Abstract
Enhancer-blocking insulators are DNA elements that disrupt the communication between a regulatory sequence, such as an enhancer or a silencer, and a promoter. Insulators participate in both transcriptional regulation and global nuclear organization, two features of chromatin that are thought to be maintained from one generation to the next through epigenetic mechanisms. Furthermore, there are many regulatory mechanisms in place that enhance or hinder insulator activity. These modes of regulation could be used to establish cell-type specific insulator activity that is epigenetically inherited along a cell and/or organismal lineage. This review will discuss the evidence for epigenetic inheritance and regulation of insulator function.
Keywords: Insulators, Epigenetic Inheritance, Chromatin, Nuclear organization
Introduction
Chromatin organization in eukaryotic cells is regulated on both local and global levels by a highly complex system of DNA elements and the proteins and RNAs that interact with them. Insulators are regulatory DNA elements that create boundaries in chromatin, delineating the ranges over which other regulatory influences take effect. There are two types of insulators: enhancer-blocking insulators, which prevent communication between discrete sequence elements (typically enhancers and promoters) when positioned between them, and barrier insulators, which prevent the spread of heterochromatin (Gaszner and Felsenfeld, 2006). This review will focus on enhancer-blocking insulators.
In Drosophila melanogaster there are multiple known insulators defined by their binding proteins, Suppressor of Hairy Wing [Su(Hw)], Boundary Element Associated Factors (BEAF-32A and BEAF-32B), Zeste-white 5 (Zw5), GAGA Binding Factor (GAF), and the most recently identified, Drosophila CTCF (dCTCF). However, in vertebrates the only known insulator protein is CCCTC-Binding Factor (CTCF) (Valenzuela and Kamakaka, 2006; Wallace and Felsenfeld, 2007). It remains unknown whether the vertebrate CTCF fulfills the roles of all the insulator proteins found in Drosophila, or whether other vertebrate insulator proteins are yet to be discovered.
Although previous work mainly focused on deciphering the mechanisms by which insulators block the communication between an enhancer and a promoter, the field has recently shifted to a more global view of insulators. Here we discuss what is known about the epigenetic inheritance and regulation of insulator information, areas that must be understood in order to place insulators in the context of a developmental system.
A role for insulators in nuclear organization
There are several models for how enhancer-blocking insulators disrupt communication between enhancers and promoters. We will refer to them as the promoter decoy model, the physical barrier model, and the loop domain model. The models are not necessarily mutually exclusive, so potentially any insulator could employ one or a combination of mechanisms in order to function properly. According to the promoter decoy model, an enhancer-blocking insulator recruits components of the transcription machinery that cause it to resemble a promoter at the molecular level. This resemblance allows it to compete with bona fide promoters for interaction with enhancers (Geyer, 1997). This model is consistent with the fact that enhancer DNA and insulator DNA have been observed to colocalize (Yoon et al., 2007; Zhu et al., 2007). However, interaction between insulators and promoters themselves has also been observed (Yoon et al., 2007), which could indicate that an alternative mechanism is responsible for both observations. Additionally, promoter decoy is unlikely to be the only mechanism underlying enhancer-blocking function, mainly because it does not explain the directional nature of the enhancer-blocking effect – that is, why an enhancer would be trapped by an insulator located in the same direction as the promoter, and not by an insulator the same distance away, but in the other direction. The physical barrier model of insulator function proposes that a molecular signal coming from the enhancer, such as a transcribing RNA polymerase complex (Kong et al., 1997; Tuan et al., 1992), simply “runs into” the insulator complex and is unable to progress further. Consistent with this mechanism is the observation that insertion of a transcriptional terminator, the lacO/R complex, between an enhancer and a promoter decreases enhancer function (Ling et al., 2004). Furthermore, when the chicken β-globin 5′ HS4 (cHS4) insulator was placed between the human HS2 enhancer and its target globin gene, RNA polymerase II accumulated at the insulator and reduced amounts at the gene promoter, suggesting that the insulator blocks polymerase progression from the enhancer to the promoter (Zhao and Dean, 2004). However, this might not be a universal mechanism of insulation, as insulators located within introns can silence downstream enhancers without truncating the gene product, implying that, at least in some cases, transcription complexes can pass through functional insulators (Geyer and Corces, 1992). The basic premise of the loop domain model is that insulator sites interact with each other and/or with other nuclear structures to form chromatin loops. This idea is straightforward and well supported, and will be discussed in detail below. However, the question of precisely how loop formation interferes with enhancer function remains an active area of study.
A large body of evidence indicates that insulators form chromatin loops. Pairing of specific insulator sites to form individual loops has been demonstrated, including the scs/scs’ insulator pair that flanks the 87A7 heat shock puff in Drosophila polytene chromosomes. The scs insulator is bound by the protein Zw5, whereas the scs’ insulator is bound by BEAF. Chromosome conformation capture (3C) experiments showed that the two sites colocalize, and coimmunoprecipitation assays demonstrasted that the two proteins interact, thus providing a mechanism for anchoring the loop (Blanton et al., 2003). Similarly, in fly lines with two insertions of the gypsy retrotransposon, which contains a Su(Hw) insulator, the chromosomal insertion sites colocalize in the nucleus in a Su(Hw)-dependent manner significantly more frequently than in fly lines lacking one or both of the insertions (Gerasimova et al., 2000). Furthermore, loops anchored by Su(Hw) insulators have been directly visualized in salt-extracted nuclei using fluorescence in situ hybridization (Byrd and Corces, 2003). Functional evidence supporting the idea that insulators form loops is provided by a study demonstrating that a loop can act as an insulator (Ameres et al., 2005). HeLa cells were transfected with a plasmid containing an enhancer and a promoter. The enhancer was flanked by arrays of tet operators, and the cells were also transfected with a plasmid expressing a protein that first binds the tetO arrays and then dimerizes. Presumably, this process should lead to interaction between the tetO arrays and looping out of the enhancer. Indeed, silencing of enhancer-driven expression was observed, but the effect could be reversed by conditions that caused the dimerizing protein to dissociate from the tetO arrays. Additionally, an endogenous example of functional loop formation has been found at the H19 imprinting control region (ICR) in mice, which binds CTCF in an allele-specific manner (Kurukuti et al., 2006; Murrell et al., 2004; Yoon et al., 2007). Yoon et al. suggest that the CTCF-bound ICR interacts with enhancer and promoter elements, whereas Kurukuti et al. suggest that the CTCF-bound ICR interacts with other DNA regulatory elements in the region, a matrix attachment region (MAR3) and a differentially methylated region (DMR1). Although these studies disagree on the specific interactions that are necessary for insulator function, both conclude that the CTCF-bound ICR is involved in loop formation that is necessary to prevent enhancer communication with the Igf2 promoter.
In addition to forming loops by interacting with one another, insulators might form loops by tethering chromatin to various nuclear structures. In mammalian cells, CTCF interacts with nucleophosmin, a protein that is enriched at the nucleolus. Also, transgenic loci with intact CTCF binding sites localize to the nucleolus more often than transgenes harboring mutated CTCF binding sites. Together, these results suggest that CTCF insulators form chromatin loops by tethering chromatin to the surface of the nucleolus (Yusufzai et al., 2004). Studies like this one, coupled with the observation that there are numerous endogenous insulator sites spread throughout the genomes of various species, imply that insulators might play a major role in global nuclear organization. This idea is further supported by observations in Drosophila diploid cells. In this model system, there are thousands of Su(Hw) binding sites distributed throughout the genome, but rather than the expected diffuse pattern of localization, immunofluorescence analyses detect a much smaller number of distinct foci, ranging from less than ten to a few dozen (Gerasimova and Corces, 1998). This finding has been interpreted to mean that the sites cluster together, organizing the entire mass of chromatin into rosette-like structures. Furthermore, most of these clusters are located at the nuclear periphery (Gerasimova et al., 2000), where there are hints that they might interact with the nuclear lamina (Capelson and Corces, 2005). This view is reinforced by a new study in human fibroblasts, which demonstrates that CTCF is found at the boundaries of many lamin-associated domains throughout the genome (Guelen et al., 2008). As peripheral vs. central positioning of genes in the nucleus generally seems to be nonrandom, an additional layer of nuclear organization might be orchestrated by insulators. Recently, dCTCF was found in the same clusters as Su(Hw) in diploid cells. Interestingly, on polytene chromosomes, Su(Hw) and dCTCF do not seem to colocalize. However, both proteins colocalize with a third protein, CP190, which is necessary for both Su(Hw) and dCTCF insulator function, thus suggesting cooperation between various classes of Drosophila insulators (Gerasimova et al., 2007; Mohan et al., 2007). If different types of insulators have variations in their properties, then interactions between these elements could permit an even finer level of control over the genome.
The role of insulators in epigenetic inheritance
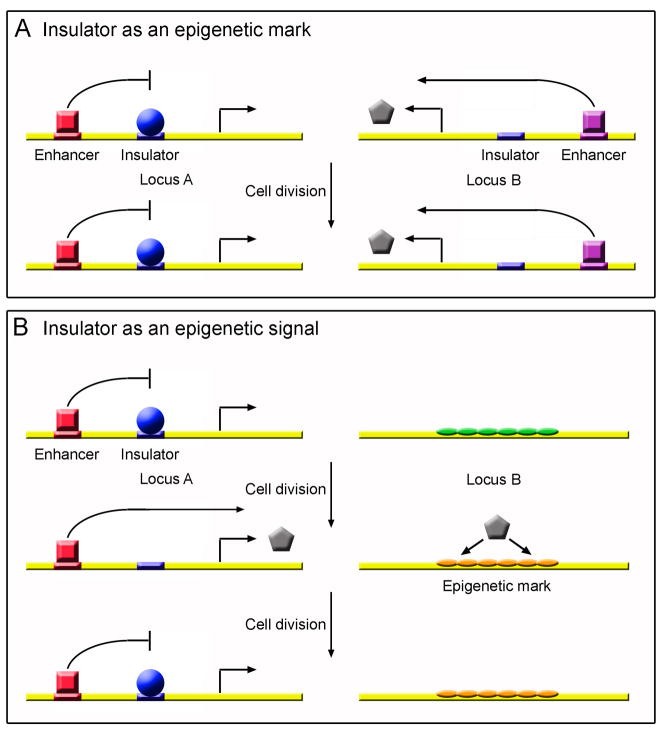
Epigenetic information is not contained in the DNA sequence, but it is nevertheless inherited from one cell generation to the next, and in some cases from one organismal generation to the next. An epigenetic system has two components: the signal that triggers an epigenetic change, and the mark that maintains the change across cell divisions, more or less stably, unless and until another signal arises to countermand it (Figure 1). If nuclear organization, as established by insulators, contains any intrinsic information, at any point during development, rather than merely responding to information encoded elsewhere, then insulators must have some connection to an epigenetic system. Although this area of inquiry has not yet been heavily researched, mounting evidence suggests that insulators can act as both an epigenetic mark and an epigenetic signal.
Figure 1.
The role of insulators in epigenetic memory. (A) Insulator protein binding could be an epigenetic mark that is maintained through cell division. Locus A and Locus B represent two different genomic loci that contain insulator elements. Locus A is bound by an insulator protein that prevents transcription, whereas locus B is unbound and therefore able to produce a gene product (pentagon). If insulator protein binding acts as an epigenetic mark, binding or lack of binding is remembered after cell division. (B) An insulator could participate in an epigenetic signal through regulating a gene that establishes an epigenetic mark. In this situation, loss of insulator protein binding leads to production of a gene product (pentagon) that establishes an epigenetic mark, here in Locus B. The epigenetic mark is maintained through cell division regardless of the state of the insulator. Therefore, the insulator participates in the establishment of the epigenetic mark, but is not the mark itself. Squares – enhancer binding proteins; Circles – insulator proteins; Pentagons – gene products whose expression is regulated by insulators; Ovals – epigenetic marks.
Early studies of Mod(mdg4), a member of the Su(Hw) insulator complex, showed a strong paternal effect on position effect variegation (PEV) (Dorn et al., 1993). PEV was assayed using the In(1)wm4h rearrangement in which the white gene is placed in close proximity to heterochromatin, leading to a variegated eye phenotype owing to varying amounts of heterochromatin spreading, and, therefore, varying amounts of white gene expression. In this system, mutations that enhance heterochromatin formation lead to a stronger white-eye phenotype, whereas mutations that perturb heterochromatin formation lead to a stronger red-eye phenotype. Mutations in the mod(mdg4) gene act as enhancers of PEV. Interestingly, male progeny, from a mod(mdg4)neo129 heterozygous father, harboring two wild-type copies of the mod(mdg4) gene show enhanced PEV. This effect is maintained through at least eleven generations of wild-type male progeny, but is not observed in offspring from a mod(mdg4)neo129 heterozygous mother. This paternal effect is also seen in eight other mod(mdg4) mutations (Buchner et al., 2000). These findings suggest that Mod(mdg4) is necessary to transmit epigenetic information, in this case, probably on the Y chromosome. However, such studies cannot determine whether Mod(mdg4) is involved in the epigenetic signal that establishes an epigenetic mark on the Y chromosome that then affects PEV, or if Mod(mdg4) is directly involved in the maintenance of the epigenetic mark that if lost in one fly generation is inefficiently re-established.
When considering mod(mdg4) mutations, it is important to note that this gene encodes at least 27 different isoforms generated by alternative splicing, and only one of these, called Mod(mdg4)2.2 or Mod(mdg4)67.2, is known to be involved in insulator function. The mutations analyzed in the PEV study mentioned above are all located within the common region of these isoforms, and, therefore, Mod(mdg4)2.2 function cannot be isolated. However, the mod(mdg4)u1 allele, which specifically affects the C-terminal region of Mod(mdg4)2.2, has also been implicated in epigenetic inheritance. This mutation inhibits insulator function when a Su(Hw) insulator is placed between an enhancer and the yellow gene promoter (Gerasimova et al., 1995). A small percentage of heterozygous male progeny from a mod(mdg4)u1 homozygous mother and a wild-type father show disrupted insulator function, whereas the reverse cross, between a wild-type mother and mod(mdg4)u1 homozygous father, leads to wild-type insulator function (Gerasimova and Corces, 1998). Therefore, there is a maternal effect that is retained in the adult fly, indicating that the protein is necessary early on in development to set up a state that is preserved through many cell divisions. As insulator function itself is the read-out in this situation, the insulator must be part of the epigenetic mark. The presence of the protein later on in development, after initiation of zygotic transcription, is not sufficient to restore normal insulator function, suggesting that Mod(mdg4)2.2 is involved in the establishment of an early embryonic epigenetic mark.
For something to act as an epigenetic mark, it must have some means of retaining its information content despite all disruptions to chromatin. Such disruptions include transcription, DNA replication, and chromatin compaction/decompaction during mitosis. Although much in this area of inquiry remains unclear, evidence is beginning to emerge indicating that insulators meet all three of these criteria. Proper function of the Drosophila Su(Hw) insulator is compatible with transcription through the insulator site, as insertion of Su(Hw) binding sites into an intron of the yellow gene results in silencing of downstream enhancers without disruption of the gene product (Geyer and Corces, 1992). In fact, the transcriptional machinery might pass through Su(Hw) insulator complexes quite often as Su(Hw) consensus sites are frequently found within introns (Ramos et al., 2006), although whether all of these sites are active insulators remains to be determined. Additionally, mapping of Drosophila CTCF and human CTCF binding sites via chromatin immunoprecipitation shows that a number of sites are located within introns (Barski et al., 2007; Holohan et al., 2007; Kim et al., 2007), suggesting that coexistence with transcription might be a common property of insulators, although it is also possible that these CTCF sites play a different role in transcription.
There are hints that insulators are also functional during DNA replication. The Drosophila chorion gene loci are amplified approximately 80-fold during oogenesis, but when randomly inserted at other genomic sites, the level of amplification decreases dramatically. Flanking the amplified locus with Su(Hw) insulators protects it against this position effect variegation (Lu and Tower, 1997), whereas inserting an insulator between two required regulatory elements abolishes amplification (Lu et al., 2001). These results suggest that the insulators might interact with replication control mechanisms in a manner similar to that by which they interact with transcriptional control mechanisms. It is plausible, therefore, that insulators could be involved in delineating separate replication domains with distinct replication timing and regulation, a role that would presumably entail maintenance of insulator function throughout S phase. It remains unclear, though, precisely what happens when the insulator site itself is replicated.
During mitosis, however, there is more direct evidence that insulator proteins might help to maintain epigenetic information. For instance, the Drosophila insulator protein BEAF remains on chromosomes during mitosis (Hart et al., 1999), but does not appear to be required for the mechanics of mitosis, suggesting that it might act as a placeholder to retain the molecular memory of active insulator sites (Gilbert et al., 2006). Additionally, in HeLa cells, CTCF has been observed on mitotic chromosomes; an examination of a subset of known interphase CTCF binding sites revealed that the same sites are also bound during mitosis (Burke et al., 2005). Another study verified this finding in a different cell line (Rubio et al., 2008). On the contrary, two additional studies reported the opposite finding that CTCF does not remain bound to chromatin during mitosis (Komura et al., 2007; Wendt et al., 2008). The maintenance of binding is critical if CTCF truly acts as an epigenetic mark; however, this idea is still controversial in the field.
A more subtle clue suggesting that insulators could be involved in the maintenance of epigenetic states comes from studies of the relationship between CTCF and the related protein BORIS (Brother of the Regulator of Imprinted Sites). BORIS contains a zinc finger domain with sequence nearly identical to that of CTCF, and binds at the same loci, but it is only normally expressed in the male germline at the developmental stage during which DNA methylation marks are erased, a stage in which CTCF expression is dramatically downregulated (Loukinov et al., 2002). Although indirect, this finding could indicate that if the absence of CTCF is important for epigenetic marks to be reset, then its presence could play a role in their maintenance.
To gain insight into the mechanisms through which insulator function is epigenetically maintained, we can look at other proteins that are involved in epigenetic inheritance. TrxG and PcG proteins are known to be involved in cellular memory of chromatin states (Grimaud et al., 2006). Mutations in mod(mdg4) enhance trxG mutations and suppress PcG mutations, and trxG mutations enhance the mod(mdg4) paternal effect described above. More importantly, the maternal effect of the mod(mdg4)u1 mutation is also enhanced by trxG mutations and suppressed by a PcG mutation, indicating that the epigenetic inheritance of insulator activity is affected by cellular memory proteins (Gerasimova and Corces, 1998). Therefore, TrxG and PcG proteins and insulator proteins are interdependent on one another for proper function. Although there is no evidence for direct interaction between these groups of proteins, they are all thought to be involved in large-scale nuclear organization. This implies that part of the information that is epigenetically retained from one generation to the next is the physical organization of chromatin in the nucleus, and each of these proteins must be functional for this process to occur properly.
Insulators modulate cell-type specific nuclear organization
If nuclear organization contains epigenetic information, then it can be inferred that it is cell-lineage specific. Additionally, nuclear organization and transcriptional regulation are known to be intimately related. The fact that gene expression patterns define cell identity also implies that nuclear organization is cell-type specific. Therefore, elements, such as insulators, that are involved in the establishment of nuclear organization and transcriptional regulation must be regulated between cell types.
Here we focus on what is known about genome-wide cell-type specific insulator information. However, the function of specific insulators which are known to be regulated between different cell types, such as the CTCF insulator at the ICR in vertebrates and the insulators found in the Drosophila Abd-B regulatory region, will be discussed in greater detail in a later section. Looking globally, CTCF binding sites in the human genome were recently mapped using chromatin immunoprecipitation followed by microarray analysis (Barski et al., 2007; Kim et al., 2007). The analysis of two different cell types revealed differences in a small but reproducible percentage of these sites, indicating that CTCF binding to DNA is partially cell-type specific. However, when a small number of Su(Hw) binding sites were analyzed in different Drosophila tissues little or no variation was observed (Adryan et al., 2007; Parnell et al., 2006). This difference could indicate that the Su(Hw) insulator is only involved in static nuclear architecture, although other Drosophila insulators might dictate cell-type specific organization. Alternatively, there might be cell-type specific changes in Su(Hw) binding to DNA that could not be resolved when comparing whole tissues that contain many different cell types. A third possibility is that Su(Hw) insulator regulation of nuclear organization occurs through the regulation of interactions between insulator proteins involved in loop formation and not at the level of DNA binding. In support of this third possibility, during heat shock, which causes the heat shock genes to be activated and transcription in the rest of the genome to be turned off, Su(Hw) insulator bodies dissociate, but insulator proteins remain bound to DNA (Gerasimova et al., 2000). Therefore, the massive change in gene expression patterns that follows heat shock is accompanied by reorganization of the protein-protein interactions between insulator proteins indicating that cell-type specific regulation of Su(Hw) insulators might occur largely at the level of insulator clustering.
The nuclear lamina is involved in cell-type specific nuclear organization and gene expression in Drosophila. As the Su(Hw) insulator interacts with the nuclear lamina, an interaction that is necessary for proper insulator function, insulators might be involved in this process of cell-type specific nuclear organization (Capelson and Corces, 2005; Gerasimova et al., 2000). In vitro differentiation prompts a small number of genes to alter their association with the nuclear lamina, and these nuclear rearrangements are accompanied by changes in gene expression (Pickersgill et al., 2006). Although the role of insulators in this context has not been examined, activation or inactivation of insulators could cause the chromatin to associate with or disassociate from the nuclear lamina, leading to the observed changes in nuclear arrangement and gene expression. Recently, lamina-associated domains were mapped throughout the entire human genome, which revealed a strong enrichment for CTCF sites within 5–10kb of these domains (Guelen et al., 2008). Although this analysis was limited to one cell type and therefore does not provide insight into the role of the nuclear lamina or CTCF in cell-type specific nuclear organization, it does suggest conservation of the functional interaction between nuclear lamina and insulator proteins in vertebrates. More thorough analysis is required to determine whether insulators are in fact involved in cell-type specific nuclear organization.
The regulation of insulator activity
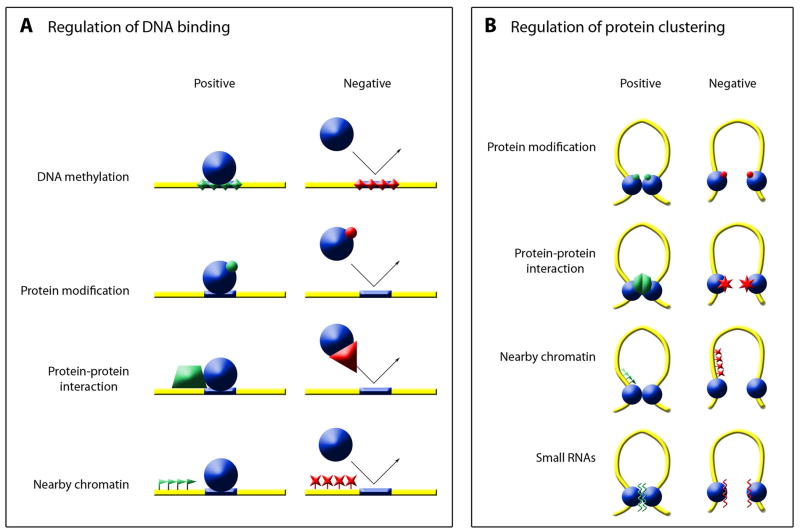
Given that the DNA sequences to which insulator proteins bind are present in all cells, but that the functionality of specific insulator sites in specific cell types exhibits some variability, it follows that there must be some means of regulating insulator protein behavior. This regulation could take place either once in a lineage as part of a differentiation event, with the effect being stably maintained by epigenetic mechanisms as discussed above, or as part of an ongoing system of control that is continually reimposed (or permitted to lapse) in every cell generation. Evidence exists for a variety of molecular mechanisms affecting insulator protein behavior at multiple stages involved in insulator function (Figure 2).
Figure 2.
Regulation of insulator activity. (A) Insulator function could be regulated on the level of DNA-binding activity. Regulation could be positive or negative and could occur by various molecular mechanisms, including DNA modification, protein modification, protein-protein interaction, and effects of nearby chromatin. (B) Insulator function could be regulated on the level of protein-protein interaction. Again, regulation could be positive or negative. Molecular mechanisms could include protein modification, protein-protein interaction, effects of nearby chromatin, and interaction with small RNAs.
A basic requirement for any kind of functional insulator protein complex is DNA-binding activity. One method of modulating DNA-binding activity is covalent modification of the DNA at the binding site. The classic example of this type of regulation is differential methylation of the CTCF binding site between the H19 and Igf2 genes (Bell and Felsenfeld, 2000; Hark et al., 2000). In this case, when CTCF is bound and the insulator is functional, only H19 is expressed; this occurs on the maternal chromosome. On the paternal chromosome, the binding site is methylated and CTCF is unable to bind, rendering the insulator inactive; only Igf2 is expressed. This methylation mark is imposed during male germ cell development and stably maintained thereafter. Another means of regulation is covalent modification of the proteins involved. It appears that ubiquitination might play a role here, as the E3 ubiquitin ligase dTopors can affect the Su(Hw) insulator in Drosophila (Capelson and Corces, 2005). Mutations in this gene reduce insulator function, whereas dTopors overexpression can rescue insulator phenotypes that already have reduced function and impaired DNA binding. A point mutation that abolishes ubiquitin ligase activity also abolishes the rescue effect, suggesting that the enzymatic activity is important; however, the identity of the enzyme’s target in this process remains unknown. A third strategy for regulating DNA binding is protein-protein competition. The relationship between BEAF and the transcriptional regulator DREF is a likely candidate for this form of regulation (Hart et al., 1999). Both proteins bind the same DNA motif, but they do not interact with each other, and electrophoretic mobility shift assays show that only one at a time can bind to a specific site. On polytene chromosomes, they appear to overlap at many points, suggesting that some copies of the chromosome might be bound by BEAF whereas others are bound by DREF. The functional significance of this relationship, if any, has not been determined.
Given that the prevailing models of insulator function suggest that insulator sites interact with one another to form chromatin loops, regulation of insulator activity at the level of protein-protein interactions required to form these loops is also likely to be important. A probable example of this type of regulation is modification of CTCF by poly(ADP-ribosyl)ation, or PARlation (Yu et al., 2004). Chromatin immunoprecipitation experiments in murine cells indicate that the PAR mark and CTCF overlap at numerous points in the genome, and in vitro mobility shift assays demonstrate that PARlated CTCF retains its DNA-binding ability. Together these results suggest that a large fraction of endogenous CTCF binding sites contain PARlated protein. In this case, there are hints that the modification is a positive regulator of insulator function, as treatment with 3-aminobenzamide, an inhibitor of PARlation, results in loss of insulation. The idea that PARlation regulates CTCF insulation by promoting interactions between insulator sites has not been shown conclusively but appears likely, as PARlation seems to promote protein-protein interactions in other contexts (Reale et al., 2000). Another case of insulator regulation by modulation of protein-protein interaction is the modification of Mod(mdg4)2.2 and CP190 in the Drosophila Su(Hw) insulator complex by addition of SUMO (Small Ubiquitin-like Modifier) (Capelson and Corces, 2006). As is the case for PAR, SUMO colocalizes with insulator proteins at a number of sites throughout the genome, as demonstrated via immunostaining of polytene chromosomes. However, unlike PAR, SUMO appears to be a negative regulator of insulator activity, as mutations in the SUMOylation pathway result in strengthened insulator function. Furthermore, the overexpression of SUMOylation machinery components disrupts the appearance of punctate insulator bodies in diploid nuclei from third instar larvae. This finding strongly suggests that SUMOylation regulates insulator activity by interfering with the ability of insulator sites at different locations to interact with one another. The RNAi machinery also appears to play a role in regulating interactions between insulator proteins (Lei and Corces, 2006). This role does not seem to be a straightforward one, however, as different components of the machinery affect insulator function in opposite ways. Based on loss-of-function and overexpression studies, the helicase Rm62 opposes, whereas Argonaute proteins facilitate, insulator activity. The interaction of insulator proteins with chromatin appears to be unaffected in any of these conditions, again indicating that regulation occurs at the level of interaction between insulator sites, rather than of proteins binding DNA at each site. The interaction between Rm62 and CP190 is RNA-dependent, which brings up an interesting possibility. In addition to determining simply whether or not a particular insulator site participates in interactions with other insulators, one can also imagine that there might be modes of regulation that specify the set of insulator sites that participate in each cluster. Potentially, the sequence specificity afforded by the inclusion of RNA in an insulator complex might provide a means of accomplishing this goal.
In addition to the regulatory mechanisms described above, there are other factors that affect insulator activity for which the functional details are still unknown. One example comes from the Drosophila Abdominal-B (Abd-B) locus. The Abd-B control region consists of a series of developmentally regulated enhancers, silencers and insulators. How the correct enhancer targets the Abd-B gene in a given cell type is an area of great interest. Interestingly, one of the insulators in this region, Fab-7, consists of multiple sub-elements that exert their insulator function either at different points during development or with different enhancer-promoter combinations (Schweinsberg et al., 2004). This finding reveals the potential for specified insulator activity that might elucidate some of the developmentally regulated gene expression at this locus. Additionally, multiple insulators within the Abd-B control region have been shown to exhibit insulator bypass, meaning that insulator activity is abrogated when two insulators are placed near one another due to insulator pairing (Gruzdeva et al., 2005; Kyrchanova et al., 2007; Kyrchanova et al., 2008; Rodin et al., 2007). Although insulator bypass has not been shown at the endogenous locus, it is possible that this mechanism plays a role in targeting specific enhancer elements to the promoter in the correct cell-type at the correct developmental time-point. Another part of the explanation for the developmentally regulated gene expression of the Abd-B gene comes from Promoter Targeting Sequences (PTS) that have been found 3′ to two of the insulators, Fab-7 and Fab-8, and allow enhancers to overcome both Drosophila CTCF and Su(Hw) enhancer-blocking activity in transgene assays (Chen et al., 2005; Zhou and Levine, 1999). These PTSs target an enhancer to one, and only one, promoter in a given cell lineage, suggesting that these sequences stabilize enhancer-promoter complexes (Lin et al., 2003). Perhaps these enhancer-promoter complexes interfere with insulator loop formation and in this way abrogate insulator activity.
Another example of nearby sequences affecting insulator activity is thyroid hormone response elements (TREs), which are often found in close proximity to CTCF binding sites in vertebrates (Arnold et al., 1996; Awad et al., 1999; Perez-Juste et al., 2000). Analysis of the relationship between these two elements suggests that CTCF enhancer-blocking activity is reduced by both the TRE found upstream of the chicken lysozyme gene and the TRE found upstream of the human c-myc gene in the presence of thyroid hormone (Lutz et al., 2003). CTCF remains bound to the DNA in the presence of hormone, indicating that TREs must inactivate CTCF.
Additional modulation of CTCF activity comes from various interaction partners (Wallace and Felsenfeld, 2007). Whereas Kaiso is thought to negatively regulate CTCF enhancer-blocking activity, CHD8 enhances this function (Defossez et al., 2005; Ishihara et al., 2006). Recent studies indicate that cohesin also interacts with CTCF. These reports demonstrated that CTCF recruits cohesin to chromatin and that it is necessary for proper insulator function at both transgenic and endogenous loci (Parelho et al., 2008; Rubio et al., 2008; Stedman et al., 2008; Wendt et al., 2008). Although the effect of cohesion on CTCF association with DNA is unclear due to conflicting data from these studies, Kaiso and CHD8 are not thought to have an effect. Therefore, at least some CTCF interacting partners must affect insulator activity through an alternative mechanism, such as promoting or hindering loop formation.
The vast number of regulatory mechanisms that exist to modulate insulator function implies that insulator activity is dynamic. Separate mechanisms have been identified for the regulation of insulator protein binding to DNA and protein-protein interactions leading to loop formation. These strategies could be used for different degrees of regulation. Interference at the level of DNA binding could be a more permanent form of regulation, whereas interference at the level of loop formation could be more transient as the proteins could remain present and poised for activation. Thus, for example, a cell could respond quickly to changes in its immediate metabolic needs and its environment by altering its loop organization, while still retaining its differentiated identity via DNA binding patterns. However, more work is needed to understand when and how the cell uses these forms of insulator regulation.
Conclusions
At least some enhancer-blocking insulators seem to participate in epigenetic inheritance, and many exhibit regulation of insulator activity. Each insulator site contains the potential for regulation on many levels, including proteins binding to the insulator DNA, modifications to these proteins, or nearby elements modulating activity of insulator complexes. As enhancer-blocking insulators are found throughout the Drosophila and vertebrate genomes, a wealth of potentially cell-type specific information exists that can be maintained throughout a cell and/or organismal lineage. It is still unknown, however, how widely these forms of regulation are used by a cell, and, therefore, it is unclear how much of the insulator activity in a given genome is cell-type specific.
Although much work has been done to understand insulator function, the mechanisms through which they function are still not well understood. It is also unclear whether the main function of insulators is to act locally through enhancer-blocking, if their function in cis-regulation of transcription simply reflects their role in global nuclear organization, or whether insulators play an active role in both transcriptional regulation and nuclear organization. If their role in nuclear organization is functional, it is possible that insulator-dependent nuclear organization of chromatin contains epigenetic information that is important for cell differentiation. Regulation of this organization by controlling insulator activity at specific genomic locations might therefore be an important mechanism for the establishment and maintenance of specific cell fates during development in higher eukaryotes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome biology. 2007;8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. The EMBO journal. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Burcin M, Kaiser B, Muller M, Renkawitz R. DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic acids research. 1996;24:2640–2647. doi: 10.1093/nar/24.14.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, Lutz M, Neiman PE, Collins SJ, Renkawitz R, Lobanenkov VV, Filippova GN. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. The Journal of biological chemistry. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & development. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, Renkawitz R. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. The EMBO journal. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. The Journal of cell biology. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Molecular cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. The EMBO journal. 2006;25:1906–1914. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Lin Q, Zhou J. Multiple Promoter Targeting Sequences exist in Abdominal-B to regulate long-range gene activation. Developmental biology. 2005;286:629–636. doi: 10.1016/j.ydbio.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Defossez PA, Kelly KF, Filion GJ, Perez-Torrado R, Magdinier F, Menoni H, Nordgaard CL, Daniel JM, Gilson E. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. The Journal of biological chemistry. 2005;280:43017–43023. doi: 10.1074/jbc.M510802200. [DOI] [PubMed] [Google Scholar]
- Dorn R, Krauss V, Reuter G, Saumweber H. The enhancer of position-effect variegation of Drosophila, E(var)3-93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature reviews. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Molecular cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Molecular cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Current opinion in genetics & development. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & development. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Gilbert MK, Tan YY, Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–1375. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–383. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF Genomic Binding Sites in Drosophila and the Organisation of the Bithorax Complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Molecular cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura J, Ikehata H, Ono T. Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc Natl Acad Sci U S A. 2007;104:15741–15746. doi: 10.1073/pnas.0702363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Molecular and cellular biology. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nature genetics. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wu D, Zhou J. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development (Cambridge, England) 2003;130:519–526. doi: 10.1242/dev.00227. [DOI] [PubMed] [Google Scholar]
- Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. The Journal of biological chemistry. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Tower J. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Molecular and cellular biology. 1997;17:2202–2206. doi: 10.1128/mcb.17.4.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhang H, Tower J. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes & development. 2001;15:134–146. doi: 10.1101/gad.822101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R. Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. The EMBO journal. 2003;22:1579–1587. doi: 10.1093/emboj/cdg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, Renkawitz R. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. The EMBO journal. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Molecular and cellular biology. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Juste G, Garcia-Silva S, Aranda A. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. The Journal of biological chemistry. 2000;275:1307–1314. doi: 10.1074/jbc.275.2.1307. [DOI] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature genetics. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–2349. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale A, Malanga M, Zardo G, Strom R, Scovassi AI, Farina B, Caiafa P. In vitro induction of H1-H1 histone cross-linking by adenosine diphosphate-ribose polymers. Biochemistry. 2000;39:10413–10418. doi: 10.1021/bi992977q. [DOI] [PubMed] [Google Scholar]
- Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. New properties of Drosophila fab-7 insulator. Genetics. 2007;177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. The EMBO journal. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nature genetics. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Molecular cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic acids research. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]