Abstract
Aminergic G protein-coupled receptors (GPCRs) have been a major focus of pharmaceutical research for many years. Due partly to the lack of reliable receptor structures, drug discovery efforts have been largely ligand-based. The recently determined X-ray structure of the β2-adrenergic receptor offers an opportunity to investigate the advantages and limitations inherent in a structure-based approach to ligand discovery against this and related GPCR targets. Approximately 1 million commercially available, “lead-like” molecules were docked against the β2-adrenergic receptor structure. On testing of 25 high-ranking molecules, 6 were active with binding affinities <4 μM, with the best molecule binding with a Ki of 9 nM (95% confidence interval 7–10 nM). Five of these molecules were inverse agonists. The high hit rate, the high affinity of the most potent molecule, the discovery of unprecedented chemotypes among the new inhibitors, and the apparent bias toward inverse agonists among the docking hits, have implications for structure-based approaches against GPCRs that recognize small organic molecules.
Keywords: docking, GPCR, inverse agonists, library bias, ligand design
Heterotrimeric guanine nucleotide-binding protein (G protein)-coupled receptors (GPCRs) are a large family of membrane receptors spanning the intra- and extracellular spaces with 7 transmembrane helices. Among receptors involved in cellular signaling, GPCRs are the family most frequently targeted by drugs (1, 2). One of the earliest successes in using GPCRs as therapeutic targets involved the group of receptors responding to aminergic hormones like epinephrine. Catecholamines such as epinephrine and norepinephrine are recognized by adrenergic GPCRs, which can be divided into 2 subfamilies (α and β), differing in ligand specificity, the tissues in which they are expressed, and downstream signal processing (3). The β2-adrenergic receptor (β2AR) is primarily found in smooth muscle tissue and agonists against this receptor are used to treat asthma and preterm labor (4–6).
Despite the lack of crystal structures, adrenergic GPCRs have been successfully targeted for drug discovery by using ligand-based analoging. Over the past 55 years (7), this has resulted in 155 drugs, 32 of which target the β2-adrenergic receptor (8). With the recent determination of the crystal structure of this receptor (9, 10), we were interested to see how well the atomic model performed as a template for ligand discovery, using docking screens of large chemical libraries. For this task, the molecular docking program DOCK3.5.54 was used (11–14). Like other widely used docking programs (15–17), DOCK screens small-molecule libraries for compounds that complement a binding site. Multiple orientations of each organic molecule are sampled, each in multiple low-energy conformations. Also, like most docking methods, DOCK ranks molecules based on polar, steric, apolar, and solvation terms. The particulars of how these are calculated differ among the different programs (DOCK3.5.54 uses a physics-based scoring function); none of them can reliably predict binding affinities, although many have proven successful at distinguishing low-likelihood ligands from those more likely to bind.
At the simplest level, the success of such a docking screen against the X-ray structure might be reflected in the hit rates emerging from it and the potency of the hits. More subtly, one might ask how strongly the hits are biased toward chemotypes previously explored by the extensive medicinal chemistry efforts against this and related targets, or whether the structure is capable of recognizing new chemotypes not explored by ligand-driven approaches. In addition, because the β2AR was crystallized in complex with an inverse agonist, it is important to determine whether the structural approach is biased toward finding inverse agonists or whether we would also identify neutral antagonists or even partial agonists and agonists, a point that has also been raised by Bouvier and colleague (18).
To investigate these questions, we docked a library of close to one million commercially available “lead-like” molecules against the X-ray crystal structure of β2AR, ranking them based on calculated complementarity to the receptor. From among the top-ranking 0.05% of the library, 25 molecules were selected based on their fit to β2AR and their novelty, and these compounds were tested experimentally for binding. Compounds with substantial affinity were further characterized for efficacy in an in vitro G protein-coupling assay. Here, we consider the resulting hit rate, affinity, and novelty of these compounds, as well as their origins in terms of the druggability of this target and the composition of a putatively unbiased library docked against it.
Results
The Binding Site of β2AR.
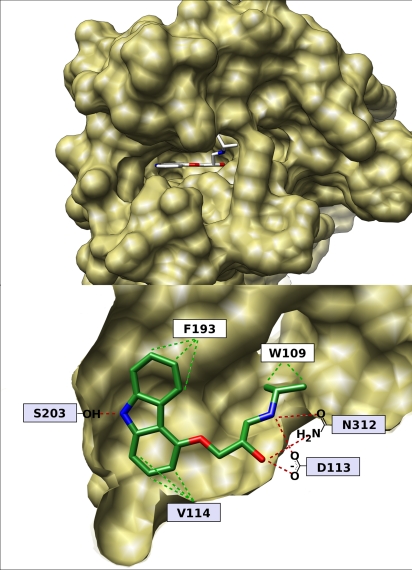
The epinephrine binding site of β2AR presents a pocket almost ideally suited to docking. It is a narrow and deep cleft that is largely concealed from solvent (Fig. 1Upper). Because the cleft is narrow, ligands can interact with both walls, offering the possibility for van der Waals contacts that span at least one principal dimension of the site. Most of the residues lining the site are hydrophobic, which contributes to potential affinity, whereas a few polar residues allow for strong directional constraints through electrostatic interactions. Most notably among these latter residues, Asp-113 [3.32 in Ballesteros–Weinstein notation (19), given as superscript hereafter] forms a salt bridge with most known ligands. Located at the back of the pocket is Ser-2035.42, which hydrogen bonds with carazolol, the ligand present in the X-ray structure. Also Gln-3127.39, adjacent to Asp-1133.32, is known to accept hydrogen bonds from the β-hydroxy-amine motif, which is prevalent among β-adrenergic modulators (Fig. 1 Lower).
Fig. 1.
The binding site of β2AR. (Upper) Top view of the binding cleft with bound carazolol. (Lower) Side view of the binding pocket, the proximal portion of the protein has been removed. Shown are some key interacting residues with their approximate placement. Red and green dashed lines indicate polar and hydrophobic contacts, respectively. Residues in a light-blue box are essential for agonist and antagonist binding (10).
Docking the ZINC Database.
The 972,608 molecules of the lead-like subset of the ZINC database (20) were docked into the carazolol-bound structure of β2AR. DOCK found poses without major clashes for 919,549 of these molecules. These were sampled in, on average, 14,930 orientations and, for each of these, 30,834 conformations (4.6·108 poses, on average, for each of the 919,549 molecules fit into the site). The time of calculation was 3.9 h on 468 CPUs. The molecules were scored for receptor complementarity based on the sum of their van der Waals [using the AMBER potential (12)] and electrostatic interaction energies [using ligand probe charges in an electrostatic potential calculated by DelPhi (13, 21, 22)], corrected for ligand desolvation (adapted from ref. 14).
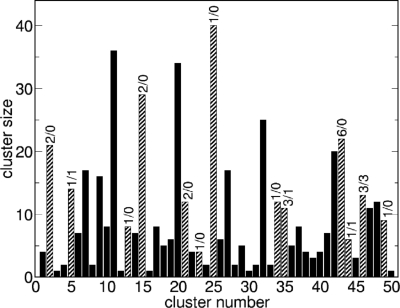
Based on their docking scores, the compounds were ranked from best to worst fitting, and all of the compounds prioritized for experimental testing were picked from among the top-ranking 500 molecules, or 0.05% of the docked library. In addition, these molecules were inspected visually for features not captured in the docking calculation, such as chemical diversity, actual commercial availability, and an overall balance between polar and nonpolar complementarity to the binding site. This is our common practice in prosecuting docking screens, and these criteria are consistent with previous studies. Twenty-five molecules were ultimately selected for experimental testing. To evaluate how these 25 represented the chemotypes present among the top 500 ranking molecules, we clustered these latter molecules using FCFP4 fingerprints with an average of 10 molecules per cluster and a maximum intracluster distance of 0.7 (23). This yielded 50 clusters; the 25 compounds chosen for testing fell into 13 of these, whereas what turned out to be the 6 true ligands originated from four of them (Fig. 2).
Fig. 2.
Bar graph showing the sizes for the 50 clusters into which the top ranked 500 docked molecules fell. The cluster number on the x axis is arbitrary. Numbers on top of each bar indicate the number of compounds tested (first number) and the number of active compounds (second number). Clusters containing compounds that were tested are striped, the others filled. Compound 1 is in cluster 5; compounds 2, 3, and 4 are in cluster 46; compound 5 is in cluster 44; and compound 6 in cluster 35.
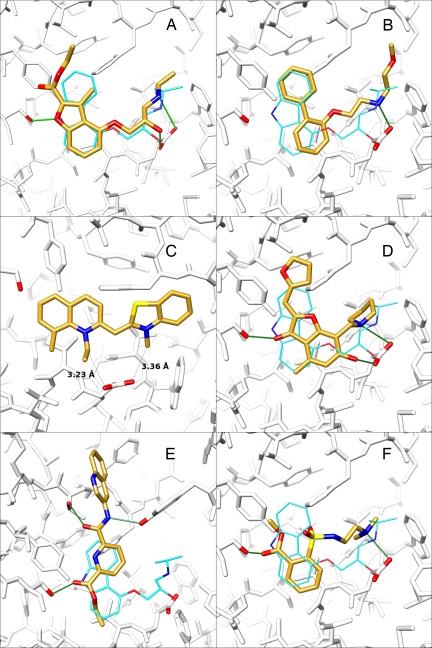
On closer inspection of these 25 selected molecules, we noticed that mutual topological resemblance, similar binding modes or physicochemical properties, would allow us to group them into 4 classes that captured common themes in their interaction with the receptor. We would like to emphasize that this classification was done a posteriori and that class membership was not a necessary condition for a molecule to be selected; it was merely a useful way to think about the compounds. “Classic” compounds featured most of the pharmacophoric characteristics of known antagonists, i.e., an aromatic ring system and an aliphatic amino group (typically charged at neutral pH). These compounds' docked poses usually overlapped well with the crystallographic pose of carazolol. Second, “bridge” compounds contained an aromatic ring system that overlapped with the location of the carbazole moiety of carazolol, but did not interact with Asp-1133.32. Instead, they connected the 2 walls of the binding site by hydrogen bonding with Thr-1955.34 and Tyr-3087.35, respectively. Third, “sulfonamide” compounds shared a common aromatic ring–sulfonamide–aliphatic nitrogen motif, with binding modes that resembled that of carazolol. Finally, the fourth class contained molecules with cationic nitrogens in ring scaffolds that have not previously been described among adrenergic ligands, or indeed among ligands for aminergic GPCRs. For the 25 compounds tested, the number in each class was 12, 9, 2, and 2, respectively, which in part reflects their relative abundance in the top 500, but also their commercial availability (Fig. 3).
Fig. 3.
Predicted binding modes of selected ligands and nonbinders identified in this study. In A–F, carazolol is depicted with cyan carbon atoms (when shown), the molecules identified in this study with golden carbon atoms, hydrogen bonds as green sticks, and residues Asp-1133.32 and Ser-2035.42 with red oxygens. Moreover, in E, Thr-1955.34 and Tyr-3087.35 are emphasized with red oxygens. (A) Overlay of the computed binding mode of 1 with the crystallographic binding mode of carazolol. (B) Binding mode of 2. The poses of 3 and 4 are very similar. (C) Predicted binding mode of 5. The distances between the alkyl substituents and the respective closest oxygen of Asp-1133.32 are shown as dashed lines. (D) Overlay of the computed binding mode of 6 with carazolol. (E) Overlay of the computed binding mode of the highest-ranking compound from the bridge class (nonbinder) with the binding mode of carazolol. (F) The binding mode of the highest-ranking sulfonamide compound. Not shown is the hydrogen bond interaction of the sulfonamide function with Asn-2936.55 because this residue has been removed for clarity.
Competition Assay and Similarity to Known Antagonists.
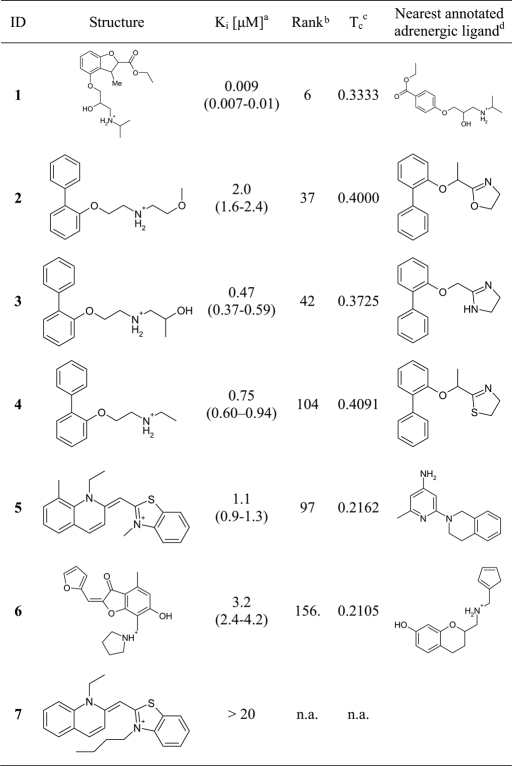
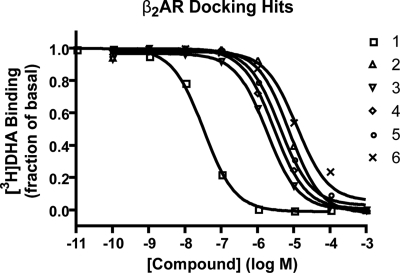
All 25 compounds were tested in a radioligand displacement assay at 20 μM, and 8 molecules measurably displaced 3H-dihydroalprenolol (DHA). Of these, the 6 compounds with percentage of displacement >10% were further characterized to obtain full DHA competition binding curves (Table 1 and Fig. 4). The measured percentage of displacement values obtained were: 100 (compound 1), 56 (compound 2), 85 (compound 3), 79 (compound 4), 67 (compound 5), and 39 (compound 6). All six had Ki values of <4 μM, with the most potent compound, 1, binding with a Ki of only 9 nM (95% confidence interval 7–10 nM). For this compound, only a racemic mixture was available for experiments. Because adrenergic receptors typically prefer one chiral form for compounds similar to 1, it is likely that the affinity of one of the enantiomers is even higher. Although docking cannot reliably rank order ligands by affinity, we note that 1 has the highest rank in DOCK score of all tested compounds, ranking 9th of the ≈1 million compounds.
Table 1.
Structures and experimental and calculated data for the six hits and one control compound
The six primary hits and the negative control compound. Membranes containing the wild-type human β2AR were prepared from Sf9 insect cells expressing the recombinant protein as described previously (10).
aAffinity values are based on 3 experiments done in parallel; numbers in parentheses represent the 95% confidence intervals.
bRank of the compound in the docking calculation.
cHighest Tanimoto similarity versus the subset of the adrenergic ligands of the WOMBAT (24) database using ECFP4 (23) fingerprints.
dThe compound of the WOMBAT database (24) most similar to the respective hit.
Fig. 4.
Dose–response curves of compounds 1–6. Ki values are calculated based on IC50 values and the Kd of DHA (0.16 nM).
The 6 confirmed ligands originated from 2 classes. Four of them, including compound 1, belong to the classic category: their docking geometry corresponds to that of carazolol and more broadly to what is expected for a β2AR ligand. Fig. 3 shows, as examples of canonical hits, the predicted poses for 1 in A and 2 in B. Compound 1, in particular, overlaps almost perfectly with carazolol and maintains all of its polar interactions. There are 2 main differences, one being the smaller aromatic ring system of compound 1 and the other one the ethoxycarbonyl substituent on this ring system. The latter is not involved in any obvious polar interactions in the docked pose. It can be speculated, however, that the side chain of Ser-2045.43 will rotate to form a hydrogen bond with the carbonyl oxygen of compound 1. As discussed below, compound 1 appears to be the most efficacious inverse agonist yet discovered for β2AR, but how this relates to these interactions remains unclear, because inverse agonism is convoluted with conformational changes distal from the orthosteric site.
The remaining 2 ligands (5 and 6) contain cyclic cations that are predicted to interact with Asp-1133.32 in manners previously unknown for adrenergic ligands. Compound 5 bears a constitutive positive charge and does not feature a hydroxy group. Because of this positive charge, which is dislocated between the 2 nitrogen atoms, the alkyl substituents on them are positively polarized. In the docking pose, these alkyl substituents form charge–charge interactions with Asp-1133.32 at 3.2 Å and 3.4 Å, respectively (Fig. 3C). Similarly, compound 6 interacts with Asp-1133.32 through the N-pyrrolidinomethylene substituent and the hydroxy group that is in the ortho position to it on the benzofuran moiety (Fig. 3D). Whereas this interaction resembles the classic aliphatic amine-hydroxyl motif in its spatial arrangement, it has not been described as a ligand chemotype.
The similarities and dissimilarities of these new ligands may be quantified by comparing their topological fingerprints with those of the 8,053 adrenergic ligands of the WOrld of Molecular BioAcTivity (WOMBAT) database (24) (see Materials and Methods and Table 1, column 5). Consistent with their categorization, the 4 classic molecules broadly resemble known adrenergic ligands. This can be seen by simply comparing them visually with their nearest neighbors among the WOMBAT ligands (Table 1) or, quantitatively, by calculating their Tanimoto coefficients to these nearest neighbors (their Tanimoto coefficients, based on ECFP4 fingerprints, range from 0.33 to 0.41). On the other hand, compounds 5 and 6 have low Tanimoto similarity values to any of the compounds in the database, and by visual inspection do not resemble their nearest neighbor in that database. These 2 molecules, although certainly cationic, seem to be novel chemotypes not previously explored. At the same time, it must be admitted that the most novel chemotypes to emerge from the docking, the bridge and sulfonamide compounds, were all nonbinders (Fig. 3 E and F).
Testing the Binding Mode of the Aryl Cations.
To test the predicted binding mode of 5, nine conservative derivatives were docked with an eye to identifying those that could not be fit in the bound geometry. From these calculations, the N-butyl derivative 7 (Table 1 and supporting information (SI) Fig. S1) appeared to clash with β2AR in a geometry that would otherwise maintain the key hydrophobic interactions between the quinoline moiety and residues Val-1143.33, Phe-2896.51 and Phe-2906.52 (Fig. 1 Lower). Consistent with this view, 7 does not detectably displace bound radioligand on experimental testing.
Ligand Efficacy.
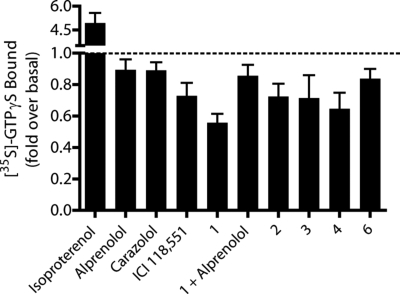
In the functional assay, 5 of the 6 ligands behaved as inverse agonists (Fig. 5); the activity of the sixth ligand, compound 5, could not be classified because of nonspecific effects on Gαs, even though its affinity was clear by ligand displacement assay. It is interesting to note that compound 1 exhibits inverse agonist activity as good or better than ICI 118551, heretofore the strongest and most potent inverse agonist for β2AR. The specificity of the inverse agonism of compound 1 was confirmed by blocking this activity with alprenolol. This behavior of the compounds is consistent with the binding site adopting a conformation adapted to the inverse agonist with which it was crystallized, carazolol, a point to which we will return.
Fig. 5.
Efficacy screen of compounds selected from competition binding. Purified wild-type β2AR and Tet-Gαs were reconstituted and stimulation of [35S]-GTPγS binding to Tet-Gαs was measured in the presence of various ligands. Isoproterenol, ICI 118551, alprenolol, and carazolol were tested at a concentration of 10 μM, all others at 100 μM. Compounds 1–4 and 6 show inverse agonist profiles. Data are shown as mean ± SEM of 3–4 independent experiments done in triplicate. Statistical analysis (Student's t test) shows significant differences (emphasized by ** if P < 0.01 and * if P < 0.05) for: bar 1, alprenolol and compound 1 (**, P = 0.0026); bar 2, carazolol and compound 1 (*, P = 0.0149); bar 3, compound 6 and compound 1 (**, P = 0.0082); bar 4, compound 1 and compound 1 + alprenolol (*, P = 0.0152).
Library Bias.
We wished to investigate how biased the docking library might be toward GPCR ligands. The lead-like subset of the ZINC database (20) contains 972,608 molecules and we compared each of these with the annotated ligands of the WOMBAT database (24) using the Similarity Ensemble Approach (25). At an E-value (confidence level) of 1·10−10, 103,563 molecules or 10.6% have an assignment to a GPCR. This compares with 41,182 (4.2%), 22,198 (2.3%), and 9,041 (0.9%) molecules that were assigned to kinases, proteases, and ligand-gated ion channels (LGICs), respectively. Comparing β2AR ligands specifically with those of an enzyme we have studied in the lab, β-lactamase TEM-1, there were 864 molecules in the ZINC subset with E-values of <1·10−10 for β2AR, but only 12 for TEM-1.
Discussion
The determination of the structure of β2AR allows us to investigate receptor-based discovery in an area long dominated by ligand-based approaches. Consistent with the druggability of this target, the β2AR binding site is well organized to bind organic molecules, partially explaining the frequency of the docking hits and their potency. An example of the latter is compound 1, which at 9 nM is among the most potent ligands to emerge from an unbiased (but see below) docking screen. Perhaps more compellingly, this compound is among the most effective inverse agonists, if not the most effective, known. A second contribution is the bias in our screening libraries toward aminergic chemotypes. The combination of potent new ligands that resemble known antagonists and structures dissimilar to previously explored ligands reflects the joint role of structural druggability and library bias, both of which are necessary to the success of a screening campaign. The β2AR seems unusually favored by both criteria.
Three features contribute to the druggability of the β2AR site and our ability to exploit it by docking. First, it is a deep and narrow pocket, leaving potential binders with only a limited choice of binding modes, i.e., mainly along a plane going down the center of the cleft. This substantially reduces the orientation space that has to be searched by the docking program. Moreover, the depth and narrowness of the pocket renders it comparatively “dry” on ligand binding. Hence, there are few possibilities for water-mediated hydrogen bonds and solvation, traditionally energy terms that are difficult to calculate correctly, making errors that arise from these terms less grave. Second, there are only a few polar and charged groups, and they are placed relatively far apart. Key among them are Asp-1133.32 at one end of the pocket and Ser-2035.42 at the other, an arrangement that emphasizes the longitudinal binding mode. This situation makes it possible for even comparatively simple organic molecules to complement all polar groups, whereas the spatial arrangement makes for a strong constraint on ligand orientation. Last, the binding site offers many hydrophobic interactions, most of them aromatic. These interactions will allow for many favorable van der Waals interactions and hence a high ligand binding affinity.
Library bias also played a role in the results of this screen. Unlike new genomic targets, or even many antibiotic targets, for which screening libraries may contain few likely ligands (26), we find that available chemical space is strongly biased toward chemotypes recognized by aminergic GPCRs. Thus, 10.6% of the commercially available molecules in the subset of the ZINC database used in this study resemble GPCR ligands, compared with only 2.3% for protease inhibitors, 4.2% for kinase-like ligands, 0.9% for LGIC-like ligands, and an even smaller proportion that resembles biogenic molecules such as natural products or metabolites. It is therefore not surprising that the most potent molecules to emerge from the docking calculation resemble adrenergic ligands. Intriguingly, a similar dominance by adrenergic-like ligands was also observed in the only other structure-based screen against this target of which we are aware, the elegant study by Topiol and colleagues (27).
Still, a principal goal of structure-based docking is to discover novel chemotypes; compounds 5 and 6 are examples of substantial departures from previously explored scaffolds. Whereas these molecules lack the affinity of the best antagonist described, the fact that such novel chemotypes are recognized supports the idea that new scaffolds remain to be discovered even for targets as intensely studied as aminergic GPCRs. The lack of measurable affinity for the negative control, compound 7, is consistent with the docking pose for its binding counterpart, compound 5, although this is not conclusive proof for the predicted conformation.
The observation that the docking-derived hits are inverse agonists is consistent with the use of the carazolol-bound conformation of β2AR in the screen, but it might also reflect the lack of agonists in the database we used. Epinephrine and related agonists, many of which are relatively small, were not part of the ZINC subset that we docked, which was restricted to lead-like molecules (see Materials and Methods). In retrospective docking screens of known agonists contained in the WOMBAT database, however, several were docked with scores that would have ranked them among the top 500 molecules of the million screened. Similarly, de Graaf and Rognan were able to successfully retrieve both agonists and antagonists when they docked to the β2AR structure, using Interaction Fingerprint Scoring (28) for the ranking (29). Additionally, after changing the rotameric states of the side chains of only 2 residues (Ser-2045.43 and Ser-2075.46), it was possible to selectively retrieve agonists in the docking. Thus, whereas the dominance of the docking hits by inverse agonists, often highly efficacious ones, is intriguing, it does not itself rule out the discovery of agonists from this structure, which remains a topic of ongoing study.
Comparing the docking approach to ligand-based methods, which are immune to the details of crystallographic conformation, it is clear that they will identify molecules to which this structure-based screen is opaque. Conversely, such methods, because they depend on ligand similarity, are unlikely to find genuine departures, such as compounds 5 and 6. In short, ligand-based methods will continue to guide drug design against GPCRs in areas where the available structures of these flexible targets are uninformative. However, the structures now emerging enable the discovery of novel chemotypes, unimaginable by inference-based methods, that complement the structure of the binding site. Based on the structural druggability of aminergic GPCRs, and the general bias in our libraries, this structure-based approach promises to be a fruitful avenue for ligand discovery against these pharmacologically important targets.
Materials and Methods
Protein Preparation for Docking.
The structure of the β2AR in complex with carazolol [PDB ID code 2RH1 (9, 10)] was used in the docking calculations. All water, organic solvent, ligand, and lipid molecules were removed. The T4-lysozyme insertion in place of loop 3 was also removed, because it carried a net formal charge of +9 that is not present in the native receptor. Protons were assigned automatically and in such a way that the protonation states of side chains and termini corresponded to pH 7. Orientations for hydroxy groups in selected binding site residues were modified to conform to the proton positions determined by the HBUILD module in CHARMM (30).
Docking Calculations.
All calculations were performed with DOCK3.5.54 against the β2AR/carazolol complex determined by crystallography, and screening the ≈1 million compounds of the lead-like subset of ZINC (see SI Methods for details).
Similarity Calculations.
The similarity calculations were performed with Pipeline Pilot (23). As a reference database, we used a subset of the WOMBAT 2006.2 database (24) containing the molecules that had been annotated as interacting with adrenergic GPCRs. Both the database and the query molecules were generated from SMILES and compared by using ECFP4 (23) fingerprints and the Tanimoto coefficient.
Competition and Functional Assays.
All Ki values were determined based on radioligand displacement assays by using ≈60 fmol of 3H-dihydroalprenolol preincubated with the receptor. Purified unliganded β2AR and tethered-Gαs protein was used in functional assays to determine ligand efficacy (see SI Methods for details).
Library Bias.
Similarities of the library molecules to sets of known ligands of the targets in WOMBAT (24), version 2006.2, were assessed with the Similarity Ensemble Approach (SEA) (25). This method yields a predicted target for every query molecule based on the similarity of the query to the set of known ligands of the respective target. Library bias was assessed by counting the numbers of molecules with a certain target class assignment, i.e., significant similarity of a molecule with the target ligand set. The criterion for a significant assignment was an E value of <1·10−10. We then calculated cumulated predictions for targets of the GPCR, protease, kinase, and LGIC classes.
Supplementary Material
Acknowledgments.
We thank Michael Keiser for SEA calculations and Michael Keiser, Denise Teotico, and Christian Laggner for reading the manuscript. This work was supported by National Institutes of Health Grants GM59957 (to B.K.S.) and NS28471 (to B.K.K.), Swiss National Science Foundation Fellowship PBZHA-118815 (to P.K.), and National Institutes of Health Grant F32 GM082028 (to D.M.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812657106/DCSupplemental.
References
- 1.Schwartz TW, Hubbell WL. Structural biology—A moving story of receptors. Nature. 2008;455:473–474. doi: 10.1038/455473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth BL. Receptor systems: Will mining the receptorome yield novel targets for pharmacotherapy? Pharmacol Ther. 2005;108:59–64. doi: 10.1016/j.pharmthera.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Milligan G, Svoboda P, Brown CM. Why are there so many adrenoceptor subtypes? Biochem Pharmacol. 1994;48:1059–1071. doi: 10.1016/0006-2952(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 4.Bai TR. Beta2 adrenergic receptors in asthma—A current perspective. Lung. 1992;170:125–141. doi: 10.1007/BF00174316. [DOI] [PubMed] [Google Scholar]
- 5.Barnes PJ. β-Adrenoceptors in smooth muscle, nerves and inflammatory cells. Life Sci. 1993;52:2101–2109. doi: 10.1016/0024-3205(93)90725-i. [DOI] [PubMed] [Google Scholar]
- 6.Smiley R, Finster M. Do receptors get pregnant too? Adrenergic receptor alterations in human pregnancy. J Matern Fetal Med. 1996;5:106–114. doi: 10.1002/(SICI)1520-6661(199605/06)5:3<106::AID-MFM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 8.Wishart DS, et al. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 11.Kuntz ID, et al. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 12.Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem. 1992;13:505–524. [Google Scholar]
- 13.Shoichet BK, Kuntz ID. Matching chemistry and shape in molecular docking. Protein Eng. 1993;6:723–732. doi: 10.1093/protein/6.7.723. [DOI] [PubMed] [Google Scholar]
- 14.Shoichet BK, Leach AR, Kuntz ID. Ligand solvation in molecular docking. Proteins Struct Funct Genet. 1999;34:4–16. doi: 10.1002/(sici)1097-0134(19990101)34:1<4::aid-prot2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 16.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 17.Jones G, et al. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 18.Audet M, Bouvier M. Insights into signaling from the β2-adrenergic receptor structure. Nat Chem Biol. 2008;4:397–403. doi: 10.1038/nchembio.97. [DOI] [PubMed] [Google Scholar]
- 19.Ballesteros JA, Weinstein H, Stuart CS. Methods in Neurosciences. New York: Academic; 1995. pp. 366–428. [Google Scholar]
- 20.Irwin JJ, Shoichet BK. ZINC—A free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilson MK, Honig BH. Calculation of electrostatic potentials in an enzyme active-site. Nature. 1987;330:84–86. doi: 10.1038/330084a0. [DOI] [PubMed] [Google Scholar]
- 22.Wei B, et al. A model binding site for testing scoring functions in molecular docking. J Mol Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 23.BioSolveIT, SciTegic. Pipeline Pilot. San Diego, CA: Accelrys Software; 2007. Version 6.1.5. [Google Scholar]
- 24.Olah M, et al. In: Chemoinformatics in Drug Discovery. Oprea TI, editor. Weinheim, Germany: Wiley-VCH; 2005. pp. 223–229. [Google Scholar]
- 25.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 26.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 27.Sabio M, Jones K, Topiol S. Use of the X-ray structure of the β2-adrenergic receptor for drug discovery. Part 2: Identification of active compounds. Bioorg Med Chem Lett. 2008;18:5391–5395. doi: 10.1016/j.bmcl.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 28.Marcou G, Rognan D. Optimizing fragment and scaffold docking by use of molecular interaction fingerprints. J Chem Inf Model. 2007;47:195–207. doi: 10.1021/ci600342e. [DOI] [PubMed] [Google Scholar]
- 29.de Graaf C, Rognan D. Selective structure-based virtual screening for full and partial agonists of the β2 adrenergic receptor. J Med Chem. 2008;51:6620. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- 30.Brooks BR, et al. CHARMM—A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.