Abstract
Asian rice (Oryza sativa) cultivars originated from wild rice and can be divided into two subspecies by several criteria, one of which is the phenol reaction (PHR) phenotype. Grains of indica cultivars turn brown in a phenol solution that accelerates a similar process that occurs during prolonged storage. By contrast, the grains of japonica do not discolor. This distinction may reflect the divergent domestication of these two subspecies. The PHR is controlled by a single gene, Phr1; here, we report the cloning of Phr1, which encodes a polyphenol oxidase. The Phr1 gene is indeed responsible for the PHR phenotype, as transformation with a functional Phr1 can complement a PHR negative cultivar. Phr1 is defective in all japonica lines but functional in nearly all indica and wild strains. Phylogenetic analysis showed that the defects in Phr1 arose independently three times. The multiple recent origins and rapid spread of phr1 in japonica suggest the action of positive selection, which is further supported by several population genetic tests. This case may hence represent an example of artificial selection driving the differentiation among domesticated varieties.
INTRODUCTION
Morphological and physiological changes during domestication have intrigued generations of geneticists and evolutionists. There are now a number of genes known to control phenotypic variation among domesticated cultivars or between cultivars and their wild progenitors (Purugganan et al., 2000; Nesbitt and Tanksley, 2002; Olsen and Purugganan, 2002; Clark et al., 2004; Wang et al., 2005; Li et al., 2006; Sweeney et al., 2006). In addition to illuminating the physiological mechanisms, which are often highly relevant to agriculture, examination of these genes also sheds light on the underlying forces driving the evolution of phenotypes during domestication (Wang et al., 2005).
Among domesticated plants and animals, Asian cultivated rice (Oryza sativa) is of particular interest as it comprises two subspecies, indica and japonica, which differ in various morphological, physiological, and life history traits (Oka, 1988). These two subspecies are also partially reproductively isolated and provide a unique opportunity to study divergence associated with speciation. Races and subspecies are attractive subjects for speciation studies as they are at the incipient stage (Ting et al., 2004).
Although the two subspecies of rice differ broadly in >40 characters (Kato et al., 1928; Oka, 1958), the indica- and japonica-type cultivars are generally distinguished by four traits: resistance to KClO3, tolerance to cold, hair length of glume tips, and the phenol reaction (PHR) (Oka, 1953; Morishima and Oka, 1960). These characteristics have been suggested to be germane to rice domestication (Oka and Chang, 1962; Chang, 1976). Of particular interest to us is PHR, which reflects grain reaction to phenol treatment (Oka, 1953; Morishima and Oka, 1960). The grains, especially hulls, of the indica-type cultivars show positive PHR by turning brown after being soaked in phenol solution, whereas those of the japonica type are PHR-negative and their color remains unchanged (Oka, 1953). Phenol treatment presumably accelerates the browning of rice grains that happens under normal storage conditions.
While both indica and japonica are generally golden-hulled at harvest, the bran and unpolished or coarse grains of indica rice darken gradually during storage. Such a process may resemble the discoloration of wheat (Triticum aestivum), which is catalyzed by polyphenol oxidases (PPOs) (Anderson and Morris, 2001; Simeone et al., 2002). As the white color of grains is preferred by consumers, wheat breeding has aimed to reduce or eliminate the grain discoloration. In rice, previous studies have shown that PHR is controlled by a single Mendelian gene, Phenol reaction 1 (Phr1; historically Ph), located on the long arm of chromosome 4 (McCouch et al., 1988; Saito et al., 1991; Tanksley et al., 1993; Lin et al., 1994). However, the nature of Phr1 and the mechanism underlying PHR remain to be elucidated. In this study, we report the cloning, molecular characterization, transformation rescue, and evolutionary analysis of Phr1 in rice.
RESULTS
Cloning and Confirmation of Phr1
To map and clone Phr1, we constructed a large mapping population derived from cross between PHR-positive indica cv MH63 and PHR-negative japonica cv CJ06 (Figure 1). The cross produced a total of 5589 F2 plants (PHR positive: 4203; PHR negative: 1386) with a segregation ratio of ∼3:1 (χ2 = 0.06; P > 0.75), consistent with the previous conclusion that a single nuclear recessive gene controls the negative PHR. Based on the previous mapping results that Phr1 was on chromosome 4 (McCouch et al., 1988), we developed two new PCR-based molecular markers, S100 and S115, located on both sides of Phr1 in the rice genetic linkage map (Chen et al., 2002; see Supplemental Table 1 online). We subsequently screened all PHR-negative F2 plants with them.
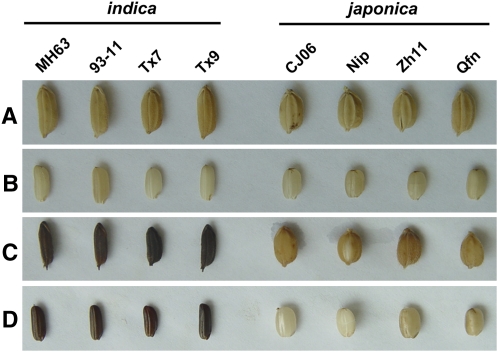
Figure 1.
PHR of Grains of Asian Cultivated Rice Subspecies.
Rice subspecies shown are as follows (from left to right): indica cv MH63, 93-11, Tx7 and Tx9, and japonica cv CJ06, Nipponbare (Nip), Zh11 and Qfn.
(A) The natural color of hulls.
(B) The color of brown rice.
(C) The PHR of hulls.
(D) The PHR of brown rice.
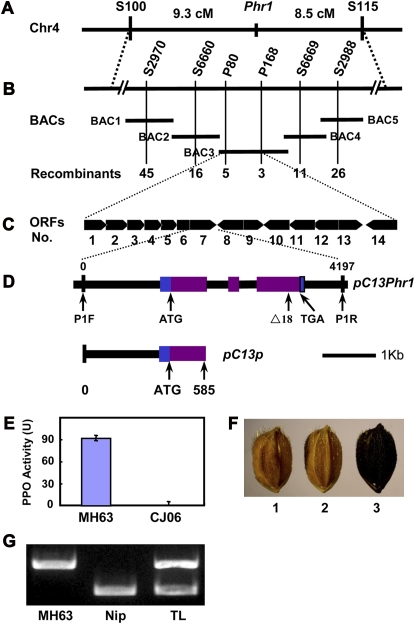
The Phr1 gene was mapped to the interval between markers S100 and S115 with genetic distances of 9.3 and 8.5 centimorgan (cM) to Phr1, respectively (Figure 2A). To fine-map Phr1, we obtained 66 recombinants between S100 and Phr1 and 40 recombinants between Phr1 and S115. With six more markers developed for this study, the breakpoints can be finely delineated (Figure 2B; see Supplemental Table 1 online). The Phr1 locus was pinpointed to an 88-kb interval between markers P80 and P168 on a single BAC clone (OSJNBa0053K19; Figure 2B). Within this DNA segment, 14 open reading frames (ORFs) have been predicted (Figure 2C) (Feng et al., 2002). Among them, OSJNBa0053K19.18 (referred to as K18 hereafter) is highly similar to plant PPO genes (Cary et al., 1992; Chevalier et al., 1999; Constabel et al., 2000; Gooding et al., 2001; Demeke and Morris, 2002). The similarity in protein sequence ranges from 43 to 68%, and the highest is found with wheat PPO.
Figure 2.
Map-Based Cloning and Confirmation of Phr1.
(A) The Phr1 locus was mapped on the long arm of chromosome 4 (Chr4) between the markers of S100 and S115.
(B) Phr1 was further localized in a single BAC clone within an interval between the markers P80 and P168. BAC1, OSJNBa0058K23; BAC2, OSJNBa0085C12; BAC3, OSJNBa0053K19; BAC4, OSJNBa0060E08; BAC5, OSJNBa0089N06. Numbers indicate the number of recombinants identified from 1386 F2 phr1 mutant plants.
(C) Predicted ORFs highlighted with arrows.
(D) The Phr1 structure and the complementation construct pC13Phr1. The start codon (ATG) and the stop codon (TGA) are indicated. Purple boxes stand for the coding sequence, blue for 5′ and 3′ untranslated regions, and black lines between boxes for introns. The mutation site in japonica cultivar Nipponbare is also indicated. Structure of the control plasmid pC13p, which contains the promoter region and a truncated Phr1 gene that encodes the first 195 amino acid residues.
(E) The comparison of PPO activities between MH63 and CJ06. One unit of PPO activity was defined as the amount of the enzyme that gives a change in absorbance of 0.001 per min.
(F) Complementation test. PHR-negative Nipponbare (Nip) (1) becomes PHR-positive when transformed with the plasmid pC13Phr1 (3), but transformed with the control vector (2) is shown the same as Nip.
(G) Confirmation of transgenic lines by specific PCR amplification of the Phr1 fragments with (bottom band) or without 18-bp deletion (top band). The DNA template was extracted from MH63, Nip, and transgenic lines (TL).
In higher plants, PPOs have been proposed to be responsible for the browning of damaged kernels, fruits, or vegetables, which may be response for disease resistance (Nicolas et al., 1994; Thipyapong et al., 1995; Gooding et al., 2001; Demeke and Morris, 2002; Li and Steffens, 2002). Therefore, we PCR amplified and sequenced the corresponding K18 sequences from MH63 and CJ06 varieties with primers designed based on the sequence of AK108237 given in GenBank (see Supplemental Table 1 online). DNA sequence comparison revealed an 18-bp deletion (Δ18) in this ORF in CJ06 compared with MH63 (Figure 2D), and the same deletion was also found in Nipponbare (PHR-negative, japonica type) compared with GLA (PHR-positive, indica type) (see Supplemental Figure 1 online). Moreover, the relative PPO activity is high in MH63 grains but nearly undetectable in CJ06 (Figure 2E). These results strongly suggested that K18 is very likely to be the Phr1 gene.
To test the prediction that k18 is the Phr1 gene, a complementation test was conducted. Transformation of the plasmid pC13Phr1 that contains the entire 93-11 Phr1 gene, including 1411-bp 5′ upstream and 454-bp downstream sequences (Figure 2D), succeeded in rescuing the negative PHR phenotype of Nipponbare, whereas the control vector pC13p (Figure 2D) containing a truncated Phr1 failed (Figure 2F). The authenticity of the transgenic plants was assured by the specific amplification of the transgene (Figure 2G). These results confirmed that the K18 is indeed the Phr1 gene and the 18-bp deletion (Δ18) is responsible for the loss-of-function phenotype of the japonica rice Nipponbare.
Phr1 Encodes a PPO
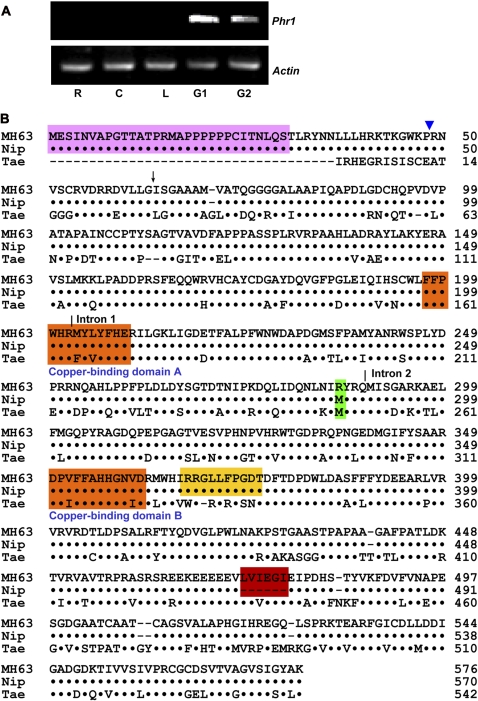
The steady state level of Phr1 mRNA was not detectable by RNA gel blot analysis; nevertheless, by RT-PCR, we could detect Phr1 expression in grains at the early flowering stage (G1) and at the mature stage (G2) (Figure 3A). An earlier study had also reported Phr1 expression in leaf tissue when the plants were under stress (Nobuta et al., 2007).
Figure 3.
Phr1 Expression Patterns and Alignment of Phr1 Protein Sequences of MH63 and Nipponbare with Wheat PPO.
(A) Phr1 expression pattern revealed by RT-PCR using total RNA isolated from roots (R), culms (C), leaves (L), and panicles 7 and 21 d after flowering (G1 and G2, respectively). Amplification of actin cDNA was used to ensure that approximately equal amounts of cDNA were loaded.
(B) Alignment of Phr1 protein sequences of MH63 and Nipponbare with wheat (T. aestivum, abbreviated as Tae) PPO (GenBank AAS00454). A dot stands for an identical amino acid residue and a dash for a gap. Putative N-terminal signal peptide and copper binding domains (A and B) are indicated in pink and orange, respectively. Black arrow indicates the predicted cleavage site of N-terminal signal peptide, and vertical lines show the intron positions. The amino acid residues encoded by the 18- and 29-bp deletion in japonica cultivars are highlighted in red and yellow, respectively. The blue triangle indicates the position of the 1-bp insertion in Tx36, a japonica cultivar.
The sequenced RT-PCR product, which was amplified from the indica rice MH63, confirmed the high similarity of cDNA sequence of Phr1 with LOC_Os04g53300 (The Institute for Genomic Research). The latter encodes a protein of polyphenol oxidase with 570 amino acid residues. Alignment of the Phr1 cDNA with its genomic DNA revealed that Phr1 contains two introns (Figure 3B; see Supplemental Figure 1 online). As a member of the tyrosinase family, the deduced amino acid sequence of Phr1 contains two putative copper binding domains (Figure 3B) (Steffens et al., 1994; Klabunde et al., 1998). Phr1 also contains a putative thylakoid-targeting sequence at its NH2 terminal, which is rich in hydroxyl amino acid residues as in other plant PPOs (Constabel et al., 2000). In addition, an NH2-terminal transit peptide of 30 amino acid residues is predicted (Figure 3B) (Keegstra, 1989; De Boer et al., 1991).
Therefore, Phr1 appears to encode an ∼62.6-kD precursor protein, which is processed into a mature PPO of ∼56.3 kD after removal of the NH2-terminal signal peptide. The deduced Phr1 amino acid sequence shows 68% identity to wheat PPO (GenBank AAS00454), indicating that Phr1 is a rice homolog of the wheat PPO (Figure 3B). BLASTX searches for Phr1 homologs identified three Phr1-like (Phr1L) proteins encoded in the rice genome. Sequence alignment analysis showed that Phr1L1 and Phr1L2 are truncated PPOs (see Supplemental Figure 2 online). It also showed that, among the 13 conserved amino acid residues in the copper binding domain B of the Phr1L3 protein, five residues have been changed (see Supplemental Figure 2 online). Most importantly, we did not detect PPO enzyme activity in the three phr1 mutation lines (Table 1). These results indicate that Phr1 is the sole source of PPO activity. Since none of the three paralogs can be functionally interchangeable with Phr1, we did not pursue these Phr1-like genes.
Table 1.
Summary of Phr1 Variations and PHR Phenotypes
| Taxon | Genome | Genotype | No. of Lines Genotyped | No. of Lines Sequenced | PHR Phenotypea |
|---|---|---|---|---|---|
| japonica (n = 35) | AA | Δ18 | 32 | 11 | − |
| Δ29 | 2 | 2 | − | ||
| Ins-1bp | 1 | 1 | − | ||
| indica (n = 20) | AA | + | 19 | 6 | + |
| Δ18/+ | 1 | 1 | + | ||
| O. rufipogon (n = 523) | AA | + | 517 | 21 | + |
| Δ18 or 18/+ | 3 | 3 | na | ||
| Δ29 or 29/+ | 3 | 3 | na | ||
| O. nivara | AA | + | 10 | 2 | + |
| O. barthii | AA | + | 67 | 2 | +b |
| O. logistaminata | AA | + | 5 | 0 | na |
| O. glaberrima (Africa cultivar) | AA | + | 1 | 1 | + |
| O. meridonalis | AA | + | 16 | 0 | na |
| O. glumaepatula | AA | + | 81 | 0 | na |
| O. panctata | BB | + | 2 | 0 | na |
| O. officinalis | CC | + | 2 | 1 | + |
| O. alta | CCDD | + | 2 | 1 | + |
| O. latifolia | CCDD | + | 1 | 0 | na |
| O. granulata | GG | + | 1 | 0 | na |
Genotypes are indicated with “+” for alleles without molecular lesions, “Δ18” for alleles bearing an 18-bp deletion, “Δ29” for alleles bearing a 29-bp deletion, “Ins-1bp” for alleles bearing a 1-bp insertion, and “Δ18/+” and “Δ29/+” for heterozygous sites. PHR phenotypes with plus and minus signs for positive and negative phenol reactions, respectively. na, not available.
Phenotype of sequenced lines.
One of two lines shows positive PHR phenotype, and another is not available.
Phr1 and Grain Discoloration during Storage
We hypothesized that functional Phr1 contributes to the discoloration of hulls and coarse grains of indica-type cultivars during prolonged storage and that the nonfunctional Phr1thus resulted in nondiscoloration of japonica grains. To determine whether this hypothesis is correct, we transferred the functional indica Phr1 gene into the PHR-negative Nipponbare rice and compared the discoloration of their grains during storage. Although no color difference between transformed and nontransformed grains could be observed at the mature grain stage, the color of transgenic hulls after harvest became darker gradually during storage (Figures 4A to 4C), indicating that Phr1 is responsible for the discoloration of PHR-positive hulls. It should be pointed out that the coarse grains of transgenic rice also show an apparent discoloration during storage (Figures 4D to 4F) as well as a strong PHR-positive reaction (Figure 4G). We further suggest that Δ18 is the predominant, if not the sole, lesion responsible for the loss of PPO activity in Nipponbare. In Figures 5 to 7 (see below) and Supplemental Figure 3 online, four sets of observations are given to support this suggestion.
Figure 4.
Discoloration of Transgenic Grains during Storage.
(A) to (C) The comparison of the hull color between untransformed Nip (Nip) and transgenic lines of Nip transformed with the functional Phr1 gene (TL) after storage for 1 month (A), 6 months (B), and 12 months (C).
(D) to (F) The comparison of the coarse grain color between untransformed Nip and transgenic lines of Nip transformed with the functional Phr1 gene after storage for 1 month (D), 6 months (E), and 12 months (F).
(G) The PHRs of coarse grains of untransformed Nip and transgenic Nip.
Figure 5.
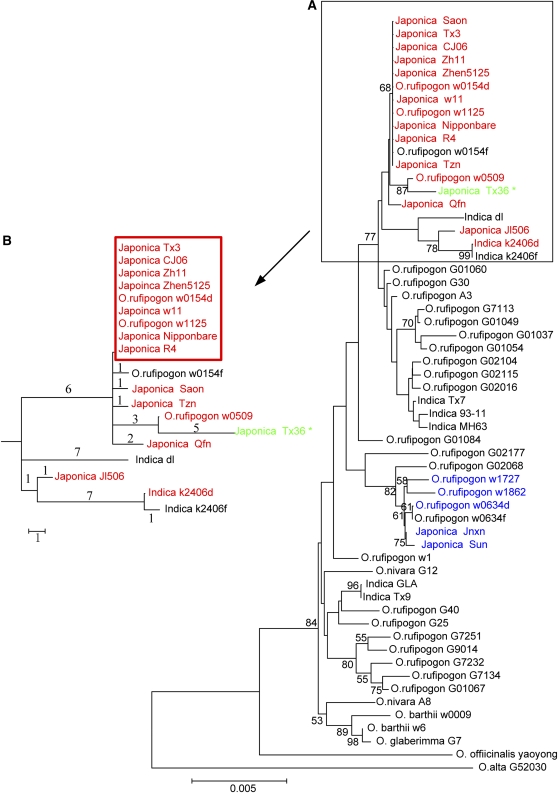
The Genealogy of Phr1 Alleles in Cultivars and Their Wild Relatives.
(A) The main panel (large tree, right) shows the optimal tree obtained using the neighbor-joining method with 1000 bootstraps. The bootstrap values are indicated at nodes with at least 50% support. The H18 (Δ18-bearing) haplotypes are labeled red, and the H29 haplotypes are labeled blue. The only 1-bp insertion line is labeled green. Note the strong genealogical clustering of these colored labels. The boxed cluster consists mainly of H18s, with a few non-Δ18-bearing haplotypes embedded within.
(B) In the inset, we zoom in on this cluster using the parsimonious haplotype cladogram. On each branch, the number of nucleotide changes is given. The suffixes -d and -f appended to accession names are abbreviations for deletion and full-length, respectively.
Figure 7.
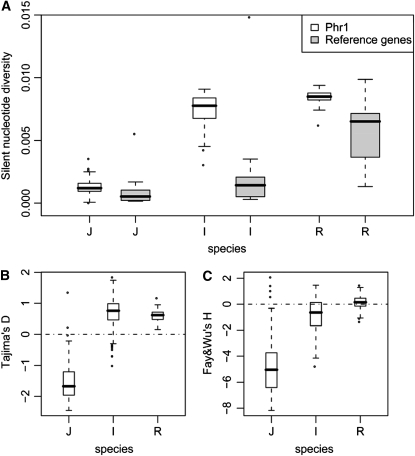
Population Genetic Tests on the Phr1 Sequences for All Three Populations.
(A) Nucleotide diversity (π) at silent sites by the box-and-whisker diagrams for Phr1 (white) and reference genes (gray). The variance is estimated by the bootstrapping procedure. For comparison, the corresponding estimates for 10 reference genes are also given.
(B) and (C) Tajima's D (B) and Fay and Wu's H (C) tests for the Phr1 sequences. The dashed lines indicated the expected values of 0 for Tajima's D and Fay and Wu's H under the standard neutral model. Both D and H statistics for Phr1 are significantly negative in japonica (P < 0.05) but much higher in indica and O. rufipogon by the bootstrapping procedure described in Methods. Significantly negative D and H indicate an excess of very low and very high frequency variants, respectively.
Multiple Independent Mutations of Phr1 in japonica Cultivars
To measure the occurrence of Phr1 deficiency among rice cultivars, we first genotyped 35 japonica and 20 indica lines using the molecular marker pSTS18, which specifically detects Δ18 (see Supplemental Table 1 online). All 20 indica lines examined are PHR-positive, although one line appears to be heterozygous for both the wild-type and Δ18 alleles. Curiously, although all 35 japonica lines examined are negative in PHR, only 32 bear the Δ18 mutation. We therefore sequenced the Phr1 alleles from the three lines that do not carry Δ18. Interestingly, all three lines carry frame-shift mutations in the Phr1 coding region, of which two carry a 29-bp deletion (Δ29) and one has a 1-bp insertion (Table 1; see Supplemental Figure 1 online).
We further surveyed the distribution of the Δ18 and Δ29 alleles with PCR markers pSTS18 and pSTS29 (see Supplemental Table 1 online) in a large panel of wild rice lines and the African cultivar Oryza glaberrima (Table 1; see Supplemental Table 2 online). Outside of O. sativa, the deletions exist only in Oryza rufipogon, but at very low frequencies. Among 523 O. rufipogon lines, the Δ18 and Δ29 deletions were observed only three times each, accounting for 0.5% of the collection. Among all other species (with 188 lines altogether), neither allele was found. These deletions are thus very rare in nature. Importantly, the Phr1 deletion correlates perfectly with the PHR-negative phenotype for the accessions where both genotype and phenotype data are available.
The Recent Origin of the Loss-of-Function Mutations in Phr1
The three apparently independent origins of Phr1 loss-of-function mutations raise several interesting questions about the evolution of the PHR-negative phenotype. Did the independent mutations all emerge during domestication or did they preexist in the wild rice? If the former, were the mutations selected strongly, and why were they favored in one subspecies only? If the latter, how old are these mutations and how are these polymorphisms maintained in the wild species?
To answer these questions, we sequenced the Phr1 alleles from 14 japonica, seven indica, 27 O. rufipogon, and seven other wild species lines (Table 1). Lines bearing deletions were preferentially included for sequencing. The haplotypes that bear the Δ18 and Δ29 deletions were referred to as H18s and H29s, respectively (both in plural form denoting multiple different haplotypes sharing the same deletion). The genealogy of all haplotypes is given in Figure 5. An obvious feature of the genealogy is that H18s and H29s are both clustered, indicating recent origins of H18s and H29s, respectively. The origins are sufficiently recent that most of the H18s, for example, have not had time to recombine with other haplotypes.
For the H18s cluster (Figure 5A, in red), four features are noteworthy. First, the H18s in O. rufipogon are all similar (ranging from 0 to 3 differences between lines); by contrast, the japonica lines are more diverse (differing by 3 to 10 bp among lines; Figure 5B). If Δ18 had existed in O. rufipogon prior to domestication, one would have expected this mutation to be more common and more diverse in wild rice. Since H18s in O. rufipogon are very rare (0.5%; Table 1) and are a subset of H18s in japonica (as shown in the genealogy of Figure 5), it seems likely that Δ18 emerged during domestication and that the presence of the deletion in O. rufipogon reflects introgression from the crop into wild populations.
Second, the genealogical cluster of H18s (labeled red in Figure 5A) harbors a few alleles that do not bear the Δ18 deficiency (i.e., O. rufipogon w0154f, indica dl, and indica K2406f). Similarly, the H29 cluster (labeled blue) contains a non-H29 sequence, O. rufipogon w0634f. One explanation might be that Δ18 first occurred on a common H18-like haplotype and some of these ancestral haplotypes that did not bear Δ18 have persisted until now. However, a closer inspection of the genealogical pattern of Figure 5A and the zoom-in picture of the H18s cluster in the haplotype tree (Figure 5B) suggest a different explanation. In this haplotype tree, three of the four non Δ18-bearing haplotypes are embedded among H18 types, and each is most closely related to another H18 haplotype. A more parsimonious explanation may be that these non-Δ18-bearing alleles were originally H18s themselves but have subsequently lost the Δ18 deficiency by recombination with a rice strain carrying a functional Phr1 allele. Because Δ18 is at the very 3′ end of the Phr1 sequence (see Supplemental Figure 1 online), single crossover is sufficient to remove this deletion without introducing many other changes to the haplotype. In other words, an H18 haplotype could lose Δ18 (and, by definition, would no longer be considered H18) but retains its genealogical relationship with other H18 haplotypes. The samples not labeled red in Figure 5B (one O. rufipogon and two indica lines) may be such examples. We also note that all japonica lines retain Δ18 (except for the 1-bp insertion line; see below), and the loss of Δ18 was observed only in the other two taxa.
Third, among japonica lines surveyed, all have either a large in-frame deletion (Δ18) or frame-shift (a 1-bp insertion or a 29-bp deletion) indel in Phr1, and they have all lost PHR activity. The contrast with indica and O. rufipogon, in which such mutations are rather uncommon, demands an explanation (see below). The 1-bp insertion in the coding region, observed only once, is particularly noteworthy (Table 1; see Supplemental Figure 1 online). If Δ18 and this 1-bp insertion arose independently, one would have to assume that, by coincidence, both occurred on a H18-like haplotype (Figures 5A and 5B). However, this haplotype is relatively uncommon in O. rufipogon. The haplotype on which the 1-bp insertion occurred is deep with the H18s genealogical cluster (Figure 5B). A more interesting scenario is as follows: an H18 haplotype underwent two changes: the gain of the 1-bp insertion and then the loss of Δ18. If this is indeed the case, this japonica haplotype would have remained nonfunctional throughout its evolution.
Fourth, the H29s cluster, characterized by another frame-shift mutation, Δ29 (Figure 5A, labeled in blue) is very young, with <1 bp of nucleotide changes per kilobase since the emergence of Δ29. There has not been sufficient time for H29s of O. rufipogon, which is not a strictly selfing species, to experience recombination with other haplotypes (see Supplemental Results online for details). Furthermore, Δ29 is only one-tenth as common in O. rufipogon as it is in japonica. While three H29s from O. rufipogon are shown in Figure 5A, the frequency of H29s in that species is only 2.5/517, vis-à-vis the frequency of 2/32 in japonica (Table 1). All these observations suggest that Δ29 is a relatively recent mutation that probably originated in japonica.
Positive Selection on the Δ18 Mutation: Signature of Differentiation
Given that all japonica lines suffer a lesion in Phr1, we ask if the spread of such lesions was driven by positive selection (i.e., was adaptive specifically to the environment of japonica). We first consider the differentiation of the Phr1 gene across populations in this section. Sites of unusually strong differentiation between populations are often suggested to be under taxon-specific selection (Hara et al., 1964; Akey et al., 2004; Beaumont, 2005).
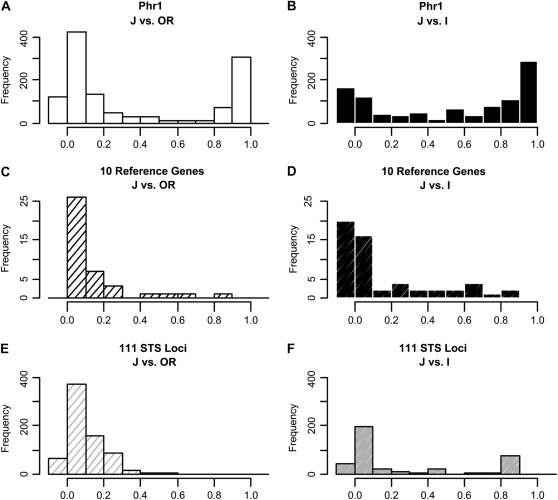
We ask whether the Δ18 region is unusually differentiated between japonica and either indica or O. rufipogon. We first calculated the extent of population differentiation for each variant site in the Phr1 gene among the three taxa. In Figure 6, we show the distribution of the Fst statistic (Weir and Cockerham, 1984) between japonica and the other two populations. High Fst values close to 1 indicate strong genetic differentiation among populations, and low Fst near 0 indicate homogeneity.
Figure 6.
Distributions of the Fst Statistic in Phr1 vis-á-vis 10 Reference Genes and Randomly Chosen Gene Fragments.
(A) Fst of Phr1 between japonica and O. rufipogon.
(B) Fst of Phr1 between japonica and indica.
(C) Fst of the 10 reference genes between japonica and O. rufipogon.
(D) Fst of the 10 reference genes between japonica and indica.
(E) Fst of randomly chosen gene fragments referred to as STS from Caicedo et al. (2007) between japonica and O. rufipogon.
(F) Fst of the same STS between japonica and indica. Phr1 sequences from each population were resampled following a multinomial distribution with 100 permutations (see Methods).
The Fst profiles of Phr1 ([A] and [B]) show a peak near 1, which is distinctive from the two reference data sets ([C] and [D] for reference genes; [E] and [F] for the STS loci).
For comparison, we used two different data sets. The first set contains 10 reference genes for which we have collected polymorphism data from the three taxa ourselves (Tang et al., 2006) (see Methods for the bootstrapping procedure). In the second set, we retrieved randomly chosen DNA fragments from the data of Caicedo et al. (2007). The two data sets differ in the lines used and in genes chosen. In Tang et al. (2006), the lines are mostly so-called elite rice, whereas Caicedo et al. (2007) included many land races. The sequences from Tang et al. (2006) are all well-annotated genes, whereas those from Caicedo et al.'s collection are random DNA fragments.
We shall compare japonica and O. rufipogon first. Phr1 is shown to have more high Fst sites than other genes, and the high peak near Fst = 1 in Figure 6A can be attributed mainly to Δ18. This peak is absent in the two reference data sets (Figures 6C and 6E). Thus, the strong differentiation observed around Δ18 is unusual for japonica and O. rufipogon. Differentiation between japonica and indica is expected to be more complex because both taxa have been influenced by domestication (population bottlenecks, selection, and hitchhiking; Lu et al., 2006). In all three sets of data, the indica-japonica differentiation is larger than the japonica–O. rufipogon comparison (Figures 6A versus 6B, 6C versus 6D, and 6E versus 6F; all with P < 0.001 by the one-sided Kolmogorov-Smirnov test). The Fst profile of Phr1 is even more distinctive from the two reference sets (Figures 6A versus 6C or 6E, and 6B versus 6D or 6F; all with P < 10−6 by the one-sided Kolmogorov-Smirnov test). The results suggest that Phr1 is unusually differentiated between indica and the other two taxa when compared with other parts of the genome.
Positive Selection on the Δ18 Mutation: Level and Pattern of Polymorphism
If Phr1 is driven by positive selection, there should be a signature of a selective sweep in japonica, but not in indica or O. rufipogon. We thus compared the within-population polymorphism for the three taxa. The signature can be observed in three forms, shown below.
First, the level of polymorphism near Phr1 should be reduced relative to other parts of the genome in japonica, and this reduction should be seen in japonica only. In Figure 7A, it can be seen that the level of polymorphism in Phr1 in japonica is indeed much lower than that in either indica or O. rufipogon. Measured against the average polymorphism of the 10 reference genes from the same taxa (Tang et al., 2006), the level of Phr1 polymorphism in japonica remains significantly lower than in the other two taxa (P < 0.001 by bootstrapping; see Methods). Note that Phr1 in non-japonica taxa is more polymorphic than the 10 reference genes. As reported in Tang et al. (2006), the rice genome is a mosaic of regions of very low to very high polymorphism, and the 10 reference genes were chosen from regions of normal polymorphism. The high-polymorphism genes can be 6 to 10 times as variable as the ten reference genes (Tang et al., 2006).
Second, in the gene suspected to have undergone a recent selective sweep, not only is the level of polymorphism expected to be reduced but the reduction should exhibit an excess of very low frequency and very high frequency mutant sites (Fay and Wu, 2000; Przeworski et al., 2001; Zeng et al., 2006). This trend is often summarized by two different statistics: Tajima's D (Tajima, 1989) and Fay and Wu's H (Fay and Wu, 2000; Zeng et al., 2006). Significantly negative D and H indicate an excess of very low and very high frequency variants, respectively. In japonica, both statistics for Phr1 are indeed significantly negative by the bootstrapping procedure described in Methods (Figures 7B and 7C). Furthermore, both D and H are much higher in indica and O. rufipogon. In neither of the two is D or H significantly different from 0.
The result that both D and H tests show the same trend is important for inferring positive selection (Figures 7B and 7C). Between the two tests, Fay and Wu's H is considered a more specific test for positive selection (Zeng et al., 2006, 2007). However, deep population subdivision with migration may sometimes lead to false positive by the H test (Przeworski et al., 2001). Indeed, Caicedo et al. (2007) found many loci in domesticated rice to be associated with a significantly negative H value. Although these authors suggested pervasive positive selection during domestication, they did not reject gene flow between the two subspecies as a major contributing factor.
Recently, Zeng et al. (2006, 2007) pointed out that the joint test of D and H, referred to as the DH compound test, is robust against most demographic factors (see Supplemental Results online for a brief explanation). The expected D or H value at the neutral equilibrium is 0 (or very close to 0) but, after a recent selective sweep, both values would often be significantly less than 0 (Tajima, 1989; Fay and Wu, 2000). When both are significantly negative, the compound statistic, DH (Zeng et al., 2006), will also be highly significant. DH has the added advantage of being insensitive to demographic influences. In japonica, but not in indica or O. rufipogon, DH is significantly negative (see Supplemental Results online). According to the simulations of Zeng et al. (2006, 2007), the most likely explanation for the simultaneous significance of D and H in japonica is positive selection.
DISCUSSION
The negative PHR phenotype in japonica cultivars is associated with functional loss of Phr1, a consequence of indels in the coding regions. While PPOs exist as a large family of functionally redundant genes in tomato (Solanum lycopersicum) and potato (Solanum tuberosum) (Thygesen et al., 1995; Thipyapong et al., 1997), the redundancy appears absent in rice. In Figure 2, it can be seen that indels in the Phr1 gene alone can nullify PPO activity.
Since rice cultivars are selfers and their wild progenitors often outcross, the impact of this breeding structure on various aspects of our observations, including coalescence time and genetic hitchhiking, is further considered in the supplemental material online. The overall population genetic patterns of Phr1 alleles suggest that the loss-of-function mutations arose three times in japonica in the recent past. Based on four different population genetic tests, we conclude that at least one of them was driven to high frequency by positive selection, presumably associated with human activities.
In our study, samples of indica, japonica, and O. rufipogon lines came predominantly from within China. Hence, the interpretation of positive selection, for example, is not confounded by possible geographical differentiation. Nevertheless, the conclusion may apply to China (or at most eastern Asian) populations only. Whether it is applicable to domesticated rice in general will have to await further studies.
An interesting parallel with the Phr1 deletions in our collection of japonica lines has been reported for the six-rowed spike phenotype in barley (Hordeum vulgare) (Komatsuda et al., 2007). Each of the three independent loss-of-function mutations in the Vrs1 gene, a homeodomain-leucine zipper gene, leads to the six-rowed phenotype, which presumably increases grain production. The difference between the rice and barley systems is that the loss of Phr1 function is restricted to only one subspecies of rice. This dichotomous distribution offers a contrast and permits some insight into subspecies differentiation during domestication. For Phr1, at least one of the mutations has spread very rapidly in japonica, likely aided by positive selection associated with human activities. Some of these mutations may have been introduced into indica and O. rufipogon. Most interesting in our finding is that the introduced alleles tend to re-acquire the functional site by recombination (Figure 5).
Why, then, is this loss of function in Phr1 associated only with japonica, but not with indica, or their immediate progenitor, O. rufipogon? We tentatively propose a hypothesis based on our observations. Traits selected by humans during domestication may sometimes be those of aesthetic appeal (Sweeney et al., 2006). Grains of the cultivars of japonica subspecies are refractory to discoloration during storage, making the Phr1 gene a plausible target of artificial selection. In this sense, Phr1 may be analogous to Rc, a domestication-related gene required for red pericarp in rice (Sweeney et al., 2006).
The retention of PHR activity in indica, in contrast with its ubiquitous loss in japonica, demands an explanation. One might argue that the history of domestication may not be long enough for every desired mutation to emerge. However, three independent losses have occurred in japonica in a rather brief period of domestication and an indica line does carry the deletion (Table 1), potentially allowing it to spread in this subspecies. Thus, the Phr1 function is retained in indica not because of the lack of time for it to be lost, but because of other reasons. Plant PPOs have been reported to be associated with disease resistance (Thipyapong et al., 1995; Li and Steffens, 2002). It may be that agriculture in the tropical and subtropical zones still put a premium on disease resistance, much like the case in the wild. Another intriguing possibility is that Phr1 activities are needed in warmer climates to maintain seed dormancy (Gu et al., 2004, 2005). Hence, the appeal of white grains might not compensate for the cost of premature seed germination in storing indica grains under some environmental conditions.
Finally, high frequency major mutations like the Phr1 indels in japonica might be a common source of phenotypic divergence among domesticated breeds or cultivars. Indeed, a widely discussed view is the less-is-more hypothesis (Olson, 1999), which posits that domestication is accompanied by extensive gene losses. A recent survey of the molecular basis of trait differentiation between cultivars of crops (rice, wheat, etc.) has suggested that loss-of-function mutations could be associated with several known traits (Yano et al., 2000, 2004; Sasaki et al., 2002; Ueguchi-Tanaka et al., 2005). In a separate study based on the two fully sequenced rice genomes, we have also found that large in-frame indels or frame-shift indels in coding regions are unusually frequent among rice cultivars (Huang et al., 2008). In light of these observations, the result on Phr1 mutations could be seen as part of a common trend in the studies of domestication.
METHODS
Material
All seeds or DNA used in this study were collected by our own lab or provided by the International Rice Research Institute. In total, 35 japonica lines, 20 indica lines, 523 Oryza rufipogon lines, and 188 others were used. All of indica lines were collected in China. For japonica lines, three are from Japan and the rest from China. For O. rufipogon, the lines were from Bangladesh (17 lines), Burma (21 lines), Cambodia (1 line), China (146 lines), India (159 lines), Indonesia (five lines), Khmer Republic (one line), Malaya (15 lines), Nepal (two lines), New Guinea (three lines), Philippines (one line), Sri Lanka (nine lines), and Thailand (115 lines). In addition, 28 O. rufipogon lines were from uncertain locales. Hence, the geographical distribution of O. rufipogon lines is properly extensive.
For indica and japonica lines, we give the identifier for each line used in Supplemental Table 2 online. For O. rufipogon and other groups, the inference made in this study depends mainly on those lines chosen for DNA sequencing. Each sequenced line is also individually listed. The number of lines chosen for genotyping is much larger, but these lines provide little information, other than the frequencies of the two deletions; hence, these lines are not individually listed. The more interesting but rare O. rufipogon lines with deletions are identified in Supplemental Table 2 online.
Cloning of Phr1
To clone Phr1, the mapping population of 5589 F2 plants was derived from the cross between an indica variety Minghui63 (MH63) and a japonica variety Chunjiang06 (CJ06). Ten F2 seeds per line were soaked in 2% (v/v) phenol solution and observed after 5 d according to the method described previously (Oka and Chang, 1962). Rice DNA was isolated according to the method described previously (Li et al., 2003b). Based on the rice genetic map and genome sequences of Nipponbare and 93-11 (McCouch et al., 1988; Feng et al., 2002; Yu et al., 2002), the PCR-based markers were developed located on either side of Phr1 and at a genetic distance from 100 to 115 cM, respectively.
Complementation Test
A 4.2-kb 93-11 genomic DNA fragment, which contains the Phr1 coding region, the 1411-bp upstream sequence, and 454-bp downstream sequence, was inserted into the binary vector pCAMBIA1300 to generate a complementation plasmid, pC13Phr1 (Figure 2D). Primers used to construct these plasmids are listed in Supplemental Table 1 online. The control plasmid pC13p containing the 1411-bp upstream sequence and 3′ truncated Phr1 that encodes the first 195 amino acid residues was also constructed (Figure 2D). The two binary plasmids were introduced into Agrobacterium tumefaciens EHA105 by electroporation, and the japonica rice Nipponbare was transformed as reported (Hiei et al., 1994; Li et al., 2003a).
PPO Enzymatic Assays and Phr1 Expression Analysis
The PPO enzymatic activity was spectrophotometrically determined using DOPA as the substrate (Robinson and Dry, 1992). Protein concentration was determined as described by Bradford using BSA as a standard (Bradford, 1976). For RT-PCR analysis, total RNA was extracted from immature rice kernels as previous reported (Li et al., 2003b). The RT reaction was performed using 2 μg of total RNA with oligo(dT) and SuperScript III RNaseH (−) reverse transcriptase (Invitrogen) according to the manufacturer's instructions. RT primers used were synthesized based on the cDNA sequences (see Supplemental Table 1 and Supplemental Figure 1 online).
Sequence Manipulations
The nonredundant peptide sequences were searched using BLASTX (Schaffer et al., 2001). The Phr1 molecular mass was calculated using the method of Kyte and Doolittle (1982) and the Genetics Computer Group software of the University of Wisconsin (Devereux et al., 1984). The signal peptide and cleavage site were predicted according to the method described previously (Nielsen et al., 1997; Nair and Rost, 2005). The comparison of Phr1 sequences was performed with the Multalign program (Corpet, 1988).
The deletions of Δ18 or Δ29 in Phr1 sequences were surveyed by PCR amplification in a total of 55 Asian cultivars, one Africa cultivar, and 710 wild rice individuals with primers pSTS18 and pSTS29 (see Supplemental Table 1 online). We further sequenced the Δ18- or Δ29-containing Phr1 genes with the primers F217, Cxp3, Cxp5, and c5311 (see Supplemental Table 1 online) from 21 Oryza sativa cultivars (seven indica and 14 japonica), the 27 O. rufipogon lines, and randomly selected lines from one African cultivated rice O. glaberrima, two O. nivara, two O. barthii, and two more distant wild relatives with a CC or CCDD genome (one O. officinalis and one O. alta) (Table 1). Two of the 27 O. rufipogon accessions and one of the seven indicas appeared to be heterozygotes (Table 1); each of the two alleles was included in the sequence analysis. Contigs were assembled using SeqMan (DNASTAR), and multiple sequences were aligned using the ClustalX program (Thompson et al., 1997).
Phylogenetic Analyses of DNA Sequences
A phylogenetic tree of the sequenced lines was reconstructed by the neighbor-joining (NJ) method (Saitou and Nei, 1987) based on Kimura's two-parameter distances (Kimura, 1980). MEGA version 4.0 (Tamura et al., 2007) was used to perform the phylogenetic reconstruction. Bootstrap values were estimated (with 1000 replicates) to assess the relative support for each branch (Felsenstein, 1985). All positions containing alignment gaps were eliminated in pairwise sequence comparisons in NJ analyses. The NJ tree was shown rooted by the midpoint to improve clarity.
In the cluster of lines containing Δ18 (H18 lines), a finer resolution is needed for visualizing the origin of the non-H18 lines embedded in that cluster. To achieve the resolution, we used the cladistic approach on the haplotypes by means of statistical parsimony (Templeton et al., 1992) with the aid of TCS v1.21 (Clement et al., 2000). Indels were treated as a single mutation. The root of this parsimonious haplotype tree, which is a cluster of the larger NJ tree, is set at the midpoint of the longest branch. This root is the same as the one given by the larger tree that encompasses all sequences. The branch length represents the number of observed mutations on the branch. This cluster is shown separately in Figure 5B.
Sequence Resampling by Bootstrapping
For phylogenetic analysis, lines with indels were chosen preferentially for sequencing. These DNA sequences are hence biased for population genetic analysis. We hence corrected this bias by resampling according to the known frequencies of these indels. (Because indels were genotyped from larger samples, their frequencies were determined with greater accuracy.) All population genetics statistics are hence presented as distributions from 100 such resamplings. The detail of this resampling scheme is as follows.
Each population contains k = 3 mutually exclusive and exhaustive classes of genotypes, for example, wild type (G+), Δ18 (GΔ18), and Δ29 (GΔ29), with the probability distribution of P = (pwild, pΔ18, pΔ29), where pwild + pΔ18 + pΔ29 = 1. Although GΔ29 individuals were not observed in the indica sample, this genotype could have been missed due to the small sample size. Let the sampling event, X, be random for Njaponica = 14, Nindica = 7, and Nrufipogon = 27 in japonica, indica, and O.rufipogon, respectively. Then, X = (Xwild, XΔ18, XΔ29) is multinomially distributed with index k = 3 and parameter P = (pwild, pΔ18, pΔ29), i.e., X ∼ Mult (k, p), where Xwild = number of trials in which G+ occurs, XΔ18 = number of trials in which GΔ18 occurs, XΔ29 = number of trials in which GΔ29occurs, and Xwild+ XΔ18+ XΔ29 = N. To infer the distribution of each genotype in the wild populations, we simulated the multinomial sampling process for each population with 100 permutations. According to the results of genotyping survey (Table 1), Pjaponica = (1/35, 32/35, 2/35), Pindica = (19/20, 0.5/20, pΔ29), and Prufipogon = (518/523, 2.5/523, 2.5/523), respectively. The missing data of PΔ29 in indica population was arbitrarily assigned within the range from 0 to 0.5/20. Increasing the value to a maximum of 0.5/20 did not change the pattern reported in this study. We first classified the available sequences data for each population into the three genotypes of G+, GΔ18, and GΔ29 and then generated the 100 multinomially distributed random number vectors for each genotype in each population. These numbers were used as the specified sizes for sampling our available sequence data with replacement. R scripts were written to perform the simulation in R 2.6.1 environment.
Population Genetic Tests
Each set of the resampled sequences was sequentially submitted to population genetic analyses. Fst statistic (Weir and Cockerham, 1984) was calculated for each site to detect genetic differentiation among japonica with indica and O. rufipogon populations. To examine the genomic distribution of Fst, randomly chosen fragments referred to as STS were retrieved from GenBank (EF000002 to EF010509) (Caicedo et al., 2007). Alignments of O. sativa ssp indica, O. sativa ssp japonica, and the wild progenitor O.rufipogon per locus were prepared with ClustalW2 (Larkin et al., 2007) and then used to calculate the Fst statistic for each site. The alignments of STS loci range from 391 to 667 bp, with an average of 493 bp. Nucleotide diversity (Tajima, 1983) was estimated using synonymous sites and noncoding regions for each population. Tajima's D statistic (Tajima, 1989) and Fay and Wu's H statistic (Fay and Wu, 2000) were computed using the H-test program (http://www.genetics.wustl.edu/jflab/htest.html). DH test (Zeng et al., 2006, 2007) was computed using Java scripts kindly provided by Kai Zeng. To calculate the H statistic, O. barthii was used as outgroup to infer ancestral character state of the O. sativa/O. rufipogon complex. The character states of segregating sites within O. barthii were resolved by checking their counterparts in more distant O. alta and O. officinalis. In all the population genetics analyses above, alignment gaps were excluded and indels were scored as binary characters. Scripts used in this study are available upon request.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MH63 (DQ532375), Nipponbare (DQ532376), CJ06 (DQ532377), Jl506 (DQ532378), Jnxn (DQ532379), Qfn (DQ532380), R4 (DQ532381), Saon (DQ532382), Sun (DQ532383), Tx3 (DQ532384), Tx36 (DQ532385), Tzn (DQ532386), w11 (DQ532387), Zh11(DQ532388), Zhen5125 (DQ532389), dl (DQ532390), 93-11 (DQ532391), GLA (DQ532392), k2406d (DQ532393), k2406f (DQ532394), Tx7 (DQ532395), Tx9 (DQ532396), A3 (DQ532397), A8 (DQ532398), G01037 (DQ532399), G01049 (DQ532400), G01054 (DQ532401), G01060 (DQ532402), G01067 (DQ532403), G01084 (DQ532404), G02016 (DQ532405), G02068 (DQ532406), G02104 (DQ532407), G02115 (DQ532408), G02177 (DQ532409), G12 (DQ532410), G25 (DQ532411), G30 (DQ532412), G40 (DQ532413), G52030 (DQ532414), G7113 (DQ532415), G7134 (DQ532416), G7232 (DQ532417), G7251 (DQ532418), G9014 (DQ532419), w0009 (DQ532420), w0154d (DQ532421), w0154f (DQ532422), w0509 (DQ532423), w0634d (DQ532424), w0634f (DQ532425), w1 (DQ532426), w1125 (DQ532427), w1727 (DQ532428), w1862 (DQ532429), w6 (DQ532430), yaoyong (DQ532431), and G7 (DQ532432).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Phr1 cDNA Sequences of Indica-Type Rice MH63 and GLA with Japonica-Type Rice Nipponbare.
Supplemental Figure 2. Alignment of PHR1 with Its Homologous Proteins Identified in the Rice Genome.
Supplemental Figure 3. The Frequency Spectrum of Phr1 Mutations.
Supplemental Table 1. PCR-Based Molecular Markers Developed in This Study.
Supplemental Table 2. Summary of Samples for Phr1 Study.
Supplemental Data Set 1. Text File Corresponding to Alignment in Figure 3.
Supplemental Data Set 1. Text File Corresponding to Alignment in Supplemental Figure 1.
Supplemental Data Set 1. Text File Corresponding to Alignment in Supplemental Figure 2.
Supplemental Results.
Supplementary Material
Acknowledgments
We thank Zhukuan Cheng (Institute of Genetics and Developmental Biology), Chuanqing Sun (China Agricultural University), and Zixuan Wang and Yuzo Minobe (Plant Gene Center, Tsukuba, Japan) for providing some seeds or DNA samples of wild rice and cultivar lines. This work was supported by grants from the National Natural Science Foundation of China (30330040, 30500049, and 30730008/C0102) and the Ministry of Science and Technology of China (2005CB1208 and 2007CB815701).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiayang Li (jyli@genetics.ac.cn).
Online version contains Web-only data.
References
- Akey, J.M., Eberle, M.A., Rieder, M.J., Carlson, C.S., Shriver, M.D., Nickerson, D.A., and Kruglyak, L. (2004). Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.V., and Morris, C.F. (2001). An improved whole-seed assay for screening wheat germplasm for polyphenol oxidase activity. Crop Sci. 41 1697–1705. [Google Scholar]
- Beaumont, M.A. (2005). Adaptation and speciation: What can F(st) tell us? Trends Ecol. Evol. 20 435–440. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Caicedo, A.L., Williamson, S.H., Hernandez, R.D., Boyko, A., Fledel-Alon, A., York, T.L., Polato, N.R., Olsen, K.M., Nielsen, R., McCouch, S.R., Bustamante, C.D., and Purugganan, M.D. (2007). Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, J.W., Lax, A.R., and Flurkey, W.H. (1992). Cloning and characterization of cDNAs coding for Vicia faba polyphenol oxidase. Plant Mol. Biol. 20 245–253. [DOI] [PubMed] [Google Scholar]
- Chang, T.T. (1976). The origin, evolution, cultivation, dissemination and differentiation of Asian and African rices. Euphytica 25 435–441. [Google Scholar]
- Chen, M., et al. (2002). An integrated physical and genetic map of the rice genome. Plant Cell 14 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, T., de Rigal, D., Mbeguie, A.M.D., Gauillard, F., Richard-Forget, F., and Fils-Lycaon, B.R. (1999). Molecular cloning and characterization of apricot fruit polyphenol oxidase. Plant Physiol. 119 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.M., Linton, E., Messing, J., and Doebley, J.F. (2004). Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. USA 101 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, M., Posada, D., and Crandall, K.A. (2000). TCS: A computer program to estimate gene genealogies. Mol. Ecol. 9 1657–1659. [DOI] [PubMed] [Google Scholar]
- Constabel, C.P., Yip, L., Patton, J.J., and Christopher, M.E. (2000). Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiol. 124 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, D., Bakker, H., Lever, A., Bouma, T., Salentijn, E., and Weisbeek, P. (1991). Protein targeting towards the thylakoid lumen of chloroplasts: Proper localization of fusion proteins is only observed in vivo. EMBO J. 10 2765–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke, T., and Morris, F. (2002). Molecular characterization of wheat polyphenol oxidase (PPO). Theor. Appl. Genet. 104 813–818. [DOI] [PubMed] [Google Scholar]
- Devereux, J., Haeberli, P., and Smithies, O. (1984). A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J.C., and Wu, C.I. (2000). Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Feng, Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420 316–320. [DOI] [PubMed] [Google Scholar]
- Gooding, P.S., Bird, C., and Robinson, S.P. (2001). Molecular cloning and characterisation of banana fruit polyphenol oxidase. Planta 213 748–757. [DOI] [PubMed] [Google Scholar]
- Gu, X.Y., Kianian, S.F., and Foley, M.E. (2004). Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X.Y., Kianian, S.F., and Foley, M.E. (2005). Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice. Genetics 171 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Hatakeyama, T., Niida, T., Yumoto, H., Watanabe, T., Noguchi, T., Khomoto, K., and Yasuda, Y. (1964). “V-factor” of Streptomyces origin, an effective substance for the control of the bacterial leaf blight of rice plant. J. Antibiot. (Tokyo) 17 174. [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Huang, X., Lu, G., Zhao, Q., Liu, X., and Han, B. (2008). Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 148 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, S., Kosaka, H., and Hara, T. (1928). On the affinity of rice varieties as shown by fertility of hybrid plants. Bull. Sci. Fac. Agric. Kyushu Univ. 3 132–147. [Google Scholar]
- Keegstra, K. (1989). Transport and routing of proteins into chloroplasts. Cell 56 247–253. [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. [DOI] [PubMed] [Google Scholar]
- Klabunde, T., Eicken, C., Sacchettini, J.C., and Krebs, B. (1998). Crystal structure of a plant catechol oxidase containing a dicopper center. Nat. Struct. Biol. 5 1084–1090. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 105–132. [DOI] [PubMed] [Google Scholar]
- Komatsuda, T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li, L., and Steffens, J.C. (2002). Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 215 239–247. [DOI] [PubMed] [Google Scholar]
- Li, X., et al. (2003. a). Control of tillering in rice. Nature 422 618–621. [DOI] [PubMed] [Google Scholar]
- Li, Y., Qian, Q., Zhou, Y., Yan, M., Sun, L., Zhang, M., Fu, Z., Wang, Y., Han, B., Pang, X., Chen, M., and Li, J. (2003. b). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.Y., Nagamura, Y., Kurata, N., Yano, M., Minobe, Y., and Sasaki, T. (1994). DNA markers tightly linked to genes, Ph, Alk and Rc. Rice Genet. Newsl. 11 108–109. [Google Scholar]
- Lu, J., Tang, T., Tang, H., Huang, J., Shi, S., and Wu, C.I. (2006). The accumulation of deleterious mutations in rice genomes: A hypothesis on the cost of domestication. Trends Genet. 22 126–131. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Kochert, G., Yu, Z.H., Wang, Z.Y., Khush, G.S., Coffman, W.R., and Tanksley, S.D. (1988). Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76 815–829. [DOI] [PubMed] [Google Scholar]
- Morishima, H., and Oka, H.I. (1960). The pattern of interspecific variation in the genus Oryza: Its quantitative representation by statistical methods. Evolution Int. J. Org. Evolution 14 153–165. [Google Scholar]
- Nair, R., and Rost, B. (2005). Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 348 85–100. [DOI] [PubMed] [Google Scholar]
- Nesbitt, T.C., and Tanksley, S.D. (2002). Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, J.J., Richard-Forget, F.C., Goupy, P.M., Amiot, M.J., and Aubert, S.Y. (1994). Enzymatic browning reactions in apple and apple products. Crit. Rev. Food Sci. Nutr. 34 109–157. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green, P.J., Wang, G.L., and Meyers, B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25 473–477. [DOI] [PubMed] [Google Scholar]
- Oka, H.I. (1953). Phylogenetic differrentiation of the cultivated rice plant. 1. Variations in respective characteristics and their combinations in rice cultivars. Jpn. J. Breed. 3 33–43. [Google Scholar]
- Oka, H.I. (1958). Intervarietal variation and classification of cultivated rice. Indian J. Genet. Plant Breed. 18 79–89. [Google Scholar]
- Oka, H.I. (1988). Origin of Cultivated Rice. (Tokyo/Amsterdam: Japan Scientific Societies Press).
- Oka, H.I., and Chang, W.T. (1962). Rice varieties intermediate between wild and cultivated forms and the origin of the japonica type. Bot. Bull. Acad. Sinica (Taiwan) 3 109–131. [Google Scholar]
- Olsen, K.M., and Purugganan, M.D. (2002). Molecular evidence on the origin and evolution of glutinous rice. Genetics 162 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.V. (1999). When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M., Wall, J.D., and Andolfatto, P. (2001). Recombination and the frequency spectrum in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18 291–298. [DOI] [PubMed] [Google Scholar]
- Purugganan, M.D., Boyles, A.L., and Suddith, J.I. (2000). Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics 155 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, S.P., and Dry, I.B. (1992). Broad bean leaf polyphenol oxidase is a 60-Kilodalton protein susceptible to proteolytic cleavage. Plant Physiol. 99 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, A., et al. (1991). Linkage map of restriction fragment length polymorphism loci in rice. Jpn. J. Breed. 41 665–670. [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416 701–702. [DOI] [PubMed] [Google Scholar]
- Schaffer, A.A., Aravind, L., Madden, T.L., Shavirin, S., Spouge, J.L., Wolf, Y.I., Koonin, E.V., and Altschul, S.F. (2001). Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, R., Pasqualone, A., Clodoveo, M.L., and Blanco, A. (2002). Genetic mapping of polyphenol oxidase in tetraploid wheat. Cell. Mol. Biol. Lett. 7 763–769. [PubMed] [Google Scholar]
- Steffens, J.C., Harel, E., and Hunt, M.D. (1994). Polyphenol Oxidase. In Genetic Engineering of Plant Secondary Metabolism, B.E. Ellis, G.W. Kuroki, and H.A. Stafford, eds (New York: Plenum Press), pp. 276–304.
- Sweeney, M.T., Thomson, M.J., Pfeil, B.E., and McCouch, S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, T., Lu, J., Huang, J., He, J., McCouch, S.R., Shen, Y., Kai, Z., Purugganan, M.D., Shi, S., and Wu, C.I. (2006). Genomic variation in rice: Genesis of highly polymorphic linkage blocks during domestication. PLoS Genet. 2 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., Fulton, T.M., and McCouch, S.R. (1993). Genetic Maps: Locus Maps of Complex Genomes. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Templeton, A.R., Crandall, K.A., and Sing, C.F. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong, P., Hunt, M.D., and Steffens, J.C. (1995). Systemic wound induction of potato (Solanum tuberosum) polyphenol oxidase. Phytochemistry 40 673–676. [Google Scholar]
- Thipyapong, P., Joel, D.M., and Steffens, J.C. (1997). Differential expression and turnover of the tomato polyphenol oxidase gene family during vegetative and reproductive development. Plant Physiol. 113 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thygesen, P.W., Dry, I.B., and Robinson, S.P. (1995). Polyphenol oxidase in potato. A multigene family that exhibits differential expression patterns. Plant Physiol. 109 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C.T., Tsaur, S.C., Sun, S., Browne, W.E., Chen, Y.C., Patel, N.H., and Wu, C.I. (2004). Gene duplication and speciation in Drosophila: Evidence from the Odysseus locus. Proc. Natl. Acad. Sci. USA 101 12232–12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., Bomblies, K., Lukens, L., and Doebley, J.F. (2005). The origin of the naked grains of maize. Nature 436 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B.S., and Cockerham, C.C. (1984). Estimating F-Statistics for the analysis of population structure. Evolution Int. J. Org. Evolution 38 1358–1370. [DOI] [PubMed] [Google Scholar]
- Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., Bennetzen, J.L., Echenique, V., and Dubcovsky, J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Zeng, K., Fu, Y.X., Shi, S., and Wu, C.I. (2006). Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174 1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, K., Shi, S., and Wu, C.I. (2007). Compound tests for the detection of hitchhiking under positive selection. Mol. Biol. Evol. 24 1898–1908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.