Abstract
DNA structural variation (SV) comprises a major portion of genetic diversity, but its biological impact is unclear. We propose that the genetic history and extraordinary phenotypic variation of dogs make them an ideal mammal in which to study the effects of SV on biology and disease. The hundreds of existing dog breeds were created by selection of extreme morphological and behavioral traits. And along with those traits, each breed carries increased risk for different diseases. We used array CGH to create the first map of DNA copy number variation (CNV) or SV in dogs. The extent of this variation, and some of the gene classes affected, are similar to those of mice and humans. Most canine CNVs affect genes, including disease and candidate disease genes, and are thus likely to be functional. We identified many CNVs that may be breed or breed class specific. Cluster analysis of CNV regions showed that dog breeds tend to group according to breed classes. Our combined findings suggest many CNVs are (1) in linkage disequilibrium with flanking sequence, and (2) associated with breed-specific traits. We discuss how a catalog of structural variation in dogs will accelerate the identification of the genetic basis of canine traits and diseases, beginning with the use of whole genome association and candidate-CNV/gene approaches.
Recent findings revealed that DNA structural variation (SV) is common in normal rodents (Graubert et al. 2007; Guryev et al. 2008) and primates (Iafrate et al. 2004; Sebat et al. 2007; Lee et al. 2008; for review, see Freeman et al. 2006). The term SV and its subset of copy number variation (CNV), refer to alterations from 1 kb to multi-Mb. The development of high-resolution array comparative genome hybridization (aCGH) allowed the genome-wide discovery of submicroscopic copy number changes (Ishkanian et al. 2004). More recently, other approaches have allowed other types of SV (e.g., inversions) and polymorphisms to be surveyed. For example, high-density single nucleotide polymorphism (SNP) platforms can allow detection of subsets of SNPs and SVs (Conrad et al. 2006; McCarroll et al. 2006). Most recently, paired end approaches have evolved to yield massive numbers of new SVs at ultra-high resolution (although requiring significant cost and effort) (Tuzun et al. 2005; Korbel et al. 2007).
Many human CNVs appear to be old because they are in linkage disequilibrium with nearby genetic markers (Hinds et al. 2006). Others are recent or recurrent. Notably, CNVs are enriched in regions that are rearrangement hotspots, such as those rich in segmental duplications (or “low copy repeats”) (Sharp et al. 2005). In humans, more than 5000 CNVs have been identified in normal individuals, and at least several hundred are common in the population (Redon et al. 2006; Korbel et al. 2007; Wong et al. 2007; Kidd et al. 2008). Most known CNVs span genes and appear likely to affect genetic networks. Thus, while SNPs and point mutations are more frequent, CNVs seem more likely to have an impact on phenotype. CNVs can have various effects, including changes in gene expression levels, disruption of gene dosage, unmasking of recessive alleles or regulatory polymorphisms, and loss of regulatory elements (for reviews, see Lupski and Stankiewicz 2005; Reymond et al. 2007).
Disease relevance of DNA copy number alteration was already appreciated from recurrent deletions of tumor suppressors (e.g., TP53 and RB1) and amplifications of oncogenes (e.g., MYC and EGFR) in cancer. However, since the recent discovery of CNV in germline DNA, several affected genes were shown to be associated with disease: CCL3L1 with HIV/AIDS, FCGR3 with glomerulonephritis, DEFB2 with Crohn's, C4A/C4B with lupus, and PRSS1 with pancreatitis (Gonzalez et al. 2005; Aitman et al. 2006; Fellermann et al. 2006; Le Marechal et al. 2006; Fanciulli et al. 2007; Yang et al. 2007). CNV also provides material and mechanism for creating new genes. CNV thus accounts for a great deal of the genetic variation in animals and humans. For example, it was recently shown that the CNV landscape in flies is under natural selection (Emerson et al. 2008). Little is known about CNV in mammalian evolution, as only Euarchontoglires—the clade of primates and rodents—has been surveyed for CNV.
Dogs have extraordinary advantages as animal models (Lindblad-Toh et al. 2005): (1) they are numerous, (2) they share many diseases that are similar to those of humans, (3) they have a greater than fivefold reduced lifespan compared to humans, (4) they often have a high level of health care, and (5) live in an environment that is very similar to that of their owners. Humans are more closely related to mice than dogs (divergence times of 80 million years ago [Mya] and 90 Mya, respectively). However, dogs share ∼650 Mb of ancestral sequence that is absent in mice, and the sequence divergence rate of dogs is half that of mice (Lindblad-Toh et al. 2005). This suggests that a subset of human biology is more similar to that of dogs versus mice. But the main advantage of canine models is their evolutionary history. Largely based on 20 yr of studying dog and pigeon breeding, Darwin (1859) began On the Origin of Species by saying that variation under domestication is the best clue to understanding evolution. He believed that selection is the most important aspect of evolution, and that it is exaggerated in domestication. In many cases, it also offers documentation of parental breeds (or subspecies), of the selection process, and of the phenotypic changes observed. Dogs coevolved with humans for >15,000 yr (Leonard et al. 2002; Savolainen et al. 2002). However, most of the ∼350 dog breeds were created by selection of extreme behavioral and morphological traits over the last 150 yr. This presumably isolated many single “founder” mutations, and thus reduced disease heterogeneity. That is consistent with the fact that most breeds are predisposed to a few diseases. Importantly, breeds can be clustered into breed classes that share genetic markers due to common descent. Breeds are thus isolated populations likely to share disease mutations with other members of the same breed class, but flanked by genetically informative sequence (i.e., due to meiotic recombination in each breed). In other words, while broad mapping is accelerated using single breeds, fine mapping can be done in related breeds. This power has begun to be applied with success (Candille et al. 2007; Parker et al. 2007; Sutter et al. 2007). The unique genetic history of dogs thus makes them an ideal mammal in which to study the biology of CNV. Here we report the use of high resolution oligonucleotide (oligo) aCGH to map common CNVs in pedigreed dogs (commonly referred to as “purebred”). We discuss the value of further cataloguing large amounts of such variation, some of which is likely to underlie breed-specific biology and disease predisposition.
Results
aCGH of a canine lymphoma cell line
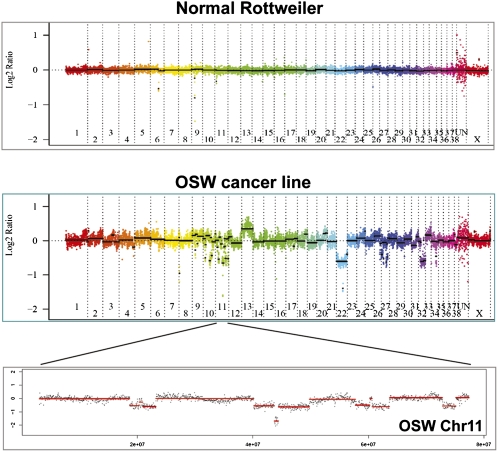
Array CGH was performed on a 385,000 spot oligo array (i.e., mean spacing of 4675 bp; NimbleGen Systems Inc.; see Methods for details of the array platform and the genome assembly used). We began our study by conducting aCGH on the well-characterized OSW T-cell lymphoma cell line from an Airedale Terrier (Kisseberth et al. 2007; Supplemental Table S1). The aCGH reference DNA for this hybridization—and for all others reported here—was from a pedigreed Boxer. Notably, OSW was previously analyzed on a low resolution BAC array, and several copy number alterations were validated (Kisseberth et al. 2007). Each spot of such BAC arrays is comprised of many fragments 0.5–2 kb in size, but our spots correspond to single unique-sequence oligos. We thus consider these two platforms to cross-validate each other (as essentially different methodologies). This comparison also confirmed the mapping of the arrayed oligos. (In depth analysis and discussion of the extensive copy number alteration present in this single lymphoma cell line are outside the scope of this work, and will be included in a second publication.) Figure 1 compares the linearized whole genome karyogram of OSW to that of a normal dog (Rottweiler), and shows a close up of all of chr 11. There is very strong correlation between the published low resolution BAC aCGH results and our largest alterations. This is most apparent for the copy number gain of nearly all of chr 13 and the one copy loss of almost all of chr 22 (cf. Fig. 1 in this study with Fig. 3 of Kisseberth et al. 2007). Kisseberth et al. (2007) additionally confirmed the DNA copy number changes of four genes within alterations detected in both studies (P16, MYC, RB1, PTEN). Beyond validating our methods, we also identified alterations previously implicated in canine lymphoma, such as the whole chromosome amplification of chr 13 (containing MYC) and loss of chr 11 and regions of chr 22 (containing RB1) and chr 26 (containing PTEN) (Hahn et al. 1994; Thomas et al. 2003; Modiano et al. 2007). The copy number alteration of some of those regions/genes, and others, are known to be associated with human lymphoma (e.g., P16 loss and MYC gain). Finally, we also identified novel candidate cancer genes (e.g., see SLITRK1 below). Our results thus support the notion that canine and human lymphoma are genetically related and suggest new candidate genes to consider in the human condition.
Figure 1.
Whole-genome DNA copy number quantification of the OSW dog lymphoma cell line by aCGH. DNA was cohybridized with a reference pedigreed Boxer (in two colors) to an oligo array with <5 kb mean spacing. Copy number is shown as log2 ratios, with gains above the midline and losses below. The top panel shows the linearized whole-genome copy number of a normal Rottweiler and the middle panel that of OSW. (There is no Y chromosome data, and the last segment before the X is the unmapped genome assembly.) Note the complete or nearly complete gain of chr 13 and losses of chr 22 and chr 32. The bottom panel shows all of OSW chr 11, with seven large single-copy losses and one small two-copy loss. The P16 tumor suppressor gene lies within the two-copy loss region.
Figure 3.
Cluster analysis. (A) Unbiased clustering of all high confidence probes altered in any of the CNV-discovery panel of dogs. Probe signals were clustered using hierarchical clustering (centroid linkage and the Pearson correlation as the distance, dChip 2005; Li and Wong 2001). CNV signal partially clusters dogs according to their breed class. Dog breeds are color-coded by breed class: Asian/ancient in red, Mastiff in blue, herding in black, and hunting in green. (Shet. Sheepdog) Shetland Sheepdog; (GS Pointer) German Shorthaired Pointer. (B) Clustering analysis of all aCGH probes for each dog. Hierarchical clustering was conducted with Genesis software (Sturn et al. 2002). All probe signals were used, except those from the X and Y chromosomes. Two of the groupings presumed to be true clusters show the shortest branch lengths: Collie and Shetland Sheepdog are both members of the Herding breed class; the two most similar dogs analyzed are two unrelated Akitas. The third pair is presumed to be a false clustering.
aCGH of pedigreed dogs
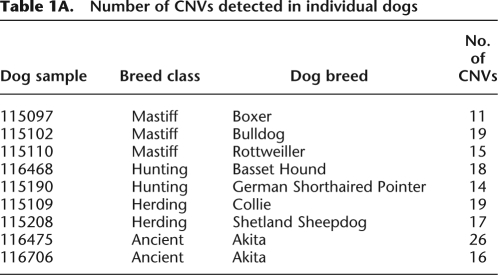
Genotyping of polymorphic canine loci has confirmed that most breeds cluster into breed classes (Parker et al. 2004, 2007). To survey CNV in normal pedigreed dogs, we assayed nine dogs that represent four main breed classes—Asian/ancient, Mastiff, Herding, and Hunting (Table 1A; Parker et al. 2004). As the reference DNA for all the aCGH assays, we used an unrelated pedigreed Boxer. We identified 155 CNVs—in 60 CNV regions (CNVrs)—at high confidence (Fig. 2; Table 1B; Supplemental Table S2). We also detected many more candidate CNVs with reduced confidence. The results of genome-wide cluster analysis of aCGH signal suggest there may be many CNVs that are true, but which are undetected or detected at lower confidence (see below). Those are likely be further resolved by higher resolution oligo arrays that are now available. We found CNVs that may be common within breed classes (Figs. 2, 3). Some other CNVs could have preceded the divergence of dog breed classes and others may have occurred independently in different breeds.
Table 1A.
Number of CNVs detected in individual dogs
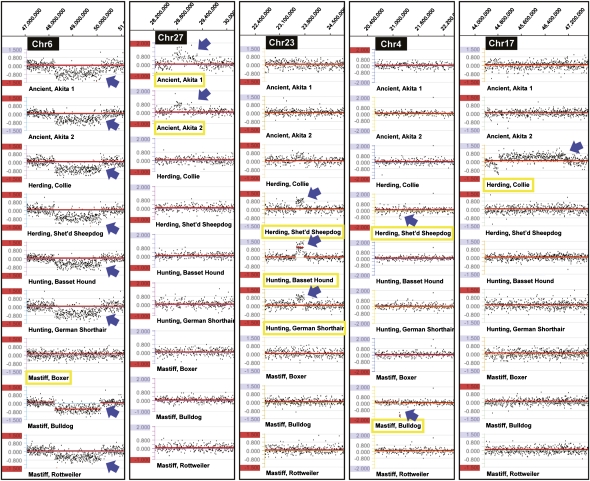
Figure 2.
Copy number variants (CNVs) in the four main classes of dog breeds. DNA from nine pedigreed dogs was cohybridized with a reference pedigreed Boxer. The first panel shows a change in all dogs but the Boxer. As the reference is another Boxer, we presume the CNV is a gain in Boxers. The second panel has a gain CNV specific to the two Akitas. The next two show CNVs shared by herding and hunting, and herding and mastif breeds. The last shows a CNV present in a single herding dog, a Collie.
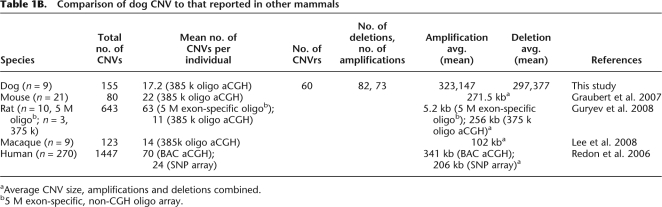
Table 1B.
Comparison of dog CNV to that reported in other mammals
aAverage CNV size, amplifications and deletions combined.
b5 M exon-specific, non-CGH oligo array.
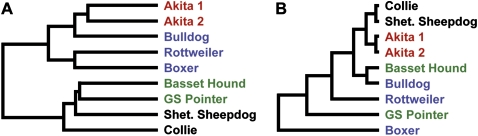
Dog phylogeny
We did not expect to find strong evidence of breed relationships in our CNV analysis. Rather, we presumed that such a signal would be obscured by CNVs that are more recent or recurrent. Surprisingly, our results suggest that a significant proportion of CNVs in modern dogs can be used to dissect evolutionary relationships. Figure 3A shows unbiased clustering of aCGH signal intensities of all probes variant in any dog. We find a trend that breeds partially cluster according to breed classes. Although this finding will have to be confirmed with more dogs, it seems unlikely that the observed pattern could be generated randomly. Moreover, the only pair of samples from the same breed not only group together, but also have the shortest branch lengths. That suggests a good portion of the identified variation has been transmitted from the founders of each class. Encouraged by those results, we tested the clustering of all genome-wide probe signals (vs. only the CNV regions as above). In Figure 3B we show the hierarchical clustering of all probes, except those on the sex chromosomes. The result shows two cluster pairs with very short branch lengths and one pair with longer branches (the remainder diverge individually, with increasing branch lengths). Notably, the two pairs with the shortest branches correspond to true clusters—the two unrelated Akitas and the two Herding class breeds. (The third pair crosses breed classes and is presumably false.) We reiterate that this clustering is based not only on the called CNV regions but on all probes on the array. The infinitesimal signal from the 60 CNVrs alone should not be detectable in the signal noise of the whole genome. The observed pattern thus hints that there may be a great deal of real CNV signal that we could not call reliably (mainly involving <50-kb CNVs and regions of low probe density). This suggests that many of our moderate confidence CNVs could be true.
Annotation and validation of CNVs
At least 56 genes overlap with our high confidence CNV regions (Supplemental Table S2), and only 16/60 CNVrs do not span annotated genes. However, the dog genome annotation is incomplete and it is likely that more genes are affected. In Supplemental Table S3 we show the Gene Ontology (GO) categories that are overrepresented in dog CNVs (and highlight those that are also in excess in humans and mice). As in humans and mice, there is overrepresentation of olfactory receptors and immunity-related genes (and others) in dogs. We also find CNV in several genes known to be involved in human disease, most notably cancer. Some genes suggest intriguing biology as well as pathophysiological relevance, such as the signaling proteins NOTCH2 and MAGI2. The latter finding is curious because MAGI2 CNV is strongly associated with human epilepsy (Marshall et al. 2008), and our CNV was discovered in the German Shorthaired Pointer, which carries a high risk for epilepsy (Slater 2007). As most of the identified CNVs span genes, we predict some of them underlie diverse breed-specific traits and disease predisposition.
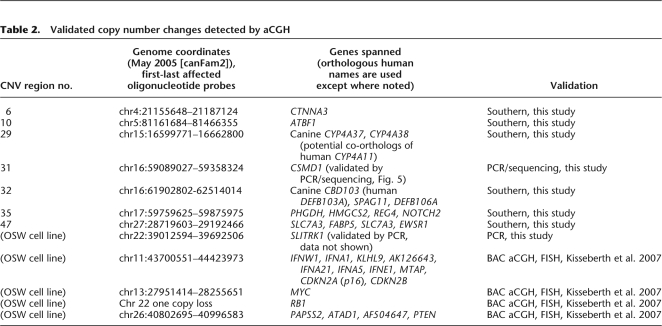
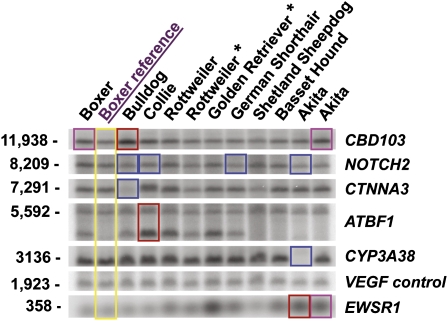
Even before we initiated validation, several criteria suggested our high confidence CNVs are true. (1) The Boxer has the fewest CNVs; this was expected because the aCGH reference is a Boxer. (2) The highest cluster similarity (see above) was seen in the only case where two dogs of one breed were tested—unrelated Akitas. (3) The cluster analysis of CNVrs tends to group breeds within their breed classes. Moreover, as we mentioned above, previous BAC aCGH and FISH analysis of the OSW cell line validated many of the large regions of copy number alteration and several specific affected genes (Kisseberth et al. 2007). We confirmed eight of the copy number changes we discovered in our aCGH survey (Table 2). Six of six variants selected for disease interest and tested by Southern blotting were validated (Fig. 4). Those were mostly chosen for relevance to cancer and neurology: CTNNA3, ATBF1, CYP4A38, CBD103, NOTCH2, and EWSR1. We equilibrated the dog samples with VEGFA, which should always be two copies (mice with copy gain or loss are not viable; Carmeliet et al. 1996; Miquerol et al. 2000). All six CNVs were validated for copy number gain or loss (breakpoint locations were not determined). Two other variants were confirmed by PCR, and one of those (Fig. 5) was resolved at sequence resolution.
Table 2.
Validated copy number changes detected by aCGH
Figure 4.
Validation of CNVs of biological interest. CNV was quantitatively tested by Southern blotting. The dogs included are the aCGH discovery set and reference, plus one Rottweiler and one Golden Retriever (asterisks). All samples were equilibrated with a known two-copy gene (VEGFA) and compared to the aCGH reference dog (in yellow). The bands corresponding to the survey dog in which a CNV was discovered are boxed—red for discovery gain, pink for intermediate gain, and blue for loss. The CNVs are labeled according to the gene of interest (other genes spanning the same variant are provided in Table 2). The size of HindIII fragments detected are provided in base pairs. Note that EWSR1 CNV gain is also present in the Golden Retriever, which was not one of the dogs in the discovery set.
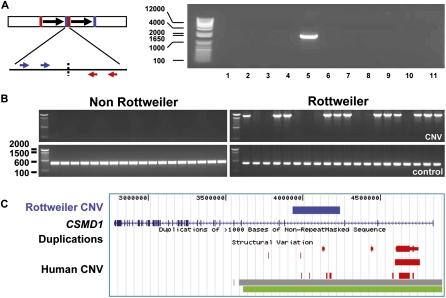
Figure 5.
Molecular definition of a Rottweiler-specific CNV. A 273-kb CNV on chr 16 was detected by aCGH only in the Rottweiler of the discovery set. (A) (Left) A schematic of the PCR assay used for validation. Each forward/reverse primer combination was used to test for tandem duplication of the aCGH CNV. (Right) The two internal primers generated a <2-kb fragment in the original Rottweiler, but not the nine other dogs. The breakpoint was determined to single nucleotide resolution by sequencing this amplicon. This confirms a head to tail orientation of the duplication. All samples are from unrelated members of pedigreed dogs (left to right): no template control, Boxer, Bulldog, Collie, Rottweiler 1, Golden Retriever, German Shorthair, Shetland Sheepdog, Basset Hound, Akita 1, Akita 2. (B) A second panel of 20 Rottweilers and 20 non-Rottweiler dogs shows the CNV is common in Rottweilers, but absent in the other breeds tested (see Methods). (C) The entire dog CNV unambiguously lies within intron 3 of CSMD1. Human CSMD1 is shown with the dog CNV mapped (blue bar) and compared to previously reported human CNVs (losses in red, gain in green, gain or loss in gray). The CNV spans several conserved elements, including four with PhastCons LOD values ≥98 (compare to the two flanking exons, with LOD = 93 and 31).
Structural variation suggestive of biological roles
A major interest in undertaking CNV discovery in dogs relates to the possibility that some of these are functional variants involved in breed-specific biology or disease predisposition. We found a candidate Rottweiler-specific microduplication in CSMD1, a putative tumor suppressor in humans. CUB and Sushi Multiple Domains 1 (CSMD1) was cloned as a candidate for the human 8p23.2 region that is frequently deleted in various epithelial tumors (Muscheck et al. 2000). This is potentially interesting because Rottweilers are at increased risk for cancer, most notably osteosarcoma. Deletion and reduced expression of CSMD1 are associated with poor prognosis in prostate and squamous cell cancers (Scholnick et al. 1996; Henshall et al. 2003; Paris et al. 2004). Both somatic point mutations and allele loss of CSMD1 are associated with cancer (Toomes et al. 2003; Richter et al. 2005; Farrell et al. 2008). We found a microduplication within CSMD1 in 14 of 22 Rottweilers (64%), but not in any of 29 non-Rottweilers (including 11 dogs in the broadly defined Mastiff breed class) (Wilcox and Walkowicz 1995). Further screening is necessary to confirm this CNV is truly Rottweiler-specific.
Figure 5 shows validation and characterization of the CSMD1 CNV identified by aCGH in the Rottweiler. We PCR-amplified through the breakpoint and sequenced it. The variant is a tandem duplication of 273 kb within the third intron of CSMD1. The dog sequence clearly maps to the same intron of the human CSMD1. Notably, the human gene is enriched for CNVs in the same third intron, as well as in the first. Analysis of the duplicated region shows that it contains several highly conserved elements. For example, we computationally confirmed the mapping of eight elements that are highly conserved in mammals—having PhastCons LOD scores > 50 (four are ≥98) (Siepel et al. 2005). We analyzed the region for PreMod regulatory modules of transcription factor binding sites that are evolutionarily conserved (Ferretti et al. 2007). Five of the eight PhastCons regions are also PreMod elements, suggesting they regulate gene expression. We found no concrete evidence of exons, genes, or miRNAs in the CNV region. Given CSMD1 may be a tumor suppressor, it is interesting that one of the PreMod elements has two possible p53 binding sites (PreMod hg17_mod108320). We did not find published support for that element, but a true p53 binding site lies 326 kb upstream of human CSMD1 (Wei et al. 2006).
We find CNV in beta defensin 103 (CBD103, ortholog of human DEFB103) is very common (Fig. 4). This is interesting because it has clear implications on immunity, feeding behavior, and coat color (see Discussion). Moreover, there is an odd genetic/epigenetic phenomenon related to that same locus and biology. Thus it will be important to molecularly define this CNV region. In Supplemental Figure S1 we show that the CNV is likely to be complex, and that the region may have sequence characteristics that make it difficult to characterize.
The OSW lymphoma cell line was previously known to have a single site where both copies of a genome region were lost (Kisseberth et al. 2007). Notably, that deleted segment contains the tumor suppressor CDKN2A (also known as p16). Our aCGH analysis of OSW confirmed that result and identified one additional two-copy loss. That second two-copy loss region (confirmed by PCR analysis, data not shown; see Supplemental Methods) is small and results in the deletion of the entire SLITRK1 gene, and otherwise only affects three candidate pseudogenes (Table 2). SLITRK1 is a transmembrane receptor enriched in the brain. It has homology with the growth-promoting secreted protein Slit in its extracellular domain and to the Trk neurotrophin receptor in its intracellular domain. Mutations in SLITRK1 are associated with psychiatric disorders, most notably Tourette's syndrome (Abelson et al. 2005). However, it was recently reported that the six members of the SLITRK gene family are expressed in hematopoietic stem cells (and are speculated to be involved in malignant hematopoiesis) (Milde et al. 2007). The two-copy loss of SLITRK1 in OSW raises the possibility that it is a tumor suppressor. We could not find published evidence of such a role in human cancer, but single cases of somatic mutations of SLITRK4 and SLITRK6 were recently reported in colorectal cancer (Wood et al. 2007).
Discussion
We have generated the first map of CNV in normal dogs. The number of CNVs we observe per animal (Table 1A,B) is very similar to the number found in mice using the murine-equivalent NimbleGen array (Graubert et al. 2007). As mice are now known to have equal or greater levels of CNV than humans, this suggests the extent of dog CNV is also comparable (She et al. 2008). Although mouse and dog NimbleGen oligo CGH arrays have similar resolution to that of human tiling BAC arrays, ∼50 kb (Ishkanian et al. 2004; Graubert et al. 2007), the latter detect far more CNVs (Table 1B). One reason for this is improved hybridization with the spotted 0.5–2 kb BAC probes (BAC-derived PCR fragment pools). A second reason for this effect is that only unique sequence is spotted in the oligo array (Sharp et al. 2007). This selectively omits—or reduces probe density in—CNV-enriched regions, such as segmental duplications and diverse repetitive sequences (Sharp et al. 2005). The increased density and improved probe design of the next generation platforms, including oligo CGH arrays, will improve this situation. It will also be important to develop a map of segmental duplications in dogs.
There is urgency to understand the contribution of CNV to natural variation and disease predisposition. However the phenotypic and genetic heterogeneity of humans makes this difficult. Dogs are especially well suited for this work: (1) they are monitored for health, as well as for breed-specific morphological and behavioral phenotypes (i.e., breed standards); (2) they have a huge range of genetic heterogeneity from intrabreed, to intrabreed class, to interbreed class; (3) they can be genetically crossed; and (4) they age much faster than humans. It seems likely that some CNVs are associated with breed standards and/or susceptibility to breed-specific disease. Notably, there are ∼75 million dogs in the United States alone. It is possible that some of the countless breeders and numerous breed clubs will join researchers to determine whether breed-specific CNVs segregate with traits or disease. For example, we validated one CNV that was only detected in Rottweilers. The CNV is present in the majority of Rottweilers and affects several highly conserved elements in an intron of CSMD1—a candidate tumor suppressor in humans. Those conserved elements are likely to be involved in gene expression, but there is no way to predict a positive or negative effect. It will be interesting to test whether that CNV is associated with Rottweiler-specific traits, such as high risk of osteosarcoma.
Another CNV of interest affects the beta defensin gene CBD103. Candille et al. (2007) recently showed that CBD103 is the long-sought-after Dominant Black (“K”) locus in dogs. The coat color of most mammals is largely determined by the ligand-receptor pair Agouti (Ag) and Melanocortin receptor 1 (MC1R). In addition, Dogs have the K locus. The top dominant allele KBlack—found in 38 dog breeds—is the product of a 3-bp deletion that removes glycine 23 (“CBD103ΔG23”). CBD103 affects coat color by binding to MC1R, and CBD103ΔG23 has two effects—increased affinity for receptor and increased extracellular levels. In addition, CBD103 binds to MC4R, which regulates feeding behavior. Thus transgenic mice overexpressing CBD103 are not only black, but also significantly smaller than littermates. We find that CBD103 has common copy number variation in multiple breeds (Fig. 4; Supplemental Fig. S1). As the physiological mechanism of CBD103ΔG23 is increased MC1R binding, it is likely that CNV of CBD103 also has phenotypic consequences. This could affect coat color, feeding behavior/metabolic regulation, and immunity. Surprisingly, the K locus also has a brindle allele that results in a pattern of whorled stripes alternating yellow and black coat color (Kerns et al. 2007). It was proposed that dog brindling is caused by clonal skin cell lineages being genetically KBlack or kyellow (Kerns et al. 2007). In this scenario, a stochastic event could set a KBlack or kyellow state in each clone, and that could be maintained epigenetically in dividing/migrating keratinocytes. The CNV observed at the K locus hints at mechanisms to explain kbrindle, including a combined structural genetic/epigenetic effect (Wilson et al. 2006).
The phylogeny of dogs is critical for the selection of breeds to be used in genetic studies. Parker et al. (2004) genotyped 96 microsatellite loci in 414 pedigreed dogs from 85 breeds. The phylogenetic tree of that data only resolved three high confidence branches—wolf, ancient/Asian dogs, and all other dogs. However, they ascertained genetic relationships to different degrees of confidence (very high to low) using Bayesian model-based clustering (Parker et al. 2004, 2007). Those studies suggested it might be difficult to solve the phylogeny of dogs due to the admixture of breeds. For example, the first genetic grouping of breeds was consistent with the historical designation of a single Mastiff class that includes subclasses of Mastiffs, Draft/Cattle dogs, and Bulldogs (Parker et al. 2004). More recently, it was proposed that Mastiffs and Bulldogs (including many terriers) group together and Draft/Cattle dogs (including mountain dogs) form another group (Parker et al. 2007). Our clustering findings hint that higher density CNV information may yield a phylogenetic tree of dog breeds. One-base-pair to 10-kb insertion/deletion polymorphisms (indels) are increasingly appreciated because they establish identity by descent with high confidence (Mills et al. 2006). Notably, small indels are being developed as genetic markers to study natural populations of dogs and wolves (Vali et al. 2008). The clustering pattern of CNVs suggests that the variation segregating with breed classes was present in the founder dogs of a breed class. As was shown for humans (Hinds et al. 2006) such CNVs should be in linkage disequilibrium with flanking sequence and would be detectable by tag SNPs. It will be important to be able to detect CNV haplotypes with the canine SNP platforms being developed for whole genome association. However, it should be noted that this is done with ∼27,000 SNPs in dogs (vs. ≥500,000 in humans) because they have an approximately sevenfold extension of linkage disequilibrium (Sutter et al. 2004). This means that current SNP platforms cannot be used to discover novel CNV. We propose the next step is to define the common CNVs, say larger than ∼10 kb, in the most popular breeds. This can already be done with second-generation platforms, such as the NimbleGen HD2 aCGH array with a greater than fivefold increased probe density (2.1 million elements on one slide) and improved oligo sequence design compared to that used here.
The availability of a catalog of CNV information will additionally enable the use of a candidate gene approach. For example, the Collie eye anomaly was recently mapped to an ∼4-centimorgan (cM) region, then fine mapped to ∼100 kb (spanning four genes) in four related breeds (Parker et al. 2007). The causative mutation was found to be a 7.8-kb intronic deletion. In the future, such studies could be vastly accelerated by high resolution CNV data. There are ∼400 identified canine diseases, and specific breeds are predisposed to a small number of conditions. That will make it straightforward to test suggestive breed-specific CNVs for particular disease associations. As was demonstrated in rats, is it also feasible to integrate genome-wide information on Quantitative Trait Loci, breed-specific CNV, and gene expression in order to identify candidate genes (Guryev et al. 2008). Another use of CNV information is in comparative genetics. For example, human genetic linkage data could implicate a genomic interval that has a suggestive CNV in a breed at high risk for a related disorder. Finally, diverse scientists will be drawn to dog models when they learn there is natural gain/loss variation in their gene of interest. Dogs are not simply animal models, but also dear companions to humans. One example of the significance of this is that 1 million dogs are treated for cancer annually in the United States. Thus, genetic advancements will also lead to translational research and improved canine health. In summary, this study shows how further canine CNV discovery can (1) impact the rapidly growing use of dog models, (2) uncover and answer basic genetic mechanisms, and (3) address dog and human diseases.
Methods
aCGH platform
We quantified DNA copy number by hybridization to the NimbleGen Systems Inc. canFam2 WG CGH oligo array platform (http://www.nimblegen.com), designed from the NCBI canine genome build of May 2005 (Lindblad-Toh et al. 2005). The array has 385,000 probes on a single slide, resulting in a median probe spacing of 4675 bp. Confident DNA CNV detection requires that alterations be covered by ∼10 oligos; thus, it has a resolution of ∼50 kb, similar to existing human tiling resolution BAC arrays (Ishkanian et al. 2004). Important for an oligo array, (1) it uses 50–75mers with generally similar melting temperatures (but see Sharp et al. 2007), and (2) it uses 2 μg of DNA without requiring complexity reduction or amplification (which generate incomplete sampling and signal noise, respectively). The probes are evenly distributed throughout the unique sequence of the genome. The methods for CGH on equivalent platforms for other species, and their analysis, are thoroughly validated and standardized (Graubert et al. 2007).
Source of genome sequence assembly and annotation
We used the May 2005 dog (Canis familiaris) whole genome shotgun (WGS) assembly v2.0 sequenced and assembled by the Broad Institute of MIT/Harvard and Agencourt Bioscience (Lindblad-Toh et al. 2005). The assembly is based on 7.5-fold shotgun coverage and has outstanding continuity. The gene annotation was largely taken from the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu). Much of the annotation was validated by manual BLAST analysis.
Source of dog DNA samples
Blood for the aCGH survey (discovery set) was collected from pedigreed dogs of volunteer contributors, under informed consent: two Boxers (one used as the reference for all other dogs), Rottweiler, Basset Hound, German Shorthaired Pointer, Collie, Shetland Sheepdog, and two Akitas. All of those dogs have Registration Certificates for their breed from the American Kennel Club (i.e., they are considered pedigreed or “purebred”). Breed classes are given in Table 1A, according to Parker et al. (2004) (see also Parker et al. 2007). The OSW (Airedale Terrier) lymphoma cell line was provided by W.C. Kisseberth (The Ohio State University College of Veterinary Medicine). The 20 Rottweiler and 20 non-Rottweiler DNAs (Fig. 4) were provided by C.A. London (The Ohio State University College of Veterinary Medicine). The 20 non-Rottweilers included the following dogs: Labrador Retriever (three), Golden Retriever (three), Shih Tzu (two), Chow Chow, Siberian Husky, Bloodhound, Pug, Collie, Fox Terrier, Dalmatian, Yorkshire Terrier, Springer Spaniel/Australian Shepherd, German Shepherd/Siberian Husky, Samoyed/Golden Retriever, and Poodle/Cocker Spaniel.
aCGH processing and analysis
Hybridization of genomic DNA to NimbleGen canFam2 WG CGH arrays (whole genome tiling of isothermal 50–75mer oligos at 4.7 kb median spacing) was performed by NimbleGen Systems, Inc. (Graubert et al. 2007). aCGH was used to detect DNA copy number. Each sample was cohybridized with the same reference DNA (taken from one normal pedigreed Boxer), one labeled with Cy3 dye and the other with Cy5. Slides were scanned and copy number was quantified from the dye fluorescence ratios. NimbleGen conducted the initial data processing, from normalization to signal calling and quantification. The high confidence calls are made according to the same criteria as used by Graubert et al. (2007). NimbleGen has an information package that describes the technology and provides measures of reproducibility, accuracy, sensitivity, and specificity (http://www.nimblegen.com/products/lit/nimblegen_grant_kit.pdf). Briefly, the normalize.qspline method from the Bioconductor package in R is used. To flag segments that deviate significantly from the baseline, a circular binary segmentation method is used (Olshen et al. 2004). We used a secondary manual method to identify false positives generated by the circular binary segmentation method and to identify other CNVs not found (i.e., false negatives). Candidate CNVs are identified by finding 11-probe segments with a log2 ratio greater than ±0.45. We removed presumptive false positives caused by single outlier probes. We conducted further analysis and visualization with SignalMap software (NimbleGen). For cluster analysis of CNVrs (Fig. 3A), all of the individual probe values within CNVrs were grouped by hierarchical clustering using centroid linkage and the Pearson correlation as the distance with dChip 2005 software (Li and Wong 2001). For genome-wide clustering (Fig. 3B), hierarchical clustering of all aCGH probes in the discovery dog panel was conducted with the Genesis software (Sturn et al. 2002). All individual probe signals were used except those on the X and Y chromosomes.
Molecular genetics
Polymerase chain reaction (PCR), DNA sequencing
PCR methods were essentially as described by the Taq polymerase manufacturer (JumpStart REDTaq, Sigma), optimizing conditions for each use. PCR primers are provided in the Supplemental Methods. PCR products were purified and directly sequenced (ABI 3130xl, The Research Institute at Nationwide Children's Hospital DNA Sequencing Core Facility).
Southern blotting
The standard methods used here have been described in precise detail (Chung et al. 2005). Probes were designed to (1) target unique sequence in HindIII fragments that did not overlap predicted breakpoints (most of which should result in a single Southern band), (2) generate fragments of different sizes to allow multiplex hybridizations, and (3) have similar melting temperatures, to result in comparable signal in multiplex format. Probes were generated by PCR (primers given in Supplemental Methods), confirmed by agarose electrophoresis, purified, and random primed-labeled with 32P. The same HindIII-digested DNA membrane was hybridized twice, in 3- and 4-plex hybridizations, and washed at high stringency. Signal was quantified by PhosphorImaging (Storm, GE Healthcare Life Sciences).
DNA samples
DNA was purified from blood using QIAamp DNA Blood Maxi Kit (Qiagen) and quantified by spectrophotometry. All DNA samples used for array hybridization were analyzed by spectrophotometry and agarose gel electrophoresis.
Acknowledgments
We thank the dog owners who volunteered to provide biological materials for this and associated studies. We thank C.A. London and W.C. Kisseberth for kindly sharing dog DNA and the OSW cell line, respectively. We thank Y.-C. Yu and H. Auer and their laboratory members for intellectual and technical support and W.C. Ray for computational assistance. We thank J.L. Rowell for technical assistance and K.L. McBride for critical reading of the manuscript. This work was supported by funding from The Research Institute at Nationwide Children's Hospital (TRINCH) to C.E.A. All samples collected for this work were acquired under informed consent, under a protocol approved by our institutional animal care and use committee.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083741.108.
References
- Abelson J.F., Kwan K.Y., O'Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Aitman T.J., Dong R., Vyse T.J., Norsworthy P.J., Johnson M.D., Smith J., Mangion J., Roberton-Lowe C., Marshall A.J., Petretto E., et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Candille S.I., Kaelin C.B., Cattanach B.M., Yu B., Thompson D.A., Nix M.A., Kerns J.A., Schmutz S.M., Millhauser G.L., Barsh G.S. A β-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C., et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Chung E.K., Wu Y.L., Yang Y., Zhou B., Yu C.Y. Curr. Protoc. Immunol. 2005. Human complement components C4A and C4B genetic diversities: Complex genotypes and phenotypes. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the origin of species. Faber and Faber; London, UK: 1859. [Google Scholar]
- Emerson J.J., Cardoso-Moreira M., Borevitz J.O., Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- Fanciulli M., Norsworthy P.J., Petretto E., Dong R., Harper L., Kamesh L., Heward J.M., Gough S.C., de Smith A., Blakemore A.I., et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C.L., Crimm H., Meeh P., Croshaw R., Barbar T.D., Vandersteenhoven J.J., Butler W., Buckhaults P. Somatic mutations to CSMD1 in colorectal adenocarcinomas. Cancer Biol. Ther. 2008;7:609–613. doi: 10.4161/cbt.7.4.5623. [DOI] [PubMed] [Google Scholar]
- Fellermann K., Stange D.E., Schaeffeler E., Schmalzl H., Wehkamp J., Bevins C.L., Reinisch W., Teml A., Schwab M., Lichter P., et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti V., Poitras C., Bergeron D., Coulombe B., Robert F., Blanchette M. PReMod: A database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–D126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.L., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., et al. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Saar K., Adamovic T., Verheul M., van Heesch S.A., Cook S., Pravenec M., Aitman T., Jacob H., Shull J.D., et al. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- Hahn K.A., Richardson R.C., Hahn E.A., Chrisman C.L. Diagnostic and prognostic importance of chromosomal aberrations identified in 61 dogs with lymphosarcoma. Vet. Pathol. 1994;31:528–540. doi: 10.1177/030098589403100504. [DOI] [PubMed] [Google Scholar]
- Henshall S.M., Afar D.E., Hiller J., Horvath L.G., Quinn D.I., Rasiah K.K., Gish K., Willhite D., Kench J.G., Gardiner-Garden M., et al. Survival analysis of genome-wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Res. 2003;63:4196–4203. [PubMed] [Google Scholar]
- Hinds D.A., Kloek A.P., Jen M., Chen X., Frazer K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Kerns J.A., Cargill E.J., Clark L.A., Candille S.I., Berryere T.G., Olivier M., Lust G., Todhunter R.J., Schmutz S.M., Murphy K.E., et al. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics. 2007;176:1679–1689. doi: 10.1534/genetics.107.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseberth W.C., Nadella M.V., Breen M., Thomas R., Duke S.E., Murahari S., Kosarek C.E., Vernau W., Avery A.C., Burkhard M.J., et al. A novel canine lymphoma cell line: A translational and comparative model for lymphoma research. Leuk. Res. 2007;31:1709–1720. doi: 10.1016/j.leukres.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S., Gutiérrez-Arcelus M., Perry G.H., Vallender E.J., Johnson W.E., Miller G.M., Korbel J.O., Lee C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum. Mol. Genet. 2008;17:1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- Le Marechal C., Masson E., Chen J.M., Morel F., Ruszniewski P., Levy P., Ferec C. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat. Genet. 2006;38:1372–1374. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- Leonard J.A., Wayne R.K., Wheeler J., Valadez R., Guillen S., Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., 3rd, Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lupski J.R., Stankiewicz P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.R., Young E.J., Pani A.M., Freckmann M.L., Lacassie Y., Howald C., Fitzgerald K.K., Peippo M., Morris C.A., Shane K., et al. Infantile spasms is associated with deletion of the MAGI2 gene on chromosome 7q11.23-q21.11. Am. J. Hum. Genet. 2008;83:106–111. doi: 10.1016/j.ajhg.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Milde T., Shmelkov S.V., Jensen K.K., Zlotchenko G., Petit I., Rafii S. A novel family of slitrk genes is expressed on hematopoietic stem cells and leukemias. Leukemia. 2007;21:824–827. doi: 10.1038/sj.leu.2404525. [DOI] [PubMed] [Google Scholar]
- Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquerol L., Langille B.L., Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- Modiano J.F., Breen M., Valli V.E., Wojcieszyn J.W., Cutter G.R. Predictive value of p16 or Rb inactivation in a model of naturally occurring canine non-Hodgkin's lymphoma. Leukemia. 2007;21:184–187. doi: 10.1038/sj.leu.2404392. [DOI] [PubMed] [Google Scholar]
- Muscheck M., Sukosd F., Pesti T., Kovacs G. High density deletion mapping of bladder cancer localizes the putative tumor suppressor gene between loci D8S504 and D8S264 at chromosome 8p23.3. Lab. Invest. 2000;80:1089–1093. doi: 10.1038/labinvest.3780114. [DOI] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Paris P.L., Andaya A., Fridlyand J., Jain A.N., Weinberg V., Kowbel D., Brebner J.H., Simko J., Watson J.E., Volik S., et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum. Mol. Genet. 2004;13:1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- Parker H.G., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker H.G., Kukekova A.V., Akey D.T., Goldstein O., Kirkness E.F., Baysac K.C., Mosher D.S., Aguirre G.D., Acland G.M., Ostrander E.A. Breed relationships facilitate fine-mapping studies: A 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1562–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Henrichsen C.N., Harewood L., Merla G. Side effects of genome structural changes. Curr. Opin. Genet. Dev. 2007;17:381–386. doi: 10.1016/j.gde.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Richter T.M., Tong B.D., Scholnick S.B. Epigenetic inactivation and aberrant transcription of CSMD1 in squamous cell carcinoma cell lines. Cancer Cell Int. 2005;5:29. doi: 10.1186/1475-2867-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P., Zhang Y.P., Luo J., Lundeberg J., Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Scholnick S.B., Haughey B.H., Sunwoo J.B., el-Mofty S.K., Baty J.D., Piccirillo J.F., Zequeira M.R. Chromosome 8 allelic loss and the outcome of patients with squamous cell carcinoma of the supraglottic larynx. J. Natl. Cancer Inst. 1996;88:1676–1682. doi: 10.1093/jnci/88.22.1676. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Itsara A., Cheng Z., Alkan C., Schwartz S., Eichler E.E. Optimal design of oligonucleotide microarrays for measurement of DNA copy-number. Hum. Mol. Genet. 2007;16:2770–2779. doi: 10.1093/hmg/ddm234. [DOI] [PubMed] [Google Scholar]
- She X., Cheng Z., Zollner S., Church D.M., Eichler E.E. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M.R. German Shorthaired Pointer National Health Committee). German Shorthaired Pointer Club of America, editor. The 2005 German Shorthaired Pointer Health Survey. 2007 http://www.gspca.org/Health/GSP_Report/GSP_HEALTH-REPORT.htm.
- Sturn A., Quackenbush J., Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Sutter N.B., Eberle M.A., Parker H.G., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N.B., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Smith K.C., Ostrander E.A., Galibert F., Breen M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br. J. Cancer. 2003;89:1530–1537. doi: 10.1038/sj.bjc.6601275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomes C., Jackson A., Maguire K., Wood J., Gollin S., Ishwad C., Paterson I., Prime S., Parkinson K., Bell S., et al. The presence of multiple regions of homozygous deletion at the CSMD1 locus in oral squamous cell carcinoma question the role of CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer. 2003;37:132–140. doi: 10.1002/gcc.10191. [DOI] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Vali U., Brandstrom M., Johansson M., Ellegren H. Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet. 2008;9:8. doi: 10.1186/1471-2156-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wilcox B., Walkowicz C. The atlas of dog breeds of the world. TFH Publications; Neptune City, NJ: 1995. [Google Scholar]
- Wilson I.M., Davies J.J., Weber M., Brown C.J., Alvarez C.E., MacAulay C., Schubeler D., Lam W.L. Epigenomics: Mapping the methylome. Cell Cycle. 2006;5:155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]
- Wong K.K., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjoblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chung E.K., Wu Y.L., Savelli S.L., Nagaraja H.N., Zhou B., Hebert M., Jones K.N., Shu Y., Kitzmiller K., et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]