Abstract
Mediator is a modular multiprotein complex required for regulated transcription by RNA polymerase (Pol) II. Here, we show that the middle module of the Mediator core contains a submodule of unique structure and function that comprises the N-terminal part of subunit Med7 (Med7N) and the highly conserved subunit Med31 (Soh1). The Med7N/31 submodule shows a conserved novel fold, with two proline-rich stretches in Med7N wrapping around the right-handed four-helix bundle of Med31. In vitro, Med7N/31 is required for activated transcription and can act in trans when added exogenously. In vivo, Med7N/31 has a predominantly positive function on the expression of a specific subset of genes, including genes involved in methionine metabolism and iron transport. Comparative phenotyping and transcriptome profiling identify specific and overlapping functions of different Mediator submodules.
Keywords: CTD, Mediator of transcriptional regulation, RNA polymerase II transcription, structure–function analysis, TFIIS
Introduction
Regulation of eukaryotic transcription by RNA polymerase (Pol) II requires the Mediator co-activator complex (Malik and Roeder, 2000; Naar et al, 2001; Bjorklund and Gustafsson, 2005; Kornberg, 2005). Mediator forms an interface between the general Pol II machinery and transcriptional activators and repressors. Mediator was identified in yeast (Kelleher et al, 1990; Flanagan et al, 1991), has been purified from various fungi, metazoans, and plant (Bäckström et al, 2007), and was detected by homology analysis in many eukaryotic genomes (Boube et al, 2002; Bourbon, 2008). Yeast Mediator has a molecular weight of 1.4 MDa and consists of 25 polypeptide subunits. Many subunit interactions have been mapped (Guglielmi et al, 2004; Larivière et al, 2006; Takagi et al, 2006). Structural information is available for subunit cyclin C (Hoeppner et al, 2005), the Med15 KIX domain (Yang et al, 2006; Thakur et al, 2008), and the subcomplexes Med7C/21 (Baumli et al, 2005), Med18/20, and Med8C/18/20 (Larivière et al, 2006, 2008) (‘C' denotes the C-terminal portion of a subunit, ‘N' denotes the N-terminal portion). Electron microscopy and biochemical analysis of yeast Mediator suggested that the subunits reside in four different modules, termed the head, middle, tail, and kinase modules (Asturias et al, 1999; Kang et al, 2001).
The Mediator modules serve various functions. The tail module binds to activators and repressors (Han et al, 2001; Jeong et al, 2001; Zhang et al, 2004; Thakur et al, 2008), the middle and head modules contact Pol II (Davis et al, 2002) and the general machinery (Kang et al, 2001; Baek et al, 2006; Larivière et al, 2006; Esnault et al, 2008), and the kinase module has inhibitory functions (van de Peppel et al, 2005; Elmlund et al, 2006). Gene deletion studies implicate the tail in regulating sporulation genes and genes for oxidative phosphorylation, the tail and middle modules in regulating low iron response and heat-shock genes, the head module in regulating conjugation genes, and the kinase module in regulating genes required during nutrient starvation (Beve et al, 2005; van de Peppel et al, 2005; Singh et al, 2006; Larivière et al, 2008). The modules may contain structurally and functionally distinct submodules, such as the Med8C/18/20 submodule of the Mediator head (Larivière et al, 2006, 2008).
Here, we show that the Mediator middle module contains a submodule formed by the N-terminal region of Med7 (Med7N) and Med31. A combination of biochemistry, X-ray crystallography, yeast phenotyping, and transcriptome analysis establishes Med7N/31 as a structurally and functionally distinct submodule that is required for activated transcription. Med31 was initially called Soh1 as it was found in a Saccharomyces cerevisiae screen for suppressors of a hyper-recombination phenotype caused by hpr1 deletion (Fan and Klein, 1994), and was later described as a Mediator subunit (Gu et al, 1999, 2002; Boube et al, 2000; Park et al, 2001; Guglielmi et al, 2004; Linder and Gustafsson, 2004). Med31 is one of the best conserved Mediator subunits and shows 28% sequence identity between yeast and human, but is not essential for yeast viability (Fan and Klein, 1994). Med31 is required for telomere maintenance (Askree et al, 2004), DNA repair (Fan and Klein, 1994), meiotic DNA processing (Jordan et al, 2007), and transposition (Nyswaner et al, 2008). The Med31 orthologue in Drosophila is a maternal-effect gene for segment specification during early Drosophila embryogenesis (Bosveld et al, 2008). We describe the Med31 structure, its interaction with the Med7N domain, and its function in vitro and in vivo on the transcriptome level.
Results
Identification and crystallization of the Med7N/31 subcomplex
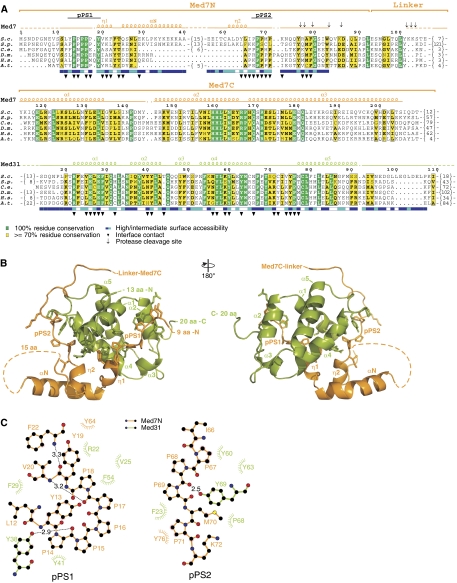
The Mediator middle module contains the subunits Med1, Med4, Med7, Med9 (Cse2), Med10 (Nut1), Med21 (Srb7), and Med31 (Soh1) (Guglielmi et al, 2004; Beve et al, 2005). To work towards the structure and function of the middle module, we purified the trimeric complex Med7/21/31 in recombinant form after bacterial co-expression of the corresponding genes and subjected it to limited proteolysis (Materials and methods). Proteolytic cleavage patterns were consistent with two flexibly linked subcomplexes, the Med7C/21 subcomplex, which we analysed earlier (Baumli et al, 2005), and a subcomplex of Med31 with the N-terminal region of Med7 (Figure 1A). We therefore co-expressed Med31 with different variants of the Med7 N-terminal region, and purified the resulting heterodimeric complexes. A complex that comprised the N-terminal region of Med7 including part of the linker (residues 1–101) was soluble, but did not crystallize, apparently due to flexibility in the proteolytically sensitive linker. A complex that lacked the entire linker and included only Med7 residues 1–83 (Med7N) crystallized. The X-ray structure of Med7N/31 was solved by selenomethionine labelling and single anomalous diffraction, and was refined to a free R-factor of 24.3% at 2.8 Å resolution (Supplementary Table I; Materials and methods).
Figure 1.
Structure of the Med7N/31 Mediator subcomplex. (A) Multiple sequence alignment and structural conservation of Med7 and Med31 from Saccharomyces cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), Caenorhabditis elegans (C.e.), Drosophila melanogaster (D.m.), Homo sapiens (H.s.) and Arabidopsis thaliana (A.t.). Secondary structure elements are indicated above the sequences (spirals, α- and 310(η)-helices; arrows, β-strands; lines, ordered but without secondary structure; dashed lines, disordered regions). Invariant and conserved residues are highlighted in green and yellow, respectively. Surface accessibility is indicated below the sequences (blue, exposed; white, buried). Cleavage sites revealed by limited proteolysis with trypsin or chymotrypsin are indicated with black arrows. The loop between Med7N helices α1 and η2 (residues 41–55) was disordered. Sequence alignments were done with MUSCLE (Edgar, 2004) and figures were prepared with ESPript (Gouet et al, 1999). (B) Ribbon model representation of the Med7N/31 crystal structure. Two views are shown that are related by a 180° rotation around the vertical axis. Med7 is depicted in orange and Med31 in green. Secondary structure elements are labelled according to (A). This and other figures were prepared with PyMol (DeLano, 2002). (C) Ligplot view (Wallace et al, 1995) of the interactions of the Med7N polyproline stretches pPS1 and pPS2 with Med31.
The Med7N/31 structure reveals novel folds
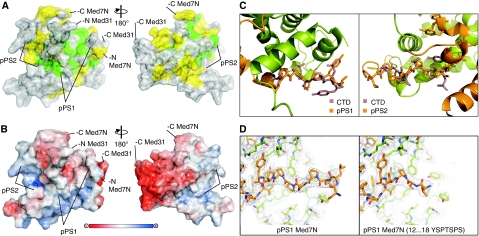
The structure shows that Med31 forms a compact right-handed four-helix bundle, whereas Med7N is extended and wraps around Med31 (Figure 1B). Both folds are novel, as neither DALI nor MSD fold searches (Holm and Park, 2000; Krissinel and Henrick, 2004) produced any hits. The Med7N/31 structure is apparently conserved among eukaryotes, as hydrophobic core residues are identical or similar from yeast to human (Figure 1A), and many residues in the Med7N–Med31 interface are also conserved. The Med7N–Med31 interface is mainly hydrophobic (Figure 1), explaining why isolated recombinant Med7N or Med31 cannot be obtained in soluble form (not shown). The Med7N/31 surface reveals several conserved, non-charged regions (Figure 2A and B) that can account for interactions of Med7N/31 with other subunits of the middle module (Guglielmi et al, 2004) or other potential binding partners.
Figure 2.
Analysis of the Med7N/31 structure. (A) Surface conservation of the Med7N/31 subcomplex. Invariant and conserved residues are highlighted in green and yellow, respectively. The surface is semitransparent to show the underlying residues in a stick model. Two views are shown that are related by a 180° rotation around the vertical axis. (B) Surface charge distribution. Red, blue, and white areas indicate negative, positive, and neutral charges, respectively. (C) The Med7N polyproline stretches adopt a CTD-like conformation. Superimposition of Med7N pPS1 and pPS2 (orange) with the Y(SEP)PT(SEP)PS peptide from the human Pin1 structure (1F8A, light purple). (D) Comparison of pPS1 in the crystal structures of Med7N/31 and Med7N/31 with pPS1 mutated to a CTD heptad repeat (mutant 6; Supplementary Table II).
Possible CTD mimicry by Med7N
Med7N contacts Med31 mainly through two polyproline stretches (pPS1 and pPS2; Figure 1A–C), which adopt a conformation similar to a left-handed type II polyproline helix (Adzhubei and Sternberg, 1993). A deletion analysis revealed that both pPS1 and pPS2 were required for the formation of a stable complex with Med31 (Supplementary Table II; constructs 2–5). pPS2 is tightly packed onto Med31, whereas pPS1 is more accessible (Figure 1C). As the C-terminal repeat domain (CTD) of Pol II is also proline rich, we compared the Med7N stretches with known CTD peptide structures. The conformation of the CTD peptide in complex with the prolyl isomerase Pin1 (PDB 1F8A; Verdecia et al, 2000) resembles the structure of Med7N pPS1, and is also distantly related to pPS2 (Figure 2C).
The CTD interacts genetically and physically with Mediator (Thompson et al, 1993; Asturias et al, 1999; Kang et al, 2001). Although med31 deletion does not suppress the cold-sensitive phenotype caused by CTD truncations (Thompson et al, 1993), it is synthetically lethal with a truncation of the CTD to only 11 intact heptad repeats (Fan et al, 1996). We therefore speculated that the CTD may bind Med31 by replacing pPS1 and/or pPS2 of Med7N under certain conditions. We could however not investigate direct CTD binding to Med31 as isolated Med31 could not be obtained in soluble form. We therefore mutated the Med7N pPS1 and pPS2 stretches such that their sequences correspond to the CTD repeat sequence consensus (Supplementary Table II), and tested whether these Med7N mutants formed stable complexes with Med31. The pPS1 mutant, but not the pPS2 mutant, formed a stable complex with Med31 (Supplementary Table II). Crystallographic analysis of the pPS1 mutant (mutant 6; Supplementary Table II) in complex with Med31 showed that four residues of the CTD heptad repeat (15-TSPS-18) were ordered and bound to Med31, whereas the preceding three residues (12-YSP-14) were flexible (Figure 2D). However, as the Med7N/31 complex also tolerated replacement of pPS1 by the reverse CTD heptad repeat sequence (mutant 10; Supplementary Table II), and as the Med7N stretches are present in organisms lacking the canonical CTD repeats (Stiller and Hall, 2002; Bourbon, 2008), a possible CTD mimicry by Med7N and CTD binding by Med31 remains hypothetical.
Med7N/31 is required for normal yeast growth in vivo
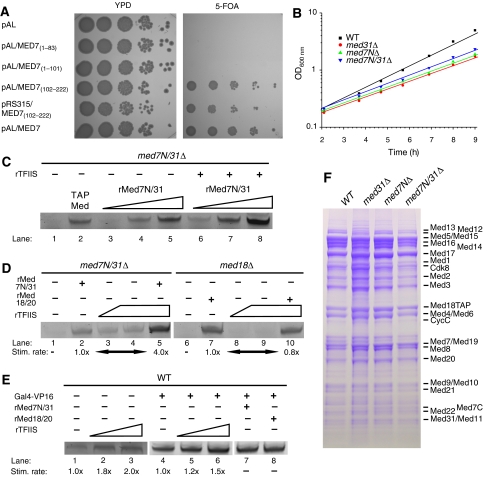
Med7 is essential for yeast viability in rich medium (Myers et al, 1998). To investigate which Med7 region is required for viability, we carried out complementation studies in yeast. We generated plasmids expressing full-length Med7, Med7N, and Med7N plus the linker part, which was not present in the Med7C/21 structure (Baumli et al, 2005) or a Med7C variant. Plasmids were introduced into a med7Δ strain rescued by a MED7-encoding URA3 plasmid. Complementation was observed with full-length MED7 or MED7C, which allows for loss of the URA3 plasmid and growth on media containing 5-fluorotic acid (5-FOA; Figure 3A). As the remaining linker residues (102–113) present in the Med7C/21 structure are not conserved, we infer that only Med7C is essential for viability, whereas Med7N and the linker are not. A strain that lacked the region encoding Med7N (med7NΔ) exhibited a slow-growth phenotype, similar to a med31Δ strain and a double deletion strain med7NΔ/med31Δ (med7N/31Δ) (Figure 3B; Supplementary Table III).
Figure 3.
Functional analysis of Med7N/31 in vivo and in vitro. (A) Yeast complementation assays. Plasmids encoding full-length MED7 or N-terminal truncations of MED7 were cloned into the SmaI/XbaI restriction sites of a pAL vector or through SalI/XbaI into a pRS315 vector. Individual plasmids were transformed into the MED7 shuffle strain (Supplementary Table III) and streaked onto 5-FOA-containing plates to shuffle out the MED7 encoding URA3 plasmid. Yeast cells carrying only MED7C (102–222) are viable, whereas the Med7 N-terminal part including part of the linker (residues 1–101) cannot rescue cell growth. (B) Mutant strains exhibit a slow-growth phenotype. Note that growth curves are on a logarithmic scale. (C) Med7N/31 is required for Gal4–VP16-activated transcription in vitro (lane 1). Transcription can be rescued by the addition of TAP-purified Mediator (0.2 pmol, lane 2) or recombinant Med7N/31 (rMed7N/31, 2–200 pmol, lanes 3–5). Addition of both rMed7N/31 (2–200 pmol) and recombinant TFIIS (rTFIIS, 20 pmol, lanes 6–8) enhances the signal. (D) A Med7N/31Δ nuclear extract that was rescued by the addition of recombinant Med7N/31 (lane 2, 200 pmol) was additionally stimulated by the addition of TFIIS (lane 5, 60 pmol). TFIIS alone can partially compensate for loss of Med7N/31 (lanes 3 and 4, 20–60 pmol). This stimulation was not observed with a med18Δ nuclear extract that was rescued by the addition of recombinant Med18/20 (rMed18/20, lanes 7–10, 20–60 pmol). (E) TFIIS addition stimulates basal and Gal4–VP16-activated transcription of wild-type (WT) nuclear extracts (lanes 2 and 3, and 5 and 6, respectively, 20–60 pmol). Recombinant Med7/31 or Med18/20 did not stimulate WT nuclear extracts. (F) Deletion of Med7N/31 or its subunits does not lead to loss of additional Mediator subunits. C-terminally TAP-tagged Med18 was purified from med31Δ/Med18-TAP, med7NΔ/Med18-TAP, and med7N/31Δ/Med18-TAP strains as described (Puig et al, 2001; Larivière et al, 2008). Copurifying proteins were separated on a 12% NuPAGE gel (Invitrogen), and bands were stained with Coomassie blue. The identity of all Mediator subunits except Med31 was confirmed by mass spectrometry (Supplementary Table V). Contaminants such as ribosomal proteins or degradation products of Mediator subunits were especially detected at lower molecular weights and do partially overlap with smaller subunits such as Med7C.
Med7N/31 is essential for transcription in vitro
To investigate whether the Med7N/31 subcomplex is required for activated transcription in vitro, we used a plasmid-based in vitro transcription assay with a HIS4 core promoter and a single upstream Gal4-binding site (Ranish et al, 1999). A nuclear extract from strain med7N/31Δ was defective in basal transcription (not shown) and in transcription activated with the Gal4–VP16 fusion protein or with a fusion protein of the Gal4 DNA-binding domain with the Gal4 activation helix (Gal4-Gal4AH) (Figure 3C, lane 1; Supplementary Figure 1, lane 1). Addition of tandem affinity purification (TAP)-purified Mediator restored transcription (Figure 3C, lane 2), providing a positive control. Transcription could also be restored by the addition of recombinant Med7N/31, demonstrating that the crystallized complex is functionally active and can act in trans without being covalently tethered to the Mediator (Figure 3C, lanes 3–5; Supplementary Figure 1, lane 2). To investigate whether loss of Med7N/31 leads to the dissociation of other subunits from Mediator in vivo, we purified Mediator from deletion strains med7NΔ, med31Δ, and med7N/31Δ with the use of a TAP tag fused to Med18 (Materials and methods). After purification from these deletion strains, the remainder of Mediator stayed intact (Figure 3F). These results show that Med7N/31 forms a peripheral subcomplex on Mediator that is required for activated transcription in our in vitro assays.
Med7N/31 is a functional Mediator submodule in vivo
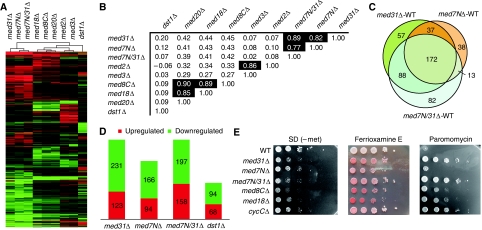
To investigate the function of the Med7N/31 subcomplex in vivo, we carried out comparative microarray-based gene expression profiling with the yeast deletion strains med31Δ, med7NΔ, and med7N/31Δ. Compared with wild-type cells, the expression profiles of the three deletion strains showed similar overall changes illustrated by high Pearson's correlations (Figure 4A–D; Table I; Supplementary Table IV). A majority of changes in mRNA levels that are induced by deletion of both Med7N and Med31 are already observed when either Med7N or Med31 is lacking from cells. This shows that Med7N/31 serves as a functional submodule of the Mediator in vivo at a majority of affected genes. Some differences between profiles for the med7NΔ, med31Δ, and med7N/31Δ strains at selected genes (Figure 4C) indicate subunit-specific functions that may result from additional protein interactions (unpublished data). As the majority of affected genes is downregulated (Figure 4D), the Med7N/31 submodule has a predominantly positive function, consistent with previous expression data on a med31Δ strain (van de Peppel et al, 2005).
Figure 4.
Transcriptome profiling analysis and phenotypic correlation. (A) Cluster diagram (Euclidean distance) of genes exhibiting significantly altered mRNA levels (at least 1.7-fold, vertical axis) for different Mediator subunit deletion strains and TFIIS (horizontal axis). Changes in mRNA levels compared with the wild-type strain are depicted in red (up), green (down), or black (no change). (B) Pearson's correlation matrix for expression profiles of the Mediator middle subunit deletion strains med31Δ, med7NΔ, med7N/31Δ, the head subunit med8CΔ, med18Δ, med20Δ, the tail subunit deletion strains med2Δ, med3Δ, and the deletion profile of the general transcription factor dst1Δ strain. (C) Venn diagram showing the overlap between the investigated med31Δ, med7NΔ, and med7N/31Δ strains. Although these three strains exhibit a very high overlap, the overlap with the dst1Δ strain was not regarded as statistically significant. (D) Number of significantly altered genes of all four investigated deletion strains, split into up- and downregulated genes. (E) Selected phenotyping analysis results. Using 10-fold serial dilutions of yeast strains spotted onto selective agar plates, we screened for phenotypes, occurring under certain growth conditions. All assays were compared with a control plate on YPD, synthetic complete (SC), or SD media plates. As typical examples, the results from the growth inhibition assay on SD (-met) plates, the siderophore uptake assay (detected using bathophenanthroline disulphonic acid (BPDS)), and the growth inhibition on paramomycin assay are depicted.
Table 1.
Summary of common biological terms over-represented in GO analysis of the differentially expressed genes in med31Δ, med7NΔ, and med7N/31Δ strains
| GO term (biological process)a | Description | med7N/31Δ submodule | |||
|---|---|---|---|---|---|
| Annotated | Upregulated | Downregulated | Expected | ||
| GO:0015891 | Siderophore transport | 9 | 7 | 0.3 | |
| GO:0000097 | Sulphur amino-acid biosynthetic process | 35 | 8 | 1.0 | |
| GO:0006555 | Methionine metabolic process | 38 | 8 | 1.1 | |
| GO:0000103 | Sulphate assimilation | 10 | 4 | 0.3 | |
| GO:0015698 | Inorganic anion transport | ||||
| – GO:0008272 | – Sulphate transport | 5 | 3 | 0.2 | |
| GO:0043419 | Urea catabolic process | 2 | 2 | 0.1 | |
| GO:0000749 | Response to pheromone during conjugation | ||||
| – GO:0000750 | – Pheromone-dependent signal transduction | 28 | 1 | 4 | 0.8 |
| GO:0015846 | Polyamine transport | 11 | 2 | 1 | 0.3 |
| GO:0007050 | Cell cycle arrest | 12 | 1 | 2 | 0.4 |
| GO:0000723 | Telomere maintenance | ||||
| – GO:0007004 | – Telomere maintenance through telomerase | 17 | 1 | 2 | 0.5 |
| – GO:0016233 | – Telomere capping | 5 | 1 | 1 | 0.2 |
| GO:0001403 | Invasive growth (sensu Saccharomyces) | 45 | 1 | 4 | 1.3 |
| GO:0032147 | Activation of protein kinase activity | 6 | 2 | 0.2 | |
| GO:0001402 | Signal transduction during filamentous growth | 7 | 1 | 1 | 0.2 |
| GO:0008645 | Hexose transport | 20 | 3 | 0.6 | |
| aGO terms are ranked according to the topGO weight algorithm (Alexa et al, 2006) exhibiting a score smaller than 0.02. | |||||
| Lower level GO terms (marked with a ‘–') are listed below the hierarchically higher GO terms. For the complete analysis including the associated genes and the analysis for the dst1Δ strain, see also Supplementary Tables IVB and C | |||||
Med7N/31 regulates a subset of genes
Gene ontology (GO) analysis of genes with significantly changed mRNA levels revealed an over-representation of genes involved in sulphate/methionine metabolism, iron homoeostasis, and transport functions (Table I; Supplementary Table IVB). A group of genes involved in telomere maintenance was also over-represented, consistent with the observation that a med31Δ strain exhibits shortened telomeres (Askree et al, 2004). The differential expression profiles for Med7N/31 were largely distinct from the previously published profiles (Larivière et al, 2008) of the tail subunit deletion strains med2Δ and med3Δ, and correlated only moderately with those of strains med8CΔ, med18Δ, and med20Δ, which lack parts of the head submodule Med8C/18/20 (Figure 4A and B). However, both the Med7N/31 and the Med8C/18/20 submodules regulate genes responsible for siderophore transport and conjugation. Thus, the Med7N/31 submodule is required not only for the regulation of a specific subset of genes but also those genes that depend on other parts of the Mediator.
To identify transcription factors that regulate the differentially expressed genes, we used the transcription factor to gene promoter mapping matrix from the YEASTRACT website (Teixeira et al, 2006), and performed Fisher's exact test with the R/Bioconductor software (Gentleman et al, 2004) (Supplementary Table IVF). This analysis revealed that the differentially expressed genes contained over-represented target genes for certain transcription factors, including Cst6, Aft1, Msn2, Msn4, Yap family factors, Met28, Met31, Met32, Hsf1, and Pdr1. Consistent with the GO analysis, Msn2, Msn4, and Yap family members are involved in stress response, Cst6 regulates genes involved in telomere maintenance, and Aft1 is required for activating the low iron response regulon through Mediator (van de Peppel et al, 2005). The mRNA levels for these factors were unaltered in the investigated strains, except for Met28 and Msn4. Met28 was downregulated for Med7N/31 deletions and upregulated for Med2/Med3 deletions (Supplementary Table IVG), explaining corresponding negative and positive responses, respectively, of target genes involved in methionine and cysteine biosynthesis (Supplementary Table IVD).
Overlapping and specific functions of Mediator submodules
To investigate the deregulation of genes induced by disruption of Med7N/31, we analysed the growth of several yeast strains with deletions in Mediator genes (Supplementary Table III) on various selective agar plates (Table II). The phenotypes of Med7N/31 deletions correlated well with results from transcription profiling. In the absence of methionine, these deletion strains grew poorly (Figure 4E), consistent with the observed downregulation of genes involved in methionine/sulphate metabolism. In the presence of siderophores, several deletion strains took up more complexed iron, as indicated by a red colour (Figure 4E), consistent with higher expression of siderophore transport genes (some likely regulated by Aft1). Sensitivities of the deletion strains towards osmotic and cell wall stress were consistent with reported impaired cell wall metabolism of Mediator subunit deletion strains of Schizosaccharomyces pombe (Linder et al, 2008). A decreased sensitivity of the med31Δ and med7N/31Δ strains for the translation inhibitor paramomycin can also be explained by impaired cellular uptake or transport (Figure 4E). Most deletion strains were further impaired in their response to temperature, oxidative, and DNA repair stresses (Table II). Also observed were inositol– phenotypes that are often indicative of defects in the general transcriptional apparatus (Hampsey, 1997).
Table 2.
Comparative phenotyping of Mediator deletion strains
| Sensitivity testa | WT | Middle | Head | Tail | Kinase | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| med31Δ | med7NΔ | med7N/31Δ | med1Δ | med9Δ | med8CΔ | med18Δ | med2Δ | med3Δ | med5Δ | cycCΔ | |||
| Siderophore uptake | SD+ferrichrome | + | +++ | ++ | ++ | + | + | +++ | +++ | ++ | ++ | − | ++ |
| SD+ferrioxamine E | + | +++ | ++ | ++ | + | + | +++ | +++ | ++ | ++ | − | ++ | |
| Lack of amino acids | SD (-met) | − | ++ | + | ++ | − | − | +++ | ++ | ++ | ++ | − | + |
| Growth on S sources | SD (-SO42−) | + | ++ | + | ++ | − | − | ++ | ++ | + | + | − | +++ |
| Growth on C sources | YP Na acetate | − | ++ | + | + | − | − | + | +++ | +++ | +++ | + | +++ |
| Growth on N sources | SD urea | − | − | + | − | − | − | ++ | ++ | ++ | + | − | + |
| Temperature | YPD 16°C | − | + | + | + | + | + | ++ | ++ | ++ | ++ | − | + |
| YPD 37°C | − | ++ | + | + | + | ++ | +++ | +++ | ++ | ++ | − | +++ | |
| Salt stress | YPD NaCl (1 M) | − | ++ | + | + | + | + | +++ | ++ | − | − | + | ++ |
| YPD KCl (1 M) | − | ++ | + | ++ | − | + | +++ | + | + | + | − | + | |
| YPD LiCl (0.4 M) | + | + | + | + | + | ++ | +++ | +++ | − | − | + | ++ | |
| Cell wall stress | YPD SDS (0.01%) | − | + | + | + | − | − | +++ | +++ | + | + | − | ++ |
| Osmotic stress | YPD glycerol (2 M) | − | ++ | ++ | ++ | + | − | +++ | +++ | + | + | − | + |
| Oxidative stress | YPD Menadione (20 mM) | − | − | ++ | + | − | − | − | ++ | + | + | − | − |
| DNA repair | YPD MMS (0.05%) | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | + | − |
| YPD HU (100 mM) | + | ++ | ++ | ++ | + | + | +++ | +++ | ++ | ++ | + | +++ | |
| Transcription | SC (-inositol) | − | ++ | ++ | ++ | − | − | +++ | +++ | ++ | ++ | − | + |
| Transcription elongation | SC (-ura) 6-AU (100 mg/ml) | ++ | + | − | − | − | − | − | + | ++ | + | + | − |
| Translation | YPD cycloheximide (0.1 mg/ml) | − | +++ | +++ | +++ | − | + | ++ | +++ | +++ | +++ | + | + |
| YPD paromomycin (2.5 mg/ml) | +++ | − | +++ | − | − | − | ++ | ++ | − | − | − | − | |
| Miscellaneous | YPD caffeine (8 mM) | + | ++ | + | + | + | + | ++ | +++ | ++ | ++ | − | ++ |
| Starvation | YP media 6 days, 30°C, then respotted on YPD | − | ++ | + | + | ND | ND | +++ | +++ | ND | ND | ND | + |
| aAll measurements are growth inhibition, except for the siderophore assay, where cellular iron content was measured. | |||||||||||||
| Growth inhibition (with the exception of the siderophore uptake assays) is indicated by relative estimates (+, moderate hypersensitivity; ++, high hypersensitivity; +++, very high hypersensitivity; −, no hypersensitivity; ND, not determined). | |||||||||||||
This comparative phenotyping of Mediator deletion strains showed that the med8CΔ and med18Δ strains exhibited the most severe growth defects (Table II), possibly reflecting an important function of these subunits in the initiation complex assembly (Ranish et al, 1999). Deletion of subunits Med1, Med5, or Med9 was less detrimental. In agreement with earlier studies (van de Peppel et al, 2005), Cdk8 behaves partially contrasting with the phenotyping assays. On the transcriptome level, analyses of Med7N/31 and Med8C/18/20 submodule deletion profiles with med2Δ and med3Δ profiles (Supplementary Table IVC and D) reveal different overlaps in enriched GO terms. Some GO terms are exclusively enriched for specific Mediator submodules (e.g. telomere capping for Med7N/31), whereas other GO terms are affected for two different submodules (e.g. siderophore transport for Med7/31 and Med8C/18/20 or serine family amino-acid catabolism with Med8C/18/20 and Med2/3), and other GO term groups are affected by all investigated mutations (e.g. response to pheromone). Such overlap in the phenotyping assays (Table II) and the GO analysis (Table I; Supplementary Table IVC and D) shows that the characterized non-essential parts of Mediator are generally required for regulating metabolic sensing, stress response, and amino-acid biosynthesis pathways.
Cooperation of Med7N/31 and TFIIS
As Med31 interacts genetically with the elongation factor TFIIS (Krogan et al, 2003; Malagon et al, 2004; Collins et al, 2007; Guglielmi et al, 2007), we tested whether Med7N/31 cooperates with TFIIS during activated transcription in vitro. Indeed, transcription in a med7N/31Δ nuclear extract that was rescued by recombinant Med7N/31 could be enhanced four-fold by the addition of recombinant TFIIS using Gal4–VP16 as an activator (Figure 3D). A stimulatory effect of TFIIS, although only about two-fold, was also seen for basal and activated transcription in wild-type (WT) extracts (Figure 3E). Addition of TFIIS to a med7N/31Δ extract resulted in a signal even in the absence of recombinant Med7N/31, although TFIIS was not limiting in the extracts, as a three-fold higher concentration did not further increase transcription (Figure 3D, lanes 3 and 4). TFIIS also stimulated transcription activated with the alternative activator Gal4–Gal4AH, although only about two-fold (Supplementary Figure 1). In contrast, transcription in a med18Δ extract was not enhanced at all by TFIIS addition (Figure 3D, lanes 8 and 9) and a med18Δ extract that was rescued by the addition of recombinant Med18/20 was not further stimulated by the addition of TFIIS (Figure 3D, lane 10). These results show that TFIIS can partially compensate for the loss of Med7N/31, and that highest transcript levels required the addition of both TFIIS and Med7N/31. These observations were specific for the med7N/31Δ extract, as this effect was not obtained in corresponding experiments with a med18Δ extract. The results are further consistent with an important role for TFIIS during initiation (Guglielmi et al, 2007; Kim et al, 2007).
To investigate whether TFIIS and Med7N/31 cooperate at specific genes in vivo, we determined the gene expression profile for a yeast strain that lacks the gene for TFIIS, dst1 (Supplementary Table IVA, B, E and F). Only 162 mRNA levels were significantly altered (at a two-fold cutoff and a false discovery rate of P<0.05; Supplementary Table IVA), consistent with a lack of a phenotype of the dst1Δ strain in rich medium (Clark et al, 1991). Pearson's correlation and GO analysis did not reveal any significant overlap between dst1Δ and med7N/31Δ profiles (Figure 4B; Supplementary Table IV). Consistently, we could not detect a direct physical interaction between TFIIS and recombinant Med7N/31 (not shown). Instead, analysis of synthetic lethality partners suggests that Med31 and TFIIS are functionally connected through the SWR1 complex that is involved in exchange of histone H2A by the variant H2A.Z (Supplementary Figure 2A). Similar correlations were also found for the Mediator subunits Med1, Med9, and Med20 (not shown). Additionally, the Mediator kinase module is a highly significant phenotypic suppressor of core Mediator subunits and TFIIS (Supplementary Figure 2B). Taken together, the functional cooperation of Med7N/31 and TFIIS is apparently not gene specific in vivo, but may generally occur during initiation, and could further be linked to Swr1-dependent histone variant exchange near transcription start sites (Santisteban et al, 2000; Zanton and Pugh, 2006; Zlatanova and Thakar, 2008).
Discussion
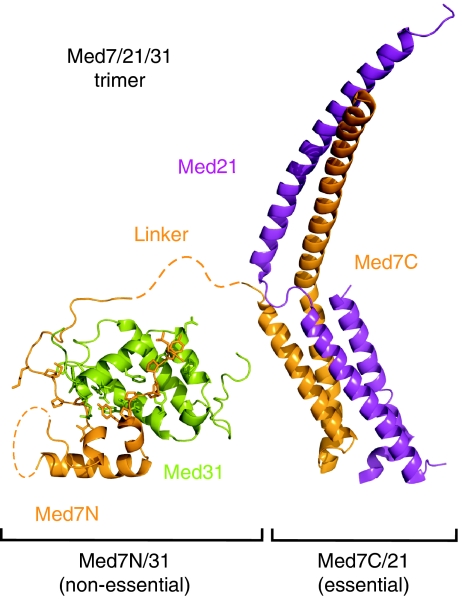
Understanding the mechanisms that eukaryotes use to regulate gene transcription through Mediator requires a detailed description of the functional substructure of this highly modular multiprotein complex. Here, we extend our previous structural and functional analyses of Mediator subcomplexes (Baumli et al, 2005; Hoeppner et al, 2005; Larivière et al, 2006, 2008) to the identification, structure, and in vitro and in vivo functional analyses of the highly conserved Med7N/31 subcomplex. The Med7N/31 structure exhibits a novel fold, and together with the previously determined Med7C/21 structure unravels the highly unusual overall structure of Med7 (Figure 5). Med7N forms a non-essential tether for the compact peripheral Med31 subunit and is flexibly linked to Med7C, which forms an extended essential heterodimer with Med21 (Figure 5). Other Mediator subunits may also show such an extended, modular, and context-dependent architecture.
Figure 5.
Structural overview of the trimeric Med7/21/31 complex architecture. Structures of the non-essential Med7N/31 submodule (this study) and the previously described essential Med7C/21 subcomplex are drawn to scale.
Intriguing features of the Med7N/31 structure are the two conserved proline-rich stretches of Med7N that bind Med31. The similarity of these stretches to the proline-rich CTD of Pol II suggests that the CTD may dynamically replace one or both stretches and bind Med31. The Med7N stretches may also detach from Med31 and bind factors that comprise a domain that recognizes polyproline peptides, such as WW, SH3, and GYF domains (Macias et al, 2002; Hesselberth et al, 2006; Kofler and Freund, 2006), or a proline isomerase. pPS1 contains the PPxY motif that can be bound by certain WW domains (Espanel and Sudol, 2001). Finally, proline-rich activators could replace one or both Med7N stretches and bind to Med31. However, insolubility of recombinant Med31 and Med7N hampered a biochemical investigation of these models. The high conservation of Med31 may reflect the putative CTD interaction, but may also stem from an alternative interaction with the polymerase or other factors.
A subsequent structure-based functional analysis investigated the cooperation of Med7N/31 with TFIIS during activated transcription, and established the subcomplex as a predominantly positive gene-regulatory submodule at the periphery of the essential middle module of Mediator. A combination of transcriptome analysis and phenotyping assays revealed the processes that are affected by a lack of intact Med7N/31, including methionine metabolism and siderophore transporter synthesis. Med7N/31 also has a role in the regulation of conjugation genes, similar to the head submodule Med8C/18/20, but in contrast to the Mediator tail subunits Med2 and Med3. Distinct phenotypes and mRNA levels were observed for mutants that do not contain an intact Med7N/31 submodule, consistent with the idea that there are gene-specific and overlapping functions associated with different Mediator submodules.
The observed overlaps in phenotyping assays and in the GO analysis of differential transcriptome data show that metabolic sensing, stress response, and certain amino-acid biosynthesis pathways are generally affected by deletion of different Mediator submodules. At present it remains difficult to decipher the cis elements and trans-acting factors that underlie the observed changes in gene expression. However, we provide evidence that Met28 and Msn4 are important transcription factors involved in Med7N/31-mediated gene regulation. Msn4 is downregulated upon Med7N/31 deletion (this study) and functions in stress response (Gasch et al, 2000; Causton et al, 2001). Met28 is downregulated upon Med7N/31 deletion but upregulated upon Med2/3 deletion, explaining the observed down- and upregulations in methionine and cysteine metabolism, respectively, as a secondary effect. Differently expressed genes were also often found to contain binding sites for Yap family transcription factors that regulate stress response (Fernandes et al, 1997). In the future, factors may be identified that use Med7N/31 as a target for gene regulation. A longer term goal remains the modelling of networks that govern gene regulation through Mediator submodules.
Materials and methods
Preparation of recombinant Med7N/31
The trimeric complex Med7/21/31His was expressed from two plasmids encoding Med7/21 and Med31His that were co-transformed into Escherichia coli BL21 (DE3) RIL cells (Stratagene). The complex was purified by Ni-NTA and anion exchange chromatography and subjected to limited proteolysis using either chymotrypsin or trypsin. The reaction was stopped by the addition of a protease inhibitor mixture, loaded onto a Superose 6 gel filtration column (GE Healthcare) and individual peaks were analysed on SDS–PAGE after TCA precipitation. The gene encoding S. cerevisiae Med31 was cloned into a pET-28b vector (Novagen) using the NdeI/NotI restriction sites, resulting in a thrombin-cleavable N-terminal hexahistidine tag. DNA encoding Med7 residues 1–83 was cloned into a pET21b vector through the EcoRI and SalI restriction sites. E. coli BL21 (DE3) RIL cells (Stratagene) were co-transformed with the two plasmids and grown in LB medium at 37°C to an optical density at 600 nm (OD600 nm) of 0.6. Expression was induced with 0.5 mM IPTG for 16 h at 18°C. Selenomethionine labelling was carried out as described (Budisa et al, 1995; Meinhart et al, 2003). For protein purification, cells were lysed using a high-pressure homogenizer (Avestin) in buffer A (50 mM Tris pH 8.0, 150 mM NaCl, and 10 mM β-mercaptoethanol). After centrifugation, imidazole was added to a final concentration of 20 mM to the supernatant and loaded onto a 3 ml Ni-NTA column (Qiagen) equilibrated with buffer A containing 20 mM imidazole. The column was washed with 20 column volumes (CVs) of buffer A containing 20 mM imidazole and eluted with buffer A containing 200 mM imidazole. Following overnight cleavage with thrombin, proteins were purified by anion exchange chromatography (Mono Q). The column was equilibrated with buffer B (50 mM Tris pH 8.0, 100 mM NaCl, and 10 mM β-mercaptoethanol), and proteins were eluted with a linear gradient of 20 CVs from 100 mM to 1 M NaCl in buffer B. After concentration, the sample was applied to a Superdex-200 size exclusion column (GE Healthcare) equilibrated with buffer C (50 mM MES pH 6.5, 150 mM NaCl, and 10 mM β-mercaptoethanol).
X-ray structure determination
For crystallization, pure Med7N/31 was concentrated to 50 mg/ml, and 5 mM DTT was added. Crystals were grown at 20°C in hanging drops over reservoirs containing 50 mM MES pH 5.6, 1.8 M Li2SO4, and 10 mM MgCl2. Crystals were harvested by gradually adding ethylene glycol to a final concentration of 10% (v/v) and were flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on a MARCCD 225 and on a PILATUS 6M detector at the Swiss Light Source (SLS), Villigen, Switzerland (native data set and mutant data set, respectively) and on a MARCCD 165 detector at the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung m.b.H. BESSY (SAD data set). Diffraction data for selenomethionine-labelled Med7N/31 was processed using XDS and XSCALE (Kabsch, 1993). Program SOLVE (Terwilliger and Berendzen, 1999) identified six selenium sites that were used for phasing. Solvent flattening, two-fold non-crystallographic symmetry averaging, and initial model building was done with RESOLVE (Terwilliger, 2003). The resulting electron density map allowed for manual building of most Med7N and Med31 using COOT (Emsley and Cowtan, 2004). The model was refined using conjugate gradient minimization in the programs CNS (Brunger et al, 1998), REFMAC (McCoy et al, 2007), and Phenix (McCoy et al, 2007). The asymmetric unit contained two Med7N/31 heterodimers that deviate only at the C termini of the proteins. The mutant structure was solved by molecular replacement using Phaser (McCoy et al, 2007) and processed accordingly. The structures and diffraction data of wild-type Med7N/31 and the mutant complex Med7 (1–83) (12.18 YSPTSPS)/Med31 (mutant 6; Supplementary Table II) have been deposited in the Protein Data Bank under accession codes 3FBI and 3FBN, respectively.
Yeast strains and growth assays
The heterozygous MED7/med7Δ strain was obtained from Euroscarf (Frankfurt, Germany) and transformed with the cognate gene clone plasmid pYCG_YOL135C (containing a URA3 marker). Diploids were sporulated and tetrads were dissected on YPD plates. To assess functionality, pAL-med71–83, pAL-med71–101, pAL-med7101–222, pAL-med71–222, or pRS315-med7101–222 were transformed into the MED7 shuffle strain. Transformants were re-streaked onto 5-FOA and YPD plates. Yeast knockout strains were obtained from Euroscarf. All strains exhibited a BY background and were, with the exception of med2Δ and med18Δ, MATa. Deletion of the N-terminal region of Med7 (residues 1–83) was obtained by homologous recombination after transforming only the C-terminal part (residues 84–222) fused to a ClonNAT marker into either WT or med31Δ strains. Additionally, a C-terminal TAP tag on Med18 was introduced using an URA3 marker into wild-type, med31Δ, med7NΔ, and med7N/31Δ strains. Phenotype analyses of Mediator subunit deletion mutants were performed from cultures grown to stationary phase. Cells were diluted to an OD600 nm of 1.0, washed, and spotted in serial dilution onto plates. Assays were mostly performed as described (Hampsey, 1997). For synthetic defined (SD) (-met) and SD (-SO42−) and for 6-azauracil plate assays, strains with methionine or uracil auxotrophy were transformed with pRS411 or pRS316 plasmids, respectively. Siderophore uptake assays were performed with SD plates containing 500 μM bathophenanthroline disulphonic acid and 10 μM of siderophores.
In vitro transcription assays
Nuclear extracts were prepared from 3 l of culture as described by the Hahn laboratory (www.fhcrc.org/labs/hahn). Plasmid transcription was performed essentially as described (Ranish and Hahn, 1991). Transcription reactions were carried out in a 25 μl volume. The reaction mixture contained 100 μg nuclear extract, 150 ng of pSH515 plasmid, 1 × transcription buffer (10 mM HEPES pH 7.6, 50 mM potassium acetate, 0.5 mM EDTA, and 2.5 mM magnesium acetate), 2.5 mM DTT, 192 μg of phosphocreatine, 0.2 μg of creatine phosphokinase, 10 U of RNase inhibitor (GE Healthcare), and 100 μM nucleoside triphosphates. For activated transcription, 150 ng of Gal4–VP16 or 200 ng of Gal4–Gal4AH was added. The reaction was incubated at room temperature for 40 min and then stopped with 180 μl of 100 mM sodium acetate, 10 mM EDTA, 0.5% sodium dodecyl sulphate, and 17 μg of tRNA/ml. Samples were extracted with phenol–chloroform and precipitated with ethanol. Transcripts were analysed by primer extension essentially as described (Ranish and Hahn, 1991). Instead of the 32P-labelled lacI oligo, 0.125 pmol of a fluorescently labelled 5′-FAM-oligo was used. Quantification was performed with a Typhoon 9400 and the ImageQuant Software (GE Healthcare).
Gene expression profiling analysis
All experiments were performed in synthetic complete medium with 2% glucose. For microarray analysis, three independent colonies were used for inoculation, and overnight cultures were diluted in fresh medium to OD600 nm=0.1 (25 ml cultures, 160 r.p.m. shaking incubator 30°C). Cells were harvested by centrifugation (4000 r.p.m., 3 min, 20°C) at OD600 nm=0.8. Total RNA was prepared after cell lysis using a mixer mill (Retsch) and subsequent purification using the RNAeasy kit (Qiagen). The total RNA preparation was treated on-column with DNase (Qiagen). All following steps were conducted according to the Affymetrix GeneChip Expression Analysis Technical Manual (P/N 702232 rev. 2). Briefly, one-cycle cDNA synthesis was performed with 1 μg of total RNA. In vitro transcription labelling was carried out for 16 h. The fragmented samples were hybridized for 16 h on Yeast Genome 2.0 expression arrays (Affymetrix), washed and stained using a Fluidics 450 station, and scanned on an Affymetrix GeneArray scanner 3000 7G. Data analysis was performed using R/Bioconductor (Gentleman et al, 2004). S. pombe probes were filtered out prior to normalization with the GCRMA algorithm (Wu et al, 2004). Linear model fitting and multiple testing correction using an empirical Bayes approach was performed using the LIMMA package (Smyth, 2004). Differentially expressed genes were defined as having an adjusted P-value smaller than 0.05 and an estimated fold change of at least 2.0 (calculated as the fold change of the average expression in the triplicate measurements). Previously published transcriptome profiles (Larivière et al, 2008) were included in further analyses by matching genes and comparing fold changes. Over-represented biological processes for genes with significant expression changes were determined using the topGO package (Alexa et al, 2006). Clustering was calculated using TIGR MeV application (Saeed et al, 2003). Microarray data were submitted to the ArrayExpress database (http://www.ebi.ac.uk/microarray) under accession number E-MEXP-1916.
Supplementary Material
Supplementary Data
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 4
Supplementary Table 5
Acknowledgments
We thank Kerstin Maier and other members of the Cramer laboratory for help. We thank Heidi Feldmann, Stephan Jellbauer, and Emanuel Clausing (Gene Center Munich) for discussions and providing materials for yeast work. We thank Georg Arnold and Thomas Fröhlich (LAFUGA at the Gene Center Munich) for mass spectrometry analysis. We thank Achim Tresch, Johannes Söding, and other members of the Gene Center computational biology groups for bioinformatics support. We thank Sake van Wageningen and Frank Holstege (Utrecht University) for providing published microarray data in R/Bioconductor format. Part of this study was performed at the BESSY (Berlin, Germany) and at the Swiss Light Source (SLS) at the Paul Scherrer Institute (Villigen, Switzerland). This study was supported by grants of the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the Transregio 5, the EU grant 3D Repertoire, contract no. LSHG-CT-2005-512028, the European Molecular Biology Organization (EMBO), the Boehringer Ingelheim Fonds, and the LMU excellent research professorship to P Cramer.
Competing interests The authors have declared that no competing interests exist.
References
- Adzhubei AA, Sternberg MJ (1993) Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol 229: 472–493 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ (2004) A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283: 985–987 [DOI] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Baek HJ, Kang YK, Roeder RG (2006) Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem 281: 15172–15181 [DOI] [PubMed] [Google Scholar]
- Baumli S, Hoeppner S, Cramer P (2005) A conserved Mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J Biol Chem 280: 18171–18178 [DOI] [PubMed] [Google Scholar]
- Beve J, Hu GZ, Myers LC, Balciunas D, Werngren O, Hultenby K, Wibom R, Ronne H, Gustafsson CM (2005) The structural and functional role of Med5 in the yeast Mediator tail module. J Biol Chem 280: 41366–41372 [DOI] [PubMed] [Google Scholar]
- Bjorklund S, Gustafsson CM (2005) The yeast Mediator complex and its regulation. Trends Biochem Sci 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Bosveld F, van Hoek S, Sibon OCM (2008) Establishment of cell fate during early Drosophila embryogenesis requires transcriptional Mediator subunit dMED31. Dev Biol 313: 802–813 [DOI] [PubMed] [Google Scholar]
- Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon HM (2000) Drosophila homologs of transcriptional Mediator complex subunits are required for adult cell and segment identity specification. Genes Dev 14: 2906–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube M, Joulia L, Cribbs DL, Bourbon H-M (2002) Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151 [DOI] [PubMed] [Google Scholar]
- Bourbon H-M (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R (1995) High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur J Biochem 230: 788–796 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Dykstra CC, Sugino A (1991) Isolation, DNA sequence, and regulation of a Saccharomyces cerevisiae gene that encodes DNA strand transfer protein alpha. Mol Cell Biol 11: 2576–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Davis JA, Takagi Y, Kornberg RD, Asturias FA (2002) Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell 10: 409–415 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM (2006) The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA 103: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M (2008) Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31: 337–346 [DOI] [PubMed] [Google Scholar]
- Espanel X, Sudol M (2001) Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem 276: 14514–14523 [DOI] [PubMed] [Google Scholar]
- Fan HY, Cheng KK, Klein HL (1996) Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics 142: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Klein HL (1994) Characterization of mutations that suppress the temperature-sensitive growth of the hpr1 delta mutant of Saccharomyces cerevisiae. Genetics 137: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K (1997) Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17: 6982–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ III, Sayre MH, Tschochner H, Kornberg RD (1991) A Mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Gu J-Y, Park JM, Song EJ, Mizuguchi G, Yoon JH, Kim-Ha J, Lee K-J, Kim Y-J (2002) Novel Mediator proteins of the small Mediator complex in Drosophila SL2 cells. J Biol Chem 277: 27154–27161 [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 3: 97–108 [DOI] [PubMed] [Google Scholar]
- Guglielmi B, Soutourina J, Esnault C, Werner M (2007) TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA 104: 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon H-M, Holstege FCP, Werner M (2004) A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133 [DOI] [PubMed] [Google Scholar]
- Han SJ, Lee JS, Kang JS, Kim YJ (2001) Med9/Cse2 and Gal11 modules are required for transcriptional repression of distinct group of genes. J Biol Chem 276: 37020–37026 [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S (2006) Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol 7: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner S, Baumli S, Cramer P (2005) Structure of the Mediator subunit cyclin C and its implications for CDK8 function. J Mol Biol 350: 833–842 [DOI] [PubMed] [Google Scholar]
- Holm L, Park J (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16: 566–567 [DOI] [PubMed] [Google Scholar]
- Jeong CJ, Yang SH, Xie Y, Zhang L, Johnston SA, Kodadek T (2001) Evidence that Gal11 protein is a target of the Gal4 activation domain in the Mediator. Biochemistry 40: 9421–9427 [DOI] [PubMed] [Google Scholar]
- Jordan PW, Klein F, Leach DRF (2007) Novel roles for selected genes in meiotic DNA processing. PLoS Genet 3: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ (2001) The structural and functional organization of the yeast Mediator complex. J Biol Chem 276: 42003–42010 [DOI] [PubMed] [Google Scholar]
- Kelleher RJ III, Flanagan PM, Kornberg RD (1990) A novel Mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA (2007) The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci USA 104: 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler MM, Freund C (2006) The GYF domain. FEBS J 273: 245–256 [DOI] [PubMed] [Google Scholar]
- Kornberg RD (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF (2003) A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Larivière L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P (2006) Structure and TBP binding of the Mediator head subcomplex Med8–Med18-Med20. Nat Struct Mol Biol 13: 895–901 [DOI] [PubMed] [Google Scholar]
- Larivière L, Seizl M, van Wageningen S, Röther S, van de Pasch L, Feldmann H, Sträßer K, Hahn S, Holstege FCP, Cramer P (2008) Structure–system correlation identifies a gene regulatory Mediator submodule. Genes Dev 22: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T, Gustafsson CM (2004) The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae mediator. J Biol Chem 279: 49455–49459 [DOI] [PubMed] [Google Scholar]
- Linder T, Rasmussen NN, Samuelsen CO, Chatzidaki E, Baraznenok V, Beve J, Henriksen P, Gustafsson CM, Holmberg S (2008) Two conserved modules of Schizosaccharomyces pombe Mediator regulate distinct cellular pathways. Nucleic Acids Res 36: 2489–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M (2002) WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 513: 30–37 [DOI] [PubMed] [Google Scholar]
- Malagon F, Tong AH, Shafer BK, Strathern JN (2004) Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics 166: 1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci 25: 277–283 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Blobel J, Cramer P (2003) An extended winged helix domain in general transcription factor E/IIE alpha. J Biol Chem 278: 48267–48274 [DOI] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev 12: 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70: 475–501 [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ (2008) Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Gim BS, Kim JM, Yoon JH, Kim HS, Kang JG, Kim YJ (2001) Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol Cell Biol 21: 2312–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Ranish JA, Hahn S (1991) The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J Biol Chem 266: 19320–19327 [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S (1999) Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev 13: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Singh H, Erkine AM, Kremer SB, Duttweiler HM, Davis DA, Iqbal J, Gross RR, Gross DS (2006) A functional module of yeast Mediator that governs the dynamic range of heat-shock gene expression. Genetics 172: 2169–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Stiller JW, Hall BD (2002) Evolution of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci USA 99: 6091–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD (2006) Head module control of Mediator interactions. Mol Cell 23: 355–364 [DOI] [PubMed] [Google Scholar]
- Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sa-Correia I (2006) The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res 34: D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr 59: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Pan S-J, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Näär AM (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452: 604–609 [DOI] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73: 1361–1375 [DOI] [PubMed] [Google Scholar]
- van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TTJP, van Leenen D, Holstege FCP (2005) Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell 19: 511–522 [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP (2000) Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol 7: 639–643 [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, Macol C, Iyer L, Tjian R, van den Heuvel S, Hart AC, Wagner G, Naar AM (2006) An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704 [DOI] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF (2006) Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev 20: 2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ (2004) A triad of subunits from the Gal11/tail domain of Srb Mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24: 6871–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A (2008) H2A.Z: view from the top. Structure 16: 166–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 4
Supplementary Table 5