Abstract
Cellular populations have been widely observed to respond heterogeneously to perturbation. However, interpreting the observed heterogeneity is an extremely challenging problem because of the complexity of possible cellular phenotypes, the large dimension of potential perturbations, and the lack of methods for separating meaningful biological information from noise. Here, we develop an image-based approach to characterize cellular phenotypes based on patterns of signaling marker colocalization. Heterogeneous cellular populations are characterized as mixtures of phenotypically distinct subpopulations, and responses to perturbations are summarized succinctly as probabilistic redistributions of these mixtures. We apply our method to characterize the heterogeneous responses of cancer cells to a panel of drugs. We find that cells treated with drugs of (dis-)similar mechanism exhibit (dis-)similar patterns of heterogeneity. Despite the observed phenotypic diversity of cells observed within our data, low-complexity models of heterogeneity were sufficient to distinguish most classes of drug mechanism. Our approach offers a computational framework for assessing the complexity of cellular heterogeneity, investigating the degree to which perturbations induce redistributions of a limited, but nontrivial, repertoire of underlying states and revealing functional significance contained within distinct patterns of heterogeneous responses.
Keywords: automated microscopy, cellular heterogeneity, image analysis
Phenotypic heterogeneity in cellular populations has been observed in diverse physiological processes (1–9), pathophysiological conditions (10, 11), and responses to therapeutics (10, 12, 13). However, interpreting cellular heterogeneity—and its changes in response to perturbations—has been an extremely challenging problem. Does heterogeneity contain biologically (or clinically) important information? If so, what level of resolution is required to extract this information?
The complexity of cellular heterogeneity is not yet well understood: At one extreme, heterogeneity may reflect biological “noise” about a single “average” phenotypic state, whereas at the other extreme, it may reflect unbounded complexity, with each cell representing a distinct state. An intermediate alternative—that we investigate here—is that cellular populations can be well described as mixtures of a limited number of phenotypically distinct subpopulations. A practical challenge is to determine the complexity of observed cellular phenotypes and develop methods for identifying patterns of cellular heterogeneity.
Here, we developed methods for characterizing patterns of spatial heterogeneity observed within cellular populations. To capture cellular heterogeneity, we made use of high-content image-based assays, an increasingly powerful approach for capturing cellular responses to perturbations and facilitating the objective quantification of cellular phenotypes (14–28). [An interesting alternative—not considered here—is to study heterogeneity captured by fluorescence-activated cell sorting (FACS), which provides increased numbers of readouts per cell, but without spatial resolution (29)]. First, we applied image-processing algorithms to extract phenotypic measurements of the activation and colocalization patterns of cellular readouts from large numbers of cells in diverse conditions. Next, we used unsupervised clustering algorithms to identify phenotypic “stereotypes” within the overall population and assign probabilities to cells belonging to subpopulations modeled on these stereotypes. Subpopulation identification is based on application of a Gaussian mixture model (GMM), which has been applied recently in the context of online phenotype discovery (30). Finally, for each population (or condition), we estimated the fraction of cells in each subpopulation and summarized the results as a probability vector. The resulting “subpopulation profiles” are human- and machine-interpretable and enable the comparison of heterogeneous responses across multiple physiological, pathological, or environmental perturbations.
To develop our approach, we analyzed existing and new image databases of drug-treated HeLa cells. Drug treatments provided reliable and well-characterized mechanistic classes of perturbations that could be varied in both time and dose. Furthermore, existing drug classifications provided a biologically motivated method for estimating a crucial property of cellular heterogeneity. Namely, whether increasing numbers of subpopulations provided improved ability to distinguish different mechanistic classes of perturbations. [Traditionally, the tradeoffs between model fit and complexity are addressed by a nonbiological criterion, such as by Bayesian Information Criterion (B.I.C.) or gap statistics (30).] Thus, drug perturbations and their classifications provided an unbiased criterion for determining the degree of complexity required to interpret patterns of cellular heterogeneity.
Results
To study the heterogeneous effects of perturbations on cells, we made use of an existing image database of HeLa cells treated with 100 drugs at 13 concentrations and stained by using a multiplexed collection of immunofluorescent markers (DNA/SC35/anillin) (25). However, this dataset did not contain temporal information and additionally lacked consistent numbers of representative drugs in several key categories of drug mechanism. Thus, we created a 25-drug dataset capturing the responses of HeLa cells to drugs at varying concentrations [low or high, as specified in supporting information (SI) Table S1] and time points (3, 6, 12, or 24 h). For this dataset, we selected 5 drugs in each of 5 known functional categories, affecting DNA replication (e.g., methotrexate), histone deacetylation (e.g., trichostatin A), microtubules (e.g., paclitaxel), the glucocorticoid receptors (e.g., dexamethasone), and topoisomerase (e.g., doxorubicin). We additionally included an “other” category containing 10 drugs of miscellaneous (e.g., dichloroacetic acid) or unspecified (e.g., green tea-extracted polyphenols) mechanism. After fixation, cells were stained with 1 of 2 sets of fluorescent markers (DNA/phospho-p38/phospho-ERK or DNA/actin/α-tubulin; see Table S2) selected as general readouts of cell signaling state (25).
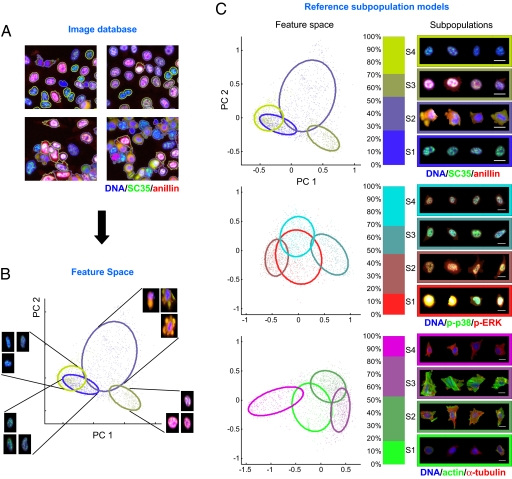
For each image, we identified individual cellular regions and extracted phenotypic features (Fig. 1 A and B). There are multiple approaches for extracting phenotypic information, including detecting either known biological phenotypes [e.g., apoptosis (31) or cell cycle phases (32) or general image properties, e.g., cell morphology and patterns of marker intensities (14, 18)]. Here, we found that a feature set designed to measure the degree of marker colocalization performed well in capturing phenotypic differences in signaling state among visually distinct cells. First, we measured ratios of marker intensities at each pixel to estimate the degree of marker colocalization. To avoid bias due to selection of a “favored” denominator, we measured all 3 possible pairs of marker ratios. Second, we quantized marker ratios into 16 bins. (Bin ranges were empirically chosen based on our data; the highest bin value was set to “infinity” to capture ratios when marker intensities in the denominator were close to background.) Third, we used 6 16 × 16 histograms to tally each of the three quantized marker ratio pairs for both cytoplasmic and nuclear subcellular regions. To down-weight low-intensity pixels, the contribution of each pixel to the histogram was weighted by its total intensity of markers. Finally, each histogram was normalized to have unit mass.
Fig. 1.
Heterogeneous cellular populations can be characterized as mixtures of subpopulations with distinct phenotypes. For convenience of visualization, results are shown for k = 4 subpopulations. (A and B) Illustration of approach. (A) HeLa cells are treated with drugs, labeled with fluorescence markers, and imaged. Individual cellular regions are identified. (B) A high-dimensional vector of phenotypic features is extracted from each cell. Cells are assigned probabilities of belonging to subpopulations that have been identified by using a GMM. Scatter plot: cells are visualized as points via feature representation and PCA reduction to two dimensions; colored cell outlines colors in A and points in B correspond to most probable subpopulation assignment; ellipses represent unit standard deviation from the mean for each subpopulation. Shown are 3 representative cells taken from various locations in each ellipse. (C) Shown are subpopulation models for different marker sets. Models are built from “reference” sets of cells sampled from multiple conditions. Scatter plots are as in B. Stacked bar graphs represent (prior) probabilities of cell membership in each subpopulation. Sample cells from within 1 standard deviation of the mean phenotype are shown at right for each subpopulation (image colors correspond to markers in order; e.g., blue/green/red corresponds to DNA/SC35/anillin). (Scale bar: 20 μm.)
Taken together, these histograms resulted in a 1,536-dimensional (equal to 6 × 16 × 16) feature vector. These high-dimensional feature vectors were subsequently reduced to ≈25 dimensions by principal-components analysis (PCA) (Materials and Methods). Although alternative choices of cellular features could certainly be substituted in the remaining analysis, we found that these ratiometric features performed well at grouping cells with visually similar signaling states in similar regions of feature space.
To enable comparisons of cellular heterogeneity across experimental conditions, a reference training set of 10,000 cells was constructed for each marker set from a uniform sampling of control and drug-treated wells (Materials and Methods). This sample represented only a small fraction (<2%; see Materials and Methods) of our overall datasets. Probabilistic clustering was then applied to this training set to characterize cell heterogeneity as stochastic variation of feature vectors around a collection of distinct phenotypes (Fig. 1 B and C). For a specified number of subpopulations k, expectation maximization (EM) clustering (33) characterized the total population as a weighted mixture of k subpopulations, each defined by a Gaussian distribution around a distinct cellular stereotype and a (prior) overall probability of subpopulation occurrence (30) (Fig. 1C, stacked bar graph). (For convenience of visualization we illustrate our approach with k = 4; the effects of varying k will be discussed subsequently.) The reference subpopulation model is thus defined by this collection of k phenotype means, covariances, and prior probabilities of occurrence.
The heterogeneity of a cell population subjected to a specific experimental condition can be estimated by using the reference subpopulation model. For each cell, the (posterior) probability that it is a member of a given subpopulation can be computed from the subpopulation model by using Bayes' rule. These probabilities for a single cell and all k subpopulations can be represented as a k-dimensional vector whose entries sum to 1. By averaging these k-dimensional vectors over a population of cells, a subpopulation profile is obtained as the expected overall proportion of each subpopulation for a given experimental condition. Replicates produced reproducible subpopulation profiles (Fig. S1), which were averaged to obtain 1 final profile per condition. Thus, the heterogeneity observed within any experimental condition can be concisely encoded by a probability distribution over the reference subpopulations.
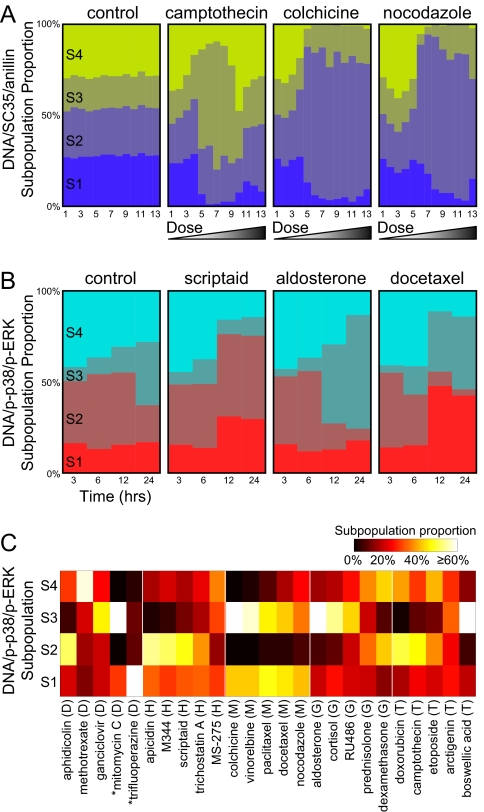
We next examined the degree to which subpopulation profiles can be used to distinguish heterogeneous responses to different drugs (Fig. 2A). We first compared subpopulation profiles for drug-treated conditions to controls. A comparison of camptothecin (topoisomerase) to controls, for example, showed a significant shift to a dominant phenotype (Fig. 2A; e.g., dose 5, population 3). Comparison of colchicine and nocodazole (both targeting microtubules) to controls showed substantial redistributions of phenotypic heterogeneity after drug treatment (Fig. 2A; e.g., dose 9, population 2). We next compared the effects of drugs to one another. We found that drugs of similar mechanism often yielded similar profiles (e.g., Fig. 2A, colchicine vs. nocodazole). Conversely, mechanistically different drugs yielded distinct profiles (e.g., Fig. 2A, camptothecin vs. colchicine).
Fig. 2.
Subpopulation profiles allow quantitative comparison of heterogeneous cellular responses. (A) Subpopulation profiles reveal dose-dependent heterogeneous responses to drugs [DNA/SC35/anillin marker set, 20-h drug exposure (25)]. Thirteen concentrations are shown, increasing from left to right, at 3-fold titration. (B) Subpopulation profiles reveal time-dependent heterogeneous responses to drugs. Subpopulation profiles for selected drugs are combined in time series (DNA/p-p38/p-ERK marker set; concentrations are given in Table S1). Profiles in A and B are based on reference models computed with k = 4 subpopulations. Colors and subpopulations ids S1-S4 on left-most images correspond to Fig. 1C. (C) Drugs of similar mechanism often induce similar subpopulation profiles. Subpopulation profiles for 25 drugs at the 24-h time point and DNA/p-p38/p-ERK marker set are displayed in a heat map and grouped by category (D, DNA replication; H, histone deacetylase; M, microtubule; G, glucocorticoid receptor; T, topoisomerase). Drugs with <200 cells per well are marked with asterisks (mitomycin C and trifluoperazine).
Dose responses typically revealed sharp transitions in subpopulation proportions as drug concentration increased, whereas untreated populations tended to remain in constant proportions (Fig. 2A). Multiphasic drug effects, such as seen for camptothecin, can be easily discerned as dramatic transitions in subpopulation proportions [Fig. 2A, second image, transitions at doses 5 and 10 in agreement with previous results (18, 25)]. The interpretation of results across time is complicated somewhat by the fact that controls did not maintain constant proportions, likely because of varying exposure times to the drug vehicle DMSO before fixation (Fig. 2B, first image). Although most drugs did not show significant differences from the controls at the 3- and 6-h time points, significant variation was observed at the 12- and 24-h time points (Fig. 2B). We note that analysis of automatically identified subpopulations revealed no significant enrichment for any specific cell cycle stage, as might be expected from our choice of pixel-based features (SI Text and Fig. S2). These observations suggest that subpopulation profiles contain information that may offer insights into drug modes of action.
We next wondered whether the significant redistributions of subpopulations were consistent across drug perturbations of similar mechanism. To assess the “information content” of observed patterns of heterogeneity, we computed subpopulation profiles for our 25 annotated drugs at the 24-h time point and grouped the results by category of mechanism (Fig. 2C). (Here, we visualized these results using a heatmap, rather than stacked bar graphs, to provide easier visual comparison among larger numbers of drugs.) Whereas DNA replication-inhibiting drugs showed high variability in their subpopulation proportions, the responses of drugs within all other categories generally showed high concordance. Rigorous assessment of the similarities and differences between drug effects requires a quantitative measure of “distance” between subpopulation profiles. When considered as probability vectors, subpopulation profiles may be viewed as information theoretic descriptions of cell population heterogeneity. Similarities among profiles can be quantitatively compared by using the Kullback–Leibler (KL) divergence (34) and visually displayed by using multidimensional scaling (MDS) plots (SI Text). We observed that profiles showed significant grouping by drug mechanism (Fig. 3A, k = 4). Thus, drugs of similar mechanism appear to induce similar patterns of cellular heterogeneity.
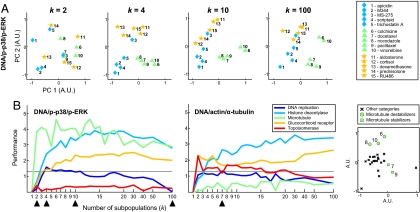
Fig. 3.
A small number of phenotypic subpopulations can distinguish drug effects by mechanism. (A) As the number of subpopulations in the reference model increases (k = 2, 4, 10, and 100), MDS plots (40) reveal relative similarities among drugs (computed by using KL divergence) within 3 mechanism categories (H, blue diamonds; M, green triangles; G, orange stars). Axes are labeled in arbitrary units. (B) Drug classification performance is computed for each value of k (and drug category; see also Table S5). Performance is measured by −log10 of the P value for a rank-sum statistic [Mann–Whitney U test (39)] obtained by comparing distributions of intra- and intercategory subpopulation profile similarity scores. Profile similarity is computed for 12- and 24-h time points of low drug concentration and combined by rank addition. Dotted lines indicate significance threshold corresponding to P = 0.05. Black arrowheads (Left) (k = 2, 4, 10, 100) correspond to MDS plots in A. Black arrowheads (Right) (k = 7) correspond to MDS plot at bottom right and uses reference model shown in Fig. S1D.
A natural question arose from our analysis: Would increasing the number of subpopulations, k, in the reference model discern finer distinctions between drug effects and thus yield a more accurate classification of drugs? In theory, increasing k allows finer resolution at which to measure cellular heterogeneity, although classification accuracy for very large values of k may fail to improve—or even degrade—because of the introduction of nonbiological separation of cellular phenotypes. We found for the DNA/p-p38/p-ERK and DNA/actin/α-tubulin marker sets that categorization performance for most drug categories sharply improved as k increased from 1 (no predictive power) to ≈5. Afterward, no significant improvement in classification was observed (Fig. 3B). In a few cases, categorization performance was poor. For example, the DNA/actin/α-tubulin marker set showed poor performance for the microtubule category (Fig. 3B Middle). However, as previously shown, this poor performance was due to the fact that the microtubule category was too coarse (18); in fact, subpopulation profiles did distinguish between microtubule-stabilizing taxanes (paclitaxel and docetaxel) and drugs that destabilize microtubules (colchicine, vinorelbine, and nocodazole) (Fig. 3B Right; Fig. S1D). Thus, a small number of subpopulations were sufficient to give accurate classification for most drug categories and both marker sets.
We extended our analysis to include the additional 10 drugs of unspecified or miscellaneous categorization (Table S1). MDS plots for the DNA/p-p38/p-ERK marker set showed these additional drugs to be interspersed with glucocorticoid receptor drugs (Fig. S3, k = 4). We hypothesized that glucocorticoid receptor ligand-like effects would be more likely to be observed for compounds more proximal to drugs in this category. Of 4 proximal compounds tested, 2 induced cytoplasmic-to-nuclear translocation of a GFP-tagged glucocorticoid receptor (35) in COS7 cells (green tea polyphenols and valproic acid), whereas 2 did not (nicotinamide and azelaic acid) (Fig. S4). Neither of the more distal compounds tested (mitoxantrone and methoxyacetic acid) induced glucocorticoid receptor translocation. Of note, the HDAC drug MS-275 closest to the glucocorticoid receptor group also induced translocation, whereas the less proximal HDAC drug trichostatin A did not (data not shown). Interestingly, these insights were derived by using a general signaling marker set, not specific to mechanisms of nuclear receptor regulatory pathways. These results suggest that analyzing patterns of cellular heterogeneity may be useful for generating mechanistic insights into modes of drug action.
Although our approach effectively captured phenotypic stereotypes common to the entire collection of drug perturbations, we wondered to what extent exceptional (i.e., rare or unmodeled) phenotypes may have been missed (30). To test for exceptional phenotypes in our dataset, we evaluated goodness of model fit for all 35 of our drugs and all treatment conditions based on the ratio of log likelihoods for drug treatment vs. the reference training data (Materials and Methods). We found that only a relatively small number of drug treatment conditions did not fit the reference model reasonably well. To ensure that results were not biased by the inclusion of all 35 drug treatments in our original model, we recomputed model fit using a new reference model based on data sampled from controls and only 5 low-concentration drug treatments, 1 from each category of mechanism (Fig. 4A).
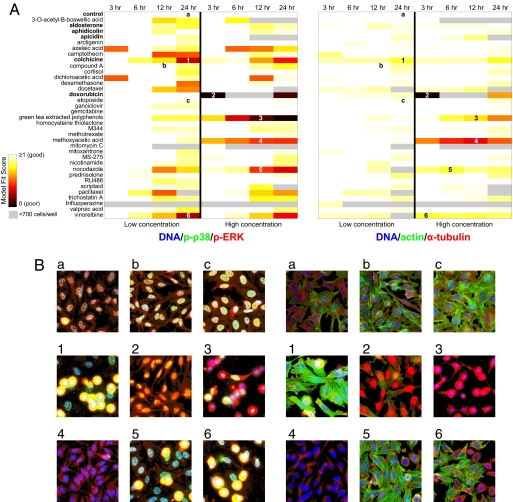
Fig. 4.
Exceptional phenotypes are detected by evaluating the fit of each drug to a reference model built by using only the low concentration of 1 drug in each functional category (control, aldosterone, aphidicolin, apicidin, colchicine, and doxorubicin) at all time points. (A) Reference models for DNA/p-p38/p-ERK and DNA/actin/α-tubulin marker sets are both computed with k = 4 subpopulations. Heat map reflects the ratio of model fit (log likelihood) of drug treatment data to training data. Four time points (3, 6, 12, and 24 h) are displayed for each of 2 marker sets, at both low and high concentrations of each drug. Gray boxes indicate poor fits due to low cell counts (<700 cells per well). (B) Shown are sample images of drug treated cells that fit the reference model well (a–c), as well as the 6 lowest scoring drugs (1–6) as indicated in A.
Examining the 6 drugs whose model fit scores were lowest revealed 2 types of exceptional phenotypes (Fig. 4B): (i) microtubule-destabilizing drugs (colchicine, nocodazole, vinorelbine) whose phenotypes were similar to each other but different from most of the remaining drugs; and (ii) 3 drugs at high concentrations (doxorubicin, green tea-extracted polyphenols, methoxyacetic acid) whose phenotypes were visually much different from those seen for any other drug treatment condition. Although colchicine-treated cells were included in the reference model, the phenotypes presented at the 24-h time point were in too low a proportion to constitute 1 of the 4 modeled subpopulations; the other exceptional phenotypes were likely not present in the reference sample. Drugs that induced poorly modeled heterogeneous response in one marker set, tended to also do so in the other marker set. Depending on the application, there are a variety of (supervised and unsupervised) strategies for updating or rebuilding a reference subpopulation model with exceptional phenotypes, such as iterative identification and merging of novel phenotypes in large-scale screens (30). Phenotypic complexity within a collection of perturbations can thus be estimated in a relative sense (i.e., the number of subpopulations required to distinguish the perturbations) or in an absolute sense (i.e., the total number of distinct subpopulations).
Discussion
Phenotypic heterogeneity is often ignored or viewed as an impediment to understanding the effects of perturbations on cellular populations. Traditional measurements of a mean and a variance typically do not capture the complexity of cellular responses to perturbations often observed in microscopy. On the other hand, cell-to-cell variation suggests a potential for limitless cellular complexity. Analysis of our datasets suggested that the effects of perturbations may be characterized largely as redistributions of a limited, but nontrivial, repertoire of underlying states. Subpopulation profiles provided an intuitive and computationally tractable method for identifying functional significance from patterns of heterogeneity. Analysis of subpopulation profiles revealed that drugs of (dis-)similar mechanism induced (dis-)similar patterns of subpopulation redistributions. Thus, our work suggests that patterns of heterogeneity can be quantified and used to extract meaningful biological information.
Our approach for grouping cells is unsupervised, and the identification of subpopulations depends on the composition of the dataset, the markers, the extracted features, and the number of subpopulations. In some cases, subpopulation phenotypes and their redistributions may be interpretable. For example, the microtubule-destabilizing drugs vinorelbine and nocodazole induced a decrease in subpopulations characterized by α-tubulin-positive staining. Our approach also identifies subpopulations whose phenotypes cannot be easily interpreted by current biological knowledge. For example, it is unclear how aldosterone induces an increase in subpopulation 3, characterized by weak diffused p-ERK staining concomitant with clear p-p38 nuclear staining. In general, the best method for interpreting subpopulations and their redistributions may be through relative comparisons with profiles of perturbations with known mechanisms.
GMMs provide a reasonable first approximation for grouping cells in high-dimensional feature space. However, certain subpopulations may still display some degree of phenotypic heterogeneity (e.g., Fig. 1C Upper, subpopulation 2). This may be due to several factors, including: Not all subpopulations will be well modeled by Gaussian distributions; some subpopulations may cover large regions of feature space and need further subdivisions; and automated grouping based on phenotypic features may not agree with visual grouping. Model parameters in future studies of heterogeneity may be optimized to improve visual tightness of identified subpopulation phenotypes.
Our analysis suggests that observed heterogeneity may reflect variation around a discrete set of phenotypic states. Our work provides a framework for future studies with larger sample sizes, greater numbers of perturbations, and higher numbers of markers to rigorously test questions about the nature of heterogeneity, including: Do unperturbed cells occupy essentially the same phenotypic space as cells with significantly perturbed pathways? And, do perturbations induce redistributions among a limited repertoire of underlying stereotypical states? We envision that the ability to identify and interpret information contained in patterns of cellular heterogeneity will provide insights into physiology and disease that may be missed by traditional population-averaged or small-cell-number studies.
Materials and Methods
Drugs and Concentrations.
The drug-dose data of 100 compounds in 13 3-fold serial dilution are as previously reported (25). The new time-course data consisted of 35 compounds at 2 concentrations (see Table S1).
Cell Culture.
For the time-course data, HeLa cells were diluted to 5,000 cells per 50 μl of media and plated onto 384-well glass-bottom plates. Plates were incubated at room temperature for 30 min to minimize edge effects in wells (36), then placed in a 37 °C/5% CO2 incubator overnight. Cells were fixed at 3, 6, 12, and 24 h after drug treatment. See SI Text for details about cell culture, drug treatment, and plate layout.
Fluorescent Markers.
The marker set for the drug-dose data (DNA/SC35/anillin) are as previously reported (25). For the time-series data, 2 sets of fluorescent markers were used: (DNA/p-p38/p-ERK) and (DNA/actin/α-tubulin). See SI Text for details about staining protocol.
Image Background Correction and Cell Segmentation.
Image background correction was done by using the ImageJ background subtraction rolling-ball algorithm (37), with rolling-ball size set to 500 pixels. Original image dimensions were 1,392 × 1,040 pixels. Cell segmentation methods used to define DNA and non-DNA (cytoplasmic) regions are as in ref. 18 and are summarized in SI Text.
Feature Extraction.
For each cell, a 1,536 dimensional feature vector was computed based on histograms of the ratios of marker intensities per pixel, in both cytoplasmic and nuclear cellular regions. See Results and SI Text.
Feature Data Sampling.
Feature data from multiple wells was drawn by using sampling with replacement, with weights assigned to each well according to the proportionate number of total cells detected within the well. The total number of cells (training data) sampled from all plates for computing PCA coordinate transformations and subpopulation reference models was 10,000 (≈2% of cells for DNA/p-p38/p-ERK and DNA/actin/α-tubulin marker sets, and ≈0.06% of cells for DNA/SC35/anillin marker set). For the DNA/p-p38/p-ERK and DNA/actin/α-tubulin marker sets, all controls and low concentration drug treatments at all 4 time points were sampled. For the DNA/SC35/anillin marker set, all wells were sampled.
PCA Coordinate Transformations.
Principal-components analysis (PCA) was used to reduce the dimensionality of feature data from 1,536 to 27 for the DNA/p-p38/p-ERK marker set, and to 26 for the DNA/actin/α-tubulin marker set. The choice of dimension was made for each marker set by randomizing the order of feature dimensions for each sampled cell and computing the eigenvalues of the resulting (randomized feature) covariance matrix. Subsequent modeling and profiling computations all used the PCA reduced data.
Reference Models.
Subpopulation reference GMM models were computed from sampled feature data by using the EM algorithm (33). Code was based on the publicly-available Netlab Toolbox. For each model, EM clustering was run 10 times, starting from a K-means clustering (38) using randomly chosen means. The final clustering with the best log-likelihood value was chosen as the subpopulation reference model. To eliminate the influence of convergence failures, each run was attempted up to 5 times with new initial conditions until convergence was reached. For each marker set, subpopulation models were computed for each number of subpopulations clusters in the set (2,3,4,…,18,19,20,30,40,50,100).
Subpopulation Profiles.
Subpopulation profiles for each drug-treatment replicate (single well) were computed by averaging the posterior probabilities for the corresponding subpopulation model for 1,000 samplings (with replacement) consisting of PCA reduced feature data from 1,000 cells per sample. Profiles for each drug-treatment condition were computed by an average of profiles for each replicate well, averaged by the number of cells observed in each well. When exactly 1 of the 2 replicate wells had <1,000 cells in total, the profile for the other well was used.
Similarity Comparison.
Individual profiles were compared by using a symmetrized version of Kullback-Leibler divergence (34). In particular, given subpopulation profiles P and Q for a model with k subpopulations, the similarity measure S(P,Q) is defined as
where
MDS Plots.
The Matlab mdscale function was used to compute data for multidimensional scaling plots (40). Kruskal's stress-1 metric (total error in similarity representation) was computed to be <0.05 for each plot in Fig. 3A, and ≈0.12 for the plot in Fig. 3B.
Identification of Exceptional Phenotypes.
For a given drug treatment condition, the model fit score was computed as the ratio of the model log likelihoods for drug treatment data vs. reference training data. Samplings of 1,000 cells were used to compute log likelihoods.
Supplementary Material
Acknowledgments.
We thank Michael Johnson (Nikon) and Jurg Rohrer (BD Biosciences), all members of the S.J.A. and L.F.W. laboratory for extremely helpful discussions and feedback, and the anonymous reviewers for their helpful suggestions. This work was supported by National Institutes of Health Grants R01 GM081549 (to L.F.W.), R01 GM071794 (to S.J.A.), K22 CA118717-01 (to E.D.M.), and P50 CA70907 UT SPORE in Lung Cancer; the Welch Foundation (I-1619 and I-1644 to L.F.W. and S.J.A.); and the University of Texas Southwestern Endowment for Scholars in Biomedical Research (to L.F.W. and to S.J.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807038105/DCSupplemental.
References
- 1.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 4.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 5.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 6.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006.0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samadani A, Mettetal J, van Oudenaarden A. Cellular asymmetry and individuality in directional sensing. Proc Natl Acad Sci USA. 2006;103:11549–11554. doi: 10.1073/pnas.0601909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 11.Zhang M, Rosen JM. Stem cells in the etiology and treatment of cancer. Curr Opin Genet Dev. 2006;16:60–64. doi: 10.1016/j.gde.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Velliste M, Murphy RF. Automated interpretation of subcellular patterns in fluorescence microscope images for location proteomics. Cytometry A. 2006;69:631–640. doi: 10.1002/cyto.a.20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad C, et al. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 2004;14:1130–1136. doi: 10.1101/gr.2383804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: A user's guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RL, et al. A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem. 2008;375:237–248. doi: 10.1016/j.ab.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–453. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 19.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 20.Mora-Bermudez F, Ellenberg J. Measuring structural dynamics of chromosomes in living cells by fluorescence microscopy. Methods. 2007;41:158–167. doi: 10.1016/j.ymeth.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 21.Neumann B, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 22.Newberg J, Murphy RF. A framework for the automated analysis of subcellular patterns in human protein atlas images. J Proteome Res. 2008;7:2300–2308. doi: 10.1021/pr7007626. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien P, Haskins JR. In vitro cytotoxicity assessment. Methods Mol Biol. 2007;356:415–425. doi: 10.1385/1-59745-217-3:415. [DOI] [PubMed] [Google Scholar]
- 24.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 25.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 26.Perrimon N, Mathey-Prevot B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics. 2007;175:7–16. doi: 10.1534/genetics.106.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 29.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 30.Yin Z, et al. Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics. 2008;9:264. doi: 10.1186/1471-2105-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plasier B, Lloyd DR, Paul GC, Thomas CR, Al-Rubeai M. Automatic image analysis for quantification of apoptosis in animal cell culture by annexin-V affinity assay. J Immunol Methods. 1999;229:81–95. doi: 10.1016/s0022-1759(99)00107-6. [DOI] [PubMed] [Google Scholar]
- 32.Wang M, Zhou X, King RW, Wong ST. Context based mixture model for cell phase identification in automated fluorescence microscopy. BMC Bioinformatics. 2007;8:32. doi: 10.1186/1471-2105-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dempster A, Laird N, Rubin D. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc. 1977;39:1–38. [Google Scholar]
- 34.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22:79–86. [Google Scholar]
- 35.Htun H, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4950. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen. 2003;8:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg S. Biomedical image processing. IEEE Comput. 1983;16:22–34. [Google Scholar]
- 38.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- 39.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 1947;18:50–60. [Google Scholar]
- 40.Borg I, Groenen P. Modern Multidimensional Scaling: Theory and Applications. New York: Springer; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.