Abstract
Activation of a number of class A G protein-coupled receptors (GPCRs) is thought to involve two molecular switches, a rotamer toggle switch within the transmembrane domain and an ionic lock at the cytoplasmic surface of the receptor; however, the mechanism by which agonist binding changes these molecular interactions is not understood. Importantly, 80% of GPCRs including free fatty acid receptor 1 (FFAR1) lack the complement of amino acid residues implicated in either or both of these two switches; the mechanism of activation of these GPCRs is therefore less clear. By homology modeling, we identified two Glu residues (Glu-145 and Glu-172) in the second extracellular loop of FFAR1 that form putative interactions individually with two transmembrane Arg residues (Arg-183(5.39) and Arg-258(7.35)) to create two ionic locks. Molecular dynamics simulations showed that binding of agonists to FFAR1 leads to breakage of these Glu-Arg interactions. In mutagenesis experiments, breakage of these two putative interactions by substituting Ala for Glu-145 and Glu-172 caused constitutive receptor activation. Our results therefore reveal a molecular switch for receptor activation present on the extracellular surface of FFAR1 that is broken by agonist binding. Similar ionic locks between the transmembrane domains and the extracellular loops may constitute a mechanism common to other class A GPCRs also.
G protein-coupled receptors (GPCRs)3 are important components of signal transduction machineries that regulate many physiological processes. They are also important as targets for therapeutic agents; a large percentage of drugs in the marketplace are GPCR ligands or modulators. Knowledge of structure-function relationships of GPCRs has been gained through many pharmacological, biochemical, and biophysical studies, and has been used extensively to enhance the discovery of GPCR ligands that have been developed into therapeutically useful agents (1–3). Knowledge of the molecular details of ligand-receptor interaction and of the mechanism of receptor activation will also likely improve efforts to identify agonists with better potency and efficacy. Tan et al. (3) have recently reported their design of agonists with higher potency and efficacy for the trace amine receptor 1 based on the rotamer toggle switch model of receptor activation that is thought to operate in a number of class A GPCRs. The rotamer toggle switch typically involves the aromatic residues Trp and Phe within transmembrane helix 6 (TMH6) of GPCRs. During agonist-mediated receptor activation or in constitutively active receptors, the dihedral angle on the side chain of these residues is predicted to be rotated compared with the inactive state and thereby triggers a movement of TMH6 away from TMH3 (e.g. Ref. 4). It is also thought that an ionic lock between an Arg residue in TMH3 and a Glu in TMH6 near the cytoplasmic surface of some GPCRs holds the receptor in the inactive conformation and that receptor activation is accompanied by breakage of the ionic bond when agonist binds; the ionic lock may also be broken by receptor mutation (e.g. Ref. 5). Although these models of receptor activation have been proposed for a number of class A GPCRs, it is not certain how generally this hypothesis can be applied across all members of this GPCR class. From the alignment of 372 sequences of human GPCRs, we noted that about 80% of GPCRs do not have the putative residues that play a role in either the rotamer toggle switch, the ionic lock, or both. For these receptors, the interaction responsible for regulating interconversion between inactive and active receptor conformations therefore remains unknown.
The free fatty acid receptor 1 (FFAR1) is a Gq-coupled, class A GPCR-activated endogenously by free fatty acids, with a preference for medium-to-long chain fatty acids (C8–12) (reviewed in Ref. 6). The receptor has been suggested to be a potential target for treatment of type 2 diabetes, as offered by the action of agonists to potentiate glucose-stimulated insulin release (reviewed in Refs. 7, 8). Several groups, including ours, have reported the discovery of novel small molecule ligands for FFAR1 (9–13). Most of these compounds were identified by high-throughput screening followed by chemical optimization (10–12). Our group has delineated the ligand-binding pocket of FFAR1 (14, 15) and used the information as a rational approach to ligand discovery by means of virtual screening (13). The mechanism of FFAR1 activation; however, remains unknown especially because this receptor does not contain either the rotamer toggle switch or the ionic lock between TMHs 3 and 6.
We have previously identified nine residues in the ligand-binding pocket of FFAR1 that are important for ligand recognition and/or receptor activation (14). In particular, two Arg residues (Arg-183(5.39)4 and Arg-258(7.35)) and an Asn residue (Asn-244(6.55)) in the TMHs coordinate the carboxylate head group of the naturally occurring agonist linoleate and the synthetic agonist GW9508. In the present study, by a collaborative effort using computational modeling and receptor mutagenesis, we report the identification of Glu-172 in the second extracellular loop (ECL2) of FFAR1 that may function together with Arg-183(5.39) and Arg-258(7.35) as locks to control activation of the receptor. Our results suggest that these ionic locks at the extracellular surface hold the receptor in an inactive state. Agonists, through interaction with the arginine residues, may break the arginine-glutamate interactions thereby allowing the receptor to adopt an active conformation. Therefore, our results have provided insights into the mechanism of activation of class A GPCRs that function in a manner not explicable by the more well-studied models.
MATERIALS AND METHODS
Construction of the Models of the Receptor-Ligand Complexes—A model of FFAR1 was constructed based on the β2-adrenergic receptor (β2AR)(PDB ID: 2rh1) using MODELLER (16) as implemented in Insight II.5 The optimization procedure, using the Discovery module of Insight II,5 consisted of 5000 steps of conjugated gradient energy minimization and 1000 steps of molecular dynamics (MD) with distance constraints for the atoms involved in interhelical hydrogen bonding in the backbone of the transmembrane domains. The interatomic distance between the atoms involved in the formation of hydrogen bonds was set at 2.5 Å, and the force of the constraints was set to 1000 kcal/mol/Å. A distance-dependent dielectric constant was applied. Linoleate and GW9508 were docked to the novel model of FFAR1 by superimposing the previously defined model of the ligand-receptor complex constructed on the basis of rhodopsin (15). The novel complexes were subjected to 1000 steps of conjugate gradient energy minimization, also performed with the Discovery module of Insight.
MD Simulations—MD simulations were conducted using CHARMM (version c33b2) (18). Simulations were performed in implicit membrane using the Generalized Born with a simple switching (GBSW) (19, 20) method implemented CHARMM. The charges for linoleate and GW9508, and dihedral angle parameters for GW9508 were obtained using Hartree-Fock/G31** calculations in Gaussian.6 Langevin simulations were performed at a temperature of 300 K with a time-step of 0.002 ps. The empty or linoleate/GW9508-occupied FFAR1 structures were first energy-minimized for 200 steps using the adopted basis set Newton Raphson (ABNR) minimizer and then subjected to 3.5-ns MD simulations. During the simulations, to preserve the helical structures of the transmembrane domains, nuclear Overhauser enhancement (NOE) restraints were applied to the distances between the backbone carbonyl O of residue n and the backbone NH of residue n+4, setting FMAX and KMAX to 200.0 and 80.0 kcal/mol/Å, respectively.
Receptor Plasmids and Site-directed Mutagenesis—A plasmid coding for wild-type FFAR1 was cloned into pcDNA3.1/hygro(+) (Invitrogen) as reported previously (14). Site-directed mutagenesis was performed using the QuikChange II XL mutagenesis kit (Stratagene). Sequences on the promoter and the full insert in all constructs were verified by sequencing (MWG Biotech).
Cell Culture and Transfection—Cells were seeded at 18,400/well in 96-well plates 1 day prior to transfection. CHO-K1 cells were maintained in F-12 medium supplemented with 10% fetal bovine serum. HEK-EM (293) cells (22) were maintained in DMEM medium supplemented with 10% fetal bovine serum. Transfection was carried out using Lipofectamine LTX (Invitrogen) according to the manufacturer's recommendation. For the reporter gene assay, the PathDetect in vivo trans-reporting system (Stratagene) was used. The trans-reporting system utilizing the ERK1/2 pathway was used in CHO-K1 cells. Cells were co-transfected per well with 48 ng of receptor/lacZ control plasmid, 48 ng of pFR-Luc plasmid, and 2 ng of pFA2-Elk1 trans-activator plasmid. For the cis-reporting system, cells were co-transfected per well with 50 ng of receptor/lacZ control plasmid and 50 ng of pAP-1-Luc plasmid.
Cytoplasmic Free Ca2+ Concentration ([Ca2+]i) Measurement—Measurement of [Ca2+]i was made by loading FFAR1-expressing HEK-EM (293) cells with Calcium4 reagent (Molecular Devices) and monitoring the changes in fluorescence intensity upon agonist stimulation with the FLIPR (Molecular Devices, Sunnyvale, CA), as described previously (14). Assays were performed in quadruplicate.
Reporter Gene Assay—Assay for reporter gene activity was carried out by washing transfected cells with the corresponding serum-free medium and incubating them in serum-free medium supplemented with 1 mm fatty acid-free BSA. The fatty acid-free BSA was used to bind endogenously released free fatty acids, in light of the report by Stoddart et al. (23). Nevertheless, further experimentation in the absence or presence of 1 nm to 3 mm fatty acid-free BSA to adsorb endogenously released FFAs showed that endogenously released FFA does not interfere with the system under study. Cells were frozen at –20 °C after a further 22-h incubation. Luciferase activity was measured following thawing and lysis of cells with 100 μl/well lysis buffer. The entire cell lysate was combined with 125 μl of reaction buffer and 25 μl of 0.4 mm luciferin (Sigma) automatically via automatic injectors in Victor3 multi-label reader (PerkinElmer Life Sciences). The composition of the lysis buffer and reaction buffer has been reported previously (24). Luminescence was measured for 3 s after shaking for 2 s.
Measurement of Expression Levels of WT, E145A, and E172A—FLAG epitope-tagged receptors were analyzed by flow cytometry in a FACS Calibur (Becton Dickinson). FLAG-receptor-expressing HEK-EM (293) cells were labeled with FITC-conjugated anti-FLAG M2 antibodies (Sigma) according to our previously described protocol (14). Levels of surface receptors were quantified as the mean fluorescence signal and normalized as a percentage of the wild-type receptor level.
RESULTS
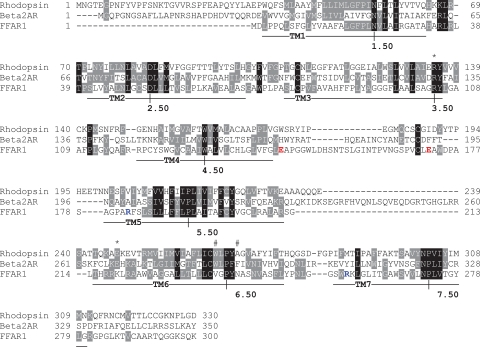
While the activation of a number of class A GPCRs is postulated to involve the rotamer toggle switch or the breakage of an ionic lock at the cytoplasmic surface, or both, we noted that FFAR1 does not contain the complement of amino acid residues required to form the putative switches (Fig. 1). At the location in FFAR1 of the putative rotamer toggle switch, a Val and Asn are present at 6.48 and 6.52, respectively, instead of aromatic acid residues. And, in the region of the putative ionic lock at the cytoplasmic surface of FFAR1, a Lys is present at 6.30 rather than a Glu, and its nearby residues are ones with hydrophobic side chains or are Arg residues. It is therefore unlikely that these residues regulate receptor activation in the same manner as described by the postulated models. It is possible, however, that a similar ionic lock may be present at a different location in the receptor.
FIGURE 1.
Sequence alignment of FFAR1, rhodopsin, and β2-adrenergic receptor. The most conserved residues in each transmembrane domain are labeled as X.50, according to the Ballesteros and Weinstein nomenclature (17). Symbols indicate positions of residues believed to be involved in the rotamer toggle switch (#) and the ionic lock (*) between the cytosolic ends of TM3 and TM6. Residues forming ionic locks between the TM helical bundle and ECL2 of FFAR1 are colored in red and blue.
We therefore performed a search for putative ionic interactions using an improved FFAR1 homology model. Our previous model was constructed in the absence of ECL2 and was based on the crystal structure of rhodopsin. Given the recent availability of a 2.4-Å structure of β2AR (25, 26), we reconstructed a new FFAR1 model based on this template because of the closer sequence identity of FFAR1 with β2AR than with rhodopsin (28% versus 16%). In the β2AR structure, the TMHs are more tilted away from the center of the receptor than in the structure of rhodopsin. Moreover, in the β2AR structure, ECL2 forms a short helix instead of the β hairpin present in rhodopsin and, unlike in rhodopsin, does not occlude the ligand-binding cavity. In this regard, the β2AR may be more representative of the typical GPCRs, which in the majority of cases are activated by diffusible ligands. The ECL2 of rhodopsin in not an ideal template for the construction of GPCR models as it is deeply buried into the transmembrane cavity and would hinder ligand docking (27). When dealing with rhodopsin-based models, significantly better results have been obtained modeling ECL2 de novo (27), removing ECL2 and expanding the ligand-binding pocket through conformational searches prior to docking (15), or guiding the docking by means of constraints based on mutagenesis data (28, 29). Because of the larger volume of the binding pocket and to the solvent-exposed conformation of ECL2, a β2AR-based homology model might be suitable for ligand docking with minimal or no optimization.
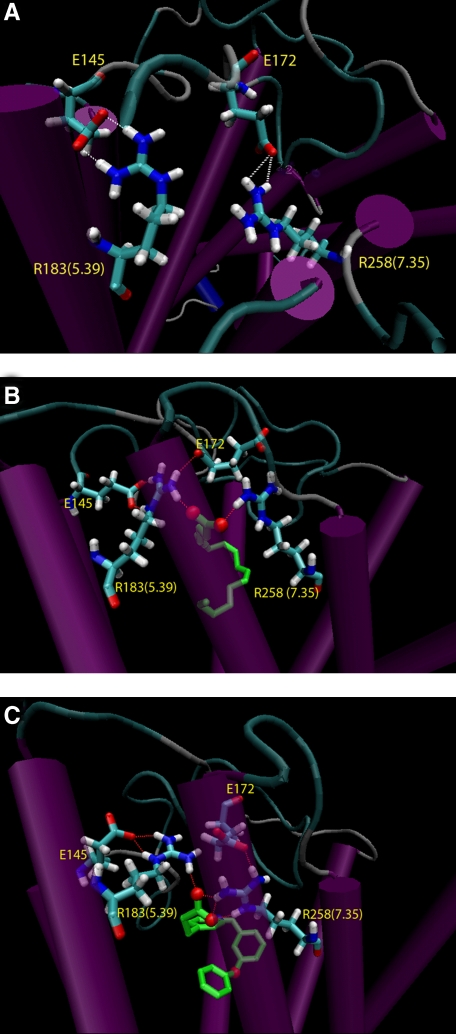
In our new FFAR1 model, the TMHs were constructed based on the β2AR, while the extracellular and intracellular loops were modeled de novo by means of the algorithm implemented in MODELLER (16). We imposed the formation of the disulfide bridge between two conserved Cys residues located in ECL2 and TMH3, and, among a total of 20 generated loop models, chose the ECL2 conformation that most closely resembled that seen in the β2AR structure, based on root-mean square deviation in the backbone of the ECL2-(170–182) portion. Notably, in our new β2AR-based model, Glu-145(ECL2) forms a putative hydrogen bond with Arg-183(5.39), and Glu-172(ECL2) forms a hydrogen bond with Arg-283(7.35) (Fig. 2A). It is noteworthy that Glu-145(ECL2) faces the binding pocket in the current model, whereas it pointed away from the transmembrane core in our previous rhodopsin-based homology model.
FIGURE 2.
Molecular models of the unoccupied FFAR1 and FFAR1 complexed with linoleate or GW9508. The salt bridges between amino acid residues Glu-145(ECL2) and Arg-183(5.39) and Glu-172(ECL2) and Arg-258(7.35) are identified in unoccupied FFAR1 (A) and in FFAR1 complexed with linoleate (B) or GW9508 (C).
MD Simulations of Unoccupied and Agonist-occupied FFAR1—Because Arg-183(5.39) and Arg-258(7.35) may form putative hydrogen bonds with Glu-145(ECL2) and Glu-172(ECL2) on the one hand, and with the carboxylate group of an agonist on the other hand (14), we wanted to examine the dynamic properties of these networks of Arg-Glu interactions in relation to agonist occupancy. To study the interactions between the two Arg-Glu pairs, we monitored the non-bonded interaction energies, including electrostatic and van der Waals components, along the MD trajectories in the unoccupied or agonist-occupied FFAR1 (Fig. 3). The simulations were carried out for 3.5 ns using Langevin dynamics with an implicit membrane model. After about 1 ns, the systems reached and maintained a stable conformation, as indicated by the root mean square deviation of the FFAR1 backbone calculated from the average structure obtained over the period of 1–3.5-ns simulations (supplemental Fig. S1). Throughout the simulations, the interaction energies between Glu and Arg residues in the unoccupied receptor remained constant, indicating that their interactions were stable and strong. Specifically, the carboxylate groups of Glu-145(ECL2) and Glu-172(ECL2) formed ionic interactions with Arg-183(5.39) and Arg-258(7.35), respectively. When FFAR1 was complexed with either linoleate or GW9508, the ligand competed via its carboxylate group for the interaction with the Arg residues. The ligand-receptor interactions remained stable and the ligands retained their initial conformations over the course of the simulations. In contrast, the interaction energy between Arg-183(5.39) and Glu-145(ECL2) became significantly weaker during the simulations (Fig. 3A). A weaker interaction energy was also observed between Arg-258(7.35) and Glu-172(ECL2) (Fig. 3B). The weaker interaction energies between the two Arg-Glu pairs in the agonist-occupied receptor suggest that the presence of agonist perturbs these ionic locks. The average structures of FFAR1 in complex with linoleate and GW9508 calculated over the period of 1–3.5-ns simulations are shown in Fig. 2 (B and C).
FIGURE 3.
Non-bonded interaction energies (van-der-Waals and electrostatic components) between two putative ionic locks. The interaction energies between Glu-145(ECL2) and Arg-183(5.39) (A) and Glu-172 and Arg-258(7.35) (B) for the unoccupied FFAR1 (black) and FFAR1 in complex with linoleate (red) or GW9508 (green) were computed along 3.5-ns trajectories. The changes in energies are expressed as percent deviation from the average energy detected for the unoccupied receptor.
Constitutive Activation of Receptor through Mutation—The results of the MD simulations suggested that the Arg-Glu might be a part of the molecular switch that governs receptor activation through an exchange of interacting partners with an agonist. We therefore hypothesized that the Arg-Glu interactions in the unliganded receptor function as locks that keep the receptor in the inactive state and agonists activate the receptor by weakening or breaking these interactions. Following this rationale, we postulated that mutations that prevent the formation of these interactions would lead to constitutive activation. We addressed this idea by constructing individual Ala mutants of Glu-145(ECL2) and Glu-172(ECL2) in FFAR1 and determined their constitutive activities using luciferase-based transcriptional reporter assays. As it has been suggested that the release of endogenous fatty acid during membrane preparation may lead to apparent constitutive activity in FFAR1 that can be reversed by incubation with BSA (23), measurement of constitutive activity in intact cells may avoid such complication. To further prevent this potential problem, cells were incubated with serum-free medium supplemented with 1 mm fatty acid-free BSA during the assay. Agonist-stimulated activities were assessed by measuring linoleate-stimulated increases in cytoplasmic free calcium concentration ([Ca2+]i) and constitutive receptor activities were assessed using a trans-reporter system, measuring the activation of ERK1/2 pathway using CHO-K1 cells, which is a more sensitive assay of constitutive signaling.
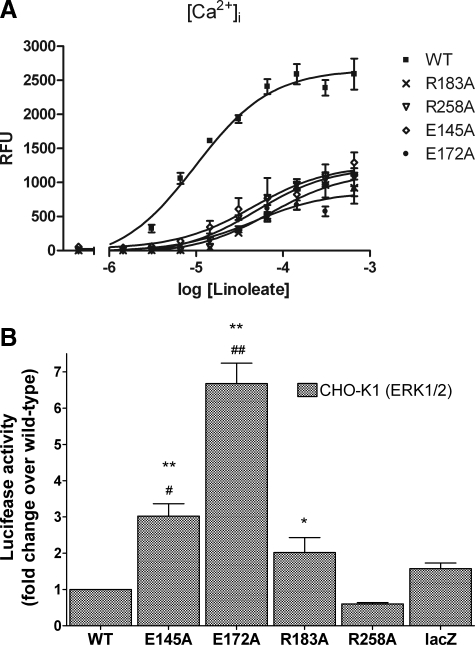
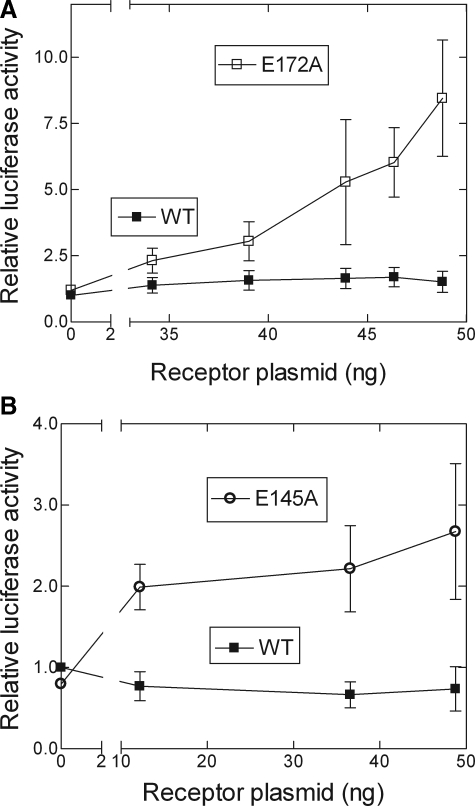
Transfection with wild-type FFAR1 confirmed that linoleate stimulated a FFAR1-dependent increase in [Ca2+]i (Fig. 4A) and showed that the receptor did not exhibit any measurable constitutive activity, as reflected by the lack of increased luciferase activity over lacZ-transfected control cells (Fig. 4B). In contrast, linoleate stimulation of cells expressing E145A or E172A caused increases in [Ca2+]i that were 48 and 33% of WT, respectively, whereas these mutant receptors exhibited increases of constitutive luciferase activities above WT of ∼3-fold for E145A and 7-fold for E172A. Mutation of Arg-183(5.39) or Arg-258(7.35) to Ala generated receptors that exhibited linoleate-stimulated signaling activities of 43 and 48% of WT, respectively. R183A exhibited a low level of constitutive activity of ∼2-fold over basal luciferase whereas R258A did not exhibit constitutive activity above that of WT. We estimated the levels of cell-surface expression of E145A and E172A using FLAG epitope-tagged receptors. FLAG-E145A was expressed at a level 173 ± 11% (mean ± S.E.) of WT and FLAG-E172A at 85 ± 5.5% of WT; we did not construct FLAG-tagged R183A or R258A. We verified the observed increases in constitutive activities of E145A and E172A in titration experiments with increasing amounts of receptor plasmid for transfection. Increasing doses of wild-type receptor resulted in background activity that was independent of plasmid level, confirming the absence of constitutive activity of wild-type FFAR1 (Fig. 5). In contrast, titrating with increasing amounts of either E145A or E172A plasmids (Fig. 5) resulted in proportionally higher luciferase activities. The measured constitutive activities were unlikely due to activation of the mutant receptors by endogenously released FFAs because the wild-type receptor showed no increased signaling over the receptor-negative control cells. Taken together, the results from this series of experiments suggest that Glu-145(ECL2), Glu-172(ECL2) and Arg-183 constrain the receptor in an inactive conformation, consistent with our hypothesis.
FIGURE 4.
Signaling activities of FFAR1 mutants. A, agonist-stimulated signaling was determined by measuring increases in [Ca2+]i in response to increasing doses of linoleate as described under “Materials and Methods.” RFU represents the relative fluorescence units of the Calcium4 reagent. Data shown are mean ± S.E. of quadruplicate samples in two experiments. B, levels of constitutive activities were determined in CHO-K1 cells by co-transfection of the receptor plasmid and an Elk1-Gal4 trans-activator element and a luciferase reporter as described under “Materials and Methods.” Data shown are mean ± S.E. after normalizing the luminescence levels relative to wild-type receptor. One-way analysis of variance performed indicated significant differences among the groups. Subsequently, Newman-Keuls multiple comparison test was performed to determine whether luciferase activity was statistically different from the lacZ (#) or wild-type group (*). ##, p < 0.001; #, p < 0.01; **, p < 0.001; *, p < 0.05.
FIGURE 5.
Increased constitutive activities of E172A (A) and E145A (B) FFAR1 mutants. The levels of constitutive activity were determined as described under “Materials and Methods.” The amount of receptor plasmid was titrated at a range of 0–48 ng by varying the amount of receptor plasmid while keeping the total amount of DNA transfected constant by supplementing the mixture with lacZ control plasmid. Data shown are means ± S.E. after normalizing the data relative to the value obtained with zero receptor plasmid in the wild-type data set.
DISCUSSION
Residues in ECL2 have been shown to be important for agonistic activity in a number of GPCRs. In the m3 muscarinic receptor, several ECL2 amino acids with different physicochemical properties have been shown to be important for agonist activity (30). The residues appear to play a role solely in receptor activation because their mutation negatively affects potency but not affinity of the agonist carbachol. A role for ECL2 in guiding thyrotropin-releasing hormone (TRH) into the transmembrane binding pocket of the TRH receptor type 1 was hypothesized based on mutagenesis and computational experiments (31, 32). The importance of ECL2 in allowing access of ligand to receptor was suggested also in the m2 muscarinic receptor in which the flexibility of the loop was reduced by creating a mutant that forms an additional disulfide bond (33). The ECL2 has also been shown to have a role in agonist and antagonist recognition in the adenosine and P2Y receptors (28, 34–36). In the 5-HT4 receptor, distinct conformational changes in the ECL2 upon binding to agonists, neutral antagonists or inverse agonists were observed in circular dichroic spectra (37). By alanine-scanning mutagenesis of ECL2 residues in V1a vasopressin receptor, several aromatic residues in the receptor were found to be important in maintaining high affinity binding to agonist and a certain class of antagonists and for potent agonistic activity (38). Furthermore, from a random mutagenesis study, residues in the ECL2 of C5a receptor were suggested to form multiple interactions that stabilize the receptor in the inactive state (39). Based on the results of all of these studies, it may be suggested that ECL2 can participate in ligand interaction and appears to contribute to receptor activation, despite differences in the specific locations and identities of the functional residues in structurally related GPCRs.
In FFAR1, we have identified two critical residues, Glu-145(ECL2) and Glu-172(ECL2), in ECL2 that play a role in regulating receptor function. By a combination of computational and biological studies, we determined that two key interactions, salt bridges between Glu-145(ECL2) and Arg-183(5.39) and between Glu-172(ECL2) and Arg-258(7.35), are responsible for constraining the receptor in an inactive state. The role of these Glu residues is supported by the decreased agonist-stimulated and increased constitutive activities of E145A and E172A and by computational simulations showing that agonists cause separation of Glu-145(ECL2) and Arg-183(5.39) as well as Glu-172(ECL2) and Arg-258(7.35). Intuitively, it may be conceived that mutation of Arg-183(5.39) and Arg-258(7.35) will cause constitutive activity similar to that caused by mutation of Glu-145 and Glu-172(ECL2). This was not found to be the case although modest constitutive activity was observed for R183A. It is worth noting that mutations of Arg3.50, the residue that forms one part of the ionic lock with Glu/Asp6.30 (see below) in other class A GPCRs, do not always cause constitutive activity and sometimes decreases constitutive signaling depending on the receptor (see Ref. 40 for review).
The results from the present study provide us with a mechanistic insight into the activation of FFAR1 and perhaps other GPCRs. For a number of them, activation is believed to involve conformational changes of aromatic residues at positions 6.48 and 6.52 and a breakage of the salt bridge formed by Arg3.50 and Glu/Asp6.30. These residues are considered molecular switches that trigger the conversion of a receptor from the inactive to the active state. A salt bridge between Arg3.50 and Glu6.30 is observed in the crystal structure of rhodopsin (41). A number of GPCRs such as the β2AR (5), α1b-adrenergic receptor (42) and 5-hydroxytryptamine receptor (43) show constitutive activity when this salt bridge is disrupted by mutation of one or both of the interacting partners. However, this activation model is not applicable to all GPCRs because the residues at these positions are not conserved among all GPCRs, suggesting the presence of alternative conformational switches apart from these well known ones. To gauge the generality of the activation model involving the aromatic toggle switch in TMH6 or the ionic lock between TMH3 and TMH6, we examined a sequence alignment of 372 human GPCRs belonging to all classes (369 sequences were derived from the analysis reported by Surgand et al. (44), while three additional sequences were derived from our previous analysis of the nucleotide and lipid receptor cluster (15)). With regard to the rotamer toggle switch, we found that 69 sequences do not feature aromatic residues at position 6.48 and 218 do not feature aromatic residues at position 6.52. In terms of the ionic lock, 90 sequences do not have a positive charge at position 3.50 of the (D/E)RY motif and 275 sequences do not have a negatively charge residue at 6.30. In FFAR1, Val and Asn are found at positions 6.48 and 6.52, while a Lys residue is in place of a negatively charged residue at position 6.30. Therefore, the two most studied molecular switches do not apply to many GPCRs, including FFAR1, and alternative molecular switches are likely to exist.
In FFAR1, we propose that a part of the activation process involves the releasing of constraints imposed by the ionic lock between Arg-183(5.39) and Glu-145(ECL2) and between Arg-258(7.35) and Glu-172(ECL2). Arg-183(5.39) and Arg-258(7.35) are located near the extracellular end of TMH5 and TMH7, respectively. Both Glu-145(ECL2) and Glu-172(ECL2) are located in ECL2, the former being two positions away from the end of TMH4 and the latter two positions away from the conserved Cys of ECL2 that forms a putative disulfide bridge. These Arg-Glu ionic interactions appear to be important for restraining the receptor in an inactive conformation because mutation of Glu-145(ECL2) and Glu-172(ECL2) leads to constitutive activation. These interactions are weakened or perhaps broken when an agonist interacts with Arg-183(5.39) and Arg-258(7.35) that along with Asn-244 function to coordinate the agonist at the head group. We suggest that this exchange of interacting partners from the receptor to the agonist for Arg-183(5.39) and Arg-258(7.35) allows the receptor to adopt an active conformation.
Charged residues are not uncommon in ECL2 of class A GPCRs, in particular, in the nucleotide and lipid receptor cluster to which FFAR1 belongs. Of 45 sequences in the nucleotide and lipid receptor cluster, 26 receptors have either a Glu or an Asp located two positions away from the Cys that forms the putative disulfide bond, forming a CX(E/D) motif (Glu-172(ECL2) in FFAR1). In contrast, it is difficult to determine the residues in other class A GPCRs that correspond to Glu-145(ECL2) as ECL2 is known to be variable both in length and sequence. Given the negatively charged nature of nucleotide and lipid agonists, the negatively charged residues of ECL2 are not involved in ligand recognition, as they would in fact exert repulsive forces on the ligands, but are likely to be the counterions of positively charged residues located within the binding pocket. In line with this hypothesis, it has been shown by molecular modeling and molecular dynamics simulations of the P2Y6 receptor that the negatively charged Asp-179 in ECL2 interacts with Arg-103(3.29) near the extracellular end of TMH3 (21). Besides interacting with Asp-179(ECL2), Arg-103(3.29) also plays a role in coordinating the phosphate group of nucleotide agonists. In an interesting analogy with FFAR1, coordination of the agonist by Arg-103(3.29) is accompanied by release of the Asp-179-Arg-103 interaction resulting in a movement of ECL2 away from the ligand binding cavity (21). In this example, a positively charged residue located in TM3, not TM7 like in FFAR1, forms an ionic interaction with a negatively charged residue located in ECL2, in the position correspondent to Glu-172 in FFAR1.
Based on the data, we propose that the breakage of the ionic lock between Glu-145(ECL2) and Arg-183(5.39) or between Glu-172(ECL2) and Arg-258(7.35) may constitute a part of the activation mechanism in FFAR1. These two ionic locks might operate independently because disruption of either one of them leads to constitutive activity. We suggest that when an agonist is attracted to the receptor via interaction with Arg-183(5.39) or Arg-258(7.35), these interactions, which function to keep the receptor inactive, become weakened and break apart. Accompanying this breakage of the ionic lock, ECL2 may become more mobile and allow the receptor to adopt a fully activated state. Two antagonists that we identified (compound 8 and 21 in Ref. 13) contain a less electronegative nitro group instead of a carboxyl group which may be unable to release the ionic lock and activate the receptor. We suggest that the knowledge gained from this study can be exploited for the design of new FFAR1 agonists and antagonists.
Analyzing the sequences of non-olfactory human GPCRs, we found that about half of them have positively charged residues in the binding pocket. We speculate that some of these residues may interact with negatively charged residues located in ECL2, which are also very common, thus forming an ionic lock that keeps the receptor in an inactive state in the absence of agonist, but that can be disrupted by agonists. The locking mechanisms would be similar to the one that we propose for FFAR1; however the specific locations of the residues within ECL2 and the TMs may be different.
Supplementary Material
Acknowledgments
We thank Drs. Stanislav Engel, Bruce Raaka, and Susanne Neumann for many helpful discussions.
This work was supported, in whole or in part, by the Intramural Research Program, NIDDK, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: GPCR, G protein-coupled receptor; MD, molecular dynamics; DMEM, Dulbecco's modified Eagle's medium; BSA, bovine serum albumin; WT, wild type; β2AR, β2-adrenergic receptor; FFAR1, free fatty acid receptor 1; TMH, transmembrane helix; ECL, extracellular loop.
Residues are identified through the indexing system of Ballesteros and Weinstein (17); see legend of Fig. 1 for details.
Insight II, Version 2001, (2006) Accelrys Inc.
Gaussian 03, Revision C. 02. (2004) Gaussian, Inc., Wallingford, CT.
References
- 1.Cristalli, G., Lambertucci, C., Marucci, G., Volpini, R., and Dal Ben, D. (2008) Curr. Pharm. Des. 14 1525–1552 [DOI] [PubMed] [Google Scholar]
- 2.Lagerström, M. C., and Schiöth, H. B. (2008) Nat. Rev. Drug. Discov. 7 339–357 [DOI] [PubMed] [Google Scholar]
- 3.Tan, E. S., Groban, E. S., Jacobson, M. P., and Scanlan, T. S. (2008) Chem. Biol. 15 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi, L., Liapakis, G., Xu, R., Guarnieri, F., Ballesteros, J. A., and Javitch, J. A. (2002) J. Biol. Chem. 277 40989–40996 [DOI] [PubMed] [Google Scholar]
- 5.Ballesteros, J. A., Jensen, A. D., Liapakis, G., Rasmussen, S. G., Shi, L., Gether, U., and Javitch, J. A. (2001) J. Biol. Chem. 276 29171–29177 [DOI] [PubMed] [Google Scholar]
- 6.Brown, A. J., Jupe, S., and Briscoe, C. P. (2005) DNA Cell Biol. 24 54–61 [DOI] [PubMed] [Google Scholar]
- 7.Costanzi, S., Neumann, S., and Gershengorn, M. C. (2008) J. Biol. Chem. 283 16269–16273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winzell, M. S., and Ahrén, B. (2007) Pharmacol. Ther. 116 437–448 [DOI] [PubMed] [Google Scholar]
- 9.Briscoe, C. P., Peat, A. J., McKeown, S. C., Corbett, D. F., Goetz, A. S., Littleton, T. R., McCoy, D. C., Kenakin, T. P., Andrews, J. L., Ammala, C., Fornwald, J. A., Ignar, D. M., and Jenkinson, S. (2006) Br. J. Pharmacol. 148 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrido, D. M., Corbett, D. F., Dwornik, K. A., Goetz, A. S., Littleton, T. R., McKeown, S. C., Mills, W. Y., Smalley, T. L., Jr., Briscoe, C. P., and Peat, A. J. (2006) Bioorg. Med. Chem. Lett. 16 1840–1845 [DOI] [PubMed] [Google Scholar]
- 11.Song, F., Lu, S., Gunnet, J., Xu, J. Z., Wines, P., Proost, J., Liang, Y., Baumann, C., Lenhard, J., Murray, W. V., Demarest, K. T., and Kuo, G. H. (2007) J. Med. Chem. 50 2807–2817 [DOI] [PubMed] [Google Scholar]
- 12.Tan, C. P., Feng, Y., Zhou, Y. P., Eiermann, G. J., Petrov, A., Zhou, C., Lin, S., Salituro, G., Meinke, P., Mosley, R., Akiyama, T. E., Einstein, M., Kumar, S., Berger, J. P., Mills, S. G., Thornberry, N. A., Yang, L., and Howard, A. D. (2008) Diabetes 57 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tikhonova, I. G., Sum, C. S., Neumann, S., Engel, S., Raaka, B. M., Costanzi, S., and Gershengorn, M. C. (2008) J. Med. Chem. 51 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sum, C. S., Tikhonova, I. G., Neumann, S., Engel, S., Raaka, B. M., Costanzi, S., and Gershengorn, M. C. (2007) J. Biol. Chem. 282 29248–29255 [DOI] [PubMed] [Google Scholar]
- 15.Tikhonova, I. G., Sum, C. S., Neumann, S., Thomas, C. J., Raaka, B. M., Costanzi, S., and Gershengorn, M. C. (2007) J. Med. Chem. 50 2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779–815 [DOI] [PubMed] [Google Scholar]
- 17.Ballesteros, J. A., and Weinstein, H. (1995) in Methods in Neurosciences (Conn, P. M., and Sealfon, S. C., eds), Academic Press, San Diego
- 18.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S., and Karplus, M. (1983) J. Comput. Chem. 4 187–217 [Google Scholar]
- 19.Im, W., Feig, M., and Brooks, C. L., III (2003) Biophys. J. 85 2900–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im, W., Lee, M. S., and Brooks, C. L., III (2003) J. Comput. Chem. 24 1691–1702 [DOI] [PubMed] [Google Scholar]
- 21.Costanzi, S., Joshi, B. V., Maddileti, S., Mamedova, L., Gonzalez-Moa, M. J., Marquez, V. E., Harden, T. K., and Jacobson, K. A. (2005) J. Med. Chem. 48 8108–8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins, A. K., and Horlick, R. A. (1998) BioTechniques 25 240–244 [DOI] [PubMed] [Google Scholar]
- 23.Stoddart, L. A., Brown, A. J., and Milligan, G. (2007) Mol. Pharmacol. 71 994–1005 [DOI] [PubMed] [Google Scholar]
- 24.Huang, W., Osman, R., and Gershengorn, M. C. (2005) Biochemistry 44 2419–2431 [DOI] [PubMed] [Google Scholar]
- 25.Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Kuhn, P., Weis, W. I., Kobilka, B. K., and Stevens, R. C. (2007) Science 318 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenbaum, D. M., Cherezov, V., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Yao, X. J., Weis, W. I., Stevens, R. C., and Kobilka, B. K. (2007) Science 318 1266–1273 [DOI] [PubMed] [Google Scholar]
- 27.Costanzi, S. (2008) J. Med. Chem. 51 2907–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzi, S., Mamedova, L., Gao, Z. G., and Jacobson, K. A. (2004) J. Med. Chem. 47 5393–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foucaud, M., Tikhonova, I. G., Langer, I., Escrieut, C., Dufresne, M., Seva, C., Maigret, B., and Fourmy, D. (2006) Mol. Pharmacol. 69 680–690 [DOI] [PubMed] [Google Scholar]
- 30.Scarselli, M., Li, B., Kim, S. K., and Wess, J. (2007) J. Biol. Chem. 282 7385–7396 [DOI] [PubMed] [Google Scholar]
- 31.Colson, A. O., Perlman, J. H., Smolyar, A., Gershengorn, M. C., and Osman, R. (1998) Biophys. J. 74 1087–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlman, J. H., Colson, A. O., Jain, R., Czyzewski, B., Cohen, L. A., Osman, R., and Gershengorn, M. C. (1997) Biochemistry 36 15670–15676 [DOI] [PubMed] [Google Scholar]
- 33.Avlani, V. A., Gregory, K. J., Morton, C. J., Parker, M. W., Sexton, P. M., and Christopoulos, A. (2007) J. Biol. Chem. 282 25677–25686 [DOI] [PubMed] [Google Scholar]
- 34.Kim, J., Jiang, Q., Glashofer, M., Yehle, S., Wess, J., and Jacobson, K. A. (1996) Mol. Pharmacol. 49 683–691 [PMC free article] [PubMed] [Google Scholar]
- 35.Moro, S., Hoffmann, C., and Jacobson, K. A. (1999) Biochemistry 38 3498–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olah, M. E., Jacobson, K. A., and Stiles, G. L. (1994) J. Biol. Chem. 269 24692–24698 [PMC free article] [PubMed] [Google Scholar]
- 37.Banères, J. L., Mesnier, D., Martin, A., Joubert, L., Dumuis, A., and Bockaert, J. (2005) J. Biol. Chem. 280 20253–20260 [DOI] [PubMed] [Google Scholar]
- 38.Conner, M., Hawtin, S. R., Simms, J., Wootten, D., Lawson, Z., Conner, A. C., Parslow, R. A., and Wheatley, M. (2007) J. Biol. Chem. 282 17405–17412 [DOI] [PubMed] [Google Scholar]
- 39.Klco, J. M., Wiegand, C. B., Narzinski, K., and Baranski, T. J. (2005) Nat. Struct. Mol. Biol 12 320–326 [DOI] [PubMed] [Google Scholar]
- 40.Rovati, G. E., Capra, V., and Neubig, R. R. (2007) Mol. Pharmacol. 71 959–964 [DOI] [PubMed] [Google Scholar]
- 41.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., Yamamoto, M., and Miyano, M. (2000) Science 289 739–745 [DOI] [PubMed] [Google Scholar]
- 42.Greasley, P. J., Fanelli, F., Rossier, O., Abuin, L., and Cotecchia, S. (2002) Mol. Pharmacol. 61 1025–1032 [DOI] [PubMed] [Google Scholar]
- 43.Shapiro, D. A., Kristiansen, K., Weiner, D. M., Kroeze, W. K., and Roth, B. L. (2002) J. Biol. Chem. 277 11441–11449 [DOI] [PubMed] [Google Scholar]
- 44.Surgand, J. S., Rodrigo, J., Kellenberger, E., and Rognan, D. (2006) Proteins 62 509–538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.