Abstract
Cytoplasmic dynein and dynactin interact to drive microtubule-based transport in the cell. The p150Glued subunit of dynactin binds to dynein, and directly to microtubules. We have identified alternatively spliced isoforms of p150Glued that are expressed in a tissue-specific manner and which differ significantly in their affinity for microtubules. Live cell assays indicate that these alternatively spliced isoforms also differ significantly in their microtubule plus end-tracking activity, suggesting a mechanism by which the cell may regulate the dynamic localization of dynactin. To test the function of the microtubule-binding domain of p150Glued, we used RNAi to deplete the endogenous polypeptide from HeLa cells, followed by rescue with constructs encoding either the full-length polypeptide or an isoform lacking the microtubule-binding domain. Both constructs fully rescued defects in Golgi morphology induced by depletion of p150Glued, indicating that an independent microtubule-binding site in dynactin may not be required for dynactin-mediated trafficking in some mammalian cell types. In neurons, however, a mutation within the microtubule-binding domain of p150Glued results in motor neuron disease; here we investigate the effects of four other mutations in highly conserved domains of the polypeptide (M571T, R785W, R1101K, and T1249I) associated in genetic studies with Amyotrophic Lateral Sclerosis. Both biochemical and cellular assays reveal that these amino acid substitutions do not result in functional differences, suggesting that these sequence changes are either allelic variants or contributory risk factors rather than causative for motor neuron disease. Together, these studies provide further insight into the regulation of dynein-dynactin function in the cell.
Cytoplasmic dynein is the major minus end-directed microtubule motor in the cell. The multisubunit protein complex dynactin binds to dynein and is required for most of its cellular activities, including spindle assembly, vesicular trafficking, and cell migration. Because of these multiple essential cellular roles, loss of either dynein or dynactin is lethal early in embryogenesis in higher eukaryotes (1, 2). However, neurons are uniquely vulnerable to more subtle defects in dynein function. Missense mutations in the genes encoding dynein or dynactin cause progressive neuronal degeneration in both mice and humans (2-5) while other cell types remain unaffected. Together, these observations suggest a key role for dynein and dynactin in neuronal maintenance and function.
The mechanisms by which dynactin facilitates dynein-driven transport are not fully clear, but microtubule binding by dynactin has been shown to enhance the processivity of the dynein motor (6, 7). The projecting arm of dynactin is composed of a dimer of the p150Glued subunit, which binds directly to dynein (8, 9). The N terminus of the p150Glued polypeptide contains a cytoskeletal-associated protein, glycine-rich (CAP-Gly)3 domain that binds directly to microtubules in a nucleotide-independent manner (10). In addition to this domain, a stretch of basic amino acids directly C-terminal to the CAP-Gly domain has been identified as a second, lower affinity microtubule-binding site. This basic domain may further enhance dynein processivity by facilitating one-dimensional diffusion of dynactin along microtubules (11).
Expression of GFP-labeled p150Glued shows tracking of growing microtubule tips in the cell (12, 13). Plus end-tracking proteins (+TIPs), including p150Glued, have been hypothesized to regulate microtubule dynamics (for a review see Ref. 14). Several of these +TIP proteins interact with microtubules through CAP-Gly domains, such as CLIP-115 and CLIP-170, while others interact with microtubules through basic/serine-rich regions, such as adenomatous polyposis coli protein (APC) and the CLASPs (14). The mechanism of plus end-tracking of p150Glued, as well as the relative contributions of the tandem CAP-Gly and basic domains in this process, are not yet understood.
Modulation of microtubule binding may serve to alter cellular function of p150Glued. Alternative splicing of DCTN1, the gene that encodes p150Glued, produces p135, a neuronal-enriched splice form that lacks the CAP-Gly and basic domains and cannot bind to microtubules (15). Also, a missense mutation in this CAP-Gly domain has been shown to cause neurodegeneration. A glycine-to-serine substitution at residue 59 of p150Glued causes a slowly progressive, autosomal dominant form of lower motor neuron disease in a human kindred (5, 16). This G59S mutation is located within the CAP-Gly domain and decreases the affinity of the mutant protein for microtubules (5, 17).
Further analysis of the DCTN1 gene has identified three heterozygous, missense mutations, M571T, R785W, and T1249I, in p150Glued in patients with amyotrophic lateral sclerosis (ALS), and one mutation, R1101K, in patients with both ALS and frontotemporal dementia (FTD) (18, 19). These substitutions occur in highly conserved residues that are located in the coiled-coiled and dynein intermediate chain (DIC)-binding regions of the protein, so they are predicted to act as either causative mutations or risk factors for human motor neuron disease. However, the effects of these mutations on the cellular function of p150Glued are not yet known.
In this study, we address the functional effects of several genetic alterations in the p150Glued subunit of dynactin. We identify naturally occurring splice forms of p150Glued that lack exon 5, exons 5 and 6, or exons 5 through 7. RT-PCR analysis of human tissues indicates that alternatively spliced isoforms of p150Glued are expressed in a tissue-specific pattern. Functional studies indicate that alternative splicing of these exons has a significant effect on microtubule binding affinity and on microtubule plus end-tracking activity in the cell. Additionally, we examine the functional consequences of sequence variants identified in patients with ALS. Together, these studies provide further insight into the regulation of dynein-dynactin function in vivo.
EXPERIMENTAL PROCEDURES
Library Screening and RT-PCR—Multiple isoforms of p150Glued were identified in a screen of a human fetal brain cDNA library (Stratagene). RT-PCR was performed using an Enhanced Avian RT-PCR kit (Sigma), poly(A)+ RNA from adult human brain, skeletal muscle and kidney (Clontech), and primers corresponding to sequences from the 5′-end of exon 3 and the 3′-end of exon 8 of the human DCTN1 gene. The PCR products were fractionated on 4% NuSieve-GTG (BMA, Rockland, ME) gel, and purified with QIAEX II Gel Extraction Kit (Qiagen) for TA cloning followed by sequence analysis.
Cell Cultures, Transfections, and Immunocytochemistry—COS7, HeLa-M and MN1 cells were maintained as described (17). Transient transfection assays were performed using Fugene-6 (Roche Applied Science) and plasmids encoding Myc-tagged or GFP-tagged forms of p150Glued. For immunofluorescence assays, COS7 and HeLa-M cells were fixed in -20 °C methanol for 10 min and MN1 cells were fixed in 4% paraformaldehyde for 20 min and then permeabilized with 0.25% Triton X-100. Immunostaining was performed using antibodies to tubulin (DM1A, Sigma or YL1/2, Serotech), p150Glued (BD Biosciences and polyclonal antibody UP502(17)), Myc (Invitrogen), Golgi protein GM130 (BD Biosciences), and TGN46 (Serotech).
Live Cell Imaging—For time-lapse imaging using OpenLab software (Improvision), cells were seeded on glass bottom dishes (World Precision Instruments) and imaged in phenol red-free Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 25 mm HEPES, 1% OxyFluor (Oxyrase), and sealed with mineral oil (Sigma). The microscope stage was maintained at 37 °C. Images were acquired at 5-s intervals for 5 min.
RNA Interference—HeLa-M cells were transfected using Lipofectamine RNAiMAX (Invitrogen) with 10 nm RNA duplexes (Dharmacon) targeted to the 3′-untranslated region of human DCTN1 (GenBank™ accession number NM_004082). Two independent RNA duplexes were used: UTR1, 5′-ccaccaccaaagguuaaguuu-3′ and UTR2, 5′-gacuucaccccuugauuaauu-3′, both of which gave similar results. Maximal p150Glued knockdown was achieved 72 h after transfection. Rescue experiments were performed by transfecting RNAi-treated cells 72 h later with the indicated DNA constructs using Fugene (Roche Diagnostics), in comparison to mock-treated controls. Overall levels of p150Glued expression were determined by quantitative Western blot of whole cell lysates; we observed no significant differences between the mean expression levels of mock-treated HeLa-M cells and cells rescued with either wild-type p150Glued or any of the specifically mutated constructs (p = 0.6 by one-way analysis of variance). We also compared the relative signal intensity at the cellular level via immunofluorescence measurements analyzed with NIH ImageJ and consistently found that expression of wild-type p150 to about 50% of the control level was sufficient to rescue Golgi morphology. We used this expression level as a standard to compare the effects of wild-type and mutant proteins. The average fluorescence intensity of p150 for each condition was: 113.5 ± 17.8 (n = 84 cells) for mock-treated cells, 56.2 ± 16.5 (n = 100 cells) for cells rescued with wild-type p150Glued, 55.5 ± 14.6 (n = 43 cells) for cells rescued with the M571T construct, 51.2 ± 15.7 (n = 60 cells) for cells rescued with the R785W construct, 48.0 ± 9.3 (n = 42 cells) for cells rescued with the R1101K construct, and 49.5 ± 13.1 (n = 44 cells) for cells rescued with the T1249I construct.
Microtubule Binding Assay—His-tagged alternatively spliced isoforms of p150Glued, spanning residues 1-333, were expressed in E. coli and purified by Ni2+-affinity chromatography. Alternatively, cDNA constructs encoding these isoforms or wild type and mutant forms of p150Glued were expressed in vitro using the TnTT7 Quick system (Promega) and clarified by centrifugation at 39,000 × g for 30 min. Either the purified recombinant or in vitro-expressed proteins were then incubated with increasing concentrations of microtubules polymerized in vitro from purified bovine tubulin (Cytoskeleton) in the presence of 0.1 mm GTP and 20 μm Taxol (Cytoskeleton) for 20 min at 37 °C. Microtubule-bound and unbound proteins were separated by centrifugation at 39,000 × g for 20 min and analyzed by SDS-PAGE and Western blot analysis. Results were quantified by densitometry using NIH ImageJ.
Dynein Intermediate Chain Pull-down Assay—His-tagged full-length recombinant dynein intermediate chain was expressed in Escherichia coli, purified by Ni2+ affinity chromatography, and then coupled to activated CH-Sepharose 4B beads (8). Myc-tagged p150Glued proteins were expressed using the TnTT7 Quick system (Promega) and clarified by centrifugation at 39,000 × g for 30 min. A fixed volume of DIC-Sepharose beads (equivalent to ∼15 μg DIC) were incubated with increasing amounts of p150Glued proteins for 1 h at 25 °C on a platform shaker at 200 rpm; bound and unbound proteins were separated by centrifugation at 1,300 × g for 2 min and analyzed by SDS-PAGE and Western blot. Results were quantified by densitometry using NIH ImageJ.
Sucrose Density Gradient Fractionation—COS7 cells transiently expressing Myc-tagged wild-type or mutant p150Glued proteins were lysed using 100 mm Na-PIPES (pH 6.8), 1 mm MgSO4, 1 mm EGTA, 25 mm NaCl, 0.5 mm dithiothreitol, 1% IGEPAL CA-630, and protease inhibitors (leupeptin, pepstatin A, N-p-tosyl-l-arginine methyl ester, and phenylmethylsulfonyl fluoride). The cell lysate was clarified by centrifugation at 38,000 × g for 10 min at 4 °C, and the resulting supernatant was layered over a 5-25% linear sucrose density gradient and centrifuged at 120,000 × g for 18 h at 4 °C. 1-ml gradient fractions were collected and analyzed by SDS-PAGE and Western blot analysis using monoclonal antibodies to Myc (Invitrogen), DIC (MAB 1618, Chemicon), and dynamitin (BD Biosciences). Results were quantified by densitometry using NIH ImageJ.
RESULTS
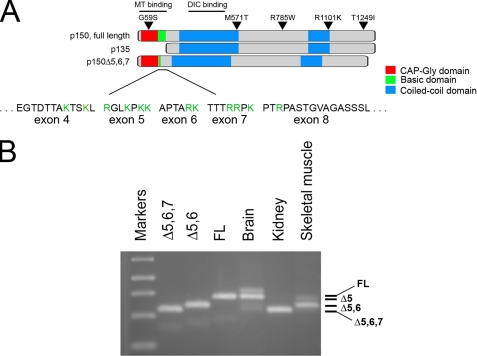
Differential Expression of Alternatively Spliced Isoforms of p150Glued—The human gene encoding p150Glued is composed of 32 exons (20). We have previously identified an alternatively spliced form of p150Glued that lacks the N-terminal CAP-Gly and basic microtubule-binding domains (Fig. 1A). This p135 isoform is expressed primarily in neurons (15). We screened a fetal human cDNA library to probe for additional alternatively spliced isoforms of p150Glued and found multiple alternatively spliced cDNAs, which differ in an N-terminal domain encoded by exons 5-7; three small exons that encode 7, 6, and 7 amino acid residues, respectively (Fig. 1A). Specifically, we identified isoforms lacking exons 5 and 6 (Δ5,6) and exons 5 through 7 (Δ5,6,7).
FIGURE 1.
Alternative splicing results in the tissue-specific expression of multiple isoforms of p150Glued. A, full-length isoform of the dynactin subunit p150Glued has a N-terminal CAP-Gly domain (red), followed by a basic domain that binds more weakly to microtubules (green), and two coiled-coil domains that mediate homodimerization (blue). These coiled-coil domains also mediate binding to the DIC and to the Arp1 subunit of dynactin. The previously characterized p135 isoform is specifically expressed in neurons and lacks both of the N-terminal microtubule-binding domains. The newly characterized Δ5, Δ5,6, and Δ5,6,7 isoforms lack much of the basic domain that forms a secondary site of association with the microtubule; basic residues in the amino acid sequences of exons 4 through 8 are highlighted in green. B, isoform expression was analyzed by RT-PCR of mRNA isolated from adult human tissues including brain, kidney, and skeletal muscle using primers spanning exons 3-8, and compared with PCR products from cDNAs corresponding to Δ5,6,7, Δ5,6, and full-length p150Glued. The identity of the bands observed by gel electrophoresis was confirmed by direct sequencing of the PCR products. A minor upper band visible in the PCR reaction from human brain mRNA was determined to be unrelated by sequence analysis.
We performed RT-PCR to compare the expression of these alternatively spliced transcripts in several human tissues, including brain, kidney, and muscle (Fig. 1B). The identity of the bands observed by gel electrophoresis was confirmed by direct sequencing of the PCR products. We noted the expression of multiple isoforms of p150Glued in adult brain, including a major band corresponding to the full-length polypeptide that includes all three exons (FL), as well as two more minor bands corresponding to isoforms lacking either exons 5 and 6 (Δ5,6) or lacking exons 5 through 7 (Δ5,6,7). In skeletal muscle we also observed the expression of multiple p150Glued isoforms, including a major band corresponding to Δ5,6 and minor bands corresponding to an isoform lacking only exon 5 (Δ5) as well as Δ5,6,7. In contrast, the predominant isoform of p150Glued expressed in kidney was Δ5,6,7, which lacks the basic domain. Together, these observations suggest tissue-specific regulation of expression of p150Glued isoforms.
Regulation of Tip Tracking Behavior by Differential Expression of p150Glued Isoforms—The region encoded by exons 5-7 spans a basic domain of the p150Glued polypeptide (Fig. 1A) that has recently been identified as possessing a weak affinity for microtubules (11). Truncation constructs of p150Glued that lack the N-terminal CAP-Gly domain but include this basic domain have been shown to exhibit one-dimensional diffusion along the microtubule, termed “skating” behavior (11). In contrast, constructs that include the N-terminal CAP-Gly domain bind more stably to the microtubule. Therefore, the additional splice forms that we identified may encode functionally distinct isoforms of the polypeptide, which differ in their relative association with the microtubule.
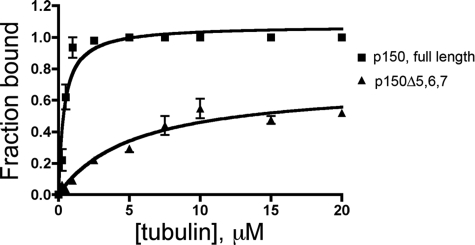
We tested whether these isoforms differ in their affinity for microtubules. As shown in Fig. 2, a comparison of the binding of the p150-FL and p150-Δ5,6,7 isoforms in a microtubule sedimentation assay indicates that the loss of the basic domain significantly alters the overall affinity for microtubules. p150-FL bound to microtubules with a Kd of 0.40 ± 0.07 μm. p150-Δ5,6,7 bound to microtubules much more weakly; we did not reach saturation (the extrapolated Bmax was 0.7 ± 0.1) potentially due to weak binding, but we estimated a Kd of 5.3 ± 1.5 μm. Consistent with this observation, we have found that a recombinant construct of p150Glued spanning the CAP-Gly domain (residues 1-107) binds to microtubules with a significantly lower affinity than a longer construct (1-333) that also includes the basic domain (data not shown).
FIGURE 2.
Alternatively spliced isoforms of p150Glued differ significantly in their binding affinity for microtubules. Microtubule binding affinities for the p150-FL and p150-Δ5,6,7 isoforms were measured in co-sedimentation assays with increasing concentrations of polymerized tubulin, and indicate that the basic domain encoded by exons 5, 6, and 7 contributes significantly to the overall affinity of p150Glued for the microtubule. Fitting the data to the equation [y = (Bmax×x)/(Kd+x)] by nonlinear regression yields a Kd of 0.40±0.07 μm for p150-FL; an estimated Kd of 5.3 ± 1.5 μm was obtained for p150-Δ5,6,7.
Cellular assays have demonstrated that GFP-labeled constructs of p150Glued can dynamically track growing microtubule plus ends (12, 13), but we have previously observed that the dynamic plus end localization of p150Glued is cell type-specific (21). Given the cell type-specific expression of p150Glued isoforms, we hypothesized that the difference in tip tracking behavior might be due to the differential expression of alternatively spliced isoforms of the polypeptide.
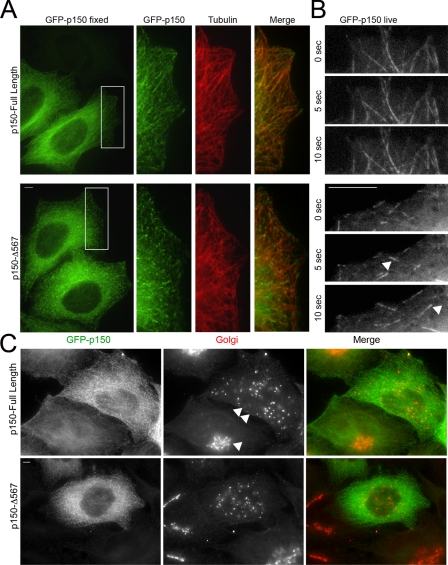
To test this hypothesis, we tagged both the p150-FL construct and the Δ5,6,7 isoform with GFP and expressed these polypeptides in HeLa cells. As shown in Fig. 3A, we saw a clear difference in the cellular localization of these constructs. P150-FL fully decorates cellular microtubules. Microtubule bundling was observed at higher expression levels, as previously noted (10). In contrast, the Δ5,6,7 isoform shows marked tip decoration at a similar expression level. Live cell assays demonstrate that the tip decoration seen in fixed cells is due to dynamic tip tracking of the alternatively spliced isoform (Fig. 3B for stills and see supplemental Movie S2). In contrast, tip tracking is not seen in dynamic assays with the p150-FL isoform (Fig. 3B and supplemental Movie S1). Similar results were obtained upon expression of these constructs in COS-7 cells (data not shown).
FIGURE 3.
Regulation of tip tracking by differential expression of p150Glued isoforms. A, HeLa cells transfected with GFP-tagged p150Glued-FL (top) or GFP-tagged p150-Δ5,6,7 (bottom) were stained with antibodies to p150Glued (UP502, green) and tubulin (red). Full-length p150Glued demonstrates lattice binding of microtubules, but p150-Δ5,6,7 localizes predominantly to microtubule tips. B, still images from a time series observing GFP-tagged p150-FL (top) or p150-Δ5,6,7 (bottom). Arrowheads point to GFP- p150-Δ5,6,7 labeling of dynamic plus ends. Frames are displayed in 5-s intervals. The full-length isoform localizes along the length of the microtubule lattice, remaining associated with the microtubule during both growth and catastrophe. In contrast, the p150-Δ5,6,7 isoform demonstrates dynamic tip-tracking behavior with growing microtubule tips. See also supplemental Movies S1 and S2. C, HeLa cells were transfected with GFP-tagged p150-FL (top) or GFP-tagged p150-Δ5,6,7 (bottom). Cells were fixed 24 h after transfection and stained with antibodies to p150Glued (UP502, green) and TGN46 (Golgi, red). Overexpression of either isoform causes Golgi disruption. The extent of Golgi disruption is dependent on expression levels; compare the relatively intact Golgi (single arrowhead) seen in a cell expressing low levels of the GFP-p150 with the dispersed Golgi (double arrowheads) in the cell expressing higher levels of GFP-p150. Scale bars, 5 μm.
Is the Microtubule Binding Domain of p150Glued Required for Dynactin Function?—The expression of distinct isoforms of p150Glued that vary in their affinity for microtubules both in vitro and in cellular assays suggests that modulation of microtubule binding may have a significant effect on dynactin function. While cytoplasmic dynein alone is a robust microtubule motor in vitro, dynactin is required for most of dynein functions in the cell. Dynactin is not required for processivity of the dynein motor, but single molecule studies have shown that dynactin enhances dynein run length along the microtubules, potentially due to the ATP-independent binding of dynactin to the microtubule (6, 7). However, in vitro studies have shown that when multiple dynein motors are bound to the same bead cargo the effects of dynactin on overall run length are negligible (22). Therefore, it has been suggested that the microtubule-binding domain of dynactin may not be required for intracellular organelle transport; studies in Drosophila S2 cells support this hypothesis (23).
We asked whether the microtubule-binding domain of p150Glued is required to maintain Golgi organization in mammalian cells. Dynactin is required to maintain a compact perinuclear Golgi in HeLa cells (17); either depletion or transient overexpression of p150Glued causes Golgi disruption. Transfection of HeLa cells with either the p150-FL or the p150-Δ5,6,7 isoforms leads to vesiculation of the Golgi (Fig. 3C). This disruption is expression-level dependent; lower levels of GFP-p150 expression do not measurably disrupt the Golgi (see cell marked with an arrowhead in Fig. 3C), but higher expression levels lead to complete dispersal (see cell marked with a double arrowhead in Fig. 3C).
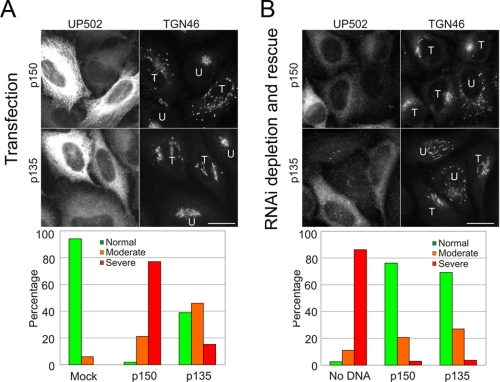
While the p150-Δ5,6,7 isoform binds to microtubules more weakly than the p150-FL polypeptide, this isoform still has an intact CAP-Gly domain. To more fully test the effects of the microtubule-binding domain, we compared the expression of p150-FL with p135, a neuronally expressed isoform that lacks both the CAP-Gly motif and the basic residues that mediate microtubule binding (Fig. 1A). As shown in Fig. 4A, overexpression of p135 does not disrupt the Golgi to the same extent as expression of p150-FL. In most cells expressing p135, the Golgi appears normal. In ∼40% of cells overexpressing p135, the Golgi appears tubulated (Fig. 4A), indicating partial rather than full disruption (24).
FIGURE 4.
The microtubule-binding domain of p150Glued is not required to maintain Golgi organization in mammalian cells. A, images of HeLa-M cells transiently transfected with either the p150-FL or p135 isoforms of dynactin. The accompanying bar graph shows the percentage of transfected cells containing either normal, moderately dispersed, or severely dispersed Golgi determined from ∼100 cells from three independent experiments for each construct. B, endogenous p150Glued was depleted from HeLa-M cells by RNAi-induced knockdown; cells were rescued by transfection with either the p150-FL or p135 isoforms of dynactin. Immunofluorescence was carried out using antibodies to p150Glued (UP502) and to TGN46 to visualize the Golgi. T, transfected cell; U, untransfected cell. Scale bar, 20 μm. The accompanying bar graph shows the percentage of transfected cells containing either normal, moderately dispersed, or severely dispersed Golgi determined from ∼100 cells from three independent experiments for each construct.
To test whether the microtubule-binding domain is required to maintain Golgi organization, we used an RNAi approach to determine if the p135 isoform can functionally substitute for full-length p150Glued in HeLa cells. Endogenous p150Glued was depleted using either of two independent siRNA oligos to the 3′-UTR (supplemental Fig. S1), leading to 70-80% knockdown of expression 72 h after transfection. Complete rescue was observed upon transfection with a full-length construct of human p150Glued that lacks the 3′-UTR and is thus resistant to the targeted siRNA (Fig. 4B).
Quantitative analysis indicates that the p135 isoform is indistinguishable from wild-type p150Glued in terms of rescuing Golgi localization following knockdown of endogenous p150Glued protein in HeLa cells (Fig. 4B). These results suggest that the microtubule-binding domain of p150Glued is not important for dynactin's role in maintaining a compact perinuclear Golgi in mammalian cells, consistent with the observation that this domain is dispensable for peroxisome transport in Drosophila (23).
Functional Analysis of Sequence Variants of p150Glued Identified in Patients with ALS—While the experiments shown in Fig. 4 indicate that the microtubule-binding domain of p150Glued is not required for normal Golgi organization in nonneuronal cells, a mutation in this domain (G59S; Fig. 1A) has a significant effect on dynactin function leading to motor neuron degeneration (5). This mutation exerts its effects through mechanisms involving both loss-of-function, and dominant gain of a toxic function (2, 17, 25). Additional allelic variants of the p150Glued polypeptide have been identified in ALS patients but not in controls (18, 19). These allelic variants, M571T, R785W, R1101K, and T1249I, affect residues that are highly conserved among the vertebrate p150Glued proteins and are therefore potential candidates as causal or risk factors for ALS. However, there is no conclusive genetic evidence linking these mutations to ALS. To test whether these new p150Glued mutations lead to functional disruption of dynein-dynactin activity, we tested the effects of these mutations in both in vitro and cellular assays.
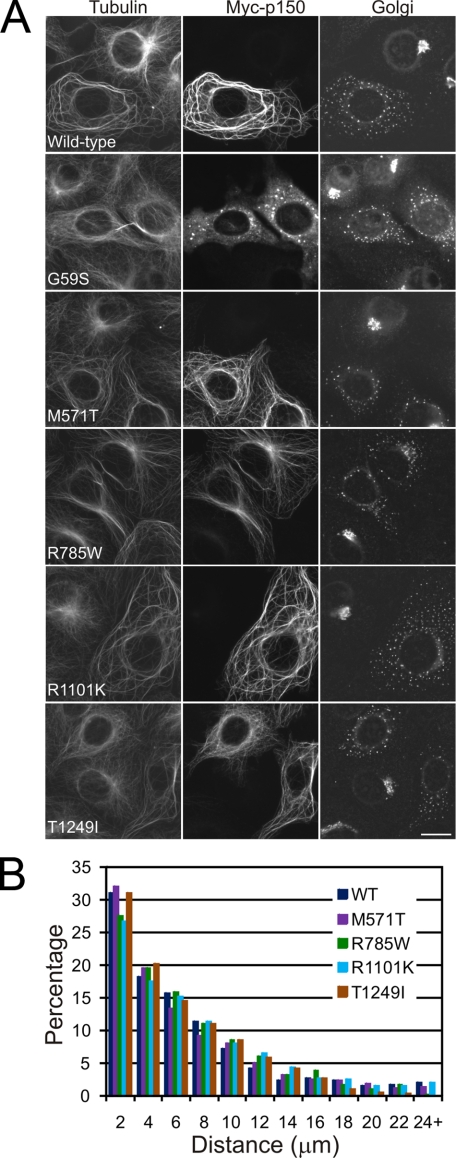
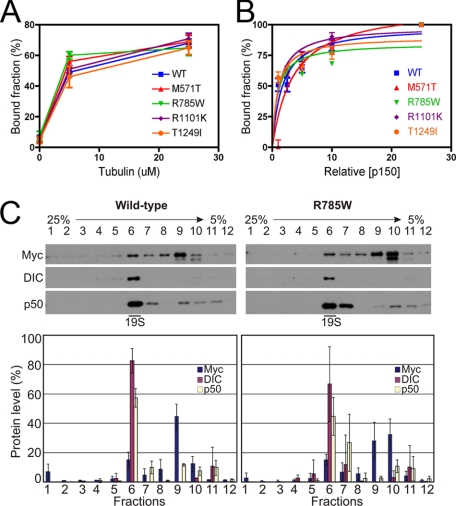
We expressed Myc-tagged constructs corresponding to each variant in both COS-7 cells (Fig. 5A) and the motor neuron-like cell line MN1 (supplemental Fig. S2), and compared the localization of these variants to wild-type full-length p150Glued and to p150Glued with the G59S mutation. The M571T, R785W, R1101K, and T1249I constructs all bound robustly to cellular microtubules, indistinguishable from wild-type p150Glued; only the G59S mutation significantly disrupts microtubule binding in this assay. Each of the Myc-tagged constructs, including the G59S construct, disrupted the Golgi in transfected cells. The dispersed Golgi staining seen in transfected cells differs significantly from the perinuclear localization in untransfected cells on the same coverslip (Fig. 5A). To quantitate these results we measured the distribution of Golgi staining as a function of distance from the nucleus in both COS7 and MN1 cells expressing Myc-tagged wild-type p150Glued, or the M571T, R785W, R1101K, and T1249I constructs. No significant differences were observed as compared with expression of Myc-tagged wild-type p150Glued (Fig. 5B and supplemental Fig. S2). These experiments suggest that these four amino acid substitutions do not perturb microtubule binding. We also verified this in microtubule sedimentation assays; as shown in Fig. 6A, no significant differences were observed in the association of these constructs with microtubules in vitro.
FIGURE 5.
Expression of wild-type and mutant p150Glued in COS7 cells. A, COS7 cells transected with either wild-type or M571T, R785W, R1101K, and T1249I Myc-tagged constructs of p150Glued were analyzed by immunofluorescence using antibodies to tubulin, the Myc tag, and TGN46 (Golgi). Note the extensive Golgi disruption in transfected cells. Scale bar, 20 μm. B, histogram showing the quantification of Golgi disruption by wild-type and mutant p150Glued proteins. The distribution of the distance of individual Golgi fragments from the nuclear surface was measured in COS7 cells expressing either wild-type or the indicated mutant p150Glued proteins. Data from ∼20 cells with measurements of >1000 individual Golgi fragments for each construct are shown.
FIGURE 6.
Comparison of microtubule and DIC binding of wild-type and mutant p150Glued. A, microtubule binding was measured in co-sedimentation assays at 0, 5, and 25 μm tubulin for in vitro translated wild-type or mutant p150Glued proteins. Data are shown as the bound fraction as a function of tubulin concentration, and are the mean ± S.D. obtained from three independent experiments. No significant differences were observed among the constructs. B, DIC binding assays were performed at increasing concentrations of p150Glued. Wild-type or mutant constructs were expressed using in vitro translation and binding to DIC-Sepharose beads was measured in pull-down assays. Data points are the mean ± S.D. obtained from three independent experiments; data were fit to the equation [y = (Bmax × x)/(Kd + x)] by nonlinear regression; no significant differences among the constructs were observed. C, sucrose density gradients of dynein-dynactin complex. Cell lysates from COS7 cells transiently expressing either wild-type or mutant p150Glued proteins (the R785W mutant is shown as a representative example) were fractionated through a 5-25% linear sucrose density gradient and the fractions subjected to immunoblot analysis using antibodies against Myc, DIC, and p50. The graphs show the relative protein amounts in each fraction as mean ± S.D. obtained from three independent experiments.
Both the M571T and R785W substitutions occur in or near the dynein-binding domain of p150Glued. Therefore we performed pull-down assays with recombinant DIC covalently bound to Sepharose beads. We noted no significant differences in the association of any of the four Myc-tagged constructs with DIC in this assay (Fig. 6B). Thus, none of the mutations have a measurable effect on the affinity of the dynein-dynactin interaction.
As noted above, the G59S mutation in p150Glued induces both a loss-of-function, and a gain-of-function, the later due to an increased tendency for the mutant protein to aggregate (17). To test whether any of ALS-associated p150Glued variants induce a similar aggregation, or whether they are less effectively incorporated into the dynactin complex, we expressed the Myc-tagged M571T, R785W, R1101K, and T1249I constructs in COS-7 cells, and then fractionated the resulting cellular lysates over sucrose density gradients. Comparisons of these gradients (Fig. 6C and data not shown) indicate comparable levels of incorporation of wild type and Myc-tagged M571T, R785W, R1101K, and T1249I constructs of human p150Glued into the endogenous dynein-dynactin complex. Additionally, both sucrose density gradient analysis and immunofluorescence analysis of transfected cells indicates that none of these mutations result in aberrant p150Glued aggregation. We also examined the effects of overexpression of wild type and Myc-tagged M571T, R785W, R1101K, and T1249I constructs of human p150Glued in the MN1 neuronal cell line (supplemental Fig. S2). Again, we observed no aggregation induced by any of the constructs, in contrast to previous observations on the formation of protein aggregates induced by the expression of the G59S mutation in this cell type (17).
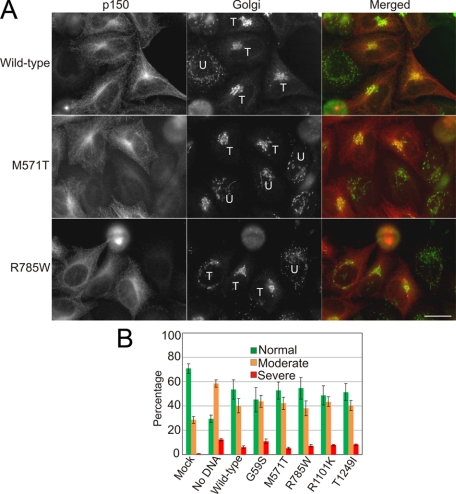
Together, these results suggest that the M571T, R785W, R1101K, and T1249I substitutions in the p150Glued polypeptide do not result in significant perturbation of dynactin function. In order to detect more subtle defects in p150Glued function, we used RNAi to specifically knockdown endogenous p150Glued in HeLa cells (supplemental Fig. S1) and examined the ability of the Myc-tagged M571T, R785W, R1101K, and T1249I constructs to rescue cellular function, using a rescue of Golgi disruption as a readout for wild-type dynein-dynactin activity in these experiments (Fig. 7). Expression levels of each of the constructs were monitored relative to expression in mock-treated controls by quantitative Western blot as well as quantitative analysis at the cellular level to ensure cells with similar expression levels were compared (as described under “Experimental Procedures”). While there were no clear-cut differences among the constructs tested, we found that the G59S construct was least effective at restoring normal Golgi morphology, consistent with a mild loss-of-function effect for this polypeptide (17). None of the ALS-associated variants were significantly different from wild type in this assay.
FIGURE 7.
Comparison of wild-type and mutant p150Glued expressed in HeLa cells following depletion of endogenous p150Glued by RNAi. A, images of HeLa-M cells transiently expressing either the wild-type or mutant p150Glued proteins (only M571T and R785W images are shown) following RNAi-induced knockdown of the endogenous p150Glued protein. Cells were fixed 24 h after plasmid transfection and immunofluorescence staining carried out using monoclonal antibodies against p150Glued and TGN46 (Golgi). T, transfected cell; U, untransfected cell. Scale bar, 20 μm. B, bar graph shows the average percentage ± S.D. of cells containing either normal, moderately dispersed, or severely dispersed Golgi determined from >300 cells from three independent experiments.
DISCUSSION
Cytoplasmic dynein and dynactin are essential for a wide range of intracellular functions in higher eukaryotes, including interphase roles of intracellular trafficking and mitotic roles in spindle assembly and checkpoint. Given these varied and important cellular functions, it is not surprising that loss of either dynein or dynactin is lethal early in development. However, it is interesting that subtle defects in dynein or dynactin caused by missense mutations in subunits of either complex result in a neuron-specific degeneration in both mice and humans.
The observation that a G59S mutation in the p150Glued subunit of dynactin results in slowly progressive neurodegenerative disease had lead to multiple studies looking for additional mutations in the polypeptide in patients with motor neuron disease. Munch et al. (18, 19) have identified three alterations in the sequence of p150Glued in patients with ALS, as well as one in a patient with ALS and FTD. Insufficient genetic data were available to causally associate the sequence variants with disease.
As all four of the mutations identified in the DCTN1 gene map to domains of interest within the p150Glued polypeptide, we tested the effects of these amino acid substitutions using a range of biochemical and cellular assays, in comparison to both wild-type and G59S forms of the polypeptide. None of these mutations appear to induce significant functional disruption of transport or trafficking in either neuronal or nonneuronal cells. Further, none of these mutations induce the formation of prominent cellular aggregates similar to those seen previously upon expression of the G59S mutation. Thus, we conclude that despite the initial identification of these variants in patients with ALS, these changes are not sufficient to cause disease. We cannot rule out the possibility that these variants may act as risk rather than causative factors, with the potential to compromise dynactin function in a subtle manner. However, in the above assays, none of these four constructs was as effective in disrupting dynactin function as the previously identified G59S mutation. One striking aspect of these studies is that expression levels of p150Glued are likely to be tightly regulated in the cell. Both depletion and overexpression of p150Glued results in a similar phenotype of Golgi disruption, and may therefore be predicted to aversely affect axonal transport (17). Mutations that alter expression levels of this protein may therefore also be considered as potential risk factors for motor neuron disease. While the ALS-associated variants tested here did not noticeably affect either protein stability or protein aggregation, these aspects may be as or more important as direct perturbations in binding domains within dynactin (25).
Using RT-PCR we identified alternatively spliced isoforms of p150Glued that do affect dynactin activity directly. Specifically, we noted that multiple isoforms of the polypeptide are expressed that differ in their microtubule binding affinity. This in turn leads to alterations in the dynamic localization of the polypeptides in live cell assays. The full-length form of p150Glued that is expressed in human brain binds along the length of cellular microtubules, and is not specifically associated with dynamically growing and shrinking microtubule plus ends. In contrast, the shorter isoform of p150Glued that lacks exons 5, 6, and 7 and is highly expressed in kidney has a significantly weaker affinity for microtubules in vitro. In the cell, this isoform dynamically tracks growing but not shrinking microtubule plus ends.
Based on these observations, we hypothesize that the differential affinity for binding to microtubules seen in vitro means that the shorter isoform will bind less strongly along the lengths of microtubules in the cell. However, the Δ5,6,7 isoform is still able to actively track with growing microtubule ends, most likely through a direct association with the plus end-binding protein EB1 (21, 26). In contrast, the full-length isoform binds more robustly along the length of the microtubule and therefore does not exhibit plus end-binding specificity. This full-length construct is also capable of direct binding to EB1 (21), but remains associated with the microtubule during both growth and catastrophe. Thus the isoform-specific alterations in microtubule binding affinity we have noted lead to a striking modulation in +Tip-tracking behavior in live cell assays. As we have previously observed using in vitro assays, dynamic localization of CAP-Gly proteins to the microtubule plus end involves a balance in relative affinities, a higher affinity for either the microtubule end itself and/or other end-binding proteins such as EB1, in association with a relatively lower affinity for lateral binding to the microtubule (26). Here, we see a clear modulation of tip-tracking activity in the cell based on isoform expression.
Despite these clear functional differences both in vitro and in live cell assays, it is unclear if these isoforms have differential functions in vesicle trafficking in nonneuronal cells. Kim et al. (23) have shown in Drosophila S2 cells, and we confirm their result in HeLa cells, that the loss of the N-terminal microtubule-binding domain of p150Glued including both the CAP-Gly domain and the adjacent basic domain, does not significantly affect intracellular trafficking of organelles including peroxisomes and the Golgi. However, Kim et al. (23) noted that loss of the microtubule-binding domain of p150Glued did affect microtubule organization during cell division. The tissue-specific regulation of isoform expression that we note here may potentially correlate with overall mitotic activity, as the more stably binding full-length isoform is expressed at a high level in brain, potentially due to higher expression levels in post-mitotic neurons. Given the neuron-specific effects of the G59S mutation in p150Glued, this domain may play a critical role in some aspect of neuronal function. Recent evidence suggests a role for dynein and dynactin in maintaining microtubule polarity in the axon (27, 28); the independent microtubule-binding domain of dynactin may be critical for this function.
Given the wide range of cellular functions that depend on dynactin, it will be important to continue to characterize the regulation of this complex within the cell, including the expression of multiple functionally distinct isoforms as described here. Also, given the identification of a mutation in dynactin as causative for motor neuron disease, additional mutations in subunits of either dynein or dynactin may be found to be causative for neurodegenerative diseases such as ALS. However, it will be essential to thoroughly investigate the effects of these mutations in biochemical and cellular assays to fully understand the effects they may have on dynactin function in the cell.
Supplementary Material
This work was supported, in whole or in part, by Grants GM068591 and GM48661 from the National Institutes of Health. This work was also supported by the ALS Association (to E. L. F. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies S1 and S2.
Footnotes
The abbreviations used are: CAP-Gly, a cytoskeletal-associated protein, glycine-rich; PIPES, 1,4-piperazinediethanesulfonic acid; GFP, green fluorescent protein; UTR, untranslated region; ALS, amyotrophic lateral sclerosis; DIC, dynein intermediate chain.
References
- 1.Harada, A., Takei, Y., Kanai, Y., Tanaka, Y., Nonaka, S., and Hirokawa, N. (1998) J. Cell Biol. 141 51-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai, C., Lin, X., Chandran, J., Shim, H., Yang, W. J., and Cai, H. (2007) J. Neurosci. 27 13982-13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hafezparast, M., Klocke, R., Ruhrberg, C., Marquardt, A., Ahmad-Annuar, A., Bowen, S., Lalli, G., Witherden, A. S., Hummerich, H., Nicholson, S., Morgan, P. J., Oozageer, R., Priestley, J. V., Averill, S., King, V. R., Ball, S., Peters, J., Toda, T., Yamamoto, A., Hiraoka, Y., Augustin, M., Korthaus, D., Wattler, S., Wabnitz, P., Dickneite, C., Lampel, S., Boehme, F., Peraus, G., Popp, A., Rudelius, M., Schlegel, J., Fuchs, H., Hrabe de Angelis, M., Schiavo, G., Shima, D. T., Russ, A. P., Stumm, G., Martin, J. E., and Fisher, E. M. (2003) Science 300 808-812 [DOI] [PubMed] [Google Scholar]
- 4.Chen, X. J., Levedakou, E. N., Millen, K. J., Wollmann, R. L., Soliven, B., and Popko, B. (2007) J. Neurosci. 27 14515-14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puls, I., Jonnakuty, C., LaMonte, B. H., Holzbaur, E. L., Tokito, M., Mann, E., Floeter, M. K., Bidus, K., Drayna, D., Oh, S. J., Brown, R. H., Jr., Ludlow, C. L., and Fischbeck, K. H. (2003) Nat. Genet. 33 455-456 [DOI] [PubMed] [Google Scholar]
- 6.King, S. J., and Schroer, T. A. (2000) Nat. Cell Biol. 2 20-24 [DOI] [PubMed] [Google Scholar]
- 7.Ross, J. L., Wallace, K., Shuman, H., Goldman, Y. E., and Holzbaur, E. L. (2006) Nat. Cell Biol. 8 562-570 [DOI] [PubMed] [Google Scholar]
- 8.Karki, S., and Holzbaur, E. L. (1995) J. Biol. Chem. 270 28806-28811 [DOI] [PubMed] [Google Scholar]
- 9.Vaughan, K. T., and Vallee, R. B. (1995) J. Cell Biol. 131 1507-1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterman-Storer, C. M., Karki, S., and Holzbaur, E. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 1634-1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culver-Hanlon, T. L., Lex, S. A., Stephens, A. D., Quintyne, N. J., and King, S. J. (2006) Nat. Cell Biol. 8 264-270 [DOI] [PubMed] [Google Scholar]
- 12.Vaughan, P. S., Miura, P., Henderson, M., Byrne, B., and Vaughan, K. T. (2002) J. Cell Biol. 158 305-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan, K. T., Tynan, S. H., Faulkner, N. E., Echeverri, C. J., and Vallee, R. B. (1999) J. Cell Sci. 112 1437-1447 [DOI] [PubMed] [Google Scholar]
- 14.Akhmanova, A., and Steinmetz, M. O. (2008) Nat. Rev. Mol. Cell Biol. 9 309-322 [DOI] [PubMed] [Google Scholar]
- 15.Tokito, M. K., Howland, D. S., Lee, V. M., and Holzbaur, E. L. (1996) Mol. Biol. Cell 7 1167-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puls, I., Oh, S. J., Sumner, C. J., Wallace, K. E., Floeter, M. K., Mann, E. A., Kennedy, W. R., Wendelschafer-Crabb, G., Vortmeyer, A., Powers, R., Finnegan, K., Holzbaur, E. L., Fischbeck, K. H., and Ludlow, C. L. (2005) Ann. Neurol. 57 687-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy, J. R., Sumner, C. J., Caviston, J. P., Tokito, M. K., Ranganathan, S., Ligon, L. A., Wallace, K. E., LaMonte, B. H., Harmison, G. G., Puls, I., Fischbeck, K. H., and Holzbaur, E. L. (2006) J. Cell Biol. 172 733-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munch, C., Rosenbohm, A., Sperfeld, A. D., Uttner, I., Reske, S., Krause, B. J., Sedlmeier, R., Meyer, T., Hanemann, C. O., Stumm, G., and Ludolph, A. C. (2005) Ann. Neurol. 58 777-780 [DOI] [PubMed] [Google Scholar]
- 19.Munch, C., Sedlmeier, R., Meyer, T., Homberg, V., Sperfeld, A. D., Kurt, A., Prudlo, J., Peraus, G., Hanemann, C. O., Stumm, G., and Ludolph, A. C. (2004) Neurology 63 724-726 [DOI] [PubMed] [Google Scholar]
- 20.Tokito, M. K., and Holzbaur, E. L. (1998) Biochim. Biophys. Acta 1442 432-436 [DOI] [PubMed] [Google Scholar]
- 21.Ligon, L. A., Shelly, S. S., Tokito, M., and Holzbaur, E. L. (2003) Mol. Biol. Cell 14 1405-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallik, R., Petrov, D., Lex, S. A., King, S. J., and Gross, S. P. (2005) Curr. Biol. 15 2075-2085 [DOI] [PubMed] [Google Scholar]
- 23.Kim, H., Ling, S. C., Rogers, G. C., Kural, C., Selvin, P. R., Rogers, S. L., and Gelfand, V. I. (2007) J. Cell Biol. 176 641-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caviston, J. P., Ross, J. L., Antony, S. M., Tokito, M., and Holzbaur, E. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10045-10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier-Larsen, E. S., Wallace, K. E., Pennise, C. R., and Holzbaur, E. L. (2008) Hum. Mol. Genet. 17 1946-1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligon, L. A., Shelly, S. S., Tokito, M. K., and Holzbaur, E. L. (2006) FEBS Lett. 580 1327-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng, Y., Wildonger, J., Ye, B., Zhang, Y., Kita, A., Younger, S. H., Zimmerman, S., Jan, L. Y., and Jan, Y. N. (2008) Nat. Cell Biol. 10 1172-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satoh, D., Sato, D., Tsuyama, T., Saito, M., Ohkura, H., Rolls, M. M., Ishikawa, F., and Uemura, T. (2008) Nat. Cell Biol. 10 1164-1171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.