-secretase is regulated by multiple factors, including APP and PLD1.
An important pathological hallmark of Alzheimer disease (AD)4 is the formation of senile plaques in the brains of patients. The major components of those plaques are β-amyloid peptides (Aβ), whose accumulation triggers a cascade of neurodegenerative steps ending in formation of senile plaques and intraneuronal fibrillary tangles with subsequent neuronal loss in susceptible brain regions (1, 2). Aβ is proteolytically derived from the β-amyloid precursor protein (APP) through sequential cleavages by β-secretase (BACE1), a novel membrane-bound aspartyl protease (3, 4), and by γ-secretase, a high molecular weight complex consisting of at least four components: presenilin (PS), nicastrin (NCT), anterior pharynx-defective-1 (APH-1), and presenilin enhancer-2 (PEN-2) (5, 6). APP is a type I transmembrane protein belonging to a protein family that includes APP-like protein 1 (APLP1) and 2 (APLP2) in mammals (7, 8). Full-length APP is synthesized in the endoplasmic reticulum (ER) and transported through the Golgi apparatus. Most secreted Aβ peptides are generated within the trans-Golgi network (TGN), also the major site of steady state APP in neurons (9–11). APP can be transported to the cell surface in TGN-derived secretory vesicles if not proteolyzed to Aβ or an intermediate metabolite. At the cell surface APP is either cleaved by α-secretase to produce soluble sAPPα (12) or reinternalized for endosomal/lysosomal degradation (13, 14). Aβ may also be generated in endosomal/lysosomal compartments (15, 16). In contrast to neurotoxic Aβ peptides, sAPPα possesses neuroprotective potential (17, 18). Thus, the subcellular distribution of APP and proteases that process it directly affect the ratio of sAPPα to Aβ, making delineation of the mechanisms responsible for regulating trafficking of all of these proteins relevant to AD pathogenesis.
Presenilin (PS) is a critical component of the γ-secretase. Of the two mammalian PS gene homologues, PS1 and PS2, PS1 encodes the major form (PS1) in active γ-secretase (19, 20). Nascent PSs undergo endoproteolytic cleavage to generate an amino-terminal fragment (NTF) and a carboxyl-terminal fragment (CTF) to form a functional PS heterodimer (21). Based on observations that PSs possess two highly conserved aspartate residues indispensable for γ-secretase activity and that specific transition state analogue γ-secretase inhibitors bind to PS1 NTF/CTF heterodimers (5, 22), PSs are believed to be the catalytic component of the γ-secretase complex. PS assembles with three other components, NCT, APH-1, and PEN-2, to form the functional γ-secretase (5, 6). Strong evidence suggests that PS1/γ-secretase resides principally in the ER, early Golgi, TGN, endocytic and intermediate compartments, most of which (except the TGN) are not major subcellular sites for APP (23, 24). In addition to generating Aβ and cleaving APP to release the APP intracellular domain, PS1/γ-secretase cleaves other substrates such as Notch (25), cadherin (26), ErbB4 (27), and CD44 (28), releasing their respective intracellular domains. Interestingly, PS1/γ-secretase cleavage of different substrates seems to occur at different subcellular compartments; APP is mainly cleaved at the TGN and early endosome domains, whereas Notch is predominantly cleaved at the cell surface (9, 11, 29). Thus, perturbing intracellular trafficking of PS1/γ-secretase may alter interactions between PS1/γ-secretase and APP, contributing to either abnormal Aβ generation and AD pathogenesis or decreased access of PS1/γ-secretase to APP such that Aβ production is reduced. However, mechanisms regulating PS1/γ-secretase trafficking warrant further investigation.
In addition to participating in γ-secretase activity, PS1 regulates intracellular trafficking of several membrane proteins, including other γ-secretase components (nicastrin, APH-1, and PEN-2) and the substrate APP (reviewed in Ref. 30). Intracellular APP trafficking is highly regulated and requires other factors such as mint family members and SorLA (2). Moreover, we recently found that phospholipase D1 (PLD1), a phospholipid-modifying enzyme that regulates membrane trafficking events, can interact with PS1, and can regulate budding of APP-containing vesicles from the TGN and delivery of APP to the cell surface (31, 32). Interestingly, Kamal et al. (33) identified an axonal membrane compartment that contains APP, BACE1, and PS1 and showed that fast anterograde axonal transport of this compartment is mediated by APP and kinesin-I, implying a traffic-regulating role for APP. Increased APP expression is also shown to decrease retrograde axonal transport of nerve growth factor (34). However, whether APP indeed regulates intracellular trafficking of proteins including BACE1 and PS1/γ-secretase requires further validation. In the present study we demonstrate that intracellular trafficking of PS1, as well as that of other γ-secretase components, but not BACE1, is regulated by APP. APP deficiency promotes cell surface delivery of PS1/γ-secretase complex and facilitates PS1/γ-secretase-mediated Notch cleavage. In addition, we find that PLD1 also regulates intracellular trafficking of PS1 through a different mechanism and more potently than APP.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—Maintenance of APP/APLP2 double knock-out (APP dKO) mouse embryonic fibroblast cells and wild type control cells (35), PS1/PS2 double knock-out (PS dKO) mouse embryonic fibroblast cells and wild type control cells (35), mouse neuroblastoma N2a cells and cells stably overexpressing human APP695 (N-695) (36), and human HeLa cells and cells stably overexpressing human APP Swedish mutant (H-sw) (37) has been described. The cells were transiently transfected with control pcDNA vector or vectors expressing full-length APP, APP CTF (C99), C99 with an ER retention signal (gift from Dr. G. Thinakaran), C99 with a Golgi/TGN retention signal (gift from Dr. G. Thinakaran), APP lacking the last 57 carboxyl-terminal amino acids, wild type PLD1, catalytically inactive form (K898R) of PLD1, or Notch NΔE-Myc using Lipofectamine (Invitrogen) following the manufacturer's protocol. A pSUPER RNA interference vector (OligoEngine, Seattle, WA) containing a small hairpin RNA targeting the APP sequence CACAAGTAGATGCCTGAAC and a vector containing a scrambled control sequence were transfected to knock down APP expression. The cells were harvested for analysis 48 h after transfection.
Antibodies and Western Blot—Treated cells were lysed in Nonidet P-40 lysis buffer (1% Nonidet P-40 in phosphate-buffered saline supplemented with a protease inhibitor mixture). Equal protein amounts of lysate were analyzed and immunoblotted as indicated. Rabbit polyclonal antibodies against BACE1 (B690), PS1 NTF (Ab14), APP CTF (369), APH-1aL carboxyl terminus, PEN-2 amino terminus (PNT2), and nicastrin (716) were developed in our laboratory (9, 38–44). Mouse anti-α-tubulin antibody was from Sigma. Monoclonal antibody 9E10 recognizing the Myc tag, monoclonal antibody against γ-adaptin, and polyclonal antibody against Rab5 were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibody against Na/K ATPase was from Abcam (Cambridge, MA). Polyclonal antibodies against Bip and against the cleaved Notch/NICD were from Cell Signaling Technology (Danvers, MA).
Cell Surface Protein Biotinylation—Biotinylation was carried out following a previously described protocol (42, 45). Briefly, the cells were washed with ice-cold phosphate-buffered saline containing CaCl2 and MgCl2 and incubated at 4 °C with NHS-LC-biotin (Pierce). The cells were lysed in 1% Nonidet P-40 lysis buffer, and lysates were affinity-precipitated with streptavidin-agarose beads (Pierce). Biotinylated proteins were eluted with SDS-PAGE sample buffer (Invitrogen) and loaded directly onto gels for electrophoresis, followed by Western blot analysis with indicated antibodies.
Pulse-Chase and Biotinylation Analysis—APP dKO and wild type control cells were starved for 30 min and labeled by [35S]methionine (500 μCi/ml) for 15 min at 37 °C. After washing away [35S]methionine, the cells were chased in normal growth medium at 20 °C for 2 h to promote accumulation of labeled proteins in the TGN. The cells were then incubated for various times at 37 °C. At the end of each chase time, the cells were biotinylated at 4 °C. The cell lysates were affinity-precipitated by streptavidin-agarose beads, and biotinylated cell surface proteins were eluted with 2% SDS. After dilution, eluted PS1 NTF was immunoprecipitated using Ab14 antibody, separated on SDS-PAGE gels, and analyzed by autoradiography.
Subcellular Fractionation—APP dKO and wild type control cells were homogenized using a ball-bearing cell cracker and then centrifuged at 800 × g for 5 min. The supernatant was fractionated by sucrose density gradient as described (9, 42, 46), and equal sample volumes were resolved by SDS-PAGE, followed by immunoblotting with antibodies recognizing indicated proteins.
Immunofluorescence Microscopy—Cell immunostaining was carried out as previously described (42). Briefly, the cells were fixed, permeabilized, and incubated with the PS1 NTF antibody Ab14. After subsequent incubation with Alexa Fluor 488-conjugated secondary antibody (Invitrogen), the cells were examined using a deconvolution fluorescent microscope (Zeiss, Thornwood, NY).
RESULTS
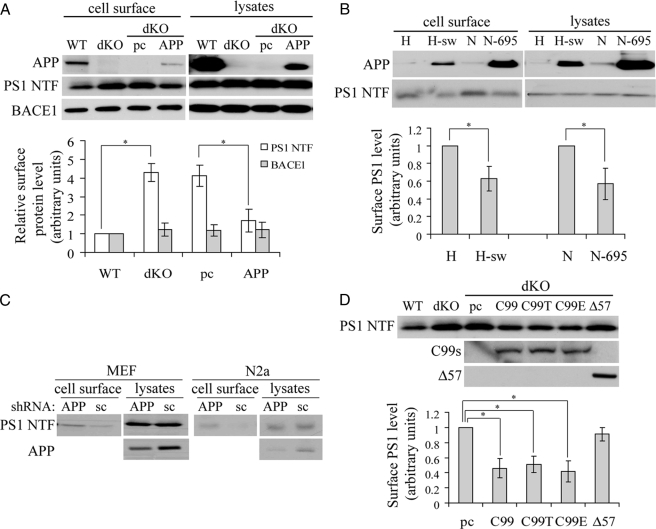
APP/APP βCTF (C99) Regulates Cell Surface Accumulation of PS1—We and others have shown that PS1 can regulate intracellular trafficking of APP (reviewed in Ref. 30). However, regulation of PS1/γ-secretase intracellular trafficking has remained elusive. One study found that axonal transport of a membrane compartment containing BACE1 and PS1 requires APP (33), implying that APP might reciprocally regulate intracellular trafficking of PS1, as well as that of BACE1. To investigate this, we compared the cell surface levels of PS1 in APP wild type (APP WT) and APP/APLP2 double knock-out (APP dKO) mouse embryonic fibroblasts. We found that although the total levels of PS1 were similar between cells, steady state levels of cell surface PS1 dramatically increased in the absence of APP/APLP2 (Fig. 1A). Such an increase was reversed by overexpressing APP in the APP dKO cells (Fig. 1A), ruling out potential clonal variation effects. In contrast, APP deficiency had no effect on cell surface levels of BACE1 (Fig. 1A). Furthermore, we found that in both HeLa cells stably expressing human APP Swedish mutations (H-sw) and N2a cells stably expressing human APP695 (N-695), steady state levels of cell surface PS1 were markedly reduced compared with those seen in control cells (Fig. 1B). Moreover, when we down-regulated APP levels in wild type mouse embryonic fibroblast and N2a cell lines using RNA interference, steady state levels of PS1 at the cell surface dramatically increased, but total PS1 levels were not affected (Fig. 1C). Taken together, these results suggest that intracellular trafficking of PS1 is regulated by APP.
FIGURE 1.
APP regulates steady state levels of PS1 at the cell surface. A, APP wild type (WT), APP/APLP2 double knock-out (dKO), and APP dKO cells transiently expressing either control pcDNA (pc) or APP were biotinylated. The cell lysates were incubated with streptavidin-agarose beads for affinity precipitation of biotinylated cell surface proteins. Biotinylated proteins and total cell lysates were subjected to SDS-PAGE and Western blot. B, HeLa cells (H), HeLa cells stably expressing APP Swedish mutations (H-sw), N2a (N), and N2a cells stably expressing APP695 (N-695) were biotinylated and affinity-precipitated, followed by SDS-PAGE and Western blot. C, mouse embryonic fibroblasts and N2a cells were transiently transfected with an APP small hairpin RNA construct to down-regulate APP or with a scrambled (sc) small hairpin RNA control construct. The cells were then subjected to biotinylation, and biotinylated cell surface proteins were precipitated with streptavidin-agarose beads for SDS-PAGE. D, APP WT, APP dKO, and APP dKO cells transiently transfected with pcDNA (pc), APP C99, C99 with a TGN retention signal (C99T), C99 with an ER retention signal (C99E), or APP lacking 57 amino acids at the carboxyl terminus (Δ57) were biotinylated. Affinity-precipitated cell surface proteins were analyzed by Western blot. Antibodies used for recognizing APP, PS1 NTF, and BACE1 were 369, Ab14, and B690, respectively. Protein levels were quantitated by densitometry and normalized to respective controls (set as 1 arbitrary unit). The data represent the means ± S.E. from three separate experiments. *, p < 0.05.
The γ-cleavage of APP requires a precedent β-cleavage to generate membrane-bound APP βCTF (C99). Because APP C99 reportedly interacts with PS1 to form a complex prior to cleavage by active PS1/γ-secretase (47, 48), we asked whether C99 regulated PS1 trafficking similarly to APP. As expected, overexpression of C99, the direct substrate of PS1/γ-secretase, rescued increased cell surface accumulation of PS1 in the APP dKO cells (Fig. 1D). Overexpression of two C99 variants containing either TGN or ER retention signals also rescued increased PS1 delivery to the cell surface. In contrast, overexpression of an APP variant lacking the carboxyl-terminal 57 amino acids did not promote rescue (Fig. 1D). These data suggest that APP or the membrane-bound APP C99 fragment may act to retain PS1 in appropriate compartments and that the intracellular domain of APP is critical for this function.
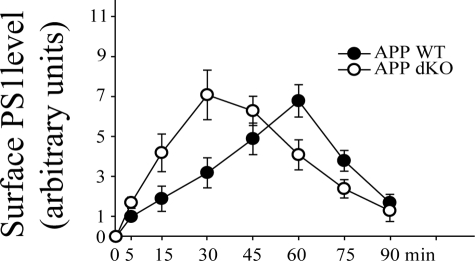
APP Deficiency Accelerates Cell Surface Delivery of PS1—We had previously found that loss of PS1 promotes budding of APP-containing vesicles from the TGN and accelerates cell surface delivery of APP (49). Therefore we compared cell surface delivery rates of PS1 in the APP WT and the APP dKO cells. As shown in Fig. 2, most [35S]methionine-labeled PS1 was delivered to the cell surface after a 60-min chase in the APP WT cells. However, in the APP dKO cells most isotope-labeled PS1 was seen at the cell surface as early as after a 30-min chase, suggesting accelerated cell surface delivery of PS1 in the absence of APP.
FIGURE 2.
APP deficiency accelerates cell surface delivery of PS1. After labeling with [35S]methionine, APP WT and APP dKO cells were incubated at 20 °C to promote accumulation of isotope-labeled protein in the TGN. The cells were then switched to 37 °C for indicated times and biotinylated at 4 °C. The cell lysates were subjected to affinity precipitation with streptavidin-agarose beads and immunoprecipitation with a PS1 antibody (Ab14 for NTF), followed by SDS-PAGE and autoradiography. PS1 levels were normalized to those of PS1 at 5 min of chase in APP WT. The data represent the means ± S.E. from three experiments.
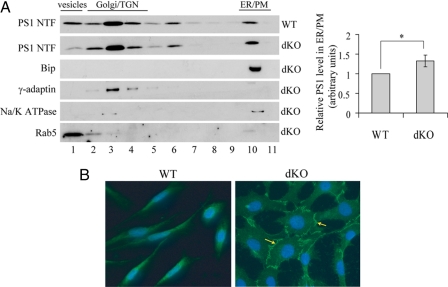
Confirmation of an Increase of Cell Surface Levels of PS1 by APP Deficiency—To corroborate that cell surface levels of PS1 are increased in the absence of APP, we performed sucrose fractionation experiments (9, 42, 46) to investigate subcellular localization of PS1. Subcellular distribution patterns of organelle markers were similar between the WT and the APP dKO cells (Fig. 3A, left panels; only dKO cells were shown). Consistent with previous reports (38, 42), PS1 was localized mainly in the Golgi/TGN and ER/plasma membrane (PM) fractions in both APP WT and dKO cells (Fig. 3A, left panels). However, the ratio of the PS1 in the ER/PM fractions relative to the total PS1 level was significantly higher in the APP dKO cells than in the WT cells (Fig. 3A, right panels). Because the sucrose fractionation assay is unable to distinguish PM from ER, we performed fluorescent immunostaining and found a much more intensive cell surface immunofluorescence of PS1 in the APP dKO cells than that in the WT cells (Fig. 3B). It is worth noting that the morphology of the APP dKO cells is quite distinct with a larger and more spreading cell body for unidentified reasons, compared with the WT fibroblasts.
FIGURE 3.
APP deficiency increases cell surface levels of PS1. A, APP WT and APP dKO cells were homogenized and fractionated by sucrose gradient sedimentation (left panels). One-ml samples from each cell line were collected from the top to the bottom of the gradient (labeled from 1 to 11). Equal sample volumes were analyzed by Western blot for PS1 NTF. Bip, γ-adaptin, Na/K ATPase, and Rab5 served as markers for ER, Golgi/TGN, PM, and vesicles, respectively. The level of PS1 NTF in each fraction was quantitated by densitometry. The ratio of PS1 NTF level in the ER/PM fractions relative to the total PS1 NTF level was analyzed and normalized to that of APP WT cells (set as 1 arbitrary unit). The data represent the means ± S.E. from three separate experiments. *, p < 0.05 (right panels). B, APP WT and dKO cells were fixed, permeabilized, sequentially incubated with anti-PS1 NTF antibody Ab14 and Alexa Fluor 488-conjugated secondary antibody, and examined with deconvolution microscope. The arrows indicate cell surface PS1.
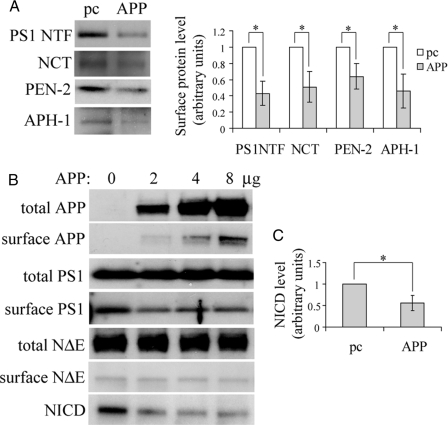
APP Regulates Cell Surface Accumulation of All γ-Secretase Components as Well as Notch Cleavage by PS1/γ-Secretase—In addition to PS1, the γ-secretase complex consists of at least three other components, NCT, APH-1, and PEN-2, which regulate each other's stability and subcellular localization (38, 42, 45). Because PS1 trafficking is regulated by APP, we asked whether APP also regulates intracellular trafficking of these factors. As expected, APP overexpression in the APP dKO cells not only reduced cell surface levels of PS1 but also reduced cell surface levels of NCT, APH-1, and PEN-2 (Fig. 4A).
FIGURE 4.
APP regulates cell surface delivery of all four γ-secretase components and modulates Notch cleavage. A, APP dKO cells were transiently transfected with pcDNA control or APP cDNA and subjected to biotinylation. Affinity-precipitated cell surface proteins were analyzed by Western blot for PS1 NTF, NCT, PEN-2, and APH-1. Protein levels were quantitated by densitometry and normalized to respective controls (set as 1 arbitrary unit). The data represent the means ± S.E. from three experiments. *, p < 0.05. B, APP dKO cells were transiently transfected with a Notch NΔE-Myc vector. After splitting equally, the cells were transfected with pcDNA (pc) or different amounts of APP cDNA and biotinylated. The cell lysates were subjected to SDS-PAGE and Western blot with antibodies against total and biotinylated (surface) APP (369), PS1 (Ab14), and NΔE (using the 9E10 anti-Myc antibody) and cleaved NΔE/NICD. C, samples transfected with 8 μg of control (APP 0 μg) or APP cDNA in B were used for comparison. After densitometry quantitation, relative levels of NICD/total NΔE were normalized to those of controls (set as 1 arbitrary unit). The data represent the means ± S.E. from three experiments. *, p < 0.05.
Another important PS1/γ-secretase substrate is Notch, whose cleavage product (NICD) serves as a transcription factor regulating several genes involved in development (50, 51). The major site for Notch cleavage is the cell surface (29). To determine whether increased cell surface levels of PS1/γ-secretase affects Notch cleavage, we overexpressed in the APP dKO cells a form of Notch1 (NΔE) lacking the ectodomain and therefore constitutively processed by PS1/γ-secretase in the absence of ligand (52). Although coexpressing APP had no effect on total and cell surface levels of Notch NΔE, NICD levels were dramatically reduced in the presence of APP (Fig. 4, B and C). Moreover, NICD levels were more highly related with cell surface levels of PS1 than with APP overexpression levels; the maximal reduction of NICD generation was achieved when cell surface levels of PS1 were maximally reduced by APP overexpression (at 2 μg), and higher APP overexpression (4 and 8 μg) could not further decrease the levels of either cell surface PS1 or NICD (Fig. 4B). These results imply that reduced Notch cleavage resulting from APP overexpression is likely due to decreased cell surface levels of PS1/γ-secretase rather than substrate competition between Notch and APP.
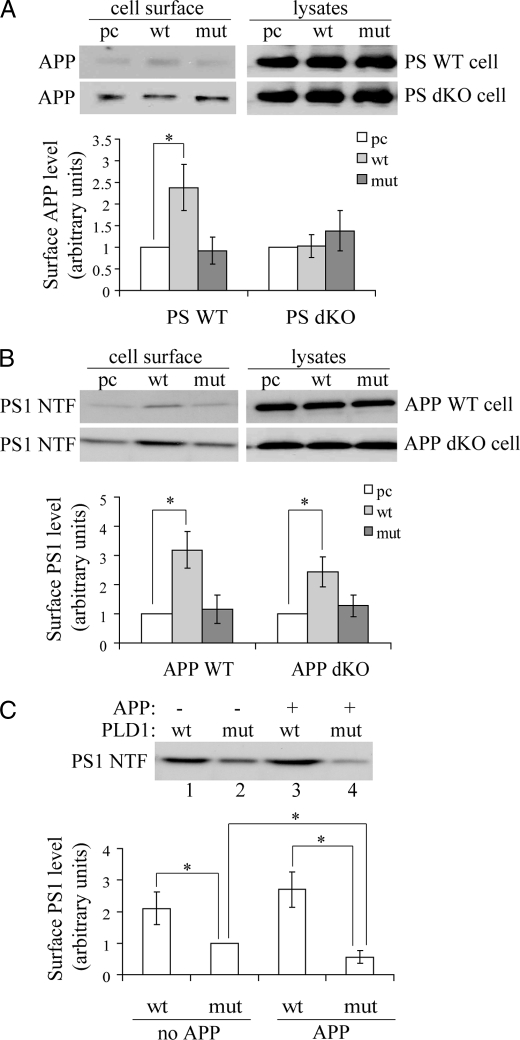
PLD1 Regulates PS1 Trafficking Differently than APP—We recently identified an interaction between PLD1 and PS1 and found that PLD1 also regulated APP trafficking but in a PS1-independent manner (31, 32). Here we confirmed that overexpression of wild type PLD1 but not its catalytically inactive K898R form promoted APP delivery to the cell surface in PS WT fibroblast cells (Fig. 5A). Although overexpressing PLD1 did not promote cell surface accumulation of APP in PS1/PS2 double knock-out (PS dKO) cells (Fig. 5A), we previously found that inhibiting PLD1 catalytic activity resulted in reduced budding of APP-containing vesicles from the TGN in both PS1 single knock-out and wild type control fibroblasts (32), implying that unchanged cell surface levels of APP in the PS dKO cells might be due to a maximal trafficking of APP-containing vesicles in the absence of PS1 so that PLD1 overexpression will not further promote APP trafficking.
FIGURE 5.
PLD1 regulates PS1 trafficking. PS wild type cells (PS WT) and PS1/PS2 double knock-out (PS dKO) cells (A) and APP WT and APP dKO cells were transiently transfected with pcDNA (pc), wild type PLD1 (wt), or a catalytically inactive form of PLD1 (K898R, mut) (B). The cells were then subjected to biotinylation and analysis of cell surface proteins. C, APP dKO cells were first transfected with control vector (-) or APP (+). After splitting equally, the cells were transfected with wt or mut PLD1 followed by biotinylation to analyze cell surface proteins. Antibodies 369 and Ab14 recognizing APP and PS1 NTF, respectively, were used for Western blot analysis. Protein levels were quantitated by densitometry and normalized to respective controls (set as 1 arbitrary unit). The data represent the means ± S.E. from three experiments. *, p < 0.05.
On the other hand, we found that overexpressing PLD1 dramatically increased cell surface levels of PS1 in both APP WT and APP dKO cells, whereas overexpressing the PLD1 mutant (K898R) had no such effects (Fig. 5B). Overexpression of wild type PLD1 also resulted in more cell surface PS1 levels than overexpression of the PLD1 mutant in both APP dKO cells transfected with control vector (-) (lanes 1 versus 2) and with APP (+) (lanes 3 versus 4) (Fig. 5C). However, unlike the interaction observed between PLD1 and PS1, we did not observe interaction between PLD1 and APP (data not shown). These results suggest that PLD1 also regulates intracellular trafficking of PS1 but through a mechanism different from that by APP. In addition, although APP overexpression reduced cell surface levels of PS1 in cells transfected with mutant PLD1 (Fig. 5C, lane 2 versus lane 4), APP overexpression did not attenuate the effects of wild type PLD1 on cell surface accumulation of PS1 (Fig. 5C, lane 1 versus lane 3), suggesting that PLD1 overexpression can override effects of APP overexpression in reducing cell surface delivery of PS1.
DISCUSSION
Since its identification, multiple roles have been proposed for APP, such as mediating signal transduction, promoting cell adhesion, and functioning in neurite outgrowth and synaptogenesis (53). We and others have also demonstrated that the APP ectodomain (sAPPα) has neuroprotective activity and prevents Tau hyperphosphorylation via suppressing overactivation of CDK5 (17, 18, 54) and that the APP intracellular domain can bind to promoter regions and regulate gene transcription (35, 55). Nevertheless, the physiological and pathological functions of APP, especially in its uncleaved full-length form, remain active areas of AD research.
Deletion of the Drosophila APP-like gene (Appl) or overexpression of human APP695 or APPL constructs in Drosophila reportedly causes abnormal axonal transport phenotypes similar to those seen in kinesin and dynein mutants (56). In addition, an axonal membrane compartment containing APP, BACE1, and PS1 has been identified, and anterograde axonal transport of that compartment is mediated by APP and kinesin-I (33). These observations suggest that APP may function as a kinesin-I membrane receptor, possibly through an indirect interaction with a kinesin light chain subunit via the adaptor JIP-1 (57), to regulate axonal transport of BACE1 and PS1 (33, 58). Consistently, it was found that APP is a major component of herpes simplex viral particles and likely mediates fast anterograde axonal transport of these particles (59, 60). Moreover, increased APP levels were recently found to markedly decrease retrograde transport of nerve growth factor and promote degeneration of forebrain cholinergic neurons in a mouse model of Down syndrome (34). These studies suggest another physiological function of APP in regulating axonal transport/protein trafficking, but this notion requires validation with additional investigation. Here we show that intracellular trafficking of PS1 is indeed regulated by APP/APP βCTF (C99) and that loss of APP increases cell surface delivery of PS1. Interestingly, our previous studies and others show that PS1 also regulates intracellular trafficking of APP and that loss of PS1 accelerates cell surface delivery of APP (reviewed in Ref. 30). Because PS1 reportedly interacts APP and APP βCTF and their proper conformation is critical for normal trafficking of membrane/secretory proteins within the secretory pathway (47, 48), interaction between APP/C99 and PS1 may be a prerequisite for proper retention of APP/PS1 in the TGN and their delivery to the cell surface.
PS1 assembles with nicastrin, APH-1, and PEN-2 to form a functional γ-secretase complex (5). These components are tightly associated, and down-regulation or targeted disruption of any one member of the complex affects maturation and/or stability of the other components, indicating that complex assembly and trafficking are highly regulated (38, 42, 45). Here, as expected and consistent with the pattern of PS1 trafficking, we found that cell surface accumulation of nicastrin, APH-1, and PEN-2 is also increased in the absence of APP. These results suggest that intracellular trafficking of the entire functional PS1/γ-secretase complex is regulated by APP. Interestingly, we found that Notch cleavage/NICD generation is reduced when APP is overexpressed in APP dKO cells (Fig. 4, B and C). This could be attributed to either substrate competition between Notch and APP for PS1/γ-secretase cleavage or decreased PS1/γ-secretase delivery to the cell surface site of Notch cleavage in the presence of APP (29). However, our results show that NICD generation is more highly related with cell surface levels of PS1 than with APP overexpression levels (Fig. 4B). Hence, although we do not completely exclude the possibility of substrate competition between APP and Notch, it is very likely that APP negatively modulates Notch cleavage/NICD generation through regulating intracellular trafficking of PS1/γ-secretase.
BACE1 is a type I transmembrane aspartyl protease and synthesized as a larger precursor, pro-BACE1, which can be modified by glycosylation (and also phosphorylation) and cleaved by a furin-like endoprotease to produce mature BACE1 (61, 62). The optimal BACE1 activity requires an acidic environment and the major cellular compartments, in various premitotic cell lines overexpressing exogenous BACE1, include early Golgi, late Golgi/early endosomes, endosomes, and the cell surface (3, 28, 63, 64). Maturation/intracellular trafficking of BACE1 is a complex process, requiring the interaction of BACE1 with a variety of different proteins such as GGA proteins that are a family of proteins involved in the recruitment and/or sorting of cargo into secretory vesicles (65–67). It has been found that BACE1 can associate with APP via the GYENPTY motif of APP (65, 68, 69), and such an interaction can be significantly reduced by sorLA, which interacts with both BACE1 and APP and regulates the trafficking/processing of the latter (2, 70, 71). However, in contrast to PS1/γ-secretase, steady state cell surface levels of BACE1 are not affected by APP (Fig. 1). Interestingly, PS1 was found to also interact with BACE1, preferably pro-BACE1, and affect BACE1 maturation (72). Therefore, although APP, PS1, and BACE1 interact with each other, their intracellular trafficking is differentially regulated.
In addition, we found that PLD1, a phospholipid-modifying enzyme known to regulate membrane trafficking events, can regulate intracellular trafficking of both APP and PS1 independent of PS1 and APP, respectively, and in a manner opposite to that by APP or PS1. We have previously found that overexpression of PLD1 can disrupt the association of γ-secretase components (31). Therefore, although PLD1 overexpression facilitates cell surface delivery of PS1, it impairs the γ-secretase activity, which results in an inhibition of γ-cleavage of APP and Notch (31). Because PLD1 is a phospholipid-modifying enzyme, it affects membrane integrity and thus may have a more general and profound effect on protein trafficking than APP or PS1 has. This notion may be supported by the observation that PLD1 overexpression overrode the effect of APP overexpression on reducing cell surface delivery of PS1 (Fig. 5C).
Cytosolic factors such as Rab11, Rab6, and Rab GDI, all of which regulate vesicular transport, have been found to interact with PS1 and/or APP (73–75), and modulation of Rab6-mediated transport affects APP processing (75). Moreover, the APP carboxyl terminus, which is conserved among APP family members (APLP1 and APLP2) and likely functionally significant (53), can interact with adaptor proteins such as Fe65, Tip60, and all three mint (X11) proteins (76–78). Mint proteins may regulate APP processing by stabilizing cellular APP, thus affecting both sAPPα and Aβ secretion (79). Interestingly, mint proteins also bind to PS1 and promote APP-PS1 interaction (64). We found that PLD1 can bind to PS1 (31) but not APP (data not shown). Together these results suggest that PS1 and APP might regulate protein trafficking via their interaction with various trafficking factors and that proper intracellular trafficking of APP and PS1/γ-secretase requires a dynamic balance among multiple regulatory pathways.
Acknowledgments
We thank Dr. Takeshi Iwatsubo for providing the APP C99 plasmids with an ER retention signal and with a Golgi/TGN retention signal.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS046673, R01 AG030197, and R01 AG021173 (to H. X.) and R01 NS054880 (to F.-F. L.). This work was also supported by grants from the Alzheimer's Association (to H. X. and F.-F. L.), the American Health Assistance Foundation (to H. X.), and National Natural Science Foundation of China Grants 30672198 and 30840036 (to Y. Z.).
Footnotes
The abbreviations used are: AD, Alzheimer disease; APLP1, APP-like protein; APP, β-amyloid precursor protein; Aβ, β-amyloid; CTF, carboxyl-terminal fragment; dKO, double knock-out; ER, endoplasmic reticulum; NCT, nicastrin; NICD, Notch intracellular domain; NTF, amino-terminal fragment; PS, presenilin; TGN, trans-Golgi network; WT, wild type; PLD, phospholipase D; PM, plasma membrane.
References
- 1.Selkoe, D. J. (1998) Trends Cell Biol. 8 447-453 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, Y. W., and Xu, H. (2007) Curr. Mol. Med. 7 687-696 [DOI] [PubMed] [Google Scholar]
- 3.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., Luo, Y., Fisher, S., Fuller, J., Edenson, S., Lile, J., Jarosinski, M. A., Biere, A. L., Curran, E., Burgess, T., Louis, J. C., Collins, F., Treanor, J., Rogers, G., and Citron, M. (1999) Science 286 735-741 [DOI] [PubMed] [Google Scholar]
- 4.Sinha, S., Anderson, J. P., Barbour, R., Basi, G. S., Caccavello, R., Davis, D., Doan, M., Dovey, H. F., Frigon, N., Hong, J., Jacobson-Croak, K., Jewett, N., Keim, P., Knops, J., Lieberburg, I., Power, M., Tan, H., Tatsuno, G., Tung, J., Schenk, D., Seubert, P., Suomensaari, S. M., Wang, S., Walker, D., Zhao, J., McConlogue, L., and John, V. (1999) Nature 402 537-540 [DOI] [PubMed] [Google Scholar]
- 5.Kimberly, W. T., LaVoie, M. J., Ostaszewski, B. L., Ye, W., Wolfe, M. S., and Selkoe, D. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6382-6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G., and Iwatsubo, T. (2003) Nature 422 438-441 [DOI] [PubMed] [Google Scholar]
- 7.Wasco, W., Bupp, K., Magendantz, M., Gusella, J. F., Tanzi, R. E., and Solomon, F. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 10758-10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasco, W., Gurubhagavatula, S., Paradis, M. D., Romano, D. M., Sisodia, S. S., Hyman, B. T., Neve, R. L., and Tanzi, R. E. (1993) Nat. Genet. 5 95-100 [DOI] [PubMed] [Google Scholar]
- 9.Xu, H., Sweeney, D., Wang, R., Thinakaran, G., Lo, A. C., Sisodia, S. S., Greengard, P., and Gandy, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3748-3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann, T., Bieger, S. C., Bruhl, B., Tienari, P. J., Ida, N., Allsop, D., Roberts, G. W., Masters, C. L., Dotti, C. G., Unsicker, K., and Beyreuther, K. (1997) Nat. Med. 3 1016-1020 [DOI] [PubMed] [Google Scholar]
- 11.Greenfield, J. P., Tsai, J., Gouras, G. K., Hai, B., Thinakaran, G., Checler, F., Sisodia, S. S., Greengard, P., and Xu, H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 742-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sisodia, S. S. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6075-6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordstedt, C., Caporaso, G. L., Thyberg, J., Gandy, S. E., and Greengard, P. (1993) J. Biol. Chem. 268 608-612 [PubMed] [Google Scholar]
- 14.Caporaso, G. L., Takei, K., Gandy, S. E., Matteoli, M., Mundigl, O., Greengard, P., and De Camilli, P. (1994) J. Neurosci. 14 3122-3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haass, C., Hung, A. Y., Schlossmacher, M. G., Teplow, D. B., and Selkoe, D. J. (1993) J. Biol. Chem. 268 3021-3024 [PubMed] [Google Scholar]
- 16.Haass, C., Hung, A. Y., Schlossmacher, M. G., Oltersdorf, T., Teplow, D. B., and Selkoe, D. J. (1993) Ann. N. Y. Acad. Sci. 695 109-116 [DOI] [PubMed] [Google Scholar]
- 17.Furukawa, K., Sopher, B. L., Rydel, R. E., Begley, J. G., Pham, D. G., Martin, G. M., Fox, M., and Mattson, M. P. (1996) J. Neurochem. 67 1882-1896 [DOI] [PubMed] [Google Scholar]
- 18.Han, P., Dou, F., Li, F., Zhang, X., Zhang, Y. W., Zheng, H., Lipton, S. A., Xu, H., and Liao, F. F. (2005) J. Neurosci. 25 11542-11552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherrington, R., Rogaev, E. I., Liang, Y., Rogaeva, E. A., Levesque, G., Ikeda, M., Chi, H., Lin, C., Li, G., Holman, K., Tsuda, T., Mar, L., Foncin, J. F., Bruni, A. C., Montesi, M. P., Sorbi, S., Rainero, I., Pinessi, L., Nee, L., Chumakov, I., Pollen, D., Brookes, A., Sansosu, P., Polinsky, R. J., Wasco, W., Da Silva, H. A. R., Haines, J. L., Pericak-Vance, M. A., Tanzi, R. E., Roses, A. D., Fraser, P. E., Rommens, J. M., and St George-Hyslop, P. (1995) Nature 375 754-760 [DOI] [PubMed] [Google Scholar]
- 20.Levy-Lahad, E., Wasco, W., Poorkaj, P., Romano, D. M., Oshima, J., Pettingell, W. H., Yu, C. E., Jondro, P. D., Schmidt, S. D., Wang, K., Crowley, A. C., Fu, Y.-H., Guenette, S. Y., Galas, D., Nemens, E., Wijsman, E. M., Bird, T. D., Schellenberg, G. D., and Tanzi, R. E. (1995) Science 269 973-977 [DOI] [PubMed] [Google Scholar]
- 21.Borchelt, D. R., Thinakaran, G., Eckman, C. B., Lee, M. K., Davenport, F., Ratovitsky, T., Prada, C. M., Kim, G., Seekins, S., Yager, D., Slunt, H. H., Wang, R., Seeger, M., Levey, A. I., Gandy, S. E., Copeland, N. G., Jenkins, N. A., Price, D. L., Younkin, S. G., and Sisodia, S. S. (1996) Neuron 17 1005-1013 [DOI] [PubMed] [Google Scholar]
- 22.Li, Y. M., Lai, M. T., Xu, M., Huang, Q., DiMuzio-Mower, J., Sardana, M. K., Shi, X. P., Yin, K. C., Shafer, J. A., and Gardell, S. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6138-6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cupers, P., Bentahir, M., Craessaerts, K., Orlans, I., Vanderstichele, H., Saftig, P., De Strooper, B., and Annaert, W. (2001) J. Cell Biol. 154 731-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovacs, D. M., Fausett, H. J., Page, K. J., Kim, T. W., Moir, R. D., Merriam, D. E., Hollister, R. D., Hallmark, O. G., Mancini, R., Felsenstein, K. M., Hyman, B. T., Tanzi, R. E., and Wasco, W. (1996) Nat. Med. 2 224-229 [DOI] [PubMed] [Google Scholar]
- 25.Haass, C., and De Strooper, B. (1999) Science 286 916-919 [DOI] [PubMed] [Google Scholar]
- 26.Chen, F., Hasegawa, H., Schmitt-Ulms, G., Kawarai, T., Bohm, C., Katayama, T., Gu, Y., Sanjo, N., Glista, M., Rogaeva, E., Wakutani, Y., Pardossi-Piquard, R., Ruan, X., Tandon, A., Checler, F., Marambaud, P., Hansen, K., Westaway, D., St George-Hyslop, P., and Fraser, P. (2006) Nature 440 1208-1212 [DOI] [PubMed] [Google Scholar]
- 27.Luo, Y., Bolon, B., Kahn, S., Bennett, B. D., Babu-Khan, S., Denis, P., Fan, W., Kha, H., Zhang, J., Gong, Y., Martin, L., Louis, J. C., Yan, Q., Richards, W. G., Citron, M., and Vassar, R. (2001) Nat. Neurosci. 4 231-232 [DOI] [PubMed] [Google Scholar]
- 28.Walter, J., Fluhrer, R., Hartung, B., Willem, M., Kaether, C., Capell, A., Lammich, S., Multhaup, G., and Haass, C. (2001) J. Biol. Chem. 276 14634-14641 [DOI] [PubMed] [Google Scholar]
- 29.Tarassishin, L., Yin, Y. I., Bassit, B., and Li, Y. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17050-17055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vetrivel, K. S., Zhang, Y. W., Xu, H., and Thinakaran, G. (2006) Mol. Neurodegener. 1 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai, D., Netzer, W. J., Zhong, M., Lin, Y., Du, G., Frohman, M., Foster, D. A., Sisodia, S. S., Xu, H., Gorelick, F. S., and Greengard, P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1941-1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai, D., Zhong, M., Wang, R., Netzer, W. J., Shields, D., Zheng, H., Sisodia, S. S., Foster, D. A., Gorelick, F. S., Xu, H., and Greengard, P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1936-1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamal, A., Almenar-Queralt, A., LeBlanc, J. F., Roberts, E. A., and Goldstein, L. S. (2001) Nature 414 643-648 [DOI] [PubMed] [Google Scholar]
- 34.Salehi, A., Delcroix, J. D., Belichenko, P. V., Zhan, K., Wu, C., Valletta, J. S., Takimoto-Kimura, R., Kleschevnikov, A. M., Sambamurti, K., Chung, P. P., Xia, W., Villar, A., Campbell, W. A., Kulnane, L. S., Nixon, R. A., Lamb, B. T., Epstein, C. J., Stokin, G. B., Goldstein, L. S., and Mobley, W. C. (2006) Neuron 51 29-42 [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Y. W., Wang, R., Liu, Q., Zhang, H., Liao, F. F., and Xu, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10613-10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, P., Li, F., Zhang, Y. W., Huang, H., Tong, G., Farquhar, M. G., and Xu, H. (2007) J. Neurochem. 100 1505-1514 [DOI] [PubMed] [Google Scholar]
- 37.Wang, R., Zhang, Y. W., Zhang, X., Liu, R., Zhang, X., Hong, S., Xia, K., Xia, J., Zhang, Z., and Xu, H. (2006) FASEB J. 20 1275-1277 [DOI] [PubMed] [Google Scholar]
- 38.Luo, W. J., Wang, H., Li, H., Kim, B. S., Shah, S., Lee, H. J., Thinakaran, G., Kim, T. W., Yu, G., and Xu, H. (2003) J. Biol. Chem. 278 7850-7854 [DOI] [PubMed] [Google Scholar]
- 39.Buxbaum, J. D., Gandy, S. E., Cicchetti, P., Ehrlich, M. E., Czernik, A. J., Fracasso, R. P., Ramabhadran, T. V., Unterbeck, A. J., and Greengard, P. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6003-6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, H., Gouras, G. K., Greenfield, J. P., Vincent, B., Naslund, J., Mazzarelli, L., Fried, G., Jovanovic, J. N., Seeger, M., Relkin, N. R., Liao, F., Checler, F., Buxbaum, J. D., Chait, B. T., Thinakaran, G., Sisodia, S. S., Wang, R., Greengard, P., and Gandy, S. (1998) Nat. Med. 4 447-451 [DOI] [PubMed] [Google Scholar]
- 41.Yan, R., Han, P., Miao, H., Greengard, P., and Xu, H. (2001) J. Biol. Chem. 276 36788-36796 [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y. W., Luo, W. J., Wang, H., Lin, P., Vetrivel, K. S., Liao, F., Li, F., Wong, P. C., Farquhar, M. G., Thinakaran, G., and Xu, H. (2005) J. Biol. Chem. 280 17020-17026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, S. F., Shah, S., Li, H., Yu, C., Han, W., and Yu, G. (2002) J. Biol. Chem. 277 45013-45019 [DOI] [PubMed] [Google Scholar]
- 44.Leem, J. Y., Vijayan, S., Han, P., Cai, D., Machura, M., Lopes, K. O., Veselits, M. L., Xu, H., and Thinakaran, G. (2002) J. Biol. Chem. 277 19236-19240 [DOI] [PubMed] [Google Scholar]
- 45.Wang, H., Luo, W. J., Zhang, Y. W., Li, Y. M., Thinakaran, G., Greengard, P., and Xu, H. (2004) J. Biol. Chem. 279 40560-40566 [DOI] [PubMed] [Google Scholar]
- 46.Gasparini, L., Gouras, G. K., Wang, R., Gross, R. S., Beal, M. F., Greengard, P., and Xu, H. (2001) J. Neurosci. 21 2561-2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia, W., Zhang, J., Perez, R., Koo, E. H., and Selkoe, D. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8208-8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waragai, M., Imafuku, I., Takeuchi, S., Kanazawa, I., Oyama, F., Udagawa, Y., Kawabata, M., and Okazawa, H. (1997) Biochem. Biophys. Res. Commun. 239 480-482 [DOI] [PubMed] [Google Scholar]
- 49.Cai, D., Leem, J. Y., Greenfield, J. P., Wang, P., Kim, B. S., Wang, R., Lopes, K. O., Kim, S. H., Zheng, H., Greengard, P., Sisodia, S. S., Thinakaran, G., and Xu, H. (2003) J. Biol. Chem. 278 3446-3454 [DOI] [PubMed] [Google Scholar]
- 50.Kopan, R., Schroeter, E. H., Weintraub, H., and Nye, J. S. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 1683-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopan, R., and Goate, A. (2000) Genes Dev. 14 2799-2806 [DOI] [PubMed] [Google Scholar]
- 52.Schroeter, E. H., Kisslinger, J. A., and Kopan, R. (1998) Nature 393 382-386 [DOI] [PubMed] [Google Scholar]
- 53.Zheng, H., and Koo, E. H. (2006) Mol. Neurodegener. 1 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mattson, M. P. (1997) Physiol. Rev. 77 1081-1132 [DOI] [PubMed] [Google Scholar]
- 55.Liu, Q., Zerbinatti, C. V., Zhang, J., Hoe, H. S., Wang, B., Cole, S. L., Herz, J., Muglia, L., and Bu, G. (2007) Neuron 56 66-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gunawardena, S., and Goldstein, L. S. (2001) Neuron 32 389-401 [DOI] [PubMed] [Google Scholar]
- 57.Sisodia, S. S. (2002) Science 295 805-807 [DOI] [PubMed] [Google Scholar]
- 58.Kamal, A., Stokin, G. B., Yang, Z., Xia, C. H., and Goldstein, L. S. (2000) Neuron 28 449-459 [DOI] [PubMed] [Google Scholar]
- 59.Satpute-Krishnan, P., DeGiorgis, J. A., and Bearer, E. L. (2003) Aging Cell 2 305-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satpute-Krishnan, P., DeGiorgis, J. A., Conley, M. P., Jang, M., and Bearer, E. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16532-16537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett, B. D., Denis, P., Haniu, M., Teplow, D. B., Kahn, S., Louis, J. C., Citron, M., and Vassar, R. (2000) J. Biol. Chem. 275 37712-37717 [DOI] [PubMed] [Google Scholar]
- 62.Creemers, J. W., Ines Dominguez, D., Plets, E., Serneels, L., Taylor, N. A., Multhaup, G., Craessaerts, K., Annaert, W., and De Strooper, B. (2001) J. Biol. Chem. 276 4211-4217 [DOI] [PubMed] [Google Scholar]
- 63.Huse, J. T., Pijak, D. S., Leslie, G. J., Lee, V. M., and Doms, R. W. (2000) J. Biol. Chem. 275 33729-33737 [DOI] [PubMed] [Google Scholar]
- 64.Lau, K. F., McLoughlin, D. M., Standen, C., and Miller, C. C. (2000) Mol. Cell Neurosci. 16 557-565 [DOI] [PubMed] [Google Scholar]
- 65.He, X., Li, F., Chang, W. P., and Tang, J. (2005) J. Biol. Chem. 280 11696-11703 [DOI] [PubMed] [Google Scholar]
- 66.Wahle, T., Prager, K., Raffler, N., Haass, C., Famulok, M., and Walter, J. (2005) Mol. Cell Neurosci. 29 453-461 [DOI] [PubMed] [Google Scholar]
- 67.Tesco, G., Koh, Y. H., Kang, E. L., Cameron, A. N., Das, S., Sena-Esteves, M., Hiltunen, M., Yang, S. H., Zhong, Z., Shen, Y., Simpkins, J. W., and Tanzi, R. E. (2007) Neuron 54 721-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Multhaup, G. (2006) Neurodegener. Dis. 3 270-274 [DOI] [PubMed] [Google Scholar]
- 69.Schmechel, A., Strauss, M., Schlicksupp, A., Pipkorn, R., Haass, C., Bayer, T. A., and Multhaup, G. (2004) J. Biol. Chem. 279 39710-39717 [DOI] [PubMed] [Google Scholar]
- 70.Spoelgen, R., von Arnim, C. A., Thomas, A. V., Peltan, I. D., Koker, M., Deng, A., Irizarry, M. C., Andersen, O. M., Willnow, T. E., and Hyman, B. T. (2006) J. Neurosci. 26 418-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersen, O. M., Reiche, J., Schmidt, V., Gotthardt, M., Spoelgen, R., Behlke, J., von Arnim, C. A., Breiderhoff, T., Jansen, P., Wu, X., Bales, K. R., Cappai, R., Masters, C. L., Gliemann, J., Mufson, E. J., Hyman, B. T., Paul, S. M., Nykjaer, A., and Willnow, T. E. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13461-13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuzuya, A., Uemura, K., Kitagawa, N., Aoyagi, N., Kihara, T., Ninomiya, H., Ishiura, S., Takahashi, R., and Shimohama, S. (2007) J. Neurosci. Res. 85 153-165 [DOI] [PubMed] [Google Scholar]
- 73.Dumanchin, C., Czech, C., Campion, D., Cuif, M. H., Poyot, T., Martin, C., Charbonnier, F., Goud, B., Pradier, L., and Frebourg, T. (1999) Hum. Mol. Genet. 8 1263-1269 [DOI] [PubMed] [Google Scholar]
- 74.Scheper, W., Zwart, R., Sluijs, P., Annaert, W., Gool, W. A., and Baas, F. (2000) Hum. Mol. Genet. 9 303-310 [DOI] [PubMed] [Google Scholar]
- 75.Scheper, W., Zwart, R., and Baas, F. (2004) Brain Res. Mol. Brain Res. 122 17-23 [DOI] [PubMed] [Google Scholar]
- 76.King, G. D., and Scott Turner, R. (2004) Exp. Neurol. 185 208-219 [DOI] [PubMed] [Google Scholar]
- 77.Rogelj, B., Mitchell, J. C., Miller, C. C., and McLoughlin, D. M. (2006) Brain Res. Rev. 52 305-315 [DOI] [PubMed] [Google Scholar]
- 78.Borg, J. P., Ooi, J., Levy, E., and Margolis, B. (1996) Mol. Cell. Biol. 16 6229-6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borg, J. P., Yang, Y., De Taddeo-Borg, M., Margolis, B., and Turner, R. S. (1998) J. Biol. Chem. 273 14761-14766 [DOI] [PubMed] [Google Scholar]