Abstract
The first bacterial genome was sequenced in 1995, and the first archaeal genome in 1996. Soon after these breakthroughs, an exponential rate of genome sequencing was established, with a doubling time of approximately 20 months for bacteria and approximately 34 months for archaea. Comparative analysis of the hundreds of sequenced bacterial and dozens of archaeal genomes leads to several generalizations on the principles of genome organization and evolution. A crucial finding that enables functional characterization of the sequenced genomes and evolutionary reconstruction is that the majority of archaeal and bacterial genes have conserved orthologs in other, often, distant organisms. However, comparative genomics also shows that horizontal gene transfer (HGT) is a dominant force of prokaryotic evolution, along with the loss of genetic material resulting in genome contraction. A crucial component of the prokaryotic world is the mobilome, the enormous collection of viruses, plasmids and other selfish elements, which are in constant exchange with more stable chromosomes and serve as HGT vehicles. Thus, the prokaryotic genome space is a tightly connected, although compartmentalized, network, a novel notion that undermines the ‘Tree of Life’ model of evolution and requires a new conceptual framework and tools for the study of prokaryotic evolution.
INTRODUCTION
Modern genomics of prokaryotes (and, generally, cellular life forms) is a rare scientific field whose birth date can be pinpointed precisely. It is natural to associate the advent of the modern era in genomics with the appearance of the first complete genome, namely, the genome of the pathogenic bacterium Haemophilus influenzae (1). Very shortly, thereafter, the second bacterial genome, that of Mycoplasma genitalium, was sequenced (2), and modern comparative genomics was born. A considerable amount of sequences from diverse organisms was available prior to these reports, but the first fully sequenced bacterial genome forever changed the state of the art in genome analysis. The availability of complete genomes (i.e. with nearly all the genetic material from the given organism sequenced as opposed to, say, 90%, so that all genes are available for analysis) is crucial to the entire enterprise of comparative genomics for at least two related but distinct, fundamental reasons: (i) some caveats notwithstanding (see below), the availability of complete genome sequences (or, more precisely, full complements of genes) provides for the possibility to identify sets of orthologs, i.e. genes that evolved from the same ancestral gene in the common ancestor of the compared genomes, (ii) comparison of complete genomes (gene sets) is the necessary condition to determine not only which genes are present in any particular genome but also which ones are absent (3, 4). The ability to delineate sets of orthologs and to pinpoint missing genes is indispensable for genome-based reconstruction of an organism's metabolism and other functional systems and for reconstructions of genome evolution.
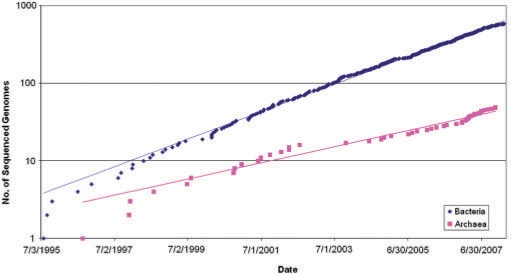
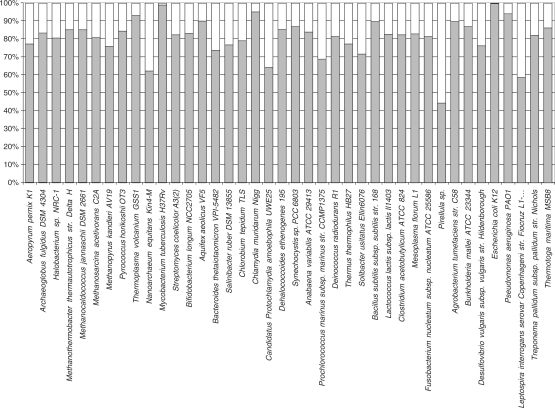
After the initial, relatively slow accumulation of bacterial and archaeal genome sequences, the rate of prokaryotic genome sequencing and public release has picked up rapidly, owing to improvements in sequencing technologies per se, and development of efficient pipelines for genome assembly and annotation (Figure 1). After the initial period of irregular growth, the accumulation of sequenced genomes of bacteria and archaea showed a remarkably good fit to exponential functions, with a doubling time of ∼20 months for bacteria and ∼34 months for archaea (Figure 1). Extrapolation suggests that the symbolic line of 1000 sequenced genomes will be crossed in March 2009, for bacteria and in April 2011, for archaea. As of this writing (10 June 2008), sequencing of the genome of any cultivable prokaryotes is considered routine, and 659 genomes of bacteria and 52 genomes of archaea have been completely sequenced (5). Moreover, inevitable biases (especially, toward medically important bacteria) in sequencing notwithstanding, these genomes are representative of the majority of recognized bacterial and archaeal phyla (Table 1). A common concern with regard to the representation of the actual prokaryotic diversity on earth in the collection of sequenced genomes is that only a small fraction of bacteria (∼0.1%) currently can be cultivated in the laboratory (6, 7). Genome sequencing of uncultivated organisms remains a major feat and so far has been successfully accomplished on very few occasions. However, recent metagenomic surveys, including very large-scale studies reported by the J. Craig Venter Institute, did not reveal abundant bacteria beyond the already known phyla and have shown that only ∼10% of the sequences in the metagenomes have no detectable homologs (8–10). The possibility, certainly, remains that major new and, perhaps, unusual groups of archaea and bacteria dwell in complex and unusual habitats. Nevertheless, it appears likely that the current collections of archaeal and bacterial genomes provide a reasonable approximation of the diversity of prokaryotic life forms on earth. This being the case, the time seems ripe to critically examine the results of bacterial and archaeal genomics.
Figure 1.
The temporal dynamics of genome sequencing for bacteria and archaea. Bacteria: doubling time ∼20 months. Archaea: doubling time ∼34 months.
Table 1.
The state of genome sequencing for the archaeal and bacterial phylaa
| Phylum | No. of genomes sequenced | Genome size range, Mb | Representative (first genome sequenced) |
|---|---|---|---|
| Archaea | |||
| Crenarchaeota | 16 | 1.3–3 | Aeropyrum pernix K1 |
| Euryarchaeota | 34 | 1.6–5.8 | Methanocaldococcus jannaschii DSM 2661 |
| Korarchaeota | 1 | 1.6 | Korarchaeum cryptofilum OPF8 |
| Nanoarchaeota | 1 | 0.5 | Nanoarchaeum equitans Kin4-M |
| Bacteria | |||
| Acidobacteria | 2 | 5.7–10.0 | Acidobacteria bacterium Ellin345 |
| Actinobacteria | 54 | 0.9–9.7 | Mycobacterium tuberculosis H37Rv |
| Aquificae | 2 | 1.6–1.8 | Aquifex aeolicus VF5 |
| Bacteriodes/Chlorobi group | 21 | 0.3–6.3 | Chlorobium tepidum TLS |
| Chlamydiae/Verrucomicrobia group | 16 | 1.0–6.0 | Chlamydia trachomatis D/UW-3/CX |
| Chloroflexi | 7 | 1.3–6.7 | Dehalococcoides ethenogenes 195 |
| Chrysiogenetes | 0 | N/A | N/A |
| Cyanobacteria | 33 | 1.6–9.0 | Synechocystis sp. PCC 6803 |
| Deinococcus–Thermus group | 4 | 2.1–3.2 | Deinococcus radiodurans R1 |
| Firmicutes (Gram-positive bacteria) | 150 | 0.6–6.0 | Mycoplasma genitalium G37 |
| Fusobacteria | 1 | 2.2 | Fusobacterium nucleatum subsp. nucleatum ATCC 25586 |
| Gemmatimonadetes | 0 | N/A | N/A |
| Nitrospirae | 0 | N/A | N/A |
| Planctomycetes | 1 | 7.1 | Rhodopirellula baltica SH 1 |
| Proteobacteria | 353 | 0.2–13.0 | Haemophilus influenzae Rd KW20 |
| Spirochaetes | 13 | 0.9–4.7 | Borrelia burgdorferi B31 |
| Synergistetes | 0 | N/A | N/A |
| Thermodesulfobacteria | 0 | N/A | N/A |
| Thermotogae | 7 | 1.8–2.2 | Thermotoga maritima MSB8 |
aThe classification is from the NCBI taxonomy as of 10 June 2008 (http://www.ncbi.nlm.nih.gov/sites/entrez?db=taxonomy).
This survey is an attempt to identify general patterns of genome organization, function and evolution that can be gleaned from the results of comparative genomics. This is a vast subject, so it is unrealistic to cover all its aspects in any depth in a relatively short article. Moreover, comparative genomics naturally feeds into the study of fundamental issues of evolution that require separate discussion. We deliberately chose a rather perfunctory style of presentation in an attempt to at least mention as many salient aspects of bacterial and archaeal genomics as possible.
SIZE AND OVERALL ORGANIZATION OF BACTERIAL AND ARCHAEAL GENOMES
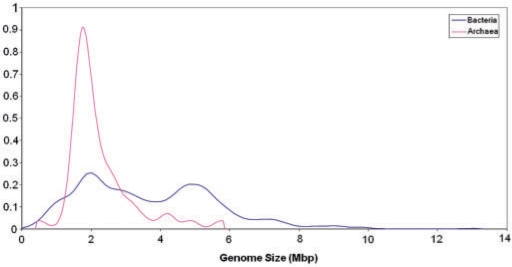
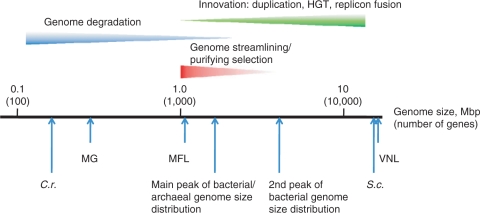
Despite the tremendous variety of life styles, as well as metabolic and genomic complexity, bacterial and archaeal genomes show easily discernible, common architectural principles. The sequenced bacterial genomes span two orders of magnitude in size, from ∼180 kb in the intracellular symbiont Carsonella rudii (11) to ∼13 Mb in the soil bacterium Sorangium cellulosum (12). Remarkably, bacteria show a clear-cut bimodal distribution of genome sizes, with the highest peak at ∼2 Mb and the second, smaller one at ∼5 Mb (Figure 2). Although there are many genomes of intermediate size, this distribution suggests the existence of two, more or less distinct classes of bacteria, those with ‘small’ and those with ‘large’ genomes [(13); the potential evolutionary forces that produced this distribution are addressed towards the end of this article]. The possibility remains that the bimodality of the bacterial genome size distribution is due to the bias of the genome sequencing efforts toward smaller genomes (such as those of symbionts and parasites) but with the growth of the genome collection, this explanation is becoming increasingly less plausible. Archaea are less diverse in genome size, from ∼0.5 Mb in the parasite Nanoarchaeum equitans (14) to ∼5.5 Mb in Methanosarcina barkeri (15) and show a sharp peak at ∼2 Mb that almost precisely coincides with the position of the highest bacterial peak, and a heavy tail corresponding to larger genomes (Figure 2). As the representation of archaeal genomes in the current databases is much less complete than the representation of bacterial genomes, it remains to be seen whether the genome size distributions in archaea and bacteria are genuinely different or the differences only reflect sequencing biases (that is, a second peak might appear in the archaeal distribution once additional, larger genomes of mesophilic archaea are sequenced). All very small (<1 Mb) genomes of bacteria and archaea belong to parasites and intracellular symbionts of eukaryotes and the only discovered archaeal parasite N. equitans that parasitizes on another archaeon, Ignicoccus hospitalis (14, 16). It appears that the minimal size of a free-living prokaryote is slightly >1 Mb, with the current record belonging to the abundant marine α-proteobacterium Pelagibacter ubique (SAR11), at ∼1.3 Mb (17).
Figure 2.
Distribution of genome sizes among bacteria and archaea. The distributions curves were obtained by Gaussian-kernel smoothing of the individual data points (276).
Notably, with the progress in genomics, it has become clear that there is no gulf in genome sizes between bacteria and archaea, viruses and eukaryotes. Indeed, the mimivirus has a genome that exceeds 1 Mb (18) and so is larger than the genomes of numerous, mostly, parasitic bacteria (and the archaeon N. equitans) and, nearly, the same size as the smallest genomes of free-living archaea and bacteria (19); such giant viruses appear to be abundant in marine habitats (20). On the other side of the genome size distribution, the smallest eukaryotic genomes, such as that of the microsporidian Encephalitozoon cuniculi (21), are substantially smaller than numerous archaeal and bacterial genomes.
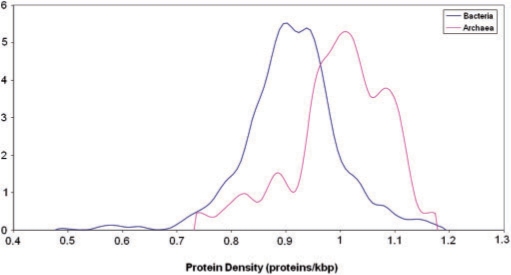
Both bacterial and archaeal genomes show unimodal and relatively narrow distributions of protein-coding gene densities, with the great majority encompassing between 0.8 and 1.2 genes per kilobase of genomic DNA (Figure 3). Notably, the archaeal distribution is significantly shifted toward higher densities compared to the bacterial distribution indicating that, on average, archaeal genomes are more compact than bacterial ones (Figure 3). Apparently, this substantial difference in gene density is a cumulative effect of small differences in characteristic protein lengths (Figure 4a) and intergenic region lengths (Figure 4b) both of which are slightly shorter in archaea than they are in bacteria.
Figure 3.
Density of protein-coding genes in bacterial and archaeal genomes. The distributions curves were obtained by Gaussian-kernel smoothing of the individual data points (276).
Figure 4.
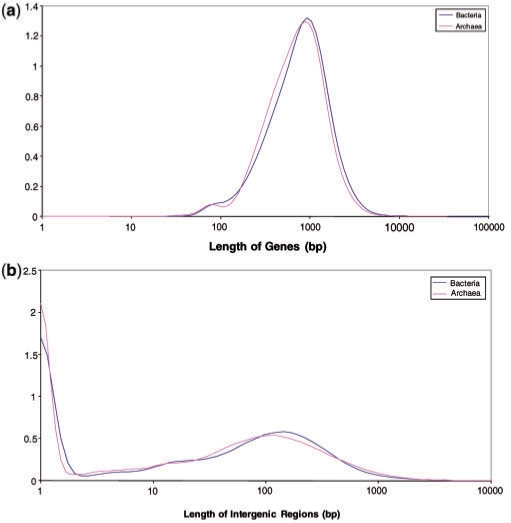
Length distributions of protein-coding genes (a) and intergenic regions (b) in bacterial and archaeal genomes. The distributions curves were obtained by Gaussian-kernel smoothing of the individual data points (276).
In accord with the general notion of genomic compactness, bacteria and archaea typically have intergenic distances that are much shorter than the characteristic lengths of the genes themselves (compare Figure 4a and b). The distributions of the lengths of intergenic regions for both archaea and bacteria (Figure 4b) are bimodal, with the first peak, at ∼0 bp, corresponding to the densely organized genome segments, primarily, within operons (see below), and the second peak, at ∼100 bp, corresponding to interoperonic regions. The tail of much longer intergenic regions (1000 bp and greater) encompasses specialized noncoding genomic segments, such as CRISPR repeats (22) and pseudogenes in certain intracellular parasitic bacteria, such as Mycobacterium leprae or Rickettsia, that appear to be in the process of extensive genome degradation via pseudogenization (22). The overwhelming majority of bacterial and archaeal proteins are encoded in uninterrupted open reading frames (ORFs), with the exceptions for a few archaeal genes that are interrupted by microintrons (23) and several split genes in archaea and bacteria that, apparently, evolved as a result of intein action (24). Furthermore, although short overlaps (a few base pairs in length) between protein-coding genes are common, there are no documented long overlaps (25).
In terms of the characteristic genome sizes and overall genome organization, bacteria do not qualitatively differ from archaea (although, as indicated above, the currently characterized archaea typically have smaller and more compact genomes), whereas both are sharply distinct from eukaryotes that span a much larger range of genome sizes, possess protein-coding genes that are, typically, interrupted by introns, and have longer intergenic regions. These features support the notion of a ‘prokaryotic principle of genome organization’ (see more below). An important practical implication of this principle is that gene prediction in sequenced archaeal and bacterial genomes is a relatively straightforward task. Considering the unity of genome organization in archaea and bacteria, in the rest of this article, we shall speak alternately of ‘archaea and bacteria’ or of ‘prokaryotes’ despite the recent objections to the use of the latter term (26); we briefly return to the legitimacy of the notion of prokaryotes toward the end of the article.
THE PROKARYOTIC GENE AND GENOME SPACES
Clusters of orthologs and classification of genes by phyletic patterns: the three classes of prokaryotic genes
One of the early and crucial generalizations of comparative genomics of prokaryotes is the readily recognizable evolutionary conservation of protein sequences encoded in the majority of the genes in each sequenced genome (27). More specifically, for a substantial majority of the genes, there are confidently identifiable orthologs in other, relatively distant bacteria and/or archaea. Orthologs are traditionally defined as genes that descend from the same ancestral gene in the common ancestor of the compared species (28). Of course, this crucial concept of evolutionary biology was originally defined in the context of evolutionary analysis of animal or plant species where the notion of the common ancestral species is unambiguous (29, 30). This is not the case in bacteria and archaea where horizontal gene transfer (HGT) is pervasive, and as the result, at least, in distant organisms, genes often have different histories (see below). Nevertheless, empirically, using the simple notion of a bidirectional best hit (BBH), it has been shown (shortly after the first complete genome sequences became available) that, for the majority of genes in any sequenced bacterial or archaeal genome, apparent counterparts (defined as orthologs, in a generalization of the original definition) were readily identifiable in other genomes (31, 32). These findings stimulated the development of the notion of clusters of orthologous genes (COGs) and methods for their identification (33, 34). Identification of COGs is a nontrivial task owing to evolutionary processes that confound orthologous relationships between genes, in particular, lineage-specific expansion of paralogous gene families that is common in archaea and bacteria (35), even if not nearly as prominent as it is in eukaryotes, and leads to coorthologous relationship between multiple paralogous genes in the compared genomes (28). Accordingly, the definition of a BBH needs to be generalized to include many-to-many (and many-to-one) relationships between genes [hence the original, rather awkward explication of COGs as Cluster of Orthologous Groups (33)]. Additional complications in the identification of orthologs stem from changes in domain architectures of proteins and differential loss of paralogous genes. Following the original COG study, a variety of increasingly sophisticated methods for identification of clusters of orthologs have been developed, some turning to explicit, genome-wide phylogenetic analysis (36–40). The latest and most comprehensive advancement in this direction is the EggNog project that relied on the COG collection as the nucleus of a new database of orthologous gene clusters including 312 bacterial and 26 archaeal genomes (41).
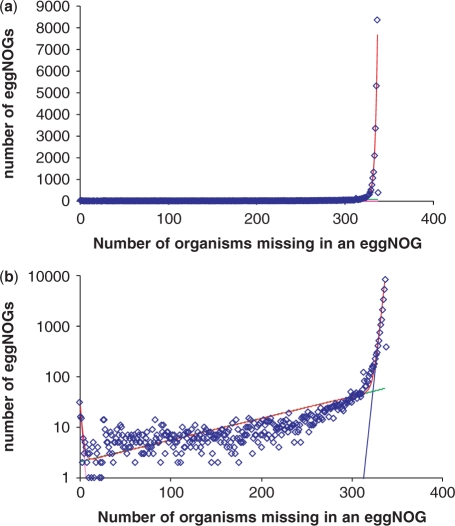
The coverage of selected archaeal and bacterial genomes in the EggNOG database is shown in Figure 5. With the notable exception of some bacteria with the largest genomes, such as Pirellula sp. and some archaea that belong to distinct, apparently, fast-evolving lineages, such as N. equitans, in most of the sequenced genomes, ∼80% of the genes (or more, in cases when closely related genomes are available) belong to clusters of orthologs. Thus, the great majority of proteins encoded in each sequenced archaeal or bacterial genome show, at least, some degree of evolutionary conservation within the explored portion of the prokaryotic gene space. However, the distribution of the clusters by the number of included organisms immediately reveals the flip side of the coin: the great majority of the clusters include only a few organisms (Figure 6a). A more detailed examination of this distribution reveals distinct structure in the prokaryotic sequence space. The distribution is, essentially, an exponential decay curve, with a rise at the left end that corresponds to the universal or nearly universal clusters. Assuming that the distribution is described by an exponent(s), the best approximation is obtained with a sum of three exponential functions (Figure 6b). The first exponent represents the conserved (universal or nearly universal) gene core (∼70 clusters), the second exponent describes the ‘shell’ of moderately common genes (∼5700 clusters), and the third exponent corresponds to the ‘cloud’ (∼24 000 clusters) that consists of genes shared by a small number of organisms. The possibility exists that the size of the cloud is somewhat inflated, i.e. some of the small clusters actually include highly diverged orthologous genes and have to be merged. However, the same overall shape of the distribution has been seen in independent studies, e.g. the recent analysis of archaeal COGs (42), suggesting that it reflects the actual structure of the prokaryotic gene space that consists of:
a miniscule fraction of highly conserved genes,
a much (two orders of magnitude) larger set of moderately conserved genes,
an even greater number of narrowly distributed genes.
This diversity of phyletic (phylogenetic) patterns (a term often used to describe the distribution of genes across organisms) reflects major trends in prokaryotic evolution, namely, extensive horizontal transfer of genes, pervasive gene loss and functional plasticity of many cellular systems (see below).
Figure 5.
Coverage of bacterial and archaeal genomes with cluster of orthologous genes. The COGs were from the EggNOG database (41), and the proteins from each genome were assigned to these clusters using a modified COGNITOR method (42).
Figure 6.
Representation of bacteria and archaea in clusters of orthologs: core, shell and cloud (a) distribution#of clusters of orthologs [from EggNOG (41)] by the number of included genomes—linear plot; (b) distribution of clusters of orthologs by the number of included genomes (semi-logarithmic plot) and approximation with three exponential functions.
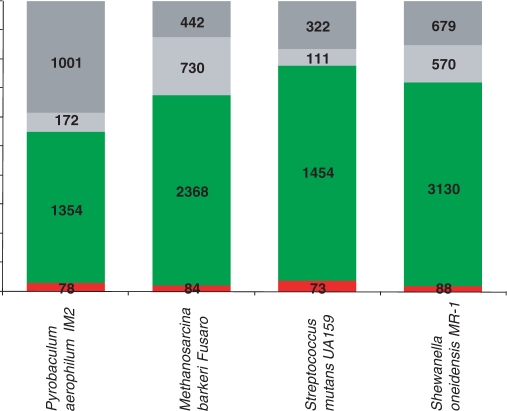
In the current databases, there is also a large number of archaeal and bacterial genes that encode protein sequences without detectable similarity to any other available protein sequences; accordingly, these genes are often denoted ORFans (43, 44). Typically, ORFans comprise 10–15% of the predicted genes in archaeal and bacterial genomes, depending on the availability of closely related genomes (Figure 7). The ORFans have also received the less flattering name ELFs, Evil Little Fellows, and it has been argued that many of them are false predictions rather than actual protein-coding genes (45). Furthermore, it has been proposed that the majority of those ORFans that are real genes were derived from bacteriophages and, accordingly, are characterized by high horizontal mobility although, occasionally, they can be recruited for a cellular function and, accordingly, fixed in a bacterial or archaeal lineage (46). Recent estimates from metagenomic surveys of bacteriophages suggest that the diversity of phage sequences is vast and remains, largely, unexplored (47). Therefore, it does seem plausible that a major fraction of bacterial and archaeal ORFans derives from the still poorly explored but, certainly, vast bacteriophage gene pool. Obviously, it is impossible to rule out and, indeed, is most likely that a fraction of the ORFans have orthologs in multiple prokaryotic genomes that avoid detection because of their rapid evolution, a possibility that is not incompatible with the origin of most ORFans in the phage gene pool.
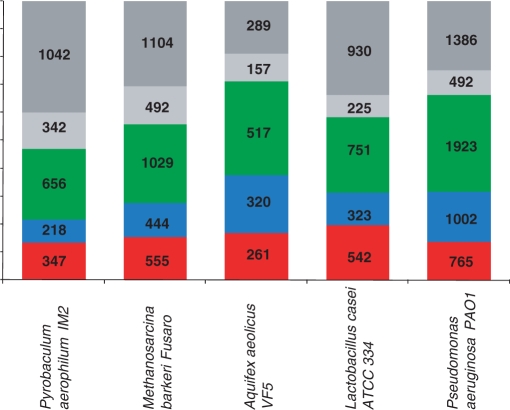
Figure 7.
Common and rare genes in selected archaeal and bacterial genomes. Red, core; green, shell; light gray, cloud; dark gray, ORFans. The assignment of the genes from each genome to one of the four classes was based on their inclusion to the core, shell or cloud EggNOGs (Figure 6); the remaining genes were classified as ORFans.
When elements of the gene space are represented as clusters of orthologs, it appears that ORFans can be reasonably merged into the ‘cloud’ of poorly represented, rare genes. This compounded ‘cloud’ obviously dominates the gene space when each cluster of orthologous genes is taken as a point. This is, however, not the case when individual genomes are considered: in each genome, the majority of the genes belong to the moderately conserved ‘shell’ (Figure 7). Of course, there is no paradox involved because, although the fraction of ‘cloud’ genes and ORFans in each genome is relatively small, they are, by definition (nearly) unique and, combined, account for the great majority of points in the gene space.
Detailed extrapolation of the expansion of the gene space with further bacterial and archaeal genome sequencing and a reliable estimate of the actual size of this space are hard to obtain, and such analysis is beyond the scope of the present article. Nevertheless, considering the vast diversity of bacteriophages revealed in metagenomic analyses, it appears most likely that the number of elements of the prokaryotic gene space will increase by orders of magnitude, and almost entirely, through expansion of the ‘cloud’.
Clustering of prokaryotes in the genome space and genome trees
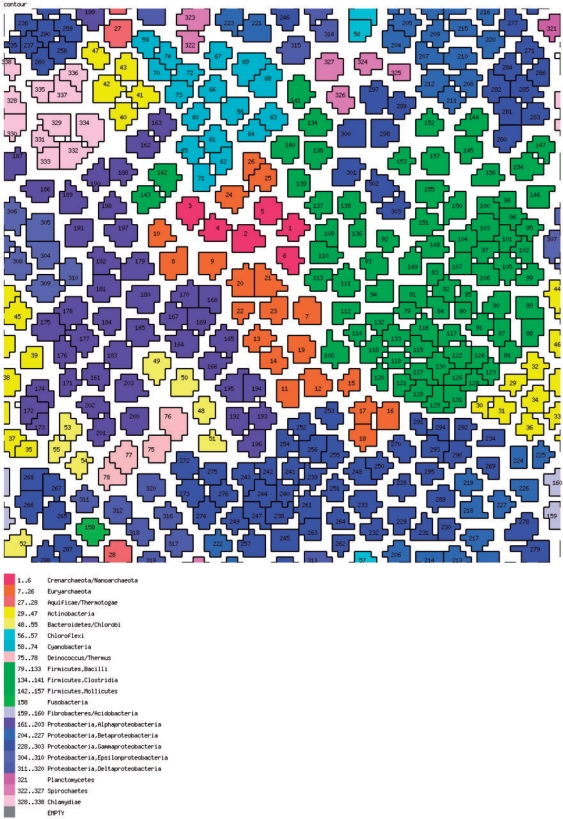
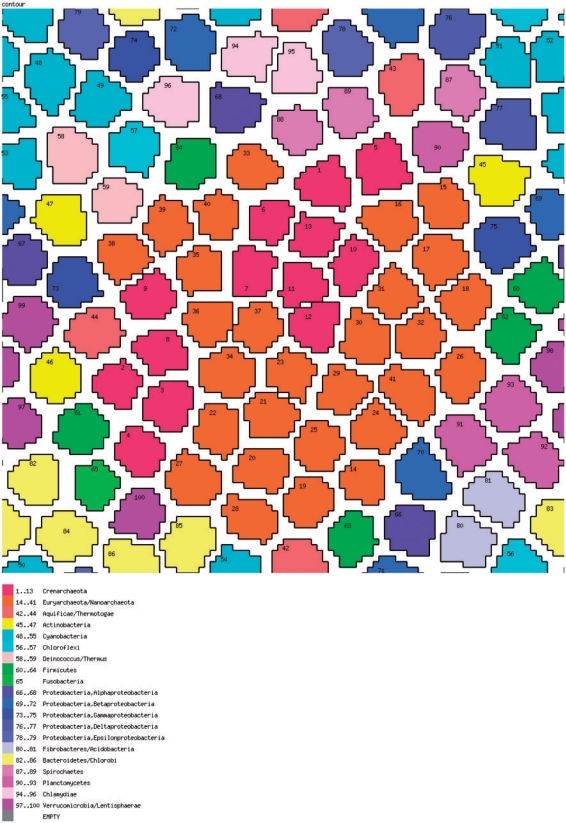
So far, we did not directly visualize the prokaryotic gene space other than in the highly abstracted form of distributions shown in Figure 6. It is easy to conceive of a more compact genome space that is conducive to simple visualization. To this end, the gene set of each organism can be conveniently represented as a vector of absence–presence in clusters of orthologs (COGs): 1 for each instance of presence of a member from the given genome in a COG, and 0 for each instance of absence. It is easy to see that these COG–genome vectors are orthogonal to phyletic patterns of COGs, i.e. phyletic patterns comprise the columns and the genome–COG vectors comprise the rows of the complete genome–COG correspondence table a fragment of which is shown in Figure 8. At this time, the number of COGs exceeds the number of genomes by, roughly, an order of magnitude, so the genome–COG vectors can be more readily compared and clustered using a variety of classification methods. We chose the self-organizing map (SOM) (48) approach to map these orthology vectors in the genome space. The SOMs are a useful and popular method to visualize a low (typically, two)-dimensional representation of high-dimensional data. Essentially, a SOM is a ‘semantic’ map where similar samples are adjacent, whereas dissimilar ones are disjointed. The SOM reveals clean separation between archaea and bacteria, and compact clustering of related genomes representing most of the major prokaryotic divisions (Figure 9). This coherence was seen not only for long recognized, firmly established groups, but also for relatively nontrivial ones such as, for instance, the Thermus–Deinococcus group. However, several larger groups were split into two or more disjointed areas, e.g. γ-proteobacteria, apparently, due to the diversity of life styles leading to dissimilar gene complements, e.g. as a result of extensive gene loss in intracellular symbionts.
Figure 8.
Genome–COG vectors. A fragment of the complete genome–COG matrix is shown. The number 1 indicates the presence and 0 indicates the absence of a gene(s) from the given genome in the given COG.
Figure 9.
The prokaryotic genome space: a SOM. The SOM was produced using a custom script that implements the Kohonen algorithm (48).
The genome–COG vectors also can be analyzed using standard phylogenetic methods and have been employed to generate ‘genome-trees’, i.e. trees that reflect the relationships between gene contents of archaea and bacteria (49). Similarly, to the message derived from the SOMs, the genome-trees seem to reveal a mixture of evolutionary and ‘biological’ signals, i.e. some of the clades reflect common aspects of the life style of the respective organisms, such as extensive gene loss in parasites (50).
THE FUNCTIONAL SPACE OF ARCHAEA AND BACTERIA
Experimental elucidation of gene functions lags far behind genome sequencing, and this gulf is unlikely to be crossed any time soon. Therefore, the central finding of comparative genomics, that the great majority of bacterial and archaeal genes belong to clusters of orthologs, is also critical for the success of functional annotation. The routine process of assignment of functions to genes in a sequenced genome involves comparison to other genomes, inclusion of genes from the new genome into preexisting clusters of orthologs and transfer of functional annotation from experimentally characterized genes to uncharacterized ones, usually, via a combination of automatic and manual procedures (51–54). Compiling information from multiple organisms progressively helps increasing annotation coverage. Additional functional information can be obtained through genome-context analysis approaches that are, also, steeped in genome comparison and rely on conservation of arrays of functionally linked genes (55, 56) (see below). Certainly, functional annotations of genomes requires extreme caution as transfer of (sometimes, incomplete or inaccurate) functional information between orthologs (not always correctly identified) from distant genomes is quite error-prone (57, 58). Functional annotation by means of comparative genomics is covered in detail in many reviews and benchmarking studies (59–61), and it is not our intention to discuss this subject in detail here. Typically, at this stage, in the evolution of prokaryotic genomics, annotation of a newly sequenced archaeal or bacterial genome goes far enough to assign 60–70% of the protein-coding genes to one of specifically defined functional categories, and another 10–15% of the genes receive a general functional prediction (typically, of biochemical activity but not biological function proper) (Figure 10). In small genomes, particularly, those of parasites, the genes that encode components of information processing systems (translation, transcription and replication) comprise a major fraction; in contrast, in larger genomes, their contribution is much smaller, whereas genes encoding metabolic and signal transduction proteins and those with other, diverse functions are prevalent (Figure 10 and see below).
Figure 10.
Distribution of predicted gene functional classes for selected archaeal and bacterial genomes. Red, information processing genes; blue, genes involved in cellular functions; green, genes involved in metabolism and transport; light gray, general prediction only; dark gray, no prediction. The function class assignment is based on the inclusion of the respective genes in COGs (34).
Today's genome annotation usually is sufficiently complete to produce the iconic illustration of numerous genomic papers, a schematic of a prokaryotic cell, with the principal metabolic pathways (and, in some cases, information processing functions as well) depicted inside and transport systems decorating the membrane [an image that, to our knowledge, was first used to depict the reconstructed biology of the spirochaete Borrelia burgdorferi (62)]. However, comparison of these in silico reconstructed cells shows, first, that they almost always contain white spots and missing links in the metabolic and transport map, and second, that the metabolic pathways and transport systems within these virtual cells are far from being the same in all bacteria or archaea (let alone across the two domains). On the contrary, remarkable biochemical diversity is a hallmark of bacterial and archaeal biology. The existence, even if not the full extent of biochemical diversity has been recognized in the pregenomic era within the confines of traditional microbiology. What has become clear with the advent of comparative genomics, is the wide spread of nonorthologous gene displacement, i.e. recruitment of unrelated genes (or distantly related, nonorthologous genes) for the same function (63). Nonorthologous displacement affects all functional classes of genes, with striking examples seen even among the most fundamental functions, such as DNA replication, where the principal replicative enzymes are nonhomologous in archaea and bacteria (64, 65). In general, however, functional diversity and nonorthologous displacement are much more prominent among proteins involved in operational (as opposed to informational) functions such as metabolism, transport and signal transduction (54, 66), which is reflected in the major differences in the distributions of the number of organisms in the respective clusters of orthologs (Figure 11). Due to nonorthologous displacement, the functional space of archaea and bacteria is not isomorphous (i.e. does not allow a one-to-one mapping) to the gene space because numerous functions correspond to more than one cluster of orthologous genes.
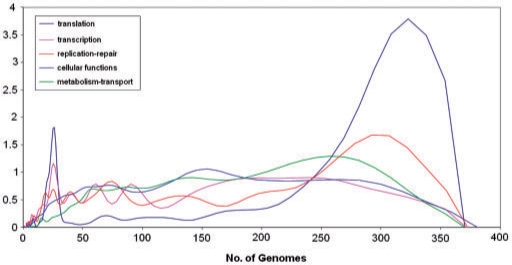
Figure 11.
Distributions of the number of organisms in clusters of orthologs for informational and operational genes. Translation, transcription and replication repair are informational function classes, and the rest are operational function classes. The distributions curves were obtained by Gaussian-kernel smoothing of the individual data points (276).
To compare the mapping of the functional space to that of the genomic space, we applied the same SOM technology to genome–function vectors, where each COG present in a particular genome is denoted by the corresponding functional category. The resulting map (Figure 12) is qualitatively different from the genomic-space map (Figure 9): archaea are, again, clearly distinct from bacteria, but the majority of bacterial phyla form multiple clusters, a pattern that seems to reflect the diversity of the functional repertoires even among rather closely related bacteria, especially, in cases of genomic degradation in parasites and symbionts.
Figure 12.
The function space of prokaryotes: a SOM. The SOM was produced using a custom script that implements the Kohonen algorithm (48).
PRINCIPLES OF PROKARYOTIC GENOME ARCHITECTURE AND ITS EVOLUTION
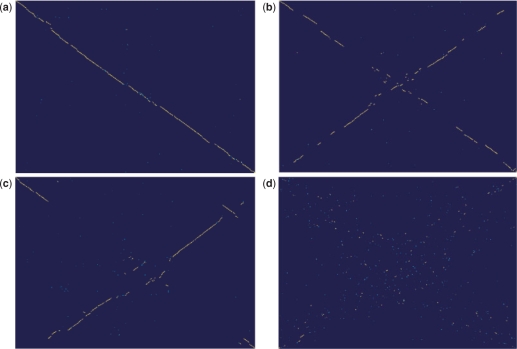
Almost immediately after the release of the first complete genome sequences, it became apparent that the gene order in bacterial and archaeal genomes is relatively poorly conserved (4,67–69), dramatically less so than genes themselves (see above). To analyze conservation of gene orders, one needs to obtain a robust set of orthologous genes between the compared genomes. Once such a set of orthologous genes is defined, it becomes straightforward to assess the gene order conservation by means of a dot-plot (one of the earliest representations of nucleotide and protein sequence similarity) where each point corresponds to a pair of orthologs. Examination of these plots reveals rapid divergence of gene order in prokaryotes (Figure 13) so that, even between closely related organisms, the chromosomal colinearity is broken at several points (Figure 13a), moderately diverged organisms show only a few extended collinear regions (Figure 13b and c), whereas for any pair of relatively distant organisms, the plot looks like the map of the night sky (Figure 13d). Disruption of synteny during evolution of bacterial and archaeal genomes typically shows a clear and striking pattern, with an X-shape seen in the dot-plots (Figure 11b and c). It has been proposed that the X-pattern is generated by symmetric chromosomal inversions around the origin of replication (70). The underlying cause of these inversions could be the high frequency of recombination in replication forks that, in the circular chromosomes of bacteria and archaea, are normally located on both sides of and at the same distance from the origin site (71).
Figure 13.
Evolution of gene order in bacteria and archaea: genomic dot-plots. (a) Colinearity with a few breakpoints between closely related bacteria: Geobacillus thermodenitrificans versus Geobacillus kaustophilus; (b) X-shaped pattern between moderately diverged bacteria: Shewanella sp. MR-4 versus Shewanella oneidensis; (c) X-shaped pattern between moderately diverged archaea: Pyrococcus horikoshii OT3 versus Pyrococcus abyssi GE5; and (d) No clear pattern between more distantly related bacteria: Streptococcus gordonii str. Challis versus Streptococcus pneumoniae R6. In each panel, the genome indicated first is plotted along the vertical axis.
Most prokaryotic genomes contain a single, bidirectional replication origin site, and this origin is a special point in the genome that defines the global genome architecture (72). By definition, a bidirectional origin is the switch point between the leading and lagging strand that in bacteria and archaea are replicated in different modes, continuous and discontinuous, respectively. In most prokaryotes, the leading and lagging strands show substantial asymmetries in nucleotide composition, gene orientation and gene content (73). A diagnostic distinction between the leading and lagging strands is the difference in GC- and AT-skews, i.e. excess of purines or pyrimidines (violation of Chargaff's second parity rule). The underlying causes of the GC/AT-skews are thought to reflect an interplay of selective and mutational forces, i.e. selection against secondary structure formation in the leading strand and differential increase of different types of mutations in single-stranded DNA (74, 75). The GC/AT-skew patterns in the leading and lagging strands of bacterial and archaeal chromosomes are consistent and significant enough to (usually) allow an accurate prediction of the origin position in an uncharacterized prokaryotic genome (76, 77). The leading and lagging strands also show asymmetric (to a widely varying degree in different genomes) distributions of genes, with a greater density of genes found on the leading strand. Moreover, a substantial majority of these genes, especially, highly expressed and/or essential ones, e.g. those coding for ribosomal RNAs and proteins, are cooriented with replication (78–81). Usually, the patterns of gene distribution are explained by different versions of the polymerase collision model that postulates selection for minimizing head-on collision between the replicating DNA polymerase and the transcribing RNA polymerase that are both more likely and more damaging than codirectional collisions (73,78,79). The exact mechanisms that affect the overall layout of bacterial chromosomes require much further analysis and cannot be discussed here in detail but the general conclusion seems clear that the mechanisms and rate of chromosomal replication are important factors that determine the genome architecture.
One of the earliest and central concepts of bacterial genetics is the operon, a group of cotranscribed and coregulated genes (82). Although enormous amount of variation on the simple theme of regulation by the Lac repressor developed by Jacob and Monod (83) has been discovered in the years since, the operon has stood the test of comparative genomics as the principle of organization of bacterial and archaeal genomes (84). Operons, particularly, those that encode physically interacting proteins, are much stronger conserved during evolution of bacterial and archaeal genomes than large-scale synteny. Comparative analysis of gene order in bacteria and archaea reveals relatively few operons that are shared by a broad range of organisms. As noticed early on, these highly conserved operons typically encode physically interacting proteins (68), a trend that is readily interpretable in terms of selection against the deleterious effects of imbalance between protein complex subunits (85). The most striking illustration of this trend is the ribosomal superoperon that includes over 50 genes of ribosomal proteins that are found in different combinations and arrangements in all sequenced archaeal and bacterial genomes (86, 87). Analysis of the ribosomal superoperon and other, smaller groups of partially conserved operons led to the notion of an überoperon (88) or a conserved gene neighborhood (89), an array of overlapping, partially conserved genes strings (known or predicted operons). In addition to the ribosomal superoperon, striking examples of conserved neighborhoods are the group of predicted overlapping operons that encode subunits of the archaeal exosomal complex (90) and the Cas genes that comprise an antivirus defense system (see also below) (89,91,92). Analysis of such large, partially conserved neighborhoods has high predictive value and can lead to the identification of novel functional systems, as in the latter two cases. The majority of genes in the überoperons encode proteins involved in the same process and/or complex but highly conserved arrangements including genes with seemingly unrelated functions exist as well, e.g. the common occurrence of the enolase gene in ribosomal neighborhoods or genes for proteasome subunits in the archaeal exosome neighborhood. The presence of these seemingly unrelated genes can be explained either by ‘gene sharing’, i.e. multiple functionalities of the respective proteins, or by ‘genomic hitchhiking’, a case when an operon combines genes without specific functional links but with similar requirements for expression (89).
The majority of operons do not belong to complex, interconnected neighborhood but instead are simple strings of two to four genes, with variations in their arrangement (69,86,89,93). Identical or similar, in terms of gene organization, operons are often found in highly diverse organisms and in different functional systems. A case in point is numerous metabolite transport operons that consist of similarly arranged genes encoding the transmebrane permease, ATPase and periplasmic subunits of the so-called ABC transporters (94). The persistence of such common operons in diverse bacteria and archaea has been interpreted within the framework of the selfish operon concept, the notion that operons are maintained not so much because of the functional importance of coregulation of the constituent genes but due to the selfish character of these compact genetic units that are prone to horizontal spread among prokaryotes (95–97) (we will return to this concept in the discussion of horizontal gene transfer subsequently).
A systematic comparison of the arrangements of orthologous genes in archaeal and bacterial genomes revealed a relatively small fraction of conserved (predicted) operons and a much greater abundance of unique directons, i.e. strings of genes transcribed in the same direction and separated by short intergenic sequences (83, 86). In benchmark studies, directons have been shown to be surprisingly accurate predictors of operons (98). Thus, the organization of archaeal and bacterial genomes seem to be governed by the operonic principle, with a small number of highly conserved operons and a much larger number of unique or rare ones. In this respect, the pattern of operon conservation is reminiscent of the distribution of clusters of orthologs that includes a small, highly conserved core, a larger, moderately conserved ‘shell’, and an expansive ‘cloud’ of (nearly) unique genes (Figure 6).
The degree of genome ‘operonization’ widely differs among bacteria and archaea; some genomes, e.g. that of the hyperthermophilic bacterium Thermotoga maritima, are almost fully covered by (predicted) operons, whereas others, such as those of most Cyanobacteria, seem to contain few operons (86, 99). What determines the extent of operonization in an organism remains unclear, although it stands to reason that this degree depends on the balance between intensity of recombination, the horizontal gene flux and selective forces that oppose disruption of operons.
PRINCIPLES OF REGULATION AND SIGNAL TRANSDUCTION IN BACTERIA AND ARCHAEA: AN OVERHAUL IN THE ERA OF COMPARATIVE GENOMICS
Bacteria and archaea possess distinct, elegantly structured systems of gene-expression regulation, and comparative genomics has dramatically changed the existing views of their organizational principles, distribution in nature and evolution. The operon concept of Jacob and Monod (82), which was introduced above as the organizing principle of the local architecture of bacterial and archaeal genome, is also the paradigm of gene expression regulation and signal transduction in these organisms (84). Under the Jacob–Monod model, the regulator (the lac-repressor in their classic study) is a sensor of extracellular or intracellular cues (in this case, the concentration of lactose) that affect the regulator protein conformation and, indirectly, the expression state of the operon (in the case of the lac-operon, the repressor binds lactose, dissociates from the operator and allows transcription). Over the 47 years that elapsed since Jacob–Monod's breakthrough, numerous variations on this subject have been discovered, including regulators that symmetrically affect transcription of adjacent divergent genes, and global regulators that regulate numerous, dispersed genes and operons, as opposed to specific regulation of a single operon under the Jacob–Monod model (100–102). The most prominent global regulators are the catabolite repressor protein (CRP) (103, 104) and the stress response (SOS) regulator LexA (105). Considering the discovery of these and other global regulators, the operon concept was amended with the notion of a regulon, a set of genes that share the same cis regulatory signal (operator) and are regulated by the same regulator protein (106, 107). Comparative genomic analysis of regulons has revealed their extreme evolutionary plasticity, with substantial differences found between regulons seen even among closely related organisms (108–110). A global transcription regulator, such as LexA, can be widespread and highly conserved in diverse bacteria but the gene composition of the LexA regulon is highly variable. The plasticity of regulons parallels the variability of genome architectures (see above) in support of the notion that regulation of gene expression and genome architecture are tightly linked in the evolution of archaea and bacteria.
In a striking contrast to the variability and plasticity of regulons, there is a remarkable unity in the architecture and structure of bacterial and archaeal transcription regulators (111–113). Typically, these regulators consist of a small molecule-binding sensor domain and a DNA-binding domain. The overwhelming majority of the DNA-binding domains are variations on the same structural theme, helix–turn–helix (113). Less common DNA-binding domains include ribbon–helix–helix and Zn-ribbon (111).
A more complex scheme of signal transduction and expression regulation that is dedicated to sensing extracellular cues is embodied in the two-component systems. Two-component systems consist of a membrane histidine kinase and a soluble response regulator between which the signal is transmitted via a phosphotransfer relay (114–116). Notably, the classical transcriptional regulators and histidine kinases share many of the same sensor (input) domain, a kinship that prompts one to consider the transcriptional regulators (one-component systems) and the two-component systems within the same, integrated framework of signal transduction and expression regulation. The one-component systems that are nearly ubiquitous and, typically, numerically dominant in bacteria and archaea are thought to be the ancestral signal transduction devices, whereas the two-component systems are likely to be a derivative, more elaborate form of signal transduction that evolved as an adaptation for environmental signaling (117).
Comparative genomics of bacteria and archaea was instrumental in the discovery of novel, previously unsuspected but, actually, common forms of signal transduction. It has been known for years that a common form of global regulation in bacteria is mediated by cAMP, with the participation of diverse adenylate cyclases (a striking case of nonorthologous gene displacement), numerous proteins containing cAMP sensors, such as the GAF domain, and the CRP, FNR and other transcription regulators also containing cAMP-binding domains (118, 119). Comparative genomic analyses revealed numerous uncharacterized proteins that contain many of the same sensor domains that are characteristic of cAMP-dependent regulators and two-component systems combined with one or two novel domains, GGDEF and EAL, so denoted after their conserved amino acid signatures (120). The genomic context of these domains and the demonstration that the GGDEF domain is a distant homolog of one of the classes of adenylate cyclases (121) has led to the hypothesis that these proteins were components of a novel signal transduction system(s). Subsequently, this system has been, indeed, discovered through the demonstration that the GGDEG domain possessed the activity of a di-GMP cyclase, whereas EAL is a cyclic di-GMP phosphodiesterase (122). The c-di-GMP-dependent signal transduction, the existence of which was not even suspected in the pregenomic era, is emerging as a major regulatory system in bacteria and archaea.
Similarly, comparative genomic analysis has convincingly shown that serine–threonine protein kinases and the corresponding phosphatases, previously conceived as staples of eukaryotic organisms, are common and diverse among archaea and bacteria (123), and appear to be another major component of the increasingly complex prokaryotic signal transduction network (124–126).
Analysis of some of the larger bacterial genomes unexpectedly revealed the presence of homologs of some of the proteins previously thought to be limited in their spread to eukaryotes and involved in such quintessentially eukaryotic signal transduction networks as programmed cell death (PCD). These proteins include proteases of the caspase superfamily, AP-ATPase family ATPases, and NACHT family GTPases, all of which are involved in various forms of plant and animal PCD (127, 128). Typically, these proteins possess complex multidomain, modular architecture, with diverse domains mediating protein–protein interactions appended to the respective catalytic domains. These predicted signaling molecules are most common in bacteria with complex developmental phases, such as cyanobacteria, actinobacteria and myxobacteria, and are present also in Methanosarcinales, so far the only group of archaea with relatively large genomes and complex morphology. A detailed investigation of the functions of these proteins remains to be performed but there are preliminary indications that, at least, in some bacteria, they might be involved in PCD (129). These findings indicate that at least some of the complex signaling networks of eukaryotes have their counterparts and putative evolutionary predecessors in bacteria. Further discussion of the implications of these findings for the evolution of eukaryotes is beyond the scope of this article but the salient point is that comparative genomics reveals the existence of previously unsuspected and unexpectedly complex signaling systems in bacteria and archaea.
The organisms with the smallest genomes, i.e. parasitic and symbiotic bacteria and the only known archaeal parasite, N. equitans, encode (virtually) no regulators, whereas in bacteria with the largest known genomes, the regulators and signaling proteins comprise a substantial portion of the gene repertoire (Figure 10). Numerous deviations from the trend notwithstanding, it has been consistently shown that the number of regulatory and signal transduction proteins that are encoded in a genome scales, roughly, as the square of the total number of genes, i.e. on average, the larger the genome, the greater is the fraction of genes dedicated to signal transduction (117,130–132) (see further discussion subsequently).
Along with the general dependence on genome size, comparative genomic analysis reveals great variation among bacteria and archaea in the complexity of their signal transduction systems that seems to reflect the organism's life style. This variation in the fraction of the genes dedicated to signal transduction was quantitatively captured in the notion of the ‘bacterial IQ’, a quotient that is proportional to the square root of the number of signal transduction proteins (given the aforementioned scaling) and inversely proportional to the total number of genes (132). The IQ reflects the ability of bacteria and archaea to respond to diverse environmental stimuli. Accordingly, the IQ values are the lowest in intracellular symbionts (parasites), are only slightly higher in organisms with compact genomes that inhabit stable environments, such as marine cyanobacteria, but are much greater in organisms from complex and changing environments, even those with relatively small genomes.
GENOMIC COMPLEXITY OF PROKARYOTES: MINIMAL GENE SETS AND THE ‘BUREAUCRATIC CEILING’ OF COMPLEXITY
All archaea and bacteria are cellular organisms that possess replicating chromosomes, the machinery for genome expression, membranes endowed with transport and energy-transforming systems, and at least a minimal metabolic circuitry. The necessity to produce and maintain all these complex systems, certainly, imposes a low bound on genomic complexity. An attempt to define a minimal gene set for a bacterial cell has been undertaken as soon as the first two bacterial genome sequences (H. influenzae and M. genitalium) became available (133). By identifying the set of orthologs and supplementing it with some more or less educated guesses on apparent instances of nonorthologous gene displacement, the minimal gene set for a bacterium growing on a rich medium (i.e. with minimal biosynthetic requirements) was estimated at ∼250 genes. Limited revisions of this estimate have been offered (134–136) drawing from more complete comparative genomic analyses, and experimental studies on knockout mutants variously defined the number of essential genes in bacteria between ∼300 and ∼700, depending on the life style (in more complex bacteria, these can be underestimates of the minimal gene set because of functional redundancy among some genes) (137–141). On the whole, it appears that the original estimate (133) was reasonable although, possibly, on the low side of a realistic minimal gene repertoire of a viable bacterium (or archaeon). In a completely unexpected development, the genome of the endosymbiont C. rudii was found to contain only ∼170 genes, which is fewer than any estimates of the minimal gene set (11, 142). However, this unusual organism lacks certain genes that are present in all other known bacteria and archaea and encode proteins that appear to be indispensable, e.g. some of the aminoacyl-tRNA synthetases. At present, the best possible explanation is that this organism imports these essential proteins from the host cell, thereby violating the apparent constraint affecting other prokaryotic parasites and symbionts, even intracellular ones (133). Thus, conceivably, Carsonella is a case of a bacterium-to-organelle transition in progress (142). The minimal complexity for a heterotrophic organism growing on a rich medium is likely to remain at approximately 250 genes. The smallest genomes of currently known free-living organisms, e.g. P. ubique, are ∼1.3 Mb in size, with ∼1100 genes (17). Considering that even these genomes contain up to 15% ORFans that are, generally, nonessential, it is reasonable to project the minimal gene set for a free-living organism to the convenient round number of approximately 1000 genes. Clearly, given the wide spread of nonorthologous gene displacement, a minimal prokaryotic gene set is not a unique combination of genes. Instead, there can be a large number of minimal organisms with diverse life styles but, roughly, the same number of genes (135).
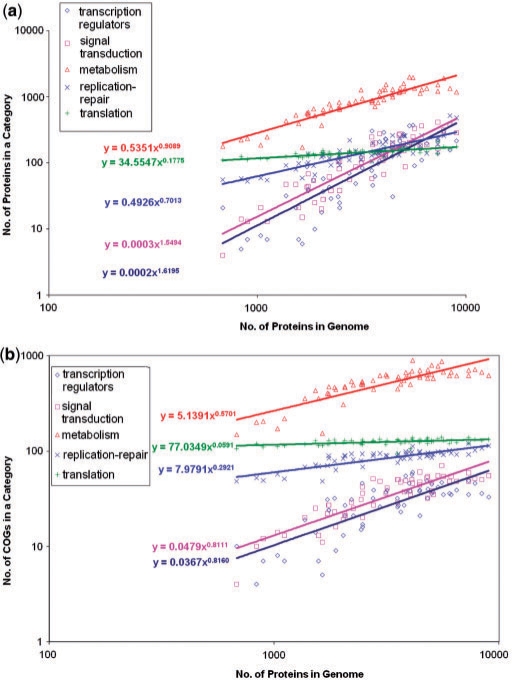
More fundamental questions, perhaps, are what determines the actual complexity of bacterial and archaeal genomes and what if anything gives the upper bound to this complexity. To address this problem, we turn to the analysis of scaling of different functional categories of genes with genome size that was already referred to in the above discussion of signal transduction systems. As first noticed (to our knowledge) by Stover et al. (143) in the course of the genome analysis of the bacterium Pseudomonas aeruginosa, investigated in detail by Van Nimwegen (130) and subsequently independently confirmed and explored by several groups (117,131,132), genes in different functional categories show dramatic differences in their dependence on the total number of genes. All broadly defined functional categories scale as a power function of the total gene number but the exponents of the power laws widely differ and reveal a distinct pattern. The numbers of genes coding for protein components of the translation system and those for proteins involved in cell division show almost no dependence on genome size (exponent close to 0); the counts of genes encoding metabolic enzymes, transporters, as well as proteins involved in DNA replication and repair are, roughly, proportional to the genome size (exponent close to 1) and, transcriptional regulators and proteins involved in signal transduction (e.g. two-component systems) have exponents close to 2, that is, scale (almost) with the square of the total number of genes, meaning that the fraction of the regulatory proteins scales (almost) linearly with the number of genes. An analysis we performed with representative sets of bacterial and archaeal genomes from diverse lineages corroborates these observations (Figure 14a). Notably, when the dependence was examined by plotting the number of orthologous clusters (COGs) in the respective categories (as opposed to individual genes), none of the categories showed an exponent greater than one (Figure 14b). Thus, the excess of regulators and signal transduction proteins in larger genomes seems to stem, primarily, from lineage-specific proliferation of families of paralogous genes (35). Van Nimwegen proposed that the ratios of the duplication rates to gene elimination rates that determine the exponents of the power laws for each class of genes are ‘universal constants’ of prokaryotic evolution (i.e. are, at least, approximately, the same in all bacterial and archaeal lineages and throughout the course of prokaryotic evolution), resulting in the observed distinct dependences for different functional classes of genes (130). This conjecture remains to be thoroughly tested by investigation of an adequate sampling of diverse prokaryotic lineages as some evidence of substantial lineage-specific differences as well as time dynamics has been reported (144).
Figure 14.
Scaling of genes in different functional categories with the total number of genes in archaeal and bacterial and genomes. (a) Data for individual protein-coding genes. (b) Data for COGs. The function class assignment is based on the inclusion of the respective genes in COGs (34).
The complexity of the translation and cell division systems seems to be almost the same in all bacteria and archaea regardless of the genome size. Presumably, these systems have undergone little evolution after the emergence of archaeal and bacterial cells, perhaps, with the exception of limited gene loss in the most degraded parasites and symbionts (145). Some metabolic proteins, in particular, those involved in the metabolism and transport of nucleotides, show a similar pattern (131), again, in agreement with their near universal conservation (135), but for most metabolic pathways, complexity grows along with the genome. Conceivably, this increasing metabolic complexity requires or, at least, strongly favors a disproportionate increase in the set of genes dedicated to regulation and signal transduction. Indeed, it appears that the architecture of the transcription regulatory network dramatically depends on the genome size. Small genomes encode a small number of transcription regulators each of which targets many binding sites on the chromosome, whereas large genomes encode many regulators with a small number of target sites each (146). In agreement with these findings, we recently observed that the degree of ‘operonization’ of bacterial and archaeal genomes significantly decreases with the increase of the genome size, that is, larger genomes seem to have smaller operons regulated by diverse transcription factors (J. Strasburger and Y.I.W., unpublished data). This increasing burden of ‘cellular bureaucracy’ (the regulators) could be at least one of the major factors that determine the maximum attainable size of bacterial and archaeal genomes. Indefinite extrapolation of the curve in Figure 14a would eventually result in the fraction of regulators exceeding 1, which is obviously absurd; of course, the actual ‘bureaucratic ceiling’ would be reached long before that point. Several approaches to estimate the upper bound on the gene number have been proposed (147). An intuitively attractive view is that the genome growth would become unsustainable around the point where more than one regulator is added per added gene. A calculation based on this criterion leads to a maximum of ∼20 000 genes in a prokaryotic genome, a reasonable value considering the currently observed genome size distribution (Figure 2) (148). Similar considerations on the optimization of prokaryotic genome size were developed from the viewpoint of ‘microeconomic principles’, that is, maximization of the ratio between the metabolic complexity (‘revenue’) and the number of regulators (‘logistic cost’) (13).
HGT: THE FORMATIVE PROCESS IN THE EVOLUTION OF PROKARYOTES
The wide spread and major importance of HGT in the evolution of archaea and bacteria might be biggest conceptual novelty brought about by comparative genomics of bacteria and archaea (31,149–153). However, no other discovery has caused so much controversy and (sometimes, acrimonious) debate during which opposite views of HGT have been expounded, from assertions of its rampant occurrence and overarching role in evolution of bacteria and archaea (150, 154) to the denial of any substantial contribution of HGT (155, 156). As such, the existence of HGT, i.e. transfer of genes between distinct organisms by means other than vertical transmission of replicated chromosomes during cell division, had been recognized long before the first genomes were sequenced(157–159). Moreover, it had been realized that, at least, under selective pressure, such as in the case of the spread of antibiotic resistance in a population of pathogenic bacteria, HGT can be rapid and extensive (160, 161). However, until extensive comparison of multiple, complete genome sequences became possible, HGT was viewed as a marginal phenomenon, perhaps, important under specific circumstances, such as evolution of resistance, but one that can be, more or less, disregarded in the study of evolution of organisms. One must remember that the very relevance of the question of the role of HGT in evolution stems from another revolution, the one brought about by Woese's demonstration that phylogenetic analysis of prokaryotic rRNA was feasible and, at least potentially, could be a reasonable depiction of evolution of bacteria and the newly discovered archaea (162).
Historically and methodologically, the problem of HGT identification and the impact of HGT on evolution of bacteria and archaea are sharply divided into the (relatively) recent transfers that typically occur between closely related organisms, and the (in many cases) ancient events that supposedly took place between distant organisms. On the ‘microscale’, HGT is common and noncontroversial. Indeed, comparisons of genomes between closely related bacterial strains provide clear-cut evidence of massive HGT. Perhaps, the most striking demonstration of the high prevalence of HGT is the discovery of pathogenicity islands, i.e. gene clusters that carry pathogenicity determinants, such as genes encoding various toxins, components of type III secretion systems, and others, in parasitic bacteria, and similar ‘symbiosis islands’ in symbiotic bacteria (163, 164). Pathogenicity islands are large genomic regions, up to 100 kb in length, and they are typically located near tRNA genes and contain multiple prophages, suggesting that the insertion of these islands is mediated by bacteriophages (165). The now classic comparative genomic analysis of the enterohemorrhagic O157:H7 strain and the laboratory K12 strain of Eschersichia coli has shown that the pathogenic strain contained 1387 extra genes distributed between several strain-specific clusters (pathogenicity islands) of widely different sizes (166). Thus, up to 30% of the genes in the pathogenic strain seem to have been acquired via a relatively recent HGT. A further, detailed analysis of individual lineages of E. coli O157:H7 has demonstrated continuous HGT, apparently, contributing to the differential virulence of these isolates (167). Furthermore, it has been convincingly demonstrated that most of the recent (estimated to occur within the last 100 million years) additions to the metabolic network of E. coli were due to HGT, often of operons encoding two or more enzymes (or transporters) of the same pathway, with limited contribution from gene duplication (168).
The pivotal contribution of HGT in the evolution of individual functional systems of prokaryotes has been revealed in many studies. Perhaps, the most spectacular results have been obtained with photosynthetic gene clusters of cyanobacteria and other photosynthetic bacteria. Phylogenetic analyses strongly suggest that these clusters are complex mosaics of genes assembled via multiple HGT events (169). Furthermore, the majority of cyanophages carry one or more photosynthetic genes, presumably utilizing them to augment the host photosynthetic machinery during infection (170). Thus, these bacteriophages are, de facto, specialized vehicles for the HGT of photosynthetic genes.
The discovery of gene transfer agents (GTAs) in several groups of bacteria and archaea seems to be of particular importance because these agents are defective derivatives of tailed bacteriophages appear to be specifically adapted to serve as generalized transducing agents that package and transfer random chromosome fragments between bacteria (171, 172). Thus, startling as this might be, it seems appropriate to view the GTAs as specialized functional devices for HGT (at least, between closely related organisms).
Apart from direct experimental demonstration and compelling genome comparisons, recent HGT is detectable through analysis of nucleotide composition, oligonucleotide frequencies, codon usage and other ‘linguistic’ features of nucleotide sequences that reveal horizontally acquired genes as compositionally anomalous for a given genome (173–175). However, horizontally transferred sequences are ameliorated at a relatively high rate as the acquired genes are ‘domesticated’ during evolution (163, 176). The molecular vehicles of HGT between closely related organisms are well (even if, probably, not completely) understood and include conjugation, bacteriophage-mediated transduction and transformation (159).
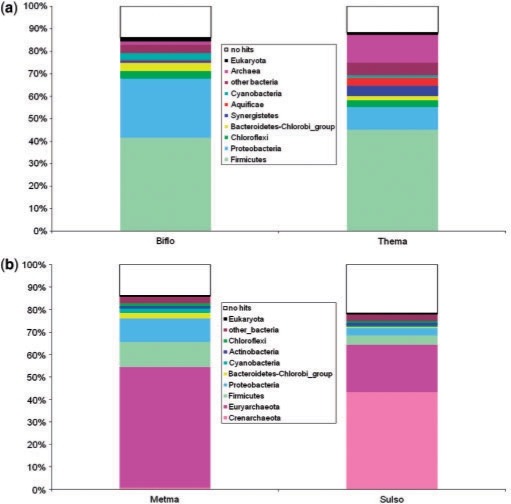
In contrast to the well-established HGT among closely related organisms, the extent of HGT across long evolutionary distances and its impact on the evolution of archaea and bacteria remains a matter of intense debate. Comparative genomics has provided ample indications of likely HGT including that between very distant organisms, in particular, archaea and bacteria. The first clear-cut indications of massive archaeal–bacterial HGT were obtained when it was shown that hyperthermophilic bacteria, namely, Aquifex aeolicus (177) and T. maritima (178), contained many more homologs of characteristic archaeal proteins than mesophilic bacteria as well as proteins with homologs both in archaea and bacteria but with much higher sequence similarity to the latter than to the former. Comparisons with mesophilic bacteria have shown that the fraction of ‘archaeal’ proteins in bacterial hyperthermophiles was much greater (with a high statistical significance) than in mesophiles (177). Subsequently, it has been shown the mesophilic archaea with relatively large genomes, Methanosarcina and halobacteria, possess many more ‘bacterial’ genes than thermophilic archaea with smaller genomes (179–181). These, admittedly, crude estimates suggest that, at least, ∼20% of the genes in an organism could have been acquired via archaeal–bacterial HGT, provided shared habitats. In Figure 15a, we compare the taxonomic breakdown of ‘best hits’ (most similar sequences in the Refseq databases detected using BLAST) for genomes of a mesophilic and a thermophilic bacteria. There is a visible and statistically highly significant excess of archaeal hits in the hyperthermophile T. maritima. Notably, this bacterium also contains a sizable fraction of proteins that are most similar to homologs from distantly related hyperthemophilic bacteria of the phylum Aquificacea, in support of the connection between the extent of apparent HGT and shared habitats. A similar comparison between a mesophilic and a hyperthemophilic archaea is even more illustrative in that the fraction of ‘bacterial’ proteins in the mesophile Methanosarcina is about threefold greater than that in the hyperthermophile Sulfolobus (Figure 15b).
Figure 15.
The taxonomic breakdown of the best database hits for proteins encoded in diverse bacterial and archaeal genomes. (a) A mesophilic bacterium, Bifidobacterium longum (Biflo), compared to a hyperthermophilic bacterium, T. maritima (Thema). (b) A mesophilic archaeon, M. mazei (Metma), compared to hyperthemrophilic archaeon, Sulfolobus solfataricus (Sulso). The best hits were obtained by processing the results of the searches of the NCBI's nonredundant protein sequence database using the BLASTP program (277).
The crucial problems with HGT between distant prokaryotes are the quality of evidence and persuasiveness of argument. The taxonomic breakdown of the results of genome-wide sequence comparisons is strongly suggestive of HGT inasmuch as widely different results are seen for different organisms (e.g. Figure 15). Nevertheless, this is not a proof of HGT, and indeed, alternative, even if not necessarily credible explanations have been duly proposed such as convergence of protein sequences in distant organisms that share similar habitats, e.g. archaeal and bacterial hyperthermophiles (182). Furthermore, it has been shown that phylogenetic analysis often does not support the conclusions on evolutionary relationships drawn from sequence similarity analysis suggesting that some of the conclusions drawn from BLAST-based comparisons could be misleading (183). Of course, it has to be kept in mind that phylogenetic analyses are themselves fraught with artifact (184), especially, when implemented on genome scale (185). Explanations rooted in methodological artifact do not readily apply to those genes that are shared exclusively by a few lineages of distant organisms (e.g. hyperthermophilic bacteria and archaea) but in such cases, the counter-argument is always ready that these genes have been lost in all other lineages.
The relationship between lineage-specific gene loss and HGT is a pervasive and formidable problem that plagues all attempts to assess the global role of HGT in the evolution of prokaryotes. The patchy phyletic patterns of numerous COGs (e.g. Figures 6 and 8) certainly testify to the dynamic character of prokaryotic evolution but the emergence of these patterns can be explained by either HGT or gene loss, or any combination thereof. The most parsimonious evolutionary scenario can be delineated if the relative rates of HGT and gene loss are known but this ratio (that undoubtedly differs between prokaryotic groups; see below) is one of the big unknowns of prokaryotic genomics. Several global reconstructions of prokaryotic evolution have been reported, all of them based on one or another version of the parsimony principle and either exploring scenarios with varying gain/loss rate ratios or attempting to estimate the optimal value of this ratio (186–188). The conclusions of these analyses are that HGT might be almost as common (188) or moderately (approximately twice) less common than gene loss during prokaryotic evolution ((186, 187) and that, accordingly, at least one HGT event was likely to have occurred during the evolution of most COGs, even within the limited sets of organisms that were analyzed. Of course, these analyses are based on gross, over-simplifying assumptions, such as uniform rates of HGT and gene loss across the prokaryotic groups, the notion that highly complex ancestral forms are unlikely, and the very concept of an underlying species tree. Although the results did not strongly depend on the species tree topology (188), the basic notion of a tree with distinct clades representing evolution of the compared organisms is indispensable for any reconstruction. The nature of ancestral organisms is hard to assess directly (although see below for a perspective on this issue) but the other two of the above fundamental have been put to test in extensive phylogenetic studies.
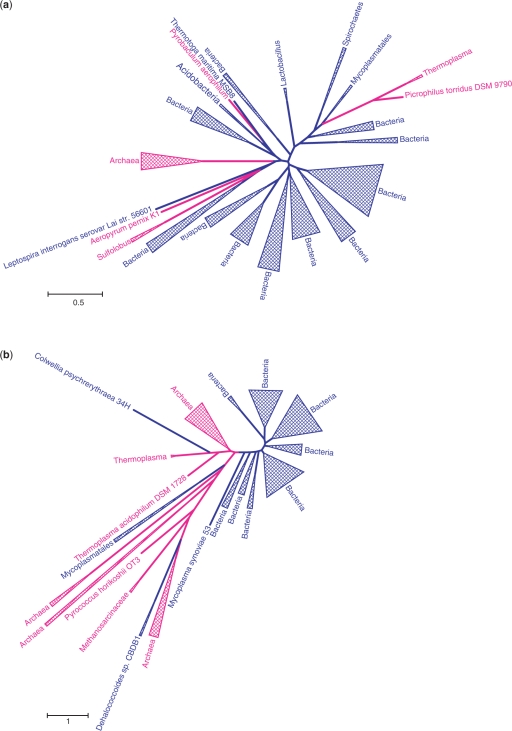
The species (organismal) tree that is supposed to depict the phylogeny of the compared organisms in their entirety is not only a key concept of evolutionary biology that descends from the original evolutionary imagery of Darwin (189) and Haeckel (190) but also a practical necessity for detecting HGT. Indeed, the most common practice of HGT detection involves identification of reliable discrepancies between the topologies of a gene tree and a species tree. The results of such a comparison are meaningful only inasmuch as the topology of the species tree can be trusted—and, of course, if this very concept is valid in the light of HGT ((154) and see below). However, the arguably most dramatic instances of HGT, those between archaea and bacteria, are more or less robust to the species tree topology inasmuch as the distinction between archaea and bacteria is not in dispute. Figure 16 shows two trees where several archaeal proteins are deeply rooted within the bacterial clade (A) or vice versa (B). Here, HGT between clades, probably, followed by subsequent HGT within the recipient clade appears to be the only sensible interpretation of the tree topology. Multiple archaeo-bacterial gene transfers have been supported by genome-wide phylogenetic analysis as well (191, 192).
Figure 16.
Two cases of readily demonstrable horizontal gene transfer between archaea and bacteria. (a) COG0030, dimethyladenosine transferase, an enzyme involved in rRNA methylation. (b) COG0206, FtsZ, a GTPase involved in cell division. Blue, bacteria; magenta, archaea. The trees were constructed using the maximum likelihood method implemented in the PhyML software (278) (WAG evolutionary model; γ-distributed site-specific rates with the shape parameter 1.0). The complete information on the analyzed sequences and the alignments are available from the authors upon request.
The validity of the species tree concept was tested by comparing phylogenetic trees for sets of several hundred single-copy COGs (i.e. those COGs that are represented by exactly one orthologous gene in each of the compared genomes) from well-characterized, widespread bacterial groups such as α-proteobacteria, γ-proteobacteria or the Bacillus–Clostridium group of Gram-positive bacteria (193–197). The results of these analyses are congruent in showing that evolution of a significant majority of these ‘simple’ COGs is compatible with a single tree topology that can be reasonably interpreted as the species tree. These findings suggest that the notion of a species tree is not without meaning, at least, when understood as a central trend of genome evolution (50). However, these analyses, in a sense, amount to a self-fulfilling prophecy because they were performed on preselected sets of genes that, indeed, might be considerably less prone to HGT than others, and within ‘shallow’ groups of bacteria in which evolution could be more tree-like than at deeper levels (198, 199). It should be noted that, by definition, in simple COGs, only the form of HGT denoted xenologous gene displacement (XGD) is possible, whereby a gene from a distant source displaces the resident ortholog (28, 179). For an essential gene, XGD is likely to require two events, first acquisition of a foreign gene and then, the loss of the native and hence is likely to be less frequent than acquisition of a new gene. Even within well-defined groups of prokaryotes, simple, one-to-one sets of orthologs include <10% genes in an average genome, and the other genes, those with patchy phyletic distributions and multiple paralogs, tend to show much higher rates of HGT (197).
Other large-scale phylogenetic analyses have aimed at reconstructing the ‘net of life’ using a variety of phylogenetic methods and, of course, relying on particular species tree topologies. A detailed discussion of such analyses is beyond the scope of this survey but the general conclusion was that, although a network graph that takes into account both vertical and horizontal connections between nodes (organisms) is, indeed, a more accurate representation of the evolution of prokaryotes than a tree, most of bacteria and archaea have experienced relatively little HGT, with only a few HGT ‘hubs’ (200) and distinct ‘highways’ of HGT that connect closely related or habitat-sharing organisms (201).
It is widely believed that ‘informational’ genes coding for proteins involved in translation, transcription and replication are much less prone to HGT than operational genes that encode metabolic enzymes, transport systems and other ‘operational’ proteins. The rationale behind this view is the complexity hypothesis according to which informational genes that, on average, are involved in a greater number of complex molecular machines whose parts are strongly coadapted and thus cannot be easily displaced with orthologs from distant organisms (xenologs) acquired via HGT (66). However, the validity of the complexity hypothesis remains uncertain as many clear-cut cases of HGT have been discovered among informational genes. Perhaps, surprisingly, these include not only most if not all aminoacyl-tRNA synthetases, enzymes that function in relative isolation (202, 203), but also many ribosomal proteins, components of the paradigmatic molecular machine, the ribosome (204, 205). On a number of occasions, HGT among translation system components involves not only XGD but also acquisition of pseudo-paralogs (205). Strong evidence of HGT has been presented also for such traditional markers of vertical phylogeny as DNA-dependent RNA polymerase subunits (206). It seems that the main difference in the modes of evolution of informational and operational genes has to do, above all, with the much lower incidence of nonorthologous gene displacement (as opposed to XGD) among informational genes (i.e. many informational functions are performed by orthologous genes in all or nearly all organisms), as reflected in the COG size distributions (Figure 11), rather than in a dramatic difference in HGT rates. Even among highly conserved informational genes including those that belong to the prokaryotic core (Figure 6), HGT seems to be common although the evolutionary scenarios are constrained by the (near) essentiality of many of these genes (207). Indeed, a large-scale analysis of phylogenetic trees for all categories of prokaryotic genes failed to reveal dramatic differences in the rates of HGT between informational and operational genes (201).
Finally, in our brief discussion of the different faces of HGT in the prokaryotic world, we must return to the selfish operon hypothesis which posits that ‘the organization of bacterial genes into operons is beneficial to the constituent genes in that proximity allows horizontal cotransfer of all genes required for a selectable phenotype’ (95). There is no contradiction between the functional and selfish aspects of operon evolution: indeed, an operon is a ‘prepackaged’ functional unit, often coming together with its own regulator, and in that capacity, operons are more likely than single genes to be fixed after HGT. Whereas the initial fixation of an operon is affected by the benefits of coregulation of functionally linked genes, their maintenance and spread through the prokaryotic world is mediated by HGT (208), an evolutionary modality that does confer on operons some (but, certainly, not all) of the properties of selfish, mobile elements. Moreover, the selfish character of operons can be seen as a way of overcoming the constraints imposed by the complexity hypothesis considering that the most common operons encode subunits of protein complexes (see above). Packaging all subunits of a complex in one operon provides for the transferability of the requisite complexity. An excellent case in point is the evolutionary history of membrane proton and sodium-translocating ATP synthases during which operons encoding multiple (up to 8) subunits of these elaborate molecular machines were repeatedly transferred between archaea and bacteria (209, 210).
So what is the take home message on the prevalence and role of HGT in the prokaryotic world? In our view, it is no longer a matter of sensible dispute that HGT is a major force in the evolution of prokaryotes that affects all aspects of bacterial and archaeal biology. Attempts to dismiss HGT as a marginal phenomenon (155, 156) seem outdated and hopeless. At the quantitative level, however, the HGT issue is far from being settled. In particular, there is a degree of tension, if not exactly a paradox, between two classes of observations: (i) there are few if any COGs that have not experienced HGT over the course of their evolution, and most, probably, have experienced multiple HGT events, but (ii) many analyses seem to reveal phylogenetic coherence in large groups of prokaryotes. There are at least three plausible, not mutually exclusive solutions to this discrepancy: (i) phylogenetic coherence is seen at limited evolutionary depths and, most importantly, in relatively small, preselected sets of COGs that are sufficiently common and ‘simple’ (no or few paralogs) to allow phylogenetic resolution and, possibly, to some extent, refractory to HGT, (ii) for the majority of COGs, the signal of vertical inheritance is stronger than the signal of HGT even if, considering the entire history of a COG, numerous HGT events are detectable, (iii) the observed phylogenetic coherence is (mostly) an illusion caused by increasingly high rates of HGT among prokaryotes with similar life styles and habitats (154). The latter idea is, probably, too sweeping to be the sole answer, but it well could be an important factor.
The subject of a truly salient debate at this time is not so much the importance and prevalence of HGT in prokaryotic evolution but, given that HGT is common and important, the legitimacy of ‘tree thinking’ in evolutionary biology of prokaryotes and the adequate formalisms and imagery for describing the process of prokaryotic evolution (211). Indeed, considering the pervasive HGT in the prokaryotic world, the very distinction between the vertical and the horizontal flows of genetic information becomes dubious (212–215). Below we return to this issue in the section on the new picture of the prokaryotic world.
THE PROKARYOTIC MOBILOME
As noted in the preceding section, hardly any COG is refractory to HGT in principle but, certainly, some genes are much more equal than others in that respect. A substantial part of the prokaryotic genetic material consists of selfish elements for which horizontal mobility is the dominant mode of dissemination and that have been aptly termed the mobilome (216). A full-fledged discussion of the mobilome requires a separate article(s) but in order to sketch an emerging coherent view of the prokaryotic world, we must briefly summarize here the salient features of this class of genetic elements. The mobilome consists of bacteriophages, plasmids, transposable elements and genes that are often associated with them and regularly become passengers such as restriction–modification (RM) and toxin–antitoxin (TA) systems. It seems natural that, inasmuch as viruses and plasmids are mobile by definition, so are the systems of defense. The mobilome is inextricably connected with the ‘main’ prokaryotic chromosomes. Viruses (bacteriophages) and many plasmids systematically integrate into chromosomes, either reversibly, in which case they often mobilize chromosomal genes or irreversibly whereby a mobile element becomes ‘domesticated’, giving rise to resident genes, initially, of the ORFan class (216, 217). It is well known since the classic experiments of Jacob and Wollman (218) that conjugative plasmids can mediate the transfer of large segments of bacterial chromosomes. The discovery of the GTAs, that seem to be specialized HGT vectors, further emphasizes the existence of regular channels of communication between the mobilome and the chromosomes.
Transfer of antibiotic resistance and secondary metabolic capabilities on plasmids are textbook examples of bacterial mobilome dynamics but the role of plasmids extends far beyond such relatively narrow biological areas (219). Actually, the boundary between chromosomes and plasmids is fuzzy (220–222). Plasmids are replicons (typically, circular but in some cases, linear) that, similarly to prokaryotic chromosomes, carry an origin site and encode at least some of the proteins involved in the plasmid replication and partitioning (223). The key proteins involved in plasmid and chromosome partitioning, in particular, ATPases of the FtsK-HerA family are homologous throughout the prokaryotic world, a fact that emphasizes common evolutionary origins and strategies of diverse prokaryotic replicons (224).
The ‘canonical’ genomes of numerous bacteria and archaea include, in addition to the ‘main’ chromosome(s), one or more relatively stable, essential, large extrachromosomal elements, often described as megaplasmids (221). Megaplasmids can be remarkably persistent during evolution. For instance, it has been shown that the single megaplasmid of Thermus thermophilus is homologous to one of the two megaplasmids of Deinococcus radiodurans and, by implication, derives from the common ancestor of these related but highly diverged bacteria (225). However, over the course of evolution of this ancient bacterial group, the megaplasmids have accumulated (relative to their size) many more differences in their gene repertoires than chromosomes. Moreover, the megaplasmids carry numerous horizontally transferred genes including genes from thermophilic organisms that apparently were acquired by the Thermus lineage and appear to be important for the thermophylic life style (225). Thus, although megaplasmids can persist in prokaryotic lineages over long evolutionary spans, they display greater genomic plasticity than chromosomes, and appear to be act as reservoirs of HGT.
All sequenced prokaryotic genomes contain traces of integration of multiple plasmids and phages (216). It is particularly notable that most of the archaeal genomes possess multiple versions of the HerA-NurA operon that encodes key component of the plasmid partitioning machinery (224). Thus, replicon fusion is likely to be a relatively common event in prokaryotes, and over the course of evolution, such fusion might have been a major factor in shaping the observed architecture of prokaryotic chromosomes.
Defense and stress response systems, in particular, RM and TA systems can be considered special parts of the mobilome. Comparative analysis of these systems shows evidence of rapid evolution and frequent HGT, and they are frequently found in plasmid and bacteriophage genomes (226). Despite their enormous molecular diversity, RM and TA systems function on the same principle: they are comprised of a toxin, a protein that destroys the chromosomal DNA (restriction enzymes), blocks translation (RNA endonuclease toxins) or kills the cell by making holes in the membrane. Cell death is prevented by specific methylation of the DNA, in the case of RM systems or by neutralization of the toxin by the antitoxin in the case of TA systems, either through toxin protein–antitoxin protein interaction or through abrogation of the translation of the toxin mRNA by the antitoxin antisense RNA. These systems possess properties of selfish elements: when the respective genes are lost from a cell, the cell typically dies either because the toxin is more stable than the antitoxin, and its activity is unleashed once the antitoxin degrades but cannot be replenished (227, 228) or because of the differential effects of dilution on the restriction and modification enzymes (229). Because of the same property of TA systems, there is strong selection for plasmids carrying TA genes that ensure plasmid ‘addiction’ by killing cells that have lost the plasmid. The currently known TA systems are likely to comprise the proverbial tips of the iceberg as bacterial and archaeal genomes carry a great variety of operons whose properties mimic those of TA operons (a pair of genes that encode small proteins and occur as a stable combination in diverse genomes and genomic neighborhoods) but that have not been experimentally characterized (K.S. Makarova, Y.I.W. and E.V.K., unpublished data).
Recently, a novel and highly unusual class of defense systems has been shown to exist in approximately half of bacteria and archaea whose genomes have been sequenced (230). This system is centered around arrays of so-called CRISPR repeats (231) and has been accordingly denoted CAS (CRISPR-Associated System) (92). The CAS systems includes ∼50 distinct gene families (91, 92) and comes across as the second largest, after the ribosomal superoperon, array of connected gene neighborhoods in prokaryotic genomes (89, 232). The CAS system protects prokaryotic cells against phages and plasmids via a ‘Lamarckian mechanism’, whereby a fragment of a phage or plasmid gene is integrated into the CRISPR locus on the bacterial chromosome and is subsequently transcribed and utilized, via still poorly characterized mechanisms, to abrogate the selfish agent's replication (233). The CAS system shows extreme plasticity, even among closely related isolates of bacteria and archaea, and strong evidence of extensive HGT (92, 230).
The selected examples discussed here point to enormous, still incompletely understood diversity of the prokaryotic mobilomeand the major contribution that the mobilomes makes to the evolution of the prokaryotic genome space.
THE NEW, DYNAMIC VIEW OF THE PROKARYOTIC WORLD AND THE DEMISE OF THE TREE OF LIFE
The ubiquity of HGT and the prominence of the prokaryotic mobilome suggest a novel, extremely dynamic picture of the prokaryotic world (Figure 17). Under this view, a Tree of Life (TOL) does not adequately represent evolution of prokaryotes (213, 214), not even in the previously envisaged form of a ‘cobweb’ of life where the main vertical flow of genetic information is complemented by functionally important but quantitatively relatively minor horizontal flow (196, 201). An image of a dynamic, weighted network graph where the nodes are genomes and edges denote gene flow between them, with the weight proportional to the intensity of the flow, is more adequate (Figure 17). In this network, it still makes sense to differentiate between vertical and horizontal gene flows. Indeed, at the microscopic level, vertical gene flow (transmission of genes to daughter cells via cell division) is readily distinguishable from HGT that constitutes gene transfer between cell via conjugation, transduction or transformation (generally, any means other than cell division). It is in the macroscopic, historical perspective that the distinction between vertical and horizontal transmission becomes conceptually dubious and practically hard to draw. Nevertheless, the network includes areas of substantial coherence of the vertical flow where the tree image is appropriate to depict coherent phylogenies of large groups of genes. Conceivably, these parts of the network, at least on average, also are characterized by intensive horizontal gene flow, emphasizing the interplay between the two directions limited applicability of genomes (154). However, on many occasions, ‘highways’ of horizontal gene flow (201), i.e. high-weight edges in the network, also connect organisms that are not tightly linked by vertical connections but coexist in the same habitats like hyperthemophilic bacteria and archaea (Figure 17).
Figure 17.
The dynamic view of the prokaryotic world. The figure is a conceptual schematic representation that is not based on specific data. The larger blue circles denote extant (solid lines) or ancestral (dashed lines) archaeal and bacterial genomes. The small red circles denote mobilome components such as plasmids or phages. Gray lines denote vertical inheritance of genes; green lines denote recent (solid) or ancient (dashed) HGT; red lines denote the permanent ongoing process of the exchange of genetic material between mobilome elements. The thickness of connecting lines reflects the intensity of gene transfer between the respective genetic elements.
Under the network vision of the prokaryotic world, archaeal and bacterial chromosomes are not envisaged as strictly defined genotypes gradually changing in time but rather as islands of temporary, relative dynamic stability that forms tightly connected (vertically and horizontally) areas of the network. The prokaryotic genome space is, obviously, not limited to chromosomes of cellular life forms but consists of a tremendous diversity of replicons including all components of the mobilome. The importance of these agents cannot be overestimated when one takes into account that metagenomic studies show that viruses are the most common entities in the biosphere, with about 10 virus particles per cell found in marine environments (47). Fusion, fission and recombination between replicons comprise the dominant mode of the genetic dynamics in the prokaryotic world. However, the notion of dynamic stability that is manifest in persistence of distinct structure in the prokaryotic world network extends also to the relationship between the genetic complements of prokaryotic cellular life forms and the mobilome. All their enormous mobility notwithstanding, selfish elements posses a core of ‘hallmark’ genes that only transiently appear in bacterial and archaeal chromosomes (234).
THE PRINCIPAL PROCESSES OF PROKARYOTIC EVOLUTION
Having formulated the notion of the dynamic prokaryotic world, we are now in a position to classify the major processes that affect evolution of prokaryotes. In doing so, one necessarily must take into account the population–genetic theory of evolution of genomic complexity that was recently expounded by Lynch (235, 236). The essence of this theory is that genetic changes leading to an increase of complexity such as duplications can be fixed only when purifying selection in a population is relatively weak, i.e. substantial complexification is possible only during population bottlenecks. Under this view, genomic complexity is not adaptive but is brought about by neutral population–genetic processes under conditions when purifying selection is (relatively) ineffective. Thus, complexification starts off as a ‘genomic syndrome’ although complex features subsequently become subject to adaptive selection. In contrast, in ‘highly successful’, large populations, purifying selection is intense, and the prevailing mode of evolution is thought to be genome streamlining (237).
The concepts of genome complexification and genome streamlining embody the ‘genome-centric’ view of evolution under which the selective pressure is a characteristic of an evolving lineage (a function of its characteristic effective population size and mutation/recombination rates) that affects the entire collectives of genes in the corresponding genomes (237). A complementary, ‘gene-centric’ perspective that is central to the description of the evolution of the mobilome elements on prokaryotic evolution considers a gene as distinct evolutionary unit that is subject to selection on its own and can compete with other genes (238).
The validity and relevance of the genome-centric perspective is supported by the observation that the distributions of sequence evolution rates across sets of orthologous genes from pairs of prokaryotic genomes have essentially the same shape within a wide range of evolutionary distances (239). In an even more direct validation of the genome-centric perspective, we have recently shown that selective pressure measured as the median ratio of nonsynonymous to synonymous substitutions is a stable characteristic of clusters of closely related prokaryotic genomes [(240); P.S. Novichkov, Y.I.W., I. Dubchak and E.V.K., unpublished data).
The relevance of the gene-centric perspective is, perhaps, most convincingly revealed by the ‘addiction’ mechanisms that lead to the retention of TA and RM modules in prokaryotic genomes through killing of the cells that lose these elements (226, 227) but is also manifest in the ‘selfish’ behavior of regular operons (97). Recently, it has been shown by mathematical modeling and computer simulation that addictive elements can spread in a bacterial population regardless of their initial concentration (241). In its extreme form, the gene-centric perspective describes evolving genomes as ‘communities’ of potentially selfish genes (241) or even as ‘ecosystems’ in which selfish genetic elements play the roles of species (242).
With the genome- and gene-centric perspectives in mind, we now can list the major evolutionary processes that shape the evolution of prokaryotic genomes (Figure 18). It seems that interaction between these six fundamental processes, along with the ‘background’ forces of purifying and positive (Darwinian) selection, is necessary and, at least, at coarse grain, sufficient, to account for prokaryotic genome evolution.
Genome streamlining under strong selection.
Neutral gene loss and genome degradation under weak selection (or neutral).
Innovation and complexification via gene duplication.
Innovation via operon shuffling.
Innovation and complexification via HGT, in particular, of partially selfish operons, a process that often leads to nonorthologous gene displacement.
Replicon fusion, propagation of mobile elements and other interactions between the relatively stable chromosomes and the mobilome.
The first four of these processes reflect the genome-centric view of evolution, whereas the remaining two relate to the gene-centric perspective. Although these processes can lead to similar and interleaved results, they are distinct, and their manifestations are discernible in comparative genomic data as discussed earlier.
Figure 18.
The principal forces of evolution in prokaryotes and their effects on archaeal and bacterial genomes. The horizontal line shows archaeal and bacterial genome size on a logarithmic scale (in megabase pairs) and the approximate corresponding number of genes (in parentheses). On this axis, some values that are important in the context of comparative genomics are roughly mapped: the two peaks of genome size distribution (Figure 2); ‘Van Nimwegen Limit’ (VNL) determined by the ‘cellular bureaucracy’ burden; the minimal genome size of free-living archaea and bacteria (MFL); the minimal genome size inferred by genome comparison [MG, (133,135,136)]; the smallest (C.r., C. rudii); and the largest (S.c., S. cellulosum) known bacterial genome size. The effects of the main forces of prokaryotic genome evolution are denoted by triangles that are positioned, roughly, over the ranges of genome size for which the corresponding effects are thought to be most pronounced.
Genome streamlining and neutral degradation are similar in their overall effect on genomes, namely, extensive loss of genes and a trend toward genome contraction but these are distinct processes as illustrated by comparison of the streamlined and degraded genomes (243). Streamlined genomes are thought to be typical of organisms that are highly abundant (i.e. evolutionarily successful) in relatively constant environments and, accordingly, should be subject to strong purifying selection, e.g. P. ubiquis (17) and cyanobacteria of the genus Prochlorococcus (244). The streamlined genomes appear to be characterized not so much by their small size (being autotrophs, these organisms cannot shed genes beyond a certain limit) as by extreme compactness and (virtual) lack of pseudogenes and integrated selfish elements. All such elements are supposed to be rapidly wiped out by the intense purifying selection that is so powerful that even short intergenic regions are contracted. In particular, P. ubiquis seems to perfectly fit this description, having no detectable pseudogenes or mobile elements, very few paralogs, and extremely shortest intergenic regions (17). However, comparative genomics of Prochlorococcus strains revealed features that might not be compatible with streamlining, namely, genomic islands (resembling pathogenicity and symbiosis islands mentioned above) containing a variety of phage-related genes (245).
Unexpectedly, the theoretically straightforward connection between the strength of selection and genome streamlining does not seem to be readily demonstrable when the selection pressure (median dN/dS) was analyzed in conjunction with other characteristics of genomes such as size, the number of protein-coding genes, and length of intergenic regions (P.S. Novichkov, Y.I.W., I. Dubchak and E.V.K., unpublished data). We found that strong selection pressure is associated with large genomes containing many genes and relatively long intergenic regions as exemplified by Figure 19 that shows the significant negative correlation between median dN/dS and the number of genes in prokaryotic genes. These definitely are not the features that are expected of streamlined genomes. Moreover, it was found that different strains of Prochlorococcus, an extremely abundant cyanobacterium with a minimal genome that is expected to evolve under a strong pressure of purifying selection, show widely different but, in all instances, moderate to high dN/dS values (P.S. Novichkov, Y.I.W., I. Dubchak and E.V.K., unpublished data). These findings emphasize the interplay between evolutionary processes that exert opposite effects on prokaryotic genomes, namely, streamlining and genome degradation that lead to genome contraction opposed to complexification and mobile element activity that favor genome expansion (Figure 18). At present, it appears that ‘pure’ streamlining is an exceptional rather than a dominant mode of prokaryotic evolution.
Figure 19.
The dependence between genome size and selection pressure in prokaryotes. The data are from the analysis of 41 alignable tight genome clusters (ATGCs) of bacteria and archaea [(240); P.S. Novichkov, Y.I.W., I. Dubchak and E.V.K., unpublished data). DN is the median of dN, and DS is the median of dS for the respective ATGC. The greater DN/DS the lower the pressure of purifying selection that affects the evolution of the genomes within an ATGC is considered to be. Rs is Spearman ranking correlation coefficient.
The genomes that apparently undergo neutral degradation, primarily, those of parasites and symbionts do not often reach a large effective population size, and hence gradually lose genes that they do not require via a ratchet-type mechanism (a gene once lost is unlikely to be regained, especially, considering the life styles of these organisms), possibly, buttressed by a deletion bias in the mutation process and exacerbated by the limited opportunities for HGT that are available to these organisms (246). Although some of these genomes are extremely small, because in parasites and symbionts many genes become dispensable, they tend to contain considerable numbers of pseudogenes and, in some cases, also sustain propagation of selfish elements. Well-characterized cases in point are Rickettsia (247, 248), Wolbachia (249), pathogenic Mycobacteria (250, 251) and some lactobacilli (252). For these organisms, the predictions of the population–genetic theory generally seem to hold in that they indeed typically have high dN/dS indicative of weak selection pressure [(253, 254) and P.S. Novichkov, Y.I.W., I. Dubchak and E.V.K., unpublished data).
As noticed earlier, organization of genes in prokaryotic genomes is highly variable, even within individual operons (69, 86). Although genome rearrangement is an intrinsically neutral process driven by recombinational events such as inversions and transpositions, it results in operon shuffling and so substantially contributes to the emergence of new operons and, accordingly, to innovation at the level of gene regulation (69, 109).
According to the population–genetic theory, the extent of innovation attainable, be it by gene duplication, by HGT or by operon shuffling, also strongly depends on an organism's effective population size that is reflected in the strength of selection (235,237,255). In a sense, innovation is the antipode of genome streamlining in that multiple duplications or genes acquired via HGT can be fixed only in small populations with a major role of drift unless the new genes confer a pronounced adaptive advantage on the organism (as is the case, e.g. with the spread of antibiotic resistance). Thus, extensive genome complexification is likely to occur only in fastidiously growing prokaryotes that inhabit complex, variable environments, where they persist as relatively small populations and/or pass through severe population bottlenecks. The results of direct analysis of selective pressure in various groups of bacteria and archaea (Figure 19) do not seem to immediately support this concept.
Gene exchange between chromosomes and the mobilome is related to and intertwined with HGT, but is nevertheless best considered a distinct phenomenon. The mobilome is a specific part of the prokaryotic world that is relatively weakly associated with the part comprising more stable chromosomes, that is, even when elements of the mobilome integrate with chromosomes, the association typically is transient. Nevertheless, lysogenic viruses of archaea and bacteria routinely integrate and occasionally mediate transduction of chromosomal genes, and plasmids (routinely, in the case of conjugative plasmids and occasionally in the case of nonconjugative ones) also can integrate and transfer chromosomal genes. Moreover, integrated viral and plasmid genes occasionally become ‘domesticated’, giving rise to ORFans that could be viewed as a genomic wasteland linking chromosomes and the mobilome. Some of the ORFans subsequently are recruited for cellular functions and leave the mobilome (46, 197). Owing to the vastness of the mobilome, these relatively weak (i.e. infrequent compared to the total number of replication cycles of selfish elements) interactions with chromosomes are crucial in shaping the chromosomal composition. Furthermore, the GTAs (171, 172), the putative devices for HGT, shed new light on the relationship between the mobilome and the chromosomes, indicating that connections between these parts of the prokaryotic world could be specifically selected for rather than just emerge sporadically.
Fusion of distinct chromosomal, plasmid and viral replicons, although even rarer than transduction, seem to make important contribution to genome evolution (256). Although here we cannot discuss the current concepts of the origins of bacterial and archaeal genomes in any detail, it is an attractive and, perhaps, not too far fetched possibility that the first prokaryotic chromosomes evolved by accretion of primordial, plasmid-like replicons (234).
It seems likely that the balance between the opposing trends of genome contraction caused by streamlining and degradation, and expansion via various routes shape are directly reflected in the size distribution of bacterial genomes, with the dominant peak shaped, primarily, by contraction and the second peak by expansion (Figure 2). However, as suggested in particular by the observation that the correlation between selection pressure and genome size in prokaryotes has the opposite sign to that predicted by the streamlining theory (Figure 19), the relationships between evolutionary processes can be complex and unexpected. Many more comparative analyses of genomes of prokaryotes with diverse genome characteristic and life styles are necessary to approach an adequate picture of the landscape of prokaryotic genome evolution.
GENOMIC SIGNATURES OF DISTINCT LIFE STYLES OF BACTERIA AND ARCHAEA
One of the greatest hopes associated with comparative genomics is the possibility, at least, in principle, to delineate ‘genomic signatures’ of distinct organismal life styles, i.e. sets of genes that are necessary and sufficient to support these lifestyles. In the current, rapidly growing collection of prokaryotic genomes, a lifestyle is often represented by multiple, diverse genomes, so the time seems ripe for studies of the genome-phenotype links to start in earnest. So far, only very modest success can be claimed. In cases where a lifestyle is linked to a well-defined biochemical pathway(s), e.g. in methanogens or photosynthetic organisms, identification of a genomic signature can be a relatively straightforward task (257, 258). Even so, for example, the analysis of the genes for proteins involved in photosynthesis illustrates the complex intertwine of lifestyle-specific and lineage-specific features. The most complete set of ‘photosynthetic’ genes was detected in cyanobacteria, whereas the other groups of photosynthetic bacteria possessed various subsets of these genes (258).
Genomic signatures of more complex phenotypes, such as thermophily or radioresistance, turned out to be much more elusive. The most effort, perhaps, has been dedicated to the quest for signs of thermophilic adaptation. Remarkably, there is a single gene that is found in all sequenced hyperthemrophilic genomes but not in any of the mesophiles, and this gene encodes a protein that is strictly required for DNA replication at extreme high temperatures, reverse gyrase (259). Moreover, the genome of a moderate thermophile T. thermophilus (strainHB27) contains a reverse gyrase pseudogene, whereas the related strain HB8 contains an intact reverse gyrase gene, demonstrating an ongoing process of reverse gyrase elimination after the probable switch from hyperthermophilic to moderate thermophilic life style (225, 260). However, search for other thermophile-specific genes yielded limited information, with no genes other than reverse gyrase showing a clean pattern of presence–absence correlated with (hyper)thermophily and only a few showing significant enrichment in thermophilic compared to mesophilic archaea and bacteria (261). Genome-wide searches for thermophilic determinants have been directed also at detecting relevant patterns of differences at the level of nucleotide and protein sequences and structures. Although these studies have revealed several suggestive distinctions of thermophilic proteins, such as higher charge density (262, 263) and overrepresentation of disulphide bridges (264), the ultimate significance of each of these features remains uncertain. The overall conclusion from these studies is that so far comparative genomics has failed to reveal ‘secrets’ of the thermophilic life style (intuitively, one would suspect that there must be major, genome-encoded differences between organisms whose optimal growth temperature exceeds 95°C and those that optimally grow at 37°C).
The story of the search for genomic correlates of extreme radioresistance and desiccation resistance might be even more illuminating. Some bacteria and archaea, of which the best characterized is the bacterium D. radiodurans, possess extreme radiation resistance that is thought to be a side effect of their adaptive desiccation resistance (265). Extensive genome analysis of D. radiodurans did not immediately reveal any unique features of the genome or of DNA repair systems that could explain the exceptional ability of this organism to survive radiation damage although homologs of plant proteins implicated in desiccation resistance and, at the time, not found in any other bacteria, have been identified (266). Deinococcus radiodurans is a model experimental system, so subsequently, transcriptomic and proteomics studies have been undertaken to characterize the response of this bacterium to high-dose irradiation (267–269). These studies have generated some excitement because substantial upregulation of several uncharacterized genes whose products were implicated in potentially relevant processes such as double-strand break repair (267). However, knockout of these genes failed to affect radiation resistance, whereas knockouts of a few genes that did not encode any recognizable domains and were not upregulated upon irradiation did render the organism radiation-sensitive (270). The recent comparative analysis of two related, radiation-resistant bacteria, D. radiodurans and D. geothermalis, failed to resolve and even further complicated the problem of genomic determinants of radioresistance (270). No genes with clear relevance to radiation resistance were discovered that would be unique to these radioresistant bacteria. Moreover, orthologs of many of the genes that are strongly upregulated in D. radiodurans upon irradiation are missing in D. geothermalis. The careful comparison of operon structure and predicted regulatory sites in the two Deinococcus genomes led to the prediction of a putative radiation-resistance regulon. However, for most of the genes that comprise this putative regulon, the relevance for radiation and desiccation resistance is uncertain. The principal determinants of radioresistance remain elusive, and there is growing evidence that important roles could belong to genes that mediate resistance in unexpected, indirect ways, e.g. through regulation of the intracellular concentrations of divalent cations that affect the level of protein damage resulting from irradiation or desiccation (271, 272).
The only possible conclusion on the current state of understanding of the genome–phenotype connections in prokaryotes is that these links are multifaceted, and that distinct sets of genes responsible for complex phenotypes are not readily identifiable despite the existence of clear signatures of certain phenotypes such as reverse gyrase in the case of hyperthermophily. The complexity of this relationship parallels the nonisomorphous mapping between the gene and functional spaces of prokaryotes discussed earlier.
ARCHAEA AND BACTERIA IN THE LIGHT OF COMPARATIVE GENOMICS: WHITHER PROKARYOTES?
The very validity of the term and concept of a prokaryote has been challenged as outdated and based on a negative definition, i.e. the absence of a eponymous organelle of the ‘higher’ organisms (eukaryotes), the nucleus (26, 273). Instead of the purportedly inadequate notion of a prokaryote, it has been proposed to classify life forms solely on the basis of phylogenetic divisions that have been derived, primarily, from rRNA trees and supported by trees for a few other (nearly) universal informational genes. The argument on the negative definition of prokaryotes has been countered by defining positive characters such as transcription–translation coupling (274). Regardless of the relative merits of these arguments, comparative genomics throws its own light on the prokaryotic problem. There is little universal conservation in terms of gene composition across archaea and bacteria, and next to none in terms of the organization of specific genes (see above). In trees built on the basis of comparisons of gene composition or conserved pairs of adjacent genes, the split between bacteria and archaea is unequivocal (50). In a stark contrast, the overall genome organization of bacteria and archaea is remarkably uniform. Some exceptions notwithstanding, this general principle of genome organization can be easily captured in a succinct description: bacteria and archaea have compact genomes with short intergenic regions so that many genes form directons that tend to function as operons. The formation of directons many of which become operons can be considered a direct consequence of genome contraction. The persistence of operons is subsequently ensured by a combination of purifying selection and frequent HGT as captured in the selfish operon concept. Thus, the uniform principle of organization of the genomes of bacteria and archaea emerges as a direct consequence of the forces operating in the evolution of these life forms, and these forces themselves are linked to their population structure. Considering this unity, we have to conclude that the concept of prokaryotes as life forms that evolve under a distinct, common mode leading to a common type of genome organization is well justified. Whether or not ‘prokaryotes’ is a good term to describe this part of the biosphere remains a debatable issue (the problem of the origin of eukaryotes from which this issues hardly can be separated is beyond the scope of this article) but, probably, one of secondary importance.
CONCLUSIONS AND PERSPECTIVE
By any account, the progress of knowledge of the prokaryotic world brought about by comparative genomics has been enormous. Many of the major trends and patterns discussed here, such as the distinction along with the similarities between archaea and bacteria, the operonic organization of bacterial genes, and the existence of HGT, have been noticed in the pregenomic era, but more as anecdotes than as general patterns. Comparative genomics allows one to actually determine how (un)common is a particular pattern, and the confidence of such inference increases with the growth of the genome collection. In the early days of genomics, a hope for a new suite of ‘laws of genomics’ has been expressed (275). Certain striking, nearly universal quantitative regularities indeed have been revealed by comparison of prokaryotic genomes. The two best candidates for ‘laws of genomics’ seem to be the scaling of different functional classes of genes with the genome (147) and the universal distribution of the evolutionary rates in orthologous gene sets (239). On the whole, however, 13 years into the comparative genomic enterprise, it seems more appropriate to speak of regularities, constraints, and perhaps, principles. Indeed, in terms of general organization, the great majority of the archaeal and bacterial genomes are notably similar, and are built according to the same, simple ‘master plan’ with wall-to-wall protein-coding and RNA-coding genes, preferentially organized in directons, typically, with a single origin of replication. Most of the arcaheal and bacterial genes are simple units, with uninterrupted coding sequence and short regulatory regions. There seems to be a nontrivial connection between gene functions and genome complexity: scaling of the number of genes of different functional classes appears to be (nearly) the same across the wide range of the available genomes, with the nearly constant, ‘frozen’ set of genes involved in translation and a steep increase in the number of regulators and signaling proteins with genome size. This increased ‘burden of bureaucracy’ is likely to be one of the important factors that set the upper limit for prokaryotic genome size and, accordingly, complexity. These regularities come as close to ‘laws of genomics’ as one can imagine although, as always in biology, there are multiple exceptions to any rule. More importantly, within these simple constraints, lie the enormous diversity and intricacy of the content and history of prokaryotic genomes.
Cases in point abound. The demonstration that the great majority of genes in each genome are not ORFans but rather have orthologs is, arguably, the very cornerstone of the genomic enterprise, which underlies all functional annotation of the sequenced genomes as well as evolutionary reconstructions. However, the flip side of the coin, namely, the patchy distribution of COGs in the gene space is no less fundamental. This distribution is the product of the major forces that shape prokaryotic evolution, namely, HGT, genes loss that often reflects genome streamlining, and nonorthologous gene displacement, which reflects the nonisomorphous mapping between the gene space and the functional space. The virtually unlimited flexibility of the architecture of prokaryotic genomes owing to extensive rearrangements, which create diverse variations on the themes of conserved operons, and the discovery of previously unsuspected signaling, regulatory and defense systems, only a few of which are briefly discussed in this article, add to the complexity of the prokaryotic genomescape that is revealed by comparative genomics.
Arguably, the most important conceptual novelty brought about by genomics is the demonstration that HGT is ubiquitous in the prokaryotic world, even as the extent of gene movement between distantly related organisms remains an issue of debate. Regardless of the further developments in these debates, the wide spread of HGT and the apparent absence of impenetrable barriers means that the prokaryotic world is a single connected gene pool, although this pool has a complex, compartmentalized structure, with its distinct parts being partially isolated from each other. Horizontal gene transfer affects different classes of genes to different extents, at least, in part, according to the complexity hypothesis, but no gene seems to be completely immune to HGT. The compartmentalization of the gene pool notwithstanding, the results of comparative genomics refute the TOL concept, at least, as applied to the prokaryotic world, as well as the notion of prokaryotic species. At best, the tree representation of genome evolution might be applicable to subsets of conserved genes from relatively close organisms. Delineation of ‘higher taxa’ of bacteria and archaea might not be a feasible project, given the erosion of the phylogenetic signal, the cumulative effect of HGT over time and the possibility that the early evolution of prokaryotes involved even more extensive HGT and could have been more akin to partially constrained sampling of the gene pool (214). From a complementary, genome-centric perspective, the results of comparative genomics indicate that the genes in any genome are far from having the same history, and it could be hard even to identify a set of genes that have a coherent history over a substantial evolutionary span. To this, it must added the a substantial fraction of most prokaryotic genomes belongs to the mobilome, the vast set of genes that come and go at striking rates and, generally, might not have any adaptive value for the organisms, even if occasionally recruited by some organisms for specific biological functions.
Taken together, these findings amount to a new, dynamic picture of the prokaryotic world that is best represented as a complex network of genetic elements, which exchange genes at widely varying rates. In this network, the distinction between the relatively stable chromosomes and the mobilome is a difference in degree (of mobility) rather than in kind. The remarkably uniform general organization of prokaryotic genomes appears to be determined by the dynamic nature of the prokaryotic genome space along with the intensive purifying selection underpinned by the large effective population size of most prokaryotes that itself is a function of extensive gene exchange.
The paradox of today's state of the art is that, despite the tremendous progress—but also owing to these advances—the emerging complexity of the prokaryotic world is currently beyond our grasp. We have no adequate language, in terms of theory or tools, to describe the workings and histories of the genomic network. Developing such a language is the major challenge for the next stage in the evolution of prokaryotic genomics.
FUNDING
DHHS (National Library of Medicine) intramural funds. Funding for open access charge: DHHS (National Library of Medicine) intramural funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The literature on prokaryotic genomics is vast, and inevitably, only a small fraction of the relevant work could be cited in this article. Moreover, we are well aware of the existence of many relevant publications from the pregenomic era that we did not have the opportunity to cite either. We sincerely apologize to all colleagues whose important contributions are not cited due to space constraints or (unfortunately but likely) our inadvertent oversight. We thank Pavel Novichkov for providing the data for Figures 13 and 19, and Kira Makarova and Sergei Maslov for useful discussions.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd [see comments] Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Mushegian AR. Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics. Curr. Opin. Genet. Dev. 1996;6:757–762. doi: 10.1016/s0959-437x(96)80032-3. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Mushegian AR, Rudd KE. Sequencing and analysis of bacterial genomes. Curr. Biol. 1996;6:404–416. doi: 10.1016/s0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- 5.Entrez Genome Project. 2008. [(accessed 10 June 2008)]. National Center for Biotechnology Information, NIH, Bethesda. Available at http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi.
- 6.Tyson GW, Banfield JF. Cultivating the uncultivated: a community genomics perspective. Trends Microbiol. 2005;13:411–415. doi: 10.1016/j.tim.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 7.DeLong EF. Microbial community genomics in the ocean. Nat. Rev. Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- 8.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 9.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 10.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 12.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 13.Ranea JA, Grant A, Thornton JM, Orengo CA. Microeconomic principles explain an optimal genome size in bacteria. Trends Genet. 2005;21:21–25. doi: 10.1016/j.tig.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, et al. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 2006;188:7922–7931. doi: 10.1128/JB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 18.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 19.Koonin EV. Virology: Gulliver among the Lilliputians. Curr. Biol. 2005;15:R167–R169. doi: 10.1016/j.cub.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 20.Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. Marine mimivirus relatives are probably large algal viruses. Virol. J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 22.Fuxelius HH, Darby AC, Cho NH, Andersson SG. Visualization of pseudogenes in intracellular bacteria reveals the different tracks to gene destruction. Genome Biol. 2008;9:R42. doi: 10.1186/gb-2008-9-2-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, Kikuchi H, Kita K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- 24.Dassa B, Amitai G, Caspi J, Schueler-Furman O, Pietrokovski S. Trans protein splicing of cyanobacterial split inteins in endogenous and exogenous combinations. Biochemistry. 2007;46:322–330. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- 25.Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan IK, Tatusov RL, Koonin EV. Purifying and directional selection in overlapping prokaryotic genes. Trends Genet. 2002;18:228–232. doi: 10.1016/s0168-9525(02)02649-5. [DOI] [PubMed] [Google Scholar]
- 26.Pace NR. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- 27.Koonin EV, Galperin MY. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr. Opin. Genet. Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- 28.Koonin EV. Orthologs, paralogs and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 29.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–106. [PubMed] [Google Scholar]
- 30.Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 31.Koonin EV, Mushegian AR, Galperin MY, Walker DR. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 32.Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS, Borodovsky M, Rudd KE, Koonin EV. Metabolism and evolution of Haemophilus influenzae deduced from a whole- genome comparison with Escherichia coli. Curr. Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 33.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zmasek CM, Eddy SR. RIO: Analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 38.Storm CE, Sonnhammer EL. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18:92–99. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Stoeckert C.J., Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 41.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siew N, Azaria Y, Fischer D. The ORFanage: an ORFan database. Nucleic Acids Res. 2004;32:D281–D283. doi: 10.1093/nar/gkh116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siew N, Fischer D. Twenty thousand ORFan microbial protein families for the biologist? Structure. 2003;11:7–9. doi: 10.1016/s0969-2126(02)00938-3. [DOI] [PubMed] [Google Scholar]
- 45.Ochman H. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 2002;18:335–337. doi: 10.1016/s0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- 46.Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards RA, Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 48.Kohonen T. Self-Organizing Maps. 2nd. Heidelberg: Springer; 1997. [Google Scholar]
- 49.Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat. Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 50.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 51.Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: from genes to genomes and back. J. Mol. Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 52.Andrade MA, Brown NP, Leroy C, Hoersch S, de Daruvar A, Reich C, Franchini A, Tamames J, Valencia A, Ouzounis C, et al. Automated genome sequence analysis and annotation. Bioinformatics. 1999;15:391–412. doi: 10.1093/bioinformatics/15.5.391. [DOI] [PubMed] [Google Scholar]
- 53.Ouzounis CA, Karp PD. The past, present and future of genome-wide re-annotation. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-comment2001. COMMENT2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin EV, Galperin MY. Sequence – Evolution- Function. Computational Approaches in Comparative Genomics. New York: Kluwer Acadademic Publication; 2002. [PubMed] [Google Scholar]
- 55.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Overbeek R, Larsen N, Pusch GD, D'Souza M, Selkov E., Jr., Kyrpides N, Fonstein M, Maltsev N, Selkov E. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 2000;28:123–125. doi: 10.1093/nar/28.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galperin MY, Koonin EV. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- 58.Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 59.Huynen MJ, Snel B. Gene and context: integrative approaches to genome analysis. Adv. Prot. Chem. 2000;54:345–379. doi: 10.1016/s0065-3233(00)54010-8. [DOI] [PubMed] [Google Scholar]
- 60.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat. Rev. Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 61.Medigue C, Moszer I. Annotation, comparison and databases for hundreds of bacterial genomes. Res. Microbiol. 2007;158:724–736. doi: 10.1016/j.resmic.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi [see comments] Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 63.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 64.Edgell DR, Doolittle WF. Archaea and the origin(s) of DNA replication proteins. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 65.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mushegian AR, Koonin EV. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- 68.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 69.Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 70.Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-research0011. RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tillier ER, Collins RA. Genome rearrangement by replication-directed translocation. Nat. Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 72.Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 73.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 74.Francino MP, Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 75.Frank AC, Lobry JR. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- 76.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frank AC, Lobry JR. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics. 2000;16:560–561. doi: 10.1093/bioinformatics/16.6.560. [DOI] [PubMed] [Google Scholar]
- 78.Nomura M, Morgan EA. Genetics of bacterial ribosomes. Annu. Rev. Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- 79.Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 80.Rocha EP, Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rocha EP, Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 82.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 83.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol. Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 86.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 87.Coenye T, Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiol. Lett. 2005;242:117–126. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 88.Lathe WC, III, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends Biochem Sci. 2000;25:474–479. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- 89.Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, Szekely LA, Koonin EV. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res. 2002;30:2212–2223. doi: 10.1093/nar/30.10.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tamames J. Evolution of gene order conservation in prokaryotes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0020. RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. , table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lawrence J. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 96.Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 97.Lawrence JG, Roth JR. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18(Suppl 1):S329–S336. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- 99.Lawrence JG. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 2003;57:419–440. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- 100.Madan Babu M, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 102.Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown CT, Callan C.G., Jr. Evolutionary comparisons suggest many novel cAMP response protein binding sites in Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:2404–2409. doi: 10.1073/pnas.0308628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 2006;62:1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- 106.Manson McGuire A, Church GM. Predicting regulons and their cis-regulatory motifs by comparative genomics. Nucleic Acids Res. 2000;28:4523–4530. doi: 10.1093/nar/28.22.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan K, Moreno-Hagelsieb G, Collado-Vides J, Stormo GD. A comparative genomics approach to prediction of new members of regulons. Genome Res. 2001;11:566–584. doi: 10.1101/gr.149301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perez-Rueda E, Collado-Vides J, Segovia L. Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput. Biol. Chem. 2004;28:341–350. doi: 10.1016/j.compbiolchem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 113.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 114.Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 115.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 116.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol. Biol. Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 117.Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lory S, Wolfgang M, Lee V, Smith R. The multi-talented bacterial adenylate cyclases. Int. J. Med. Microbiol. 2004;293:479–482. doi: 10.1078/1438-4221-00297. [DOI] [PubMed] [Google Scholar]
- 120.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 121.Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 122.Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 123.Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the ‘eukaryotic’ protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 124.Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 2002;206:1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- 125.Kennelly PJ. Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem. J. 2003;370:373–389. doi: 10.1042/BJ20021547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aravind L, Dixit VM, Koonin EV. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 128.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 129.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2004;2:643–655. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 130.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 131.Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl Acad. Sci. USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes [see comments] Proc. Natl Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koonin EV. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 136.Gil R, Silva FJ, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome [see comments] Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 138.Gerdes SY, Scholle MD, Campbell JW, Balaszi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. Experimental determination and system-level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salama NR, Falkow S. Genomic clues for defining bacterial pathogenicity. Microbes Infect. 1999;1:615–619. doi: 10.1016/s1286-4579(99)80061-6. [DOI] [PubMed] [Google Scholar]
- 141.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison C.A., III, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proc. Natl Acad. Sci. USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tamames J, Gil R, Latorre A, Pereto J, Silva FJ, Moya A. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 144.Cordero OX, Hogeweg P. Large changes in regulome size herald the main prokaryotic lineages. Trends Genet. 2007;23:488–493. doi: 10.1016/j.tig.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 145.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Molina N, van Nimwegen E. Universal patterns of purifying selection at noncoding positions in bacteria. Genome Res. 2008;18:148–160. doi: 10.1101/gr.6759507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Van Nimwegen E. In: Power Laws, Sacle-Free Networks and Genome Biology. Koonin EV, Wolf YI, Karev GP, editors. Georgetown, TX: Landes Bioscience; 2006. pp. 236–253. [Google Scholar]
- 148.Croft LJ, Lercher MJ, Gagen MJ, Mattick JS. Is prokaryotic complexity limited by accelerated growth in regulatory overhead? 2003. [(accessed 10 September 2008)]. Available at http://arxiv.org/abs/q-bio.mn/0311021.
- 149.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 150.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–M8. [PubMed] [Google Scholar]
- 151.Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 152.Brown JR. Ancient horizontal gene transfer. Nat. Rev. Genet. 2003;4:121–132. doi: 10.1038/nrg1000. [DOI] [PubMed] [Google Scholar]
- 153.Lawrence JG, Hendrickson H. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 2003;50:739–749. doi: 10.1046/j.1365-2958.2003.03778.x. [DOI] [PubMed] [Google Scholar]
- 154.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 155.Kurland CG. Something for everyone. Horizontal gene transfer in evolution. EMBO Rep. 2000;1:92–95. doi: 10.1093/embo-reports/kvd042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proc. Natl Acad. Sci. USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smith MW, Feng DF, Doolittle RF. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem. Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 158.Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu. Rev. Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 159.Bushman F. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 160.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 161.Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007;271:147–161. doi: 10.1111/j.1574-6968.2007.00757.x. [DOI] [PubMed] [Google Scholar]
- 162.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 164.Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 165.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 166.Perna NT, Plunkett G., III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 167.Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson AK, Taboada E, Gannon VP. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 169.Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE. Whole-genome analysis of photosynthetic prokaryotes. Science. 2002;298:1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 170.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stanton TB. Prophage-like gene transfer agents-novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe. 2007;13:43–49. doi: 10.1016/j.anaerobe.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 172.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 173.Medigue C, Rouxel T, Vigier P, Henaut A, Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- 174.Mrazek J, Karlin S. Detecting alien genes in bacterial genomes. Ann. NY Acad. Sci. 1999;870:314–329. doi: 10.1111/j.1749-6632.1999.tb08893.x. [DOI] [PubMed] [Google Scholar]
- 175.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes [In Process Citation] Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 177.Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 178.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 179.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Baumer S, Jacobi C, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- 182.Kyrpides NC, Olsen GJ. Archaeal and bacterial hyperthermophiles: horizontal gene exchange or common ancestry? [comment] Trends Genet. 1999;15:298–299. doi: 10.1016/s0168-9525(99)01811-9. [DOI] [PubMed] [Google Scholar]
- 183.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 184.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 185.Sicheritz-Ponten T, Andersson SG. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 2001;29:545–552. doi: 10.1093/nar/29.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 187.Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Darwin C. On the Origin of Species. London: Murray; 1859. [Google Scholar]
- 190.Haeckel E. Whitefish, MT: Kessinger Publishing; 1997. The Wonders of Life: A Popular Study of Biological Philosophy. [Google Scholar]
- 191.Gophna U, Doolittle WF, Charlebois RL. Weighted genome trees: refinements and applications. J. Bacteriol. 2005;187:1305–1316. doi: 10.1128/JB.187.4.1305-1316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 193.Novichkov PS, Omelchenko MV, Gelfand MS, Mironov AA, Wolf YI, Koonin EV. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J. Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLoS Biol. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ochman H, Daubin V, Lerat E. A bunch of fun-guys: the whole-genome view of yeast evolution. Trends Genet. 2005;21:1–3. doi: 10.1016/j.tig.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 196.Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bapteste E, Boucher Y, Leigh J, Doolittle WF. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 2004;12:406–411. doi: 10.1016/j.tim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 199.Ochman H, Lerat E, Daubin V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl Acad. Sci. USA. 2005;102(Suppl 1):6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 204.Brochier C, Philippe H, Moreira D. The evolutionary history of ribosomal protein RpS14: horizontal gene transfer at the heart of the ribosome. Trends Genet. 2000;16:529–533. doi: 10.1016/s0168-9525(00)02142-9. [DOI] [PubMed] [Google Scholar]
- 205.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. RESEARCH0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 207.Fang G, Rocha EP, Danchin A. Persistence drives gene clustering in bacterial genomes. BMC Genomics. 2008;9:4. doi: 10.1186/1471-2164-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Price MN, Huang KH, Arkin AP, Alm EJ. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res. 2005;15:809–819. doi: 10.1101/gr.3368805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hilario E, Gogarten JP. Horizontal transfer of ATPase genes—the tree of life becomes a net of life. Biosystems. 1993;31:111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- 210.Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat. Rev. Microbiol. 2007;5:892–899. doi: 10.1038/nrmicro1767. [DOI] [PubMed] [Google Scholar]
- 211.Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. Do orthologous gene phylogenies really support tree-thinking? BMC Evol. Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.O’Malley MA, Boucher Y. Paradigm change in evolutionary microbiology. Stud. Hist. Philos. Biol. Biomed. Sci. 2005;36:183–208. doi: 10.1016/j.shpsc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 213.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl Acad. Sci. USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Koonin EV. The Biological Big Bang model for the major transitions in evolution. Biol. Direct. 2007;2:21. doi: 10.1186/1745-6150-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McInerney JO, Cotton JA, Pisani D. The prokaryotic tree of life: past, present … and future? Trends Ecol. Evol. 2008;23:276–281. doi: 10.1016/j.tree.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 216.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 217.Sundin GW. Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 2007;45:129–151. doi: 10.1146/annurev.phyto.45.062806.094317. [DOI] [PubMed] [Google Scholar]
- 218.Wollman EL, Jacob F, Hayes W. Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb. Symp. Quant. Biol. 1956;21:141–162. doi: 10.1101/sqb.1956.021.01.012. [DOI] [PubMed] [Google Scholar]
- 219.Moreno E. Genome evolution within the alpha Proteobacteria: why do some bacteria not possess plasmids and others exhibit more than one different chromosome? FEMS Microbiol. Rev. 1998;22:255–275. doi: 10.1111/j.1574-6976.1998.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 220.Kolsto AB. Dynamic bacterial genome organization. Mol. Microbiol. 1997;24:241–248. doi: 10.1046/j.1365-2958.1997.3501715.x. [DOI] [PubMed] [Google Scholar]
- 221.Egan ES, Fogel MA, Waldor MK. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol. Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- 222.Slater FR, Bailey MJ, Tett AJ, Turner SL. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 2008 doi: 10.1111/j.1574-6941.2008.00505.x. [28 May 2008, Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 223.Kado CI. Origin and evolution of plasmids. Antonie Van Leeuwenhoek. 1998;73:117–126. doi: 10.1023/a:1000652513822. [DOI] [PubMed] [Google Scholar]
- 224.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Omelchenko MV, Wolf YI, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Daly MJ, Koonin EV, Makarova KS. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 2005;5:57. doi: 10.1186/1471-2148-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 228.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 229.Ichige A, Kobayashi I. Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J. Bacteriol. 2005;187:6612–6621. doi: 10.1128/JB.187.19.6612-6621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sorek R, Kunin V, Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 231.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 234.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl Acad. Sci. USA. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 237.Lynch M. Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol. 2006;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- 238.Dawkins R. The Selfish Gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- 239.Grishin NV, Wolf YI, Koonin EV. From complete genomes to measures of substitution rate variability within and between proteins. Genome Res. 2000;10:991–1000. doi: 10.1101/gr.10.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Novichkov PS, Ratner I, Wolf YI, Koonin EV, Dubchak I. ATGC: a database of orthologous genes from closely related prokaryotic genomes and a research platform for microevolution of prokaryotes. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn684. , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–1323. doi: 10.1534/genetics.105.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Le Rouzic A, Dupas S, Capy P. Genome ecosystem and transposable elements species. Gene. 2007;390:214–220. doi: 10.1016/j.gene.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 243.Marais GA, Calteau A, Tenaillon O. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica. 2008;134:205–210. doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 244.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, Chisholm SW. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 246.Dagan T, Blekhman R, Graur D. The “domino theory” of gene death: gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol. Biol. Evol. 2006;23:310–316. doi: 10.1093/molbev/msj036. [DOI] [PubMed] [Google Scholar]
- 247.Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007;23:511–520. doi: 10.1016/j.tig.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 248.Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, et al. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008 doi: 10.1093/dnares/dsn011. [28 May 2008, Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 251.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 2002;61:435–447. doi: 10.1006/tpbi.2002.1588. [DOI] [PubMed] [Google Scholar]
- 254.Mamirova L, Popadin K, Gelfand MS. Purifying selection in mitochondria, free-living and obligate intracellular proteobacteria. BMC Evol. Biol. 2007;7:17. doi: 10.1186/1471-2148-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- 256.McGeoch AT, Bell SD. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat. Rev. Mol. Cell Biol. 2008;9:569–574. doi: 10.1038/nrm2426. [DOI] [PubMed] [Google Scholar]
- 257.Bapteste E, Brochier C, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–363. doi: 10.1155/2005/859728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mulkidjanian AY, Koonin EV, Makarova KS, Mekhedov SL, Sorokin A, Wolf YI, Dufresne A, Partensky F, Burd H, Kaznadzey D, et al. The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl Acad. Sci. USA. 2006;103:13126–13131. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 260.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2007;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Makarova KS, Wolf YI, Koonin EV. Potential genomic determinants of hyperthermophily. Trends Genet. 2003;19:172–176. doi: 10.1016/S0168-9525(03)00047-7. [DOI] [PubMed] [Google Scholar]
- 262.Ladenstein R, Antranikian G. Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv. Biochem. Eng. Biotechnol. 1998;61:37–85. doi: 10.1007/BFb0102289. [DOI] [PubMed] [Google Scholar]
- 263.Greaves RB, Warwicker J. Mechanisms for stabilisation and the maintenance of solubility in proteins from thermophiles. BMC Struct. Biol. 2007;7:18. doi: 10.1186/1472-6807-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ladenstein R, Ren B. Reconsideration of an early dogma, saying “there is no evidence for disulfide bonds in proteins from archaea”. Extremophiles. 2008;12:29–38. doi: 10.1007/s00792-007-0076-z. [DOI] [PubMed] [Google Scholar]
- 265.Cox MM, Battista JR. Deinococcus radiodurans - the consummate survivor. Nat. Rev. Microbiol. 2005;3:882–892. doi: 10.1038/nrmicro1264. [DOI] [PubMed] [Google Scholar]
- 266.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl Acad. Sci. USA. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl Acad. Sci. USA. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhang C, Wei J, Zheng Z, Ying N, Sheng D, Hua Y. Proteomic analysis of Deinococcus radiodurans recovering from gamma-irradiation. Proteomics. 2005;5:138–143. doi: 10.1002/pmic.200300875. [DOI] [PubMed] [Google Scholar]
- 270.Makarova KS, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Lapidus A, Copeland A, Kim E, Land M, et al. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS ONE. 2007;2:e955. doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 272.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD, Lai B, Ravel B, Li SM, Kemner KM, et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Woese CR. There must be a prokaryote somewhere: microbiology's search for itself. Microbiol. Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Martin W, Koonin EV. A positive definition of prokaryotes. Nature. 2006;442:868. doi: 10.1038/442868c. [DOI] [PubMed] [Google Scholar]
- 275.Slonimski P, Mosse M, Golik P, Henault A, Diaz Y, Risler J, Comet J, Aude J, Wozniak A, Glemet E, Codani J. The first laws of genomics. Microbial Comp. Genomics. 1998;3:46. [Google Scholar]
- 276.Parzen E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962;33:1065–1076. [Google Scholar]
- 277.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]