Abstract
Previous data showed that the cellular proteins TIA-1 and TIAR bound specifically to the West Nile virus 3′ minus-strand stem-loop [WNV3′(−)SL] RNA (37) and colocalized with flavivirus replication complexes in WNV- and dengue virus-infected cells (21). In the present study, the sites on the WNV3′(−)SL RNA required for efficient in vitro T-cell intracellular antigen-related (TIAR) and T-cell intracellular antigen-1 (TIA-1) protein binding were mapped to short AU sequences (UAAUU) located in two internal loops of the WNV3′(−)SL RNA structure. Infectious clone RNAs with all or most of the binding site nucleotides in one of the 3′ (−)SL loops deleted or substituted did not produce detectable virus after transfection or subsequent passage. With one exception, deletion/mutation of a single terminal nucleotide in one of the binding sequences had little effect on the efficiency of protein binding or virus production, but mutation of a nucleotide in the middle of a binding sequence reduced both the in vitro protein binding efficiency and virus production. Plaque size, intracellular genomic RNA levels, and virus production progressively decreased with decreasing in vitro TIAR/TIA-1 binding activity, but the translation efficiency of the various mutant RNAs was similar to that of the parental RNA. Several of the mutant RNAs that inefficiently interacted with TIAR/TIA-1 in vitro rapidly reverted in vivo, indicating that they could replicate at a low level and suggesting that an interaction between TIAR/TIA-1 and the viral 3′(−)SL RNA is not required for initial low-level symmetric RNA replication but instead facilitates the subsequent asymmetric amplification of genome RNA from the minus-strand template.
West Nile virus (WNV) is a member of the genus Flavivirus in the family Flaviviridae (39). The WNV genome is a single-stranded positive-sense RNA of approximately 11 kb in length that has a 5′ type I cap but no 3′ poly(A) tail; contains a single, long open reading frame; and serves as the only viral mRNA. The 5′ end of the single genome open reading frame encodes the structural proteins, while the 3′ end encodes the nonstructural proteins (39). The viral replication cycle takes place in the cytoplasm of infected cells. The viral polyprotein is co- and posttranslationally cleaved by viral and cellular proteases into three structural proteins (capsid, membrane, and envelope) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5). The RNA genome serves as the template for transcription of the complementary minus-strand RNA, which in turn serves as a template for the synthesis of nascent genomic RNA. At early times after infection, equivalent low levels of viral plus- and minus-strand RNA are produced. A subsequent switch to asymmetric amplification of plus-strand RNA results in a ratio of nascent genomic RNA to minus-strand RNA of about 10:1 (13). The mechanism(s) involved in the switch to asymmetric plus-strand amplification is not currently known.
The 5′ noncoding region (NCR) of the WNV genome RNA is 96 nucleotides (nt) in length, while the length of the 3′ NCR is ∼600 nt (39). For flavivirus RNAs, evidence supporting the formation of predicted unique, conserved, terminal stem-loop (SL) structures by the 3′ nucleotides (9, 12) and the 5′ nucleotides of the genome (8, 19) and by the 3′ nucleotides of the complementary minus-strand (46) was previously obtained by structure probing. Deletion of either the 3′ or the 5′ terminal SL in flavivirus infectious clones was lethal, suggesting that these regions contain essential cis-acting elements (7, 11, 42). Data from in vitro polymerase assays done with recombinant WNV NS5 protein indicated that the 3′ terminal 230 nt of the minus-strand RNA were sufficient for de novo initiation of genomic RNA synthesis (43).
Four hamster cell proteins of about 42, 50, 60, and 108 kDa were previously reported to bind specifically to the WNV3′ (−)SL RNA (46). p42 was identified as T-cell intracellular antigen-related (TIAR) protein (37). The closely related T-cell intracellular antigen-1 (TIA-1) protein was subsequently shown to bind to the WNV3′(−)SL RNA about 10 times less efficiently than TIAR (37). Although they were first discovered in T cells, TIA-1 (2) and TIAR (31) are expressed in most types of cells and tissues and shuttle between the cytoplasm and the nucleus (1, 28, 58). These evolutionarily conserved cellular proteins are members of the RNA recognition motif (RRM) family. They are multifunctional proteins that regulate alternative splicing (17, 47, 48, 59), silence translation (18, 29, 34, 45, 56), regulate Fas-mediated apoptosis (38, 50, 52), sequester cell mRNAs in cytoplasmic stress granules (32, 33), and provide critical functions during embryonic development (4, 45). A high degree of embryo lethality prevented the establishment of a TIAR−/− mouse strain, but both the TIAR−/− and the TIA-1−/− embryo fibroblast cell lines were generated (45). WNV production was decreased by six- to eightfold in the TIAR−/− cell line but was similar to that of control cells in TIA-1−/− cells (37). An increase in the level of TIA-1 in TIAR−/− cells, which is due to the lack of downregulation by TIAR (37), is thought to partially compensate for the loss of TIAR and to be responsible for the limited effect of the loss of TIAR on virus yields. Since all attempts to knock out both proteins in mice were unsuccessful (45) and suppression of both TIA-1 and TIAR expression in chicken cells resulted in cell death (35), the effect of knockdown of both proteins on virus replication cannot be evaluated.
The reduced WNV yield observed from TIAR−/− cells, as well as previous studies showing that TIAR and TIA-1 bind specifically to the WNV3′(−)SL RNA (37) and that first TIAR and then TIA-1 colocalize with viral replication complexes in flavivirus-infected cells (21), suggests the possibility that these cell proteins play a role in viral RNA replication. As a means of more directly analyzing whether TIAR/TIA-1 facilitate genome RNA replication, the binding sites for these proteins within the WNV3′(−)SL RNA were first mapped to two short, single-stranded AU sequences, and then the functional importance of these sequences was analyzed by mutagenesis of a WNV infectious clone. The results showed that the efficiency of genomic RNA synthesis and virus production progressively decreased with decreasing in vitro binding efficiency of TIAR/TIA-1 for mutant 3′(−)SL RNAs. The reversion of several mutant RNAs that bound inefficiently to TIAR/TIA-1 in vitro supported the hypothesis that this viral RNA-cell protein interaction is not required for initial low-level symmetric viral plus- or minus-strand RNA replication but is instead needed for subsequent asymmetric amplification of genome RNA from the minus-strand template.
MATERIALS AND METHODS
Cells.
Baby hamster kidney 21 strain W12 (BHK) cells (53) were maintained at 37°C in a CO2 incubator in minimal essential medium (MEM) supplemented with 5% fetal bovine serum (Atlas) and 10 μg/ml gentamicin (Invitrogen).
Cloning, expression, and purification of recombinant TIA-1 and TIAR from Escherichia coli.
The cDNAs of the most abundant of the two isoforms of each protein, TIA-1a and TIARb, were amplified from total RNA extracted from C3H/He mouse embryo fibroblasts by reverse transcription-PCR (RT-PCR), cloned into the TA cloning vector pCR 2.1 (Invitrogen), and then subcloned into pCRT7/CT-TOPO (Invitrogen) to generate the expression vectors pTIA-1a and pTIARb. The expressed proteins contained a C-terminal (six-His) tag. All inserts were verified by restriction and sequence analyses. Recombinant TIA-1 and TIAR proteins were expressed in E. coli Rosetta (DE3) pLysS cells (Novagen), as follows. Cells were transformed with plasmid DNA (10 ng) and grown in LB medium containing carbenicillin (50 μg/ml) and chloramphenicol (34 μg/ml) to an optical density at 600 nm of 0.6 at 37°C. Protein expression was induced by the addition of 0.05 mM isopropyl-β-d-thiogalactopyranoside for 5 h with continuous shaking at 37°C. To purify proteins to near homogeneity, cell pellets from a 0.5-liter culture were resuspended in 1× equilibration/wash buffer (50 mM sodium phosphate, 300 mM NaCl [pH 7.0]) containing a protease inhibitor cocktail (complete mini, EDTA free; Roche), and then lysed with an SLM-Aminco French pressure cell (Heinemann) at 20,000 lb/in2. Clarified supernatants were loaded onto cobalt columns (Talon metal affinity resin; Clontech), the columns were washed with 1× equilibration/wash buffer containing 15 mM imidazole, and the bound proteins were eluted with 1× equilibration/wash buffer containing 150 mM imidazole. The eluted protein fractions were combined, dialyzed against storage buffer (20 mM sodium phosphate, 60 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, and 3% Ficoll 400), aliquoted, and stored at −80°C. The protein concentration was measured using a Coomassie Plus protein assay reagent kit (Pierce) with a bovine serum albumin protein standard. Proteins in eluted fractions were analyzed by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and detected by Coomassie blue staining. Recombinant eukaryotic elongation factor 1 alpha (eEF1A) containing a C-terminal six-His tag was expressed in E. coli Rosetta (DE3) pLysS cells (Novagen) and partially purified on a cobalt column as described above for TIA-1/TIAR (15).
DNA constructs used as templates for RNA synthesis.
The construction of a plasmid containing the first 75 nt of the WNV5′NCR protein (p5′NCR) was previously described (46). This plasmid was amplified by PCR, using appropriate primers with a T7 promoter (Table 1) to generate DNA templates for in vitro transcription of the WNV3′(−)SL and WNV5′(+)SL RNAs. Mutant constructs were generated using a Quik-Change II site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The sequences of the primers used to introduce mutations are shown in Table 1. DNA extracted from selected positive colonies was sequenced to confirm the presence of the introduced mutations and/or deletions. pWNV-Trun (20) was used to amplify DNA templates for in vitro synthesis of the 3′ terminal 89 nt of the WNV genome.
TABLE 1.
Sequences of primers
| Primer | Sequenceb |
|---|---|
| L1→Cs Fora | 5′GTTTGTGAGGATTAACAACGGGGGACACGGTGCGAGCTG3′ |
| L1→Cs Rev | 5′CAGCTCGCACCGTGTCCCCCGTTGTTAATCCTCACAAAC3′ |
| L2→Cs For | 5′CTTAGTAGTGTTTGTGAGGGGGGACAACAATTAACACGG3′ |
| L2→Cs Rev | 5′CCGTGTTAATTGTTGTCCCCCCTCACAAACACUACUAAG3′ |
| L3→Cs For | 5′CGCCTGTGTGAGCTGACAAACGGGGGGGGGTTTGTGAGGATTAACAAC3′ |
| L3→Cs Rev | 5′GTTGTTAATCCTCACAAACCCCCCCCCGTTTGTCAGCTCACACAGGCG3′ |
| L1+L2→Cs For | 5′GTTTGTGAGGGGGGACAACGGGGGACACGGTGCGAGCTGTTTC3′ |
| L1+L2→Cs Rev | 5′GAAACAGCTCGCACCGTGTCCCCCGTTGTCCCCCCTCACAAAC3′ |
| pΔL1 For | 5′GTTTGTGAGGATTAACAACTTACACAGTGCGAGCTGTTTCTTGGC3′ |
| pΔL1 Rev | 5′GCCAAGAAACAGCTCGCACTGTGTAAGTTGTTAATCCTCACAAAC3′ |
| ΔL2 For | 5′CTTAGTAGTGTTTGTCAACAATTAACACAGTGCG3′ |
| ΔL2 Rev | 5′CGCACTGTGTTAATTGTTGACAAACACTACTAAG3′ |
| ΔUAAL3 For | 5′GTGAGCTGACAAACGTAGTGTTTGTGAGG3′ |
| ΔUAAL3 Rev | 5′CCTCACAAACACTACGTTTGTCAGCTCAC3′ |
| pΔL1+ΔL2 For | 5′GTAGTGTTTGTCAACTTACACGGTGCGAGC3′ |
| pΔL1+ΔL2 Rev | 5′GCTCGCACCGTGTAAGTTGACAAACACTAC3′ |
| L1U16→C For | 5′GTGAGGATTAACAACAATTGACACAGTGCGAGCTG3′ |
| L1U16→C Rev | 5′CAGCTCGCACTGTGTCAATTGTTGTTAATCCTCAC3′ |
| L1A18→C For | 5′GTGAGGATTAACAACAAGTAACACAGTGCGAGCTG3′ |
| L1A18→C Rev | 5′CAGCTCGCACTGTGTTACTTGTTGTTAATCCTCAC3′ |
| L1U19→C For | 5′GTGAGGATTAACAACAGTTAACACAGTGCGAGCTG |
| L1U19→C Rev | 5′CAGCTCGCACTGTGTTAACTGTTGTTAATCCTCAC3′ |
| L1U20→C For | 5′GTGAGGATTAACAACGATTAACACAGTGCGAGCTG |
| L1U20→C Rev | 5′CAGCTCGCACTGTGTTAATCGTTGTTAATCCTCAC3′ |
| L2U25→C For | 5′GTAGTGTTTGTGAGGATTAGCAACAATTAACACAGTGCG3′ |
| L2U25→C Rev | 5′CGCACTGTGTTAATTGTTGCTAATCCTCACAAACACTAC3′ |
| L2U26→C For | 5′GTAGTGTTTGTGAGGATTGACAACAATTAACACAGTGCG3′ |
| L2U26→C Rev | 5′CGCACTGTGTTAATTGTTGTCAATCCTCACAAACACTAC3′ |
| L2A27→C For | 5′GTAGTGTTTGTGAGGATGAACAACAATTAACACAGTGCG3′ |
| L2A27→C Rev | 5′CGCACTGTGTTAATTGTTGTTCATCCTCACAAACACTAC3′ |
| L2U29→C For | 5′GTAGTGTTTGTGAGGGTTAACAACAATTAACACAGTGCG3′ |
| L2U29→C Rev | 5′CGCACTGTGTTAATTGTTGTTAACCCTCACAAACACTAC3′ |
| L1ΔU19ΔU20 For | 5′GTGAGGATTAACAACTTAACACAGTGCGAGCTG |
| L1ΔU19ΔU20 Rev | 5′CAGCTCGCACTGTGTTAAGTTGTTAATCCTCAC3′ |
| L1ΔU19ΔU20+G21A For | 5′GTGAGGATTAACAATTTAACACAGTGCGAGCTG |
| L1ΔU19ΔU20+G21A Rev | 5′CAGCTCGCACTGTGTTAAATTGTTAATCCTCAC3′ |
| L1ΔU20 For | 5′GTGAGGATTAACAACATTAACACAGTGCGAGCTG |
| L1ΔU20 Rev | 5′CAGCTCGCACTGTGTTAATGTTGTTAATCCTCAC3′ |
| L2ΔUAAUU For | 5′CTTAGTAGTGTTTGTGAGGCAACAATTAACACAGTGCG3′ |
| L2ΔUAAUU Rev | 5′CGCACTGTGTTAATTGTTGCCTCACAAACACTACTAAG3′ |
| L2ΔUAAUU+G24U For | 5′GTAGTGTTTGTGAGGAAACAATTAACACAGTGCG3′ |
| L2ΔUAAUU+G24U Rev | 5′CGCACTGTGTTAATTGTTTCCTCACAAACACTAC3′ |
| L2ΔU25 For | 5′GTAGTGTTTGTGAGGATTACAACAATTAACACAGTGCG3′ |
| L2ΔU25 Rev | 5′CGCACTGTGTTAATTGTTGTAATCCTCACAAACACTAC3′ |
| L2ΔU25ΔU26 For | 5′GTAGTGTTTGTGAGGATTACAACAATTAACACAGTGCG3′ |
| L2ΔU25ΔU26 Rev | 5′CGCACTGTGTTAATTGTTGAATCCTCACAAACACTAC3′ |
| Parental T7 For | 5′AGTAGTTCGCCTGTGTGAGC3′ |
| Parental T7 Revc | 5′[T7]CAGCTCG CACCGTGTTAATTGTTG3′ |
| pΔL1 T7 Rev | 5′[T7]CAGCTCG CACCGTGAAGTTG3′ |
| L1→Cs T7 Rev | 5′[T7]CAGCTCG CACCGTGTCCCCCGTTG3′ |
For, forward; Rev, reverse.
Substituted nucleotides are underlined.
T7 represents the sequence of the T7 promoter.
In vitro transcription of 32P-labeled and unlabeled RNA.
WNV3′(−)SL RNA, WNV3′(+)SL RNA, WNV5′(+)SL RNA, and a number of WNV3′(−)SL mutant RNAs were in vitro transcribed using a MAXIscript in vitro transcription kit (Ambion) in 20-μl reaction mixtures that contained T7 RNA polymerase (30 U), a PCR purification kit (Qiagen)-purified PCR product (1 μg), 0.8 mM [α-32P]GTP (3,000 Ci/mmol, 10 mCi/ml; GE Healthcare), 3 μM GTP, and 0.5 mM CTP, UTP, and ATP. The in vitro transcription mixture was incubated at 37°C for 2 h, and transcription was stopped by the addition of DNase I (1 U) for 15 min at 37°C. After the addition of an equal volume of 2× Gel Loading Buffer II (Ambion), the reaction mixture was heated at 95°C for 5 min. RNA transcripts were purified by electrophoresis on a 6% polyacrylamide gel containing 7 M urea. The wet gel was autoradiographed, and the 32P-labeled RNA band was excised. RNA was eluted from the gel slices by rocking overnight at 4°C in elution buffer (0.5 M NH4OAC, 1 mM EDTA, and 0.2% SDS). The eluted RNA was filtered through a 0.45-μm cellulose acetate filter unit (Millipore) to remove gel pieces, precipitated with ethanol, resuspended in water, aliquoted, and stored at −80°C. The amount of radioactivity incorporated into each RNA probe was measured with a scintillation counter (model LS6500; Beckman), and the specific activity, which was calculated as described previously (5), was routinely about 1.3 × 107 cpm/μg. Unlabeled viral RNA was in vitro transcribed as described above, except that 0.5 mM of each nucleoside triphosphate was added to the reaction mixture. After synthesis, these RNAs were purified on NucAway spin columns (Ambion), and RNA concentrations were calculated based on the UV absorbance measured at 260 nm.
Transfection of in vitro-transcribed full-length WNV genomic RNA.
Construction of the WNV infectious clone used in this study was previously described (55). Genomic RNA was synthesized from WNV infectious clone DNA (1 μg) that had been gel purified and linearized with XbaI, using a Message Maker kit (Epicentre) and SP6 RNA polymerase. BHK cells were seeded in six-well plates (2 × 105 cells per well) and grown to ∼90% confluence. The cells were washed and then transfected with 100 ng of infectious clone RNA using a 1:1 (M/M) liposome formulation of the cationic lipid 1,2-dimyristyloxypropyl-3-dimethyl-hydroxyethyl ammonium (DMRIE-C; Invitrogen) as the transfection agent in Opti-MEM (Invitrogen). After 3 h of incubation at 37°C in 5% CO2, the transfection medium was removed and replaced either with fresh MEM containing 5% fetal calf serum or MEM containing 0.5% SeaKem ME agarose (BioWhittaker Molecular Applications) and 2.5% fetal calf serum. Three serial passages were done in duplicate by transferring 100 μl of medium harvested at 72 h after transfection/infection to a fresh well in a six-well plate. Plaque assays were done as previously described (20).
Site-directed mutagenesis of the infectious clone.
Mutations were inserted into the WNV infectious clone in a shuttle vector containing the ligated 3′ and 5′ regions of the WNV genome sequence as described previously (20). Specific mutations or deletions were introduced into the first 75 nt of the WNV3′(−)SL RNA in the shuttle vector DNA, using a Quik-Change II site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The sequences of all of the mutant shuttle vectors, as well as those of the appropriate regions of the final mutant infectious clone DNAs, were checked by DNA sequencing. The primers used for site-directed mutagenesis and RNA synthesis are shown in Table 1.
Gel mobility shift assays.
32P-labeled WNV3′(−)SL RNA probe diluted in binding buffer (20 mM sodium phosphate [pH 7.0], 60 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 3% Ficoll 400, and 10 U RNasin) was denatured at 85°C for 10 min and slowly renatured (0.1°C/s) to 20°C. Reaction mixtures containing the RNA probe (2,000 cpm; 0.2 nM) and different concentrations of recombinant TIA-1 (rTIA-1; 50 to 800 nM) or rTIAR (10 to 90 nM) in a final volume of 10 μl of binding buffer plus 10 nM of the nonspecific competitor tRNA were incubated at room temperature for 30 min, and the RNA-protein complexes formed were electrophoresed on a 5% nondenaturing polyacrylamide gel at 100 V/h in 0.5× Tris-borate-EDTA buffer at 4°C. Gels were dried and visualized by autoradiography.
An estimated dissociation constant (Kd) was calculated for each protein-viral RNA interaction. Increasing amounts of partially purified rTIA-1 (50, 300, 600, and 800 nM) or rTIAR (10, 20, 60, and 90 nM) were incubated with a constant amount of the parental WNV3′(−)SL RNA probe and analyzed on nondenaturing gels. The amount of RNA-protein complex formed was quantified by dividing the amount of probe shifted by the total amount of probe in each lane. The intensities of the RNA-protein complex and free-probe bands were quantified using a Fuji BAS 1800 analyzer (Fuji Photo Film Co., Japan) and Image Gauge software (Science Lab, 98, version 3.12; Fuji Photo Film Co.). The Kd values were estimated by plotting [log (% bound RNA/% unbound RNA) + 2] against [log (protein concentration) + 1] (5, 40) using KaleidaGraph version 3.6 (Synergy Software) software. An average Kd value was determined for each interaction using Kd values calculated for three replicate binding experiments. The Kd values estimated by this protocol were very similar to the Kd values obtained for parental RNA and a representative set of viral RNAs using a more extensive set of protein concentrations (data not shown).
For competition gel shift assays, purified rTIA-1 (600 nM) and purified rTIAR (60 nM) were incubated with increasing concentrations of a nonspecific competitor, poly(I-C) (0.1 to 1 ng) or tRNA (2.5 to 20 nM), or with the specific competitor, unlabeled WNV3′(−)SL RNA (0.2 to 5 nM), for 10 min prior to the addition of 0.2 nM (2,000 cpm) of the 32P-labeled RNA probe.
Kinetics of progeny virus production.
Duplicate wells of BHK cells (80% confluence) in T25 flasks were transfected with 1 μg of parental or mutant WNV infectious clone RNA, as described above. Aliquots of culture fluid (500 μl) were harvested at 12, 24, 36, and 48 h after transfection, and 500 μl of fresh medium was replaced at each time point. The aliquots were stored at −80°C until titrated on BHK cells by plaque assay, as previously described (20). Plaque titrations were done in duplicate, and average virus titers were calculated.
Analysis of intracellular viral RNA levels by real-time quantitative RT-PCR (qRT-PCR).
BHK cells (90% confluence) in six-well plates were transfected with 200 ng of parental or mutated WNV infectious clone RNA. Total cell RNA was extracted at different times after transfection, using TRI reagent (Molecular Research Center, Inc.) according to the manufacturer's protocol. Intracellular viral genomic RNA levels were quantified as described previously (15).
Detection of intracellular viral antigen.
BHK cells (2 × 103 cells per well) were seeded on 15-mm coverslips, and 24 h later, the cells were transfected with 100 ng of WNV infectious clone RNA. Three hours after transfection, cells were fixed in 4% paraformaldehyde and then permeabilized using cold 100% methanol. The cells were washed several times in PBS and incubated with anti-WNV hyperimmune serum (kindly provided by Robert B. Tesh, University of Texas Medical Branch at Galveston, TX) at a dilution of 1:100 for 1 h at 37°C. The cells were then washed three times with cold PBS and incubated with anti-mouse immunoglobulin G antibody conjugated with rhodamine (1:300) (Santa Cruz Biotechnology). Cell nuclei were stained with 0.5 μg/ml Hoechst 33258 (Molecular Probes). After being washed, coverslips were mounted with Prolong mounting medium (Invitrogen), and the cells were viewed and photographed with a Zeiss LSM 510 confocal microscope (Zeiss, Germany) using a 100× oil immersion objective with 1× zoom. The images were merged and analyzed using Zeiss software version 3.2. The same camera settings were used for each experimental series. Relative fluorescence intensity was measured using LSM 5 (version 3.2) software in 7-μm-diameter circles in five locations within the cytoplasm of 10 representative BHK cells transfected with each viral RNA tested.
RNA secondary structure prediction.
The secondary structure of each RNA probe used in this study was predicted using Mfold version 3.1 software (60).
RESULTS
Expression, purification, and RNA binding activities of rTIA-1 and rTIAR.
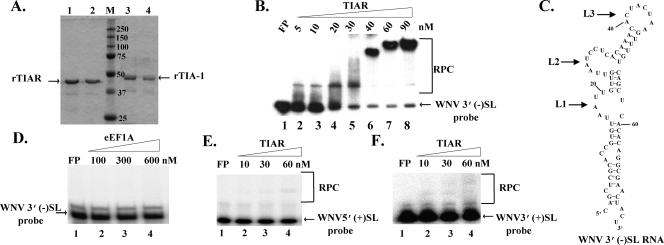
A previous study reported that both TIA-1 and TIAR bound specifically to the WNV3′(−)SL RNA (37). To map protein binding sites on the viral RNA, His-tagged rTIAR/rTIA-1 proteins were expressed in E. coli and partially purified to ∼85% homogeneity on a cobalt affinity column as described in Materials and Methods (Fig. 1A). A single band was detected for each recombinant protein by Western blot analysis using protein-specific polyclonal antibodies directed against unique C-terminal regions (data not shown). Gel mobility shift assays with the His-tagged proteins and a WNV3′(−)SL RNA probe were done as described in Materials and Methods. rTIAR bound to the probe at protein concentrations as low as 5 nM (Fig. 1B), while concentrations of ∼50 nM were required to detect rTIA-1 binding (data not shown). This 10-fold difference in binding efficiency was consistent with previous data obtained with recombinant glutathione S-transferase fusion proteins (37). Neither a control recombinant protein, eEF1A, expressed and partially purified under the same conditions as TIAR and TIA-1, nor any background protein in the partially purified TIAR/TIA-1 samples bound to the WNV3′(−)SL RNA in gel mobility shift assays (Fig. 1A, B, and D). Consistent with previous data (37), competition gel mobility shift assays done as described in Materials and Methods showed that unlabeled WNV3′(−)SL RNA, but not poly(I-C) or tRNA, competed efficiently with the WNV3′(−)SL RNA probe (data not shown). rTIAR bound minimally to two other viral RNA probes, the WNV5′(+)SL (Fig. 1E) and the WNV3′(+)SL RNA (Fig. 1F). Similar results were observed for rTIA-1 (data not shown). These data confirmed that both His-tagged TIA-1 and TIAR bind specifically and preferentially to the WNV3′(−)SL RNA. These TIAR and TIA-1 protein preparations were used to obtain all of the in vitro binding data subsequently described.
FIG. 1.
Purification and characterization of rTIA-1 and rTIAR protein. (A) rTIA-1 and rTIAR proteins were purified on cobalt affinity columns and then separated by 10% SDS-polyacrylamide gel electrophoresis. Fractions of rTIAR (lanes 1 and 2) and rTIA-1 (lanes 3 and 4) were eluted from the affinity column with 150 mM imidazole. (B) A representative gel mobility shift assay done with increasing concentrations of rTIAR and a 32P-labeled WNV3′(−)SL RNA probe and analyzed on a 5% nondenaturing polyacrylamide gel. Lanes 1, free probe (FP); 2 to 8, probe plus the indicated concentration of purified protein. The region of the gel containing RNA-protein complexes (RPC) is indicated by brackets. (C) Secondary structure predicted for the WNV3′(−)SL RNA sequence by Mfold version 3.1 software. Three loops (L1, L2, and L3) that contain short AU tracts are indicated by arrows. (D) A representative gel mobility shift assay done with increasing concentrations of recombinant eEF1A and the WNV3′(−)SL RNA probe. (E) A representative gel mobility shift assay done with increasing concentrations of rTIAR and a 32P-labeled WNV5′(+)SL RNA probe. (F) A representative gel mobility shift assay done with increasing concentrations of rTIAR and a 32P-labeled WNV3′(+)SL RNA probe.
Mapping the binding sites for TIA-1 and TIAR within the WNV3′(−)SL RNA.
Both TIA-1 and TIAR were previously reported to bind specifically to AU-rich sequences in the 3′ untranslated regions (UTRs) of a subset of cell mRNAs. The binding region in the tumor necrosis factor-α mRNA was mapped to a 39-nt AU-rich sequence (26, 36). Seven AUUUA repeats within this sequence were hypothesized to be the protein binding sites, but this was not experimentally verified. The secondary structure predicted for the WNV3′(−)SL RNA by Mfold version 3.1 was similar to the one previously determined using structure probing data by Shi et al. (46) and consists of a stem with three loops, designated L1, L2, and L3 (Fig. 1C). Each of these loops contained a short, single-stranded AU sequence.
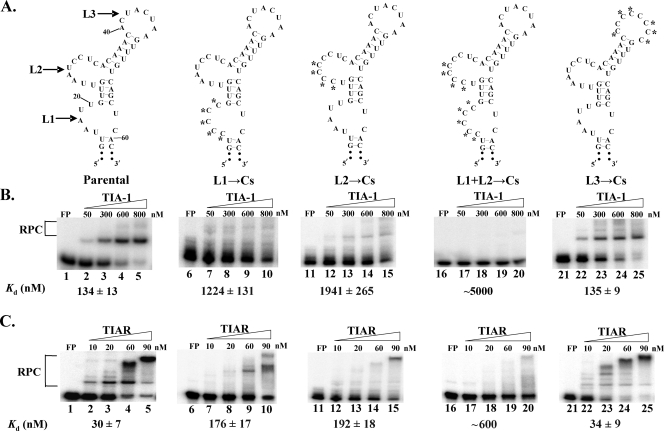
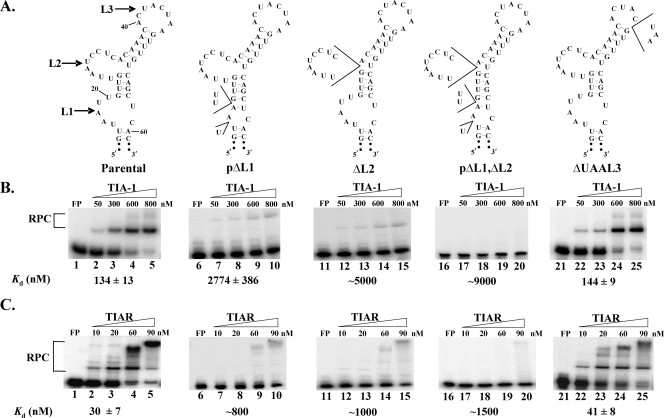
To determine whether one or more of these AU sequences are required for TIA-1 and TIAR binding to the WNV3′(−)SL RNA, substitutions (Fig. 2) or deletions (Fig. 3) were introduced into the AU sequences in each of the loops, separately or in combination, and the mutant RNAs were used as probes in gel mobility shift assays with the recombinant proteins. The optimal structures predicted by Mfold for the mutant RNAs with C substituted for A or U at positions 16 to 20 (L1→Cs), positions 26 to 29 (L2→Cs), positions 40 to 47 (L3→Cs), and positions 16 to 20 plus 26 to 29 (L1+L2→Cs) are shown in Fig. 2A. Only three A or U nucleotides could be deleted in L1 (pΔL1) or in L3 (ΔUAAL3) without altering the RNA secondary structure, while all of L2 (ΔL2) could be deleted (Fig. 3A). Gel mobility shift assay data indicated a significant decrease in the binding efficiencies of both recombinant proteins for the L1→Cs (Fig. 2B, lanes 7 to 10), L2→Cs (Fig. 2B, lanes 12 to 15), pΔL1 (Fig. 3B, lanes 7 to 10), and ΔL2 (Fig. 3B, lanes 12 to 15) mutant RNAs compared to that for the parental RNA. Both proteins bound even less efficiently to the L1+L2→Cs (Fig. 2B, Lanes 17 to 20) and pΔL1,ΔL2 (Fig. 3B, lanes 17 to 20) mutant RNAs. In contrast, the binding efficiencies of both proteins for the L3→Cs (Fig. 2B, lanes 22 to 25) and ΔUAAL3 (Fig. 3B, lanes 22 to 25) mutant RNAs were similar to that for the parental RNA. The Kd values for the RNA-protein interactions were estimated to facilitate comparisons of the binding efficiencies of the proteins for the different mutant RNAs. The Kd values were calculated as described in Materials and Methods and are shown under each gel (Fig. 2 and 3). Calculated Kd values are given for interactions that fit the curve well enough to provide a reasonable estimate. For weak interactions, where the estimated Kd value was more than five times the highest concentration of protein used in the binding assay, only a rounded number was given for the Kd value to indicate that the estimate was less precise.
FIG. 2.
Effect of C substitutions in L1, L2, or L3 of the WNV3′(−)SL RNA on in vitro rTIA-1 and rTIAR binding activity. (A) Predicted secondary structures of the WNV3′(−)SL RNA mutant probes. Substituted nucleotides are indicated by asterisks. (B) Representative gel mobility shift assays done with rTIA-1. (C) Representative gel mobility shift assays done with rTIAR. The 32P-labeled RNA probes were as follows: lanes 1 to 5, parental RNA; 6 to 10, L1→Cs RNA; 11 to 15, L2→Cs RNA; 16 to 20, L1+L2→Cs RNA; 21 to 25, L3→Cs RNA. FP, free probe. The protein concentrations used are indicated above each gel. The gels shown are representative of three replicate experiments done with each viral RNA. The estimated Kd values for the RNA-protein interactions are the averages of Kd values calculated separately from three replicate experiments. The plus-or-minus variation in Kd values is indicated.
FIG. 3.
Effect of deletions of A and U nucleotides in L1, L2, or L3 of the WNV3′(−)SL RNA on in vitro rTIA-1 and rTIAR binding activity. (A) Predicted secondary structures of WNV3′(−)SL mutant RNA probes. Only three Us in L1 and only UAA in L3 could be deleted without altering the predicted RNA secondary structure. Deleted nucleotides are indicated within wedges. (B) Representative gel mobility shift assays with rTIA-1. (C) Representative gel mobility shift assays with rTIAR. The 32P-labeled RNA probes were as follows: lanes 1 to 5, parental RNA; 6 to 10, pΔL1 RNA; 11 to 15, ΔL2 RNA; 16 to 20, pΔL1,ΔL2 RNA; 21 to 25, ΔUAAL3 RNA. The protein concentrations used are indicated above each gel. The gels shown are representative of three replicate experiments done with each viral RNA. The estimated Kd values for the RNA-protein interactions are the averages of Kd values calculated separately from three replicate experiments. The plus-or-minus variation in Kd values is indicated.
The results indicated that AU sequences in both L1 and L2 are required for efficient TIA-1 and TIAR binding to the WNV3′(−)SL RNA in vitro. However, substitution of C for A and U nucleotides in L2 resulted in a greater decrease in the binding of both proteins than substitutions in L1. The observation that TIAR bound to the C-substituted RNAs more efficiently than to the deleted RNAs is consistent with a recent report that TIAR can bind to CU-rich sequences in cell mRNAs, although TIAR binding to these sequences was less efficient than to AU-rich RNA sequences (34).
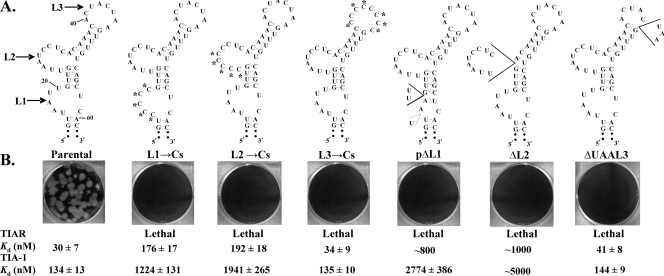
Effect of C substitution or deletion of L1 or L2 A and U nucleotides in a WNV infectious clone on progeny virus production.
Previous studies showing a reduced yield of WNV from infected TIAR knockout cells and colocalization of TIAR and TIA-1 with viral replication complexes in infected cells (21, 37) suggested that these proteins play a role in flavivirus replication. To more directly test this hypothesis, an A or U nucleotide in L1 or L2 was replaced by C or deleted in a WNV infectious clone, as described in Materials and Methods, and the resulting mutant DNA templates were used to synthesize mutant viral RNAs in vitro. BHK monolayers were transfected with parental or mutant infectious clone RNA, and the phenotype of the virus plaques produced by 72 h after transfection in agarose-overlaid transfection wells was assessed. No plaques were observed on wells transfected with any of these mutant RNAs (Fig. 4B). In addition, culture fluids harvested from nonoverlaid, duplicate wells at 72 h after transfection were assayed for virus by plaque assay and for viral RNA by RT-PCR. No plaques or viral RNA was detected in either the transfection well culture fluids nor in 72-h culture fluids from three serial blind passages in BHK cells. These mutations were designated as lethal. These results suggested that both the L1 and L2 AU sequences required for efficient TIA-1 and TIAR binding in vitro are also required for virus viability.
FIG. 4.
Effect of C substitutions or deletions in L1, L2, or L3 in a WNV infectious clone on virus production. (A) Predicted WNV3′(−)SL secondary structures of the mutant RNAs. Substituted nucleotides are indicated by asterisks. Deleted nucleotides are shown within wedges. (B) Plaques produced at 72 h after RNA transfection of BHK cells in agarose-overlaid wells are shown. The Kd values estimated from in vitro RNA binding assays (Fig. 2 and 3) are shown for each RNA.
Although deletion of 5′UAA3′ in L3 (ΔUAAL3) or replacement of all of the A or U nucleotides in L3 by Cs (L3→Cs) had no effect on the in vitro binding efficiency of TIA-1 and TIAR (Fig. 2), infectious clones with these mutations produced no detectable virus (Fig. 4). These results suggested that nucleotides in this loop are involved in other essential RNA or protein interactions as part of either the 3′(−)SL or the complementary 5′(+)SL RNA. None of the nucleotides in L3 that were mutated/deleted was previously shown to be required for the binding of the methyltransferase region of NS5 to the genome 5′(+)SL RNA, and neither of these mutant RNAs was predicted to have a significantly altered 5′(+)SL RNA structure (data not shown).
Effect of substitution or deletion of particular AU nucleotides in L1 or L2 on TIA-1 and TIAR binding in vitro and on progeny virus production.
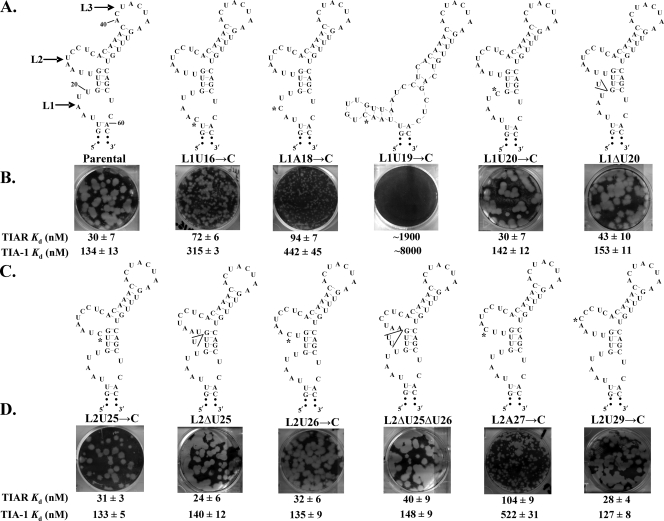
Since the substitution or deletion of all or most of the L1 or L2 AU nucleotides was lethal for the infectious clone, the viability of mutant viral RNAs with 1 or 2 nt substitutions/deletions in the 3′(−)SL was analyzed next. The in vitro binding activity for each of the mutant 3′(−)SL RNAs was assayed by gel mobility shift assay, and a Kd value was estimated for each interaction. Individual As or Us in L1 were replaced by a C to generate the L1U16→C, L1A18→C, L1U19→C, and L1U20→C mutant viral RNAs (Fig. 5A). The A or U nucleotides in L2 were changed to C to create the L2U25→C, L2U26→C, L2A27→C, and L2U29→C mutant viral RNAs (Fig. 5C). Also, a single U was deleted from either L1 or L2 to create the L1ΔU20 and L2ΔU25 mutant RNAs, respectively, and two Us were deleted from L2 to generate the L2ΔU25Δ26 mutant RNA (Fig. 5A and C). Mfold analyses of these mutant RNAs showed that only the L1U19→C mutant was predicted to form an altered secondary structure (Fig. 5A).
FIG. 5.
Effect of substitution or deletion of one or two A or U nucleotides in L1 and L2 in a WNV infectious clone on virus production. (A) and (C) Predicted WNV3′(−)SL secondary structures of the mutant RNAs. Substituted nucleotides are indicated by asterisks, and deleted nucleotides are shown within wedges. (B) and (D) Plaques produced at 72 h after RNA transfection of BHK cells in an agarose-overlaid wells are shown. The Kd values estimated from in vitro RNA binding assays (Fig. 2 and 3) are shown for each RNA.
TIAR bound to mutant RNAs that had a substitution or deletion of one or two of the 5′ terminal nucleotides of L2 (L2U25→C, L2ΔU25, L2U26→C, and L2ΔU25Δ26) and to RNAs that had a substitution or deletion of one 3′ terminal nucleotide of L1 (L1U20→C and L1ΔU20) or L2 (L2U29→C) with an efficiency similar to that observed for the parental RNA. The Kd value estimated for these interactions ranged between 24 and 43 nM (Fig. 5B). The Kd value for the interaction between TIAR and the L1U16→C RNA was higher (72 nM) (Fig. 5B). The C substitutions in the L1A18→C and L2A27→C mutant RNAs were located in the central part of either the L1 or L2 AU sequence and caused a further reduction in TIAR binding activity (Kd of about 94 nM and 103 nM, respectively) (Fig. 5A and C). A significantly lower TIAR binding efficiency (Kd of ∼1,900 nM) was observed with the L1U19→C mutant RNA that was predicted to form an altered secondary structure (Fig. 5B).
The effect of each of these single or double mutations on virus replication was next analyzed by introducing them into a WNV infectious clone. The L1U20→C, L1ΔU20, L2U25→C, L2ΔU25, L2U26→C, L2ΔU25Δ26, and L2U29→C mutant viral RNAs each had a Kd value similar to that observed for the interaction between TIAR and the parental RNA (24 to 43 nM) and produced parent-size plaques (2.5 to 3 mm in diameter) on the transfection plates (Fig. 5B and D), and no change in plaque phenotype or RNA sequence was detected during three subsequent serial passages of progeny virus, indicating that these mutant RNAs were stable. For each of these mutants, an AU sequence of at least 3 nt was preserved in one loop, while the other loop contained the full-length AU sequence. The L1U16→C mutant RNA (TIAR Kd of 72 nM) produced virus with a small-plaque phenotype (about 1 mm in diameter), whereas pinpoint-size plaques (about 0.1 mm in diameter) were produced by the L1A18→C mutant RNA (TIAR Kd of 94 nM) (Fig. 5B). A mixture of medium-size plaques (1.5 to 2 mm in diameter) and pinpoint plaques was observed on the transfection wells with the L2A27→C RNA mutant (TIAR Kd of 104 nM), indicating that this mutant rapidly reverted. Viral RNAs extracted from picked medium-size and pinpoint plaques were amplified by RT-PCR, and the resulting cDNAs were sequenced. The RNA extracted from pinpoint plaques retained the mutant C27, but this nucleotide had reverted to the parental nucleotide A in RNA extracted from medium-size plaques. The reduced size of the plaques produced by the reverted parental RNA on the transfection wells is likely due to the time needed for the revertant to be generated. After one passage of the L2A27→C virus or two serial passages of the L1U16→C or L1A18→C virus, only parent-size plaques were observed, and viral RNA extracted from these plaques had the parental sequence. No plaques were detected by 72 h after transfection of the L1U19→C mutant RNA (TIAR Kd of ∼1,900 nM) (Fig. 5B), but after one passage, parent-size plaques containing RNA with the parental sequence were detected. These results indicate that this mutant RNA underwent a level of RNA replication sufficient to generate a rescuing mutation. Three replicate experiments with this mutant RNA produced the same results.
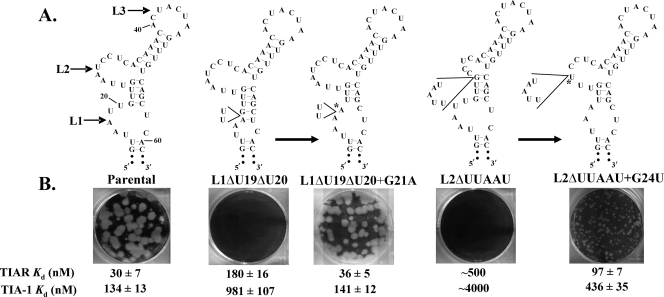
The Kd for TIAR interactions with two partially deleted mutant RNAs, L1ΔU19ΔU20 (two 3′ Us in L1 deleted) and L2ΔUUAAU (five of the 5′ A and U nucleotides in L2 deleted) were ∼180 nM and ∼500 nM, respectively (Fig. 6). No plaques were observed on wells transfected with either of these mutant RNAs, but after one passage of L1ΔU19ΔU20 culture fluid, parent-size plaques were observed. Pinpoint plaques were detected after the first passage of L2ΔUUAAU (Fig. 6B). Viral RNA extracted from the L1ΔU19ΔU20 parent-size plaques contained the second site mutation G21 to A, while viral RNA extracted from the L2ΔUUAAU pinpoint plaques contained the second site mutation G24 to U (Fig. 6A). These results indicate that both of these mutant RNAs underwent a level of RNA replication sufficient to generate a rescuing mutation.
FIG. 6.
Pseudorevertants of the L1ΔU19ΔU20 and L2ΔUUAAU mutant 3′(−)SL RNAs. (A) Predicted 3′(−)SL secondary structures of the mutant and spontaneous pseudorevertant RNAs are shown. Pseudorevertent nucleotides are indicated by asterisks. (B) Plaques produced at 72 h after RNA transfection of BHK cells on agarose-overlaid wells. The Kd values estimated from in vitro RNA binding assays are shown for each RNA strain.
Both of these spontaneous mutations altered the predicted secondary structure of the WNV3′(−)SL RNA so that three or more adjacent As or Us were again present in the loop that retained the engineered deletion (Fig. 6A). Specifically, in the L1ΔU19ΔU20+G21A revertant RNA, the spontaneous substitution at position 21 was predicted to increase the size of L1 and reduce the number of base pairs between L1 and L2 from four to three. In the L2ΔUUAAU+G24U revertant RNA, the spontaneous substitution at position 24 was predicted to reduce the size of L1 from 5 to 3 nt (5′UAA3′), to shift the other two original L1 nucleotides (5′UU3′) to the stem region between L1 and L2, to decrease the number of base pairs between L1 and L2 by 1, and to increase the size of L2 from 4 to 7 nt (5′UUUCCUC3′). The in vitro binding efficiencies of TIAR and TIA-1 for the two pseudorevertant RNAs were assayed, and the Kd value for the TIAR interaction with the L1ΔU19ΔU20+G21A RNA was 37 nM, which was similar to the Kd value for other viral RNAs that produced parent-size plaques. The Kd value for the TIAR interaction with the L2ΔUUAAU+G24U RNA was 96 nM, which was similar to the interaction with the L1A18→C RNA that also produced pinpoint plaques. An L1ΔU19ΔU20 infectious clone that was engineered to also contain the G21A mutation produced parent-size plaques, while an L2ΔUUAAU infectious clone engineered to contain the G24U mutation produced pinpoint plaques. These results indicate that the revertant phenotypes detected were not due to an unknown mutation elsewhere in the viral RNA.
Effect of mutations or deletions in the TIA-1 and TIAR binding sites in the WNV3′(−)SL RNA on the kinetics of progeny virus production.
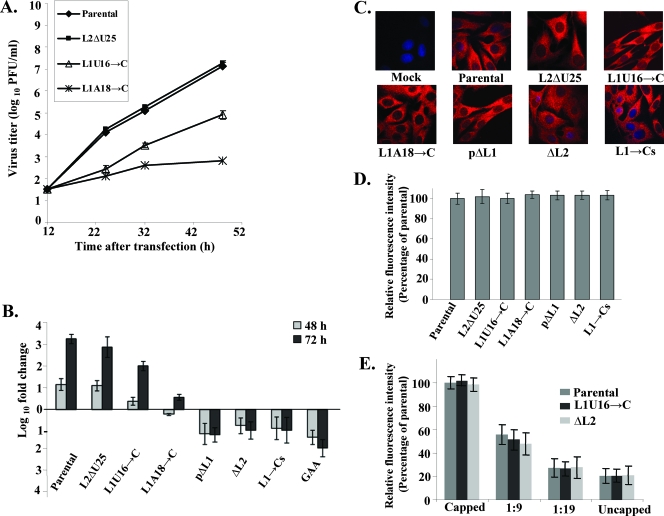
As an additional means of analyzing the effect of the L1 or L2 AU nucleotide mutations or deletions on viral growth, the kinetics of extracellular progeny virus production was assayed for selected mutant RNAs. Because the majority of the mutant viruses were observed to revert during the first passage, progeny virus production was analyzed at different times after RNA transfection. BHK cells were transfected with the L1U16→C, L1A18→C, or L2ΔU25 mutant infectious clone RNA or the parental RNA, and the titer of extracellular virus produced at different times after transfection was determined by plaque assay. The L1A18→C mutant (pinpoint plaques) produced the lowest viral yields, while the L2ΔU25 mutant (parent-size plaques) produced virus yields similar to that of the parental RNA. The L1U16→C mutant (small plaques) produced more progeny virus than the L1A18→C mutant did but significantly less virus than the parental RNA (Fig. 7A).
FIG. 7.
Effect of mutations/deletions in L1 or L2 in a WNV infectious clone on virus yield, viral RNA replication, and viral RNA translation. (A) The kinetics of virus growth in BHK cells transfected with parental or mutant infectious clone RNAs. Error bars represent the standard deviations (SD) (n = 4) in plaque titers. (B) Relative quantification of the levels of the intracellular viral genomic RNA at 48 and 72 h after viral RNA transfection of BHK cells by real-time qRT-PCR. Viral RNA levels are expressed as the log10-fold change in relative quantification units compared to the level of viral RNA (mostly input viral RNA) present at 6 h after transfection. Each RNA sample was also normalized to cellular GADPH mRNA in the same sample. (C) Immunofluorescence imaging of BHK cells transfected with parental or mutant infectious clone RNA or of mock-transfected cells. Cells were fixed and permeabilized 3 h after transfection and incubated with anti-WNV hyperimmune serum (red). Nuclear DNA (blue) was stained with Hoechst 33258 dye. (D) Relative fluorescence intensities of WNV protein in the cytoplasm of BHK cells transfected for 3 h with parental or mutant infectious clone RNA. (E) Relative fluorescence intensities of WNV protein in the cytoplasm of BHK cells transfected with either capped, uncapped, or different ratios (1:9 and 1:19) of capped to uncapped viral RNAs. Error bars represent the SD (n = 3).
Relative quantification of viral RNA replication by real-time qRT-PCR.
To determine whether the mapped TIAR/TIA-1 binding sites in L1 and L2 function as cis-acting sequences for genomic RNA synthesis, the relative amounts of genomic RNA in BHK cells were assayed at different times after transfection with parental or mutant viral RNA. The kinetics of decay of transfected input viral RNA were first assessed in cells transfected with a replication-deficient infectious clone RNA that had the conserved RNA-dependent RNA polymerase GDD motif mutated to GAA (15). Total cell RNA was extracted from BHK cells at 6, 24, 48, and 72 h after transfection of the GAA mutant RNA, and the relative amount of genomic RNA present was determined by real-time qRT-PCR using primers targeting the NS1 region. Viral RNA levels measured at 24, 48, and 72 h after transfection were expressed as a log10-fold change in relative quantification units compared to the level of viral RNA (primarily input RNA) present at 6 h after transfection. Each RNA sample was also normalized to cellular GADPH mRNA to control for sample variation. The levels of the GAA mutant RNA detected at 24 h were similar to those detected at 6 h. Thereafter, the input GAA mutant RNA levels progressively decreased with time after transfection in the absence of virus replication (Fig. 7B). In contrast, by 48 h after transfection of parental RNA, an ∼10-fold increase in genomic RNA levels was observed, and a 1,000-fold increase was detected by 72 h, indicating that viral RNA replication had occurred (Fig. 7B). Genomic RNA levels similar to those detected for the parental RNA were observed after transfection of the L2ΔU25 mutant RNA (parent-size plaques). However, after transfection of L1U16→C RNA (small plaques), only twofold and 80-fold increases in genomic RNA levels were observed by 48 and 72 h, respectively. After L1A18→C RNA (pinpoint plaques) transfection, no increase in genomic RNA levels was observed by 48 h, and about a fourfold increase was detected by 72 h after transfection (Fig. 7B). No increase in the intracellular levels of the pΔL1,ΔL2 or L1→Cs mutant genomic RNAs (the lethal phenotype) was detected at either 48 or 72 h after transfection, and the kinetics of decay of input RNA were similar to those observed for the GAA mutant RNA (Fig. 7B). The results showed that the relative efficiency of genomic RNA synthesis by the mutant RNAs correlated well with the efficiency of virus production and plaque size. The data also showed that the efficiency of genomic RNA production correlated with the relative binding efficiency of TIA-1 and/or TIAR to these RNAs in in vitro assays.
Effect of mutations in the WNV3′(−)SL RNA on viral RNA translation efficiency.
Because the sequence at the 3′ end of the minus-strand RNA is complementary to that of the 5′ end of the genomic RNA, the possibility that some of the mutations introduced might have affected the efficiency of genome translation could not be ruled out. Since in vitro translation assays for full-length flavivirus genome RNA are not available, the translation efficiency of selected mutant genomic RNAs was assayed using immunofluorescence microscopy to detect viral antigen in BHK cells 3 h after transfection of either parental or mutant infectious clone RNA, as described in Materials and Methods. About 90% of the cells in monolayers transfected with parental RNA showed bright WNV protein staining throughout the cytoplasm (Fig. 7C). Minimal fluorescence was detected in mock-transfected cells. The distribution of viral antigen and the intensity of cytoplasmic fluorescence detected after transfection of the L1U16→C (small plaques), L1A18→C (pinpoint plaques), and L2ΔU25 (parent-size plaques) mutant RNAs and the three lethal mutant RNAs, pΔL1, ΔL2, and L1→Cs, were similar to those observed after transfection of parental viral RNA (Fig. 7C and D). The high levels of fluorescence detected after transfection of 1 μg of viral RNA would have masked any small differences in translation efficiency. An additional experiment was done under conditions that reduced viral RNA translation efficiency. Cells were transfected with the same total amount of viral RNA, but the overall level of translation was reduced by replacing various amounts of the capped viral RNA with uncapped RNA. Under the capping reaction conditions used, about 80% of the in vitro-synthesized viral RNA was estimated to be capped (Ambion). The relative fluorescence intensity for the parental RNA was set at 100%, and, as shown in Fig. 7D, similar fluorescence levels were observed for two mutant RNAs. When only uncapped RNA was used, the relative intensity was about 20% (Fig. 7E). When capped and uncapped RNA were mixed at a 1:9 ratio, the fluorescence intensity observed for parental RNA was reduced by about 50% compared to that for capped viral RNA alone. The intensity was reduced by about 70% when the ratio was 1:19 (Fig. 7E). Similar reductions in fluorescence intensity were observed under the same conditions with the L1U16→C (small-plaque) and ΔL2 (lethal) mutant RNAs. These results indicated that the mutations made in the TIAR/TIA-1 binding sites within the WNV3′(−)SL, which would also have mutated the complementary 5′(+)SL, had little, if any, effect on the translation efficiency of the viral genome.
DISCUSSION
The cellular proteins TIA-1 and TIAR were previously shown to bind specifically to the WNV3′(−)SL RNA (37). Colocalization of first TIAR and then TIA-1 with flavivirus replication complexes was observed for both WNV- and dengue virus-infected cells, and anti-TIAR antibody coprecipitated viral nonstructural replication complex proteins from infected cell extracts (21). These results suggested that TIAR/TIA-1 might play a role in viral genomic RNA synthesis. In the present study, the sites on the WNV3′(−)SL RNA required for efficient in vitro binding of TIA-1 and TIAR were mapped, and the effects of mutation of these binding sites in a WNV infectious clone on virus replication were assessed. The results showed that the relative efficiency of TIAR/TIA-1 binding to the WNV3′(−)SL RNA consistently correlated with plaque size, virus yield, and genomic RNA levels (Table 2). Efficient in vitro WNV3′(−)SL RNA-TIAR interactions (Kd values ranging from 24 to 43 nM) were observed for the parental RNA and all of the mutant RNAs that produced large plaques, high virus yields, and high intracellular genomic RNA levels (e.g., the mutant L2ΔU25). Mutant RNAs with less-efficient in vitro viral 3′(−)SL RNA-TIAR interactions (e.g., the L1U16→C mutant, Kd of 72 nM) produced a small-plaque phenotype, ∼1,000-fold-lower virus yield, and ∼10-fold-lower levels of intracellular viral genomic RNA. Mutant RNAs with even less-efficient in vitro 3′(−)SL RNA-TIAR interactions (e.g., the L1A18→C mutant, Kd of 94 nM) produced pinpoint plaques, ∼15,000-fold lower virus yield, and ∼1,000-fold lower levels of intracellular genomic RNA. Mutant viral RNAs with very inefficient in vitro 3′(−)SL RNA-TIAR interactions (such as pΔL1, with a TIAR Kd of ∼800 nM; and ΔL2, with a TIAR Kd of ∼1,000 nM) did not produce detectable progeny virus. These results indicate that the TIA-1/TIAR binding sites in the viral 3′(−)SL RNA are cis acting. The 3′(−)SL RNA contains promoter elements for genome RNA synthesis. Neither TIA-1 nor TIAR binds to the 3′(+)SL or 5′(+)SL of the genomic RNA. Both of these SLs have been proposed to contain promoter elements for viral minus-strand RNA synthesis (15, 22, 41, 57).
TABLE 2.
Comparison of the efficiency of the TIAR/TIA-1-WNV 3′(−)SL RNA interaction with the efficiency of virus production and intracellular genomic RNA levels
| Viral RNAa | Estimated Kd (nM)b
|
Plaque phenotype | Virus yield (PFU/ml)c | Relative intracellular plus-strand RNA leveld | |
|---|---|---|---|---|---|
| TIAR | TIA-1 | ||||
| Parental | 30 ± 7 | 134 ± 13 | Large | 1.3 × 107 | 1 |
| L2ΔU25 | 24 ± 6 | 140 ± 12 | Large | 2.6 × 107 | 1 |
| L1U16→C | 72 ± 6 | 315 ± 3 | Small | 9.5 × 104 | 0.1 |
| L1A18→C | 94 ± 7 | 442 ± 45 | Pinpoint | 8.5 × 102 | 0.001 |
| pΔL1 | ∼800 | 2,774 ± 386 | No plaques | ND | ND |
| ΔL2 | ∼1,000 | ∼5,000 | No plaques | ND | ND |
3′(−)SL mutant RNAs are indicated.
The plus-or-minus variation in Kd is indicated.
Values shown are extracellular virus titers at 72 h after RNA transfection.
Relative amounts of intracellular plus-strand viral RNA detected 72 h after transfection of a mutant infectious clone RNA compared to the amount of plus-strand RNA detected in cells transfected with parental infectious clone RNA, which was set at 1. ND, not detected.
In infected cells, minus-strand RNA has been found only in association with double-stranded replication intermediates that are located in perinuclear membrane-associated viral replication complexes (12, 13). Colocalization of TIAR with viral replication complexes in WNV-infected BHK cells was detected starting as early as 6 h after infection, and the majority of TIAR was concentrated in this region by 24 h (21). The timing of TIAR colocalization with viral replication complexes is coincident with the switch from low-level symmetric plus- and minus-strand RNA synthesis to asymmetric amplification of plus-strand viral RNA synthesis (12). Mutant RNAs such as L1U19→C, L1ΔU19ΔU20, and L2ΔUUAAU that had 1 or 2 nt mutations/deletions were able to generate a rescuing mutation within one or two blind passages. However, even though it is likely that mutant RNAs with more than two substitutions were also able to carry out low-level symmetric RNA replication, they were not able to revert a sufficient number of the mutated nucleotides to become efficient at binding TIAR/TIA-1, and thus, these mutants were not able to amplify plus-strand RNA synthesis or produce detectable levels of virus. The generation of second-site mutations by L1ΔU19ΔU20 and L2ΔUUAAU, which significantly improved the efficiency of the interaction of each of these RNAs with TIAR/TIA-1 and resulted in virus production (Fig. 6), provided further support for the hypothesis that TIAR/TIA-1 are not required for initial low-level symmetric plus- and minus-strand synthesis but instead facilitate the amplification of plus-strand RNA synthesis.
The finding that mutations/deletions of the nucleotides in L3 had no effect on TIAR/TIA-1 binding efficiency but were lethal when engineered into the infectious clone suggest that these nucleotides are also cis acting. They may be important in the context of the 3′(−)SL and/or the complementary 5′(+)SL. Three additional cell proteins, 50, 60, and 108 kDa, were previously reported to bind specifically to the WNV 3′(−)SL RNA (37). It is possible that one or more of these cell proteins bind specifically to the 3′(−)SL RNA L3 nucleotides and play a role in plus-strand synthesis from the minus-strand template. Also, the nucleotides complementary to the 3′(−)SL L3 nucleotides are located in the top loop of the 5′(+)SL, and in this context, these nucleotides may interact with cell proteins during the initiation of translation or replication of the genome RNA. Alternatively, the nucleotides in this loop, in either the plus- and/or minus-strand RNAs, could be involved in as-yet-unidentified RNA-RNA interactions. Experiments are in progress to further investigate the function of the L3 nucleotides.
The binding sites for TIA-1 and TIAR were mapped to two single-stranded UAAUU sequences located in two adjacent loops, L1 and L2, within the WNV3′(−)SL RNA structure. The efficiency of protein binding decreased when A or U nucleotides in either L1 or L2 were deleted or replaced by C. Although the TIA-1/TIAR binding sites in the 3′ UTRs of some cell mRNAs were previously reported to be canonical AUUUA motifs, this was not experimentally confirmed, and the structural context of these motifs was not considered. The ARED mRNA database (http://rc.kfshrc.edu.sa/ared/) classifies ARE-containing mRNAs in one of five clusters based on the number of canonical AUUUA motifs present in their 3′ UTRs. The 3′ UTRs of the cell mRNAs previously reported to interact with TIAR/TIA-1 fall within cluster III (14, 18) or cluster V (30), or they have no canonical AUUUA sequences (29). TIAR was also previously reported to bind efficiently to the trailer sequence (UUUUAAAUUUU) of the Sendai virus genomic RNA (27). These data indicate that TIA-1 and TIAR can bind efficiently to RNA sequences that contain the AUUUA consensus motifs, as well as to those that do not. Variation in the binding efficiency of TIA-1/TIAR for various cell target RNAs may serve important regulatory functions for alternative splicing and translational silencing in the cell. The efficient colocalization of TIAR and TIA-1 to flavivirus replication complexes observed in infected cells suggests that the viral 3′(−)SL RNA can efficiently out-compete cell RNA targets for binding to these proteins. A recent microarray study detected TIAR binding to ∼2,500 cell mRNAs with CU-rich 3′ UTR sequences (34). The binding activity of TIAR for individual CU-rich sequences varied but was, in general, at least 135-fold lower than that for poly(U) (Kd of 1 nM). The parental WNV 3′(−)SL RNA (Kd of 30 nM) would be expected to efficiently out-compete all of the cell CU-rich 3′ UTRs for TIAR. The Kd values for the interactions between TIAR and individual ARE 3′ UTRs have not been reported.
TIA-1 and TIAR contain three RRM RNA binding domains and a C-terminal glutamine-rich auxiliary domain structurally similar to a prion domain (23, 31, 51). RRM2 was previously shown to account for the majority of the TIA-1 and TIAR binding activity to short synthetic U-rich RNAs (16) and to the WNV3′(−)SL RNA (37). An RRM contains two conserved ribonucleoprotein motifs (RNP 1 and RNP 2) (3, 49). The two RNP motifs of several RNP family proteins have been reported to interact in a sequence-specific manner with two single-stranded RNA sequences of 5 to 7 nt (10, 24, 25). The crystal structure of a complex of the RRM of the small nuclear ribonucleoprotein U1A and a 21-nt RNA hairpin from the U1 snRNA has been determined at 1.92-Å resolution (44). The β sheets of each of the U1A RNP motifs were present in an open conformation and each interacted extensively with 1 of the 7-nt internal loops (AUUGUAC and AUUGCAC) of the U1 snRNA hairpin. These two loops are located on opposite sides of the U1 snRNA hairpin and are separated from each other by a 4-bp stem. The TIA-1 and TIAR binding sites in the viral 3′(−)SL RNA are located in two adjacent loops (L1 and L2) within a hairpin structure. The AU sequences of L1 (UAAUU) and L2 (UUAAUCCUC) are in opposite orientation to each other and are separated by a 4-bp stem. Mutation/deletion of a terminal nucleotide in the L1 or the L2 AU sequences had little or no effect on either the efficiency of protein binding to the RNA in vitro or on virus production by the mutant infectious clone, with the exception of the L1U16→C (CAAUU) RNA mutant, which showed reduced protein binding efficiency and produced virus with a small-plaque phenotype (Fig. 5). These results suggest that U16 might be a critical contact nucleotide for the initiation or stabilization of the RNA-protein interaction. A single C in the middle of the AU sequence in either loop (L1A18→C [UACUU] and L2A27→C [UUCAU]) significantly reduced the efficiency of in vitro protein binding (Fig. 5) and of virus replication (pinpoint plaque phenotype). The L1 and L2 AU sequences are predicted to fit into the grooves of the TIAR/TIA-1 RNP1 and RNP2 motifs in a sequence-specific manner, and therefore, the mutation/deletion of nucleotides located in the center of these sequences would be expected to reduce the efficiency of the specific interaction of the viral RNA with the two RNPs of the TIAR/TIA-1 RRM2 domain.
The deletions or mutations introduced into L1 and L2 in the 3′(−)SL RNA had little effect on the translation efficiency of the complementary genomic RNA assayed at 3 h after transfection of BHK cells. However, the input viral RNAs used for these experiments were capped in vitro. The possibility that some of the 3′(−)SL RNA mutations might have also affected the efficiency of capping of the complementary 5′ end of the nascent mutant genomic viral RNAs synthesized in infected cells was not directly investigated. However, none of the mutations/deletions made were predicted to change any of the nucleotides or structural elements in the WNV 5′(+)SL previously reported to be important for recognition by the NS5 methyltransferase during either N-7 methylation or 2′-OH ribose methylation (19). A few of the mutations made in the 3′(−)SL RNA were predicted to alter the folding of the 5′(+)SL RNA, but these changes did not correlate with the observed viral phenotype.
Although TIAR/TIA-1 binding studies were not done with other flavivirus 3′(−)SL RNAs as part of this study, the previous observation that TIAR and TIA-1 colocalize with viral replication complexes in dengue virus-infected cells suggests that the enhancement of genomic RNA synthesis by TIAR/TIA-1 also occurs during other flavivirus infections. Flaviviruses replicate in a wide range of host species, and both TIAR and TIA-1 show a high degree of evolutionary conservation (6, 54).
Acknowledgments
This work was supported by Public Health Service research grant AI048088 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank S. V. Scherbik for technical advice and discussion of the data, W. D. Wilson and G. Gadda for assistance with the estimation of Kd values, and D. Scherbik for assistance with graphics.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Anderson, P. 1995. TIA-1: structural and functional studies on a new class of cytolytic effector molecule. Curr. Top. Microbiol. Immunol. 198131-143. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., C. Nagler-Anderson, C. O'Brien, H. Levine, S. Watkins, H. S. Slayter, M. L. Blue, and S. F. Schlossman. 1990. A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J. Immunol. 144574-582. [PubMed] [Google Scholar]
- 3.Beck, A. R., Q. G. Medley, S. O'Brien, P. Anderson, and M. Streuli. 1996. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 243829-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, A. R., I. J. Miller, P. Anderson, and M. Streuli. 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 952331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 716433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, S., and H. M. Bourbon. 1993. The developmentally-regulated Drosophila gene rox8 encodes an RRM-type RNA binding protein structurally related to human TIA-1-type nucleolysins. Nucleic Acids Res. 213699-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 841261-1268. [DOI] [PubMed] [Google Scholar]
- 8.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162290-299. [DOI] [PubMed] [Google Scholar]
- 9.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153113-121. [DOI] [PubMed] [Google Scholar]
- 10.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 131197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahour, A., A. Pletnev, M. Vazielle-Falcoz, L. Rosen, and C. J. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 20768-76. [DOI] [PubMed] [Google Scholar]
- 12.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44649-688. [DOI] [PubMed] [Google Scholar]
- 13.Cleaves, G. R., T. E. Ryan, and R. W. Schlesinger. 1981. Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 11173-83. [DOI] [PubMed] [Google Scholar]
- 14.Cok, S. J., S. J. Acton, A. E. Sexton, and A. R. Morrison. 2004. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 2798196-8205. [DOI] [PubMed] [Google Scholar]
- 15.Davis, W. G., J. L. Blackwell, P. Y. Shi, and M. A. Brinton. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 8110172-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dember, L. M., N. D. Kim, K. Q. Liu, and P. Anderson. 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 2712783-2788. [DOI] [PubMed] [Google Scholar]
- 17.Dirksen, W. P., S. A. Mohamed, and S. A. Fisher. 2003. Splicing of a myosin phosphatase targeting subunit 1 alternative exon is regulated by intronic cis-elements and a novel bipartite exonic enhancer/silencer element. J. Biol. Chem. 2789722-9732. [DOI] [PubMed] [Google Scholar]
- 18.Dixon, D. A., G. C. Balch, N. Kedersha, P. Anderson, G. A. Zimmerman, R. D. Beauchamp, and S. M. Prescott. 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong, H., D. Ray, S. Ren, B. Zhang, F. Puig-Basagoiti, Y. Takagi, C. K. Ho, H. Li, and P. Y. Shi. 2007. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 814412-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elghonemy, S., W. G. Davis, and M. A. Brinton. 2005. The majority of the nucleotides in the top loop of the genomic 3′ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology 331238-246. [DOI] [PubMed] [Google Scholar]
- 21.Emara, M. M., and M. A. Brinton. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 1049041-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filomatori, C. V., M. F. Lodeiro, D. E. Alvarez, M. M. Samsa, L. Pietrasanta, and A. V. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 202238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 155383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J. Biol. Chem. 26923074-23078. [PubMed] [Google Scholar]
- 25.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211400-407. [DOI] [PubMed] [Google Scholar]
- 26.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 2742322-2326. [DOI] [PubMed] [Google Scholar]
- 27.Iseni, F., D. Garcin, M. Nishio, N. Kedersha, P. Anderson, and D. Kolakofsky. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 215141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, K., W. Li, T. Nagayama, X. He, A. D. Sinor, J. Chang, X. Mao, S. H. Graham, R. P. Simon, and D. A. Greenberg. 2000. Expression of the RNA-binding protein TIAR is increased in neurons after ischemic cerebral injury. J. Neurosci. Res. 59767-774. [DOI] [PubMed] [Google Scholar]
- 29.Kandasamy, K., K. Joseph, K. Subramaniam, J. R. Raymond, and B. G. Tholanikunnel. 2005. Translational control of beta2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein. J. Biol. Chem. 2801931-1943. [DOI] [PubMed] [Google Scholar]
- 30.Kawai, T., A. Lal, X. Yang, S. Galban, K. Mazan-Mamczarz, and M. Gorospe. 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 263295-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami, A., Q. Tian, X. Duan, M. Streuli, S. F. Schlossman, and P. Anderson. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 898681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 1511257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1471431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, H. S., Y. Kuwano, M. Zhan, R. Pullmann, Jr., K. Mazan-Mamczarz, H. Li, N. Kedersha, P. Anderson, M. C. Wilce, M. Gorospe, and J. A. Wilce. 2007. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 276806-6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Guiner, C., M. C. Gesnel, and R. Breathnach. 2003. TIA-1 or TIAR is required for DT40 cell viability. J. Biol. Chem. 27810465-10476. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, T., C. Gueydan, G. Huez, J. J. Toulme, and V. Kruys. 1998. Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 27313781-13786. [DOI] [PubMed] [Google Scholar]
- 37.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. M. Swiderek, G. T. Moreno, and M. A. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 7611989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, W., M. Simarro, N. Kedersha, and P. Anderson. 2004. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 2410718-10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 40.Lu, C. D., J. E. Houghton, and A. T. Abdelal. 1992. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J. Mol. Biol. 22511-24. [DOI] [PubMed] [Google Scholar]
- 41.Markoff, L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 703930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nomaguchi, M., T. Teramoto, L. Yu, L. Markoff, and R. Padmanabhan. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 27912141-12151. [DOI] [PubMed] [Google Scholar]
- 44.Oubridge, C., N. Ito, P. R. Evans, and K. Nagai. 1994. Crystal structure at 1.92 angstroms resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372432-438. [DOI] [PubMed] [Google Scholar]
- 45.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 194154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, P. Y., W. Li, and M. A. Brinton. 1996. Cell proteins bind specifically to West Nile virus minus-strand 3′ stem-loop RNA. J. Virol. 706278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shukla, S., F. Del Gatto-Konczak, R. Breathnach, and S. A. Fisher. 2005. Competition of PTB with TIA proteins for binding to a U-rich cis-element determines tissue-specific splicing of the myosin phosphatase targeting subunit 1. RNA 111725-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukla, S., W. P. Dirksen, K. M. Joyce, C. Le Guiner-Blanvillain, R. Breathnach, and S. A. Fisher. 2004. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J. Biol. Chem. 27913668-13676. [DOI] [PubMed] [Google Scholar]
- 49.Siomi, H., and G. Dreyfuss. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 7345-353. [DOI] [PubMed] [Google Scholar]
- 50.Taupin, J. L., Q. Tian, N. Kedersha, M. Robertson, and P. Anderson. 1995. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 921629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67629-639. [DOI] [PubMed] [Google Scholar]
- 52.Tian, Q., J. Taupin, S. Elledge, M. Robertson, and P. Anderson. 1995. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med. 182865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaheri, A., W. D. Sedwick, S. A. Plotkin, and R. Maes. 1965. Cytopathic effect of rubella virus in BHK21 cells and growth to high titers in suspension culture. Virology 27239-241. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, R., R. Ainscough, K. Anderson, C. Baynes, M. Berks, J. Bonfield, J. Burton, M. Connell, T. Copsey, J. Cooper, et al. 1994. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 36832-38. [DOI] [PubMed] [Google Scholar]
- 55.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281294-304. [DOI] [PubMed] [Google Scholar]
- 56.Yu, Q., S. J. Cok, C. Zeng, and A. R. Morrison. 2003. Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 2781579-1584. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, B., H. Dong, Y. Zhou, and P. Y. Shi. 2008. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5′ stem-loop of genomic RNA. J. Virol. 827047-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, T., N. Delestienne, G. Huez, V. Kruys, and C. Gueydan. 2005. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 1185453-5463. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, H., R. A. Hasman, K. M. Young, N. L. Kedersha, and H. Lou. 2003. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 235959-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuker, M. 2003. M fold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]