EMBO J 28, 315–325 (2009); published online 15 February 2009
New experiments show that different combinations of translesion DNA polymerases act to bypass lesions in mammalian cells, depending on the type of DNA damage. Bypass of most lesions tested was dependent on REV3L (polζ) and at least one additional DNA polymerase. The data fit a model whereby DNA polymerases work sequentially to bypass adducts in DNA.
Mammalian genomes encode 15 different DNA polymerases. Why are there so many? One reason, it appears, is that some of these are specialized to allow bypass of particular types of damaged bases in DNA. Although endogenous and environmentally induced DNA damage is primarily removed by DNA repair mechanisms (including nucleotide excision repair, base excision repair, mismatch repair, and various forms of strand break repair), some damage may remain, which can block progression of the normal DNA replication machinery. Translesion DNA synthesis (TLS) enables cells to tolerate damaged DNA without repairing it.
The most-studied TLS enzymes from human and mouse cells belong to the ‘Y family' of DNA polymerases, and include POLH (polη), POLI (polι), POLK (polκ), and REV1. These DNA polymerases are able to insert bases opposite DNA lesions at the expense of low-fidelity mutagenic replication. These enzymes also have low processivity, incorporating only a few nucleotides before dissociating from the template–a characteristic that may help allow a higher fidelity replicative polymerase to take over as soon as possible.
It has been recognized for some time that polζ, an enzyme in the ‘B family' of DNA polymerases, is an exceptionally important player in TLS. In the yeast Saccharomyces cerevisiae, DNA polymerase ζ (polζ) has the catalytic subunit Rev3 and an auxiliary subunit Rev7. Yeast rev3 mutants show moderate UV sensitivity and a frequency of UV-induced mutation that is an order of magnitude lower than wild type (Lemontt, 1971). REV3L is the mammalian homologue of Rev3 and it is thought to have a similar function in DNA damage-induced mutagenesis in mammalian cells, as antisense and shRNA suppression of REV3L reduce the level of mutagenesis induced by UV light and several other DNA-damaging agents (Li et al, 2002; Diaz et al, 2003). Purified polζ from yeast is able to efficiently extend from a mismatched base, and from primer termini following insertion of a base opposite a lesion in DNA. An in vitro experiment with synthetic DNA showed that after mammalian POLI inserted a base opposite an abasic site or a thymine-thymine 6-4 photoproduct (TT 6-4 PP), yeast polζ could extend the primer terminus (Johnson et al, 2000). Mammalian REV3L (353 kDa) is twice the size of yeast Rev3, and there is no information about its biochemical properties. Until now, there has been no direct evidence to support a TLS model involving multiple specialized DNA polymerases in mammalian cells.
Previously, experiments to discern the function of specialized DNA polymerases have used one of two approaches. One way is to treat DNA polymerase-defective cells with radiation or chemicals to induce multiple lesions in their genomes. A second technique employs single lesions in a synthetic DNA template, and purified DNA polymerases. The new work from Zvi Livneh and colleagues (Shachar et al, 2009) is a significant advance because it combines these two approaches. DNA, containing defined sites of damage, was introduced into mammalian cells to test the role of specific specialized DNA polymerases.
Shachar et al at the Weizmann Institute of Science in Israel used a quantitative TLS assay system. Plasmids were constructed carrying a defined DNA lesion at a specific site in a short single-stranded ‘gap'. Cultured mammalian cells were transfected with a gapped plasmid containing a lesion and encoding kanamycin resistance (kanR), together with a control gapped plasmid without a lesion encoding chloramphenicol resistance (cmR). The recipient cells carried disruptions of specific TLS DNA polymerase genes, or had TLS polymerase gene expression suppressed by siRNA technology. Following an incubation period, closed circular plasmids were extracted from the mammalian cells and transformed into a TLS-defective Escherichia coli strain. The ratio of kanR/cmR colonies revealed the extent of gap repair. The DNA sequence of the bypassed region of the plasmids was analysed in individual kanR colonies to determine whether the TLS was mutagenic.
Using this assay, the authors found three combinations of TLS reactions in mammalian cells, depending on the particular DNA lesion. A thymine-thymine cyclobutane pyrimidine dimer (TT CPD) was bypassed rapidly and accurately in a process dependent on POLH and independent of polζ. In a second mechanism, TLS of an intrastrand GG adduct formed by cisplatin utilized POLH and polζ, whereas a (+)-trans-benzopyrene diol epoxide-N2-G adduct used POLK and polζ. These events involved the participation of both a Y family DNA polymerase and polζ in a process that was accurate and moderately rapid. A third type of reaction was observed for the bypass of an abasic site, a 4-hydroxyequilenin-C adduct, or a TT 6-4 PP. TLS of these adducts depended on polζ and another DNA polymerase not yet identified, and it was slow and relatively mutagenic. These findings provide direct evidence that multiple specialized DNA polymerases mediate TLS in mammalian cells, and they emphasize the critical role of polζ. One interpretation is that bypass usually consists of an insertion step by a Y-family DNA polymerase, and extension by polζ (Figure 1).
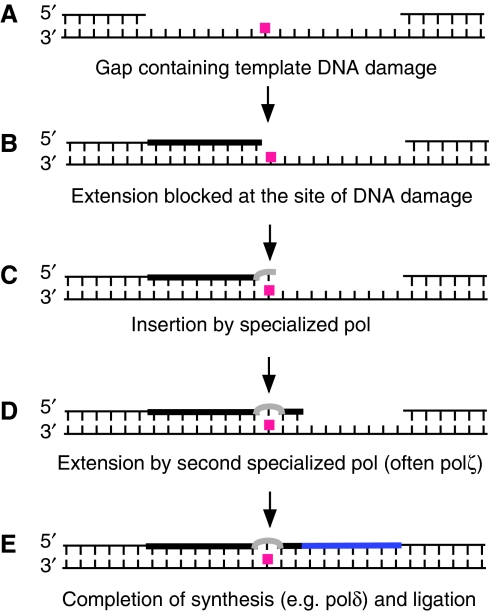
Figure 1.
Working model for the bypass of a site of DNA damage by the action of multiple DNA polymerases. Shachar et al. (2009) constructed plasmids containing a gap, each with a different type of single DNA template lesion near the centre of the gap (A). In mammalian cells, several steps are necessary for complete gap filling. Replicative DNA polymerases are normally stalled at sites of damage (B), and a specialized DNA polymerase such as POLH, POLI, POLK or polζ inserts a base (or bases) opposite the adduct (C). The polymerase selected depends on the type of DNA lesion. Extension of this aberrant terminus may require another specialized DNA polymerase, often polζ. The length of tracts synthesized by polζ in vivo is not yet known (D). For the bypass of a TT CPD, polζ is not necessary. Some post-replication repair gap filling may occur in G2 phase, and some lesion bypass may take place during S phase. If bypass happens in S phase or for filling of long gaps in vivo, switching back to a replicative DNA polymerase is necessary (E).
Consistent with previous work in budding yeast, genetic studies indicate that yeast Rev3 is involved in TLS of an AP site and a TT 6-4 PP, but not in the bypass of a TT CPD (Nelson et al, 2000). Yeast Rad30 (the homologue of POLH) can insert a base opposite a 6-4 PP but cannot extend it. The full bypass reaction requires yeast polζ (Johnson et al, 2001).
Future work with this system could enable a systematic cataloguing of the lesions in DNA that can be bypassed, and the DNA polymerases responsible. It will be important to know which lesions are bypassed in S-phase and which during gap filling in G2 phase, after DNA replication is completed (Waters and Walker, 2006). Work with other genetic mutations will help define the mechanisms of switching between DNA polymerases, a reaction that is currently proposed to involve post-translational modifications, including monoubiquitination of the sliding clamp PCNA protein at stalled DNA replication forks (Kannouche and Lehmann, 2004). Although Shachar et al. emphasize ‘two-polymerase mechanisms', some of the reactions are most likely to involve more than two DNA polymerases. For instance, some complete bypass reactions involve the replicative enzymes polδ or polɛ (McCulloch et al, 2004). Switching back to a replicative DNA polymerase may involve deubiquitination of PCNA (Zhuang et al, 2008).
References
- Diaz M, Watson NB, Turkington G, Verkoczy LK, Klinman NR, McGregor WG (2003) Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the Hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase zeta. Mol Cancer Res 1: 836–847 [PubMed] [Google Scholar]
- Johnson RE, Haracska L, Prakash S, Prakash L (2001) Role of DNA polymerase ζ in the bypass of a (6-4) TT photoproduct. Mol Cell Biol 21: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Lehmann AR (2004) Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle 3: 1011–1013 [PubMed] [Google Scholar]
- Lemontt JF (1971) Mutants of yeast defective in mutation induction by ultraviolet light. Genetics 68: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang H, McManus TP, McCormick JJ, Lawrence CW, Maher VM (2002) hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res 510: 71–80 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA (2004) Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res 32: 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol 37: 549–554 [DOI] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reißner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC (2006) The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci USA 103: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ (2008) Proc Natl Acad Sci USA 105: 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]