Abstract
Since its discovery in the early 1990’s, cortactin has emerged as a key signaling protein in many cellular processes, including cell adhesion, migration, endocytosis, and tumor invasion. While the list of cellular functions influenced by cortactin grows, the ability of cortactin to interact with and alter the cortical actin network is central to its role in regulating these processes. Recently, several advances have been made in our understanding of the interaction between actin and cortactin, providing insight into how these two proteins work together to provide a framework for normal and altered cellular function. This review examines how regulation of cortactin through post-translational modifications and interactions with multiple binding partners elicits changes in cortical actin cytoskeletal organization, impacting the regulation and formation of actin-rich motility structures.
Keywords: cortactin, actin, Arp2/3, N-WASp, motility
Introduction
Cell motility and directional migration are essential for a wide range of cellular processes and disease states, including wound healing, vascular disease, osteoporosis, mental retardation, chronic inflammatory disease, as well as tumor invasion and metastasis (Ridley 2006). The ability of a cell to migrate in response to external stimuli involves several coordinated steps: Formation of lamellipodia at the leading edge, attachment of the lamella to the substratum, forward translocation of the cell body, detachment of adhesions and subsequent retraction of the cell rear (reviewed in (Ridley et al. 2003)). Similarly, invasion requires altered cellular adhesion properties, degradation of the adjacent extracellular matrix, and directional cell migration. The complicated processes of cell migration and invasion require dynamic regulation of the cortical actin cytoskeleton, utilizing a network of signaling proteins that coordinate regulated changes in the actin architecture. One of the key molecules involved in cortical actin regulation is the adaptor protein cortactin. Cortactin is a multi-domain protein that was first identified as a Src substrate (Kanner et al. 1990; Wu et al. 1991). Subsequently, cortactin has been shown to play an essential role in many actin-based cellular processes, encompassing cell migration and invasion (Yamaguchi and Condeelis 2007), axon guidance (Knoll and Drescher 2004), neuronal morphogenesis (Martinez et al. 2003; Gray et al. 2005), and tumor cell metastasis (Li et al. 2001). Several recent reviews have highlighted cortactin’s role in a variety of cellular processes, including cancer progression, invasion and metastasis, and endocytosis (Daly 2004; Ayala et al. 2006; Cosen-Binker and Kapus 2006; Orth and McNiven 2006; Weaver 2006; Buday and Downward 2007; Yamaguchi and Condeelis 2007; Weaver 2008). In this review we focus on the structure/function relationship between cortactin and actin, and what is known about cortactin and the formation and regulation of dynamic actin-based structures and its role in the formation of motility-related membrane protrusions.
Mechanisms of actin assembly
The cortical actin cytoskeleton is a complex and dynamic structure that participates in key aspects of cellular homeostasis. In response to extracellular stimuli, specific intracellular signaling cascades produce various actin-based protrusions of the plasma membrane, including lamellipodia, filopodia, dorsal waves, and invadopodia/podosomes. Lamellipodia formation can occur in response to treatment with EGF, leading to the activation of several kinase families, including Src, Erk, and PI3K (Yamaguchi and Condeelis 2007). PI3K in turn activates Rac, a member of the Rho family GTPases, which regulates the signaling pathway involved in barbed end generation and dendritic actin nucleation (Yamaguchi and Condeelis 2007). In addition to phosphorylating cortactin, Src mediates the phosphorylation of several downstream effector molecules, facilitating the activation of Arp2/3 complex and actin nucleation (Yamaguchi and Condeelis 2007). Stimulation with PDGF activates PI3K and Src, as well as PAK1, PKA, and Abl kinases (Buccione et al. 2004). The concerted action of these signaling proteins serves to aid in the formation of circular dorsal ruffles (CDRs), invadopodia, and lamellipodia. Regardless of the signaling pathway, initiation of actin polymerization remains the consistent factor responsible for initiating the formation of membrane protrusions required for motility and invasion, allowing the cell to react and adapt to a variety of environmental cues.
A critical step in dynamic remodeling of the cortical actin cytoskeleton is activation of the actin-related protein (Arp)2/3 complex. The Arp2/3 complex consists of seven subunits (Arp2, Arp3, and ARPC1-5) that function together to nucleate and initiate new actin filaments. Arp2/3 binds to the side of a preexisting F-actin (mother) filament and nucleates production of a new (daughter) filament at a 70° angle (Machesky et al. 1999; Higgs and Pollard 2001). Arp2/3-nucleated branched actin networks in vitro are very similar in appearance to the cortical actin network present in lamellipodia (Svitkina and Borisy 1999), and subsequent work has demonstrated a requirement of Arp2/3 activity in lamellipodia formation (Mullins et al. 1998; Bailly et al. 2001). Newly branched actin filaments elongate by addition of actin monomers to their barbed ends, and are terminated by the binding of capping protein (CP) to the barbed end. The addition of capping protein ensures that the newly produced filaments are short and rigid, with polymerization concentrated near the leading edge to produce protrusive force (Condeelis 2001). Activation of Arp2/3 results in significant conformational change of several complex members, allowing all seven subunits of the complex to interact with the mother filament (Blanchoin et al. 2000a). Spatially, Arp2/3 is positioned at the pointed end of the newly forming filament (Pollard 2007). The exposed surfaces of the Arp2 and Arp3 subunits assume a conformation similar to that of an F-actin barbed end, and these proteins function to mimic the first two G-actin subunits in daughter filaments (Beltzner and Pollard 2004)(reviewed in (Pollard 2007)). Subsequent network disassembly and actin filament depolymerization involves the inhibition of PAK and LIMK. Kinase inhibition results in ADF/cofilin dephosphorylation by the phosphatase slingshot, promoting ADF/cofilin-mediated severing and disassembly of actin filaments at the network rear, replenishing the actin monomer pool for subsequent rounds of polymerization (Lappalainen and Drubin 1997; Theriot 1997; Bamburg 1999; Carlier et al. 1999; Chen et al. 2000; Niwa et al. 2002). In addition to the Arp2/3 complex, several other proteins have been shown to initiate actin nucleation, including formins, spire and Cordon blue (reviewed in (Watanabe and Higashida 2004; Baum and Kunda 2005; Pollard 2007; Winckler and Schafer 2007)). The action of these proteins differs from Arp2/3 in that they nucleate straight actin filaments.
Cortactin structure and subcellular localization
Cortactin is a 63–65kDa protein initially characterized as a tyrosine phosphorylated substrate in v-Src-transformed chick embryo fibroblasts (Kanner et al. 1990; Wu et al. 1991). Cortactin was also independently identified as downstream target of FGFR-mediated c-Src activation (Zhan et al. 1993) and as a gene amplified and overexpressed in tumor cells with chromosome 11q13 amplification (Schuuring et al. 1993). When analyzed by SDS-PAGE, cortactin migrates as an 80/85kDa doublet that cannot be attributed to post-translational modification of the protein (Wu and Parsons 1993). The reason for the incongruity between the calculated MW and the observed Mr in SDS-PAGE experiments is currently unclear, but may involve an altered ability to bind SDS inherent in the primary sequence and/or spontaneous reacquisition of secondary structural elements.
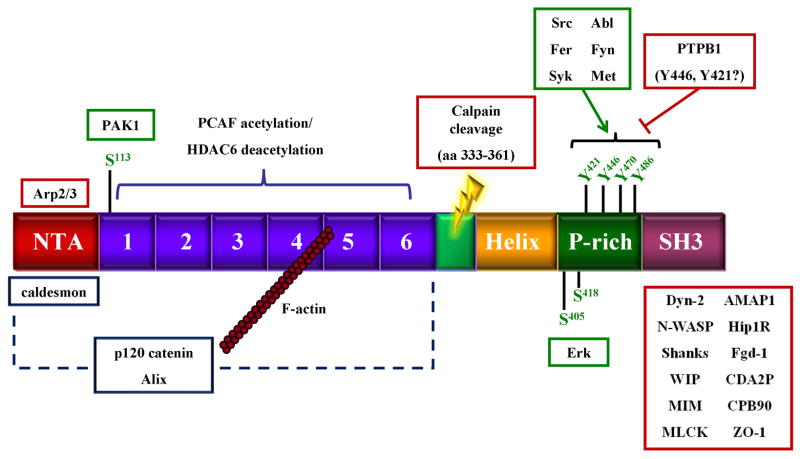
Equilibrium sedimentation and deep etch electron microscopy has determined that cortactin is a monomeric rod-shaped protein ~220Å in length, thinner than the diameter of an actin filament (Weaver et al. 2002). This may represent the “open” form of the protein, since recent work indicates that cortactin can assume an alternative confirmation that is partially globular (Cowieson et al. 2008) (see below). Based on primary sequence analysis cortactin is composed of several distinct domains that either bind other proteins or are sites of post-translation modification (Fig. 1) (Wu et al. 1991). In the murine form, these regions include an amino terminal acidic domain (NTA) (amino acids 1–84). The NTA domain contains a conserved DDW region (amino acids 20–22) that is responsible for interaction with Arp3 and subsequent activation of the Arp2/3 complex (Weed et al. 2000; Higgs and Pollard 2001). Carboxyl terminal to the NTA domain is a series of 6 complete and one partial tandemly repeating segments (amino acids 85–326) termed cortactin repeats that comprise the actin binding region (ABR). The cortactin repeats region directly binds F-actin, with maximal binding activity centered on the fourth repeat. Following the repeats region is a α-helical domain (site of calpain cleavage (Huang et al. 1997b; Perrin et al. 2006)) and a proline-rich region (PRR; amino acids 327–494) enriched in sites of tyrosine and serine phosphorylation. The extreme C-terminus contains an Src homology (SH)3 domain (amino acids 495–542) that interacts with proline-rich binding sequences in several cortactin interacting partners. These include WIP (Kinley et al. 2003), N-WASP (Weaver et al. 2002), MLCK (Dudek et al. 2002), and dynamin 2 (McNiven et al. 2000) (reviewed in (Cosen-Binker and Kapus 2006)).
Figure 1.
Domain structure of cortactin and associated binding proteins. Specific cortactin domains are described in the text. Proteins outlined in red represent binding partners with known interaction sites. Proteins outlined in blue represent known binding partners with undetermined binding sites. Proteins outlined in green represent kinases known to phosphorylate cortactin at the indicated sites.
In serum-starved or unstimulated cells, cortactin is normally present in the cytoplasm as a non-phosphorylated protein (Head et al. 2003). Upon growth factor stimulation (EGF, FGF, or PDGF), activation of Rac1 leads to cortactin relocalization from internal cytoplasmic regions to the cortical actin network (Zhan et al. 1993; Zhan et al. 1994; Head et al. 2003). In addition to Src, cortactin is also phosphorylated by the cytoplasmic tyrosine kinases Fyn, Fer, cMet, c-Abl, Syk ((Gallet et al. 1999; Kapus et al. 2000; Crostella et al. 2001; Huang et al. 2003) Boyle et al. 2007). The serine/threonine kinases PAK1 and MAPK also directly phosphorylate cortactin (Campbell et al. 1999; Webb et al. 2006). The actions of these kinases enable cortactin to function in enhancing actin polymerization and branched actin network formation through its interaction with the actin nucleating complex Arp2/3, as well as the recruitment and positioning of other signaling proteins through SH2 and SH3-mediated interactions as described below.
Cortactin interactions with Arp2/3 and N-WASp
The formation of motility-associated plasma membrane protrusive structures requires the choreographed actions of specific actin effector proteins responsible for the proper spatio-temporal regulation of actin polymerization. Stabilization of branch points in the actin cytoskeletal network is essential for efficient membrane protrusion and directed cell motility. Arp2/3 branch points are inherently metastable, debranching spontaneously in vitro over time (Blanchoin et al. 2000b). Binding of cortactin to Arp2/3 at F-actin branch points stabilizes the branch juncture, inhibiting filament debranching and network breakdown (Weaver et al. 2001).
Cortactin is able to directly influence actin polymerization by serving as a nucleation promoting factor (NPF) for Arp2/3 (Uruno et al. 2001; Weaver et al. 2001). NPFs are a class of proteins that can activate Arp2/3 through direct binding, resulting in a conformational change in the Arp2/3 complex that facilitates de novo actin nucleation and filament elongation as described above (Welch and Mullins 2002). The Wiskott-Aldrich syndrome protein (WASP) family of proteins, including WASP and N-WASp, and the related WAVE/Scar proteins also serve as NPFs for Arp2/3 (Pollard 2007). Cortactin functions as an NPF for Arp2/3 through its ability to bridge Arp3 with ARPC2, 4 and 5, stabilizing the active conformation of the Arp2/3 complex and facilitating the addition of actin monomers to the Arp2 and Arp3 subunits on the side of the mother filament (Weaver et al. 2002; Pollard 2007) (Fig. 2A). Interestingly, the NPF function of cortactin requires both the NTA and the actin binding region (ABR) of cortactin, demonstrating the importance of interaction with both F-actin and Arp2/3 for efficient actin polymerization (Weaver et al. 2001).
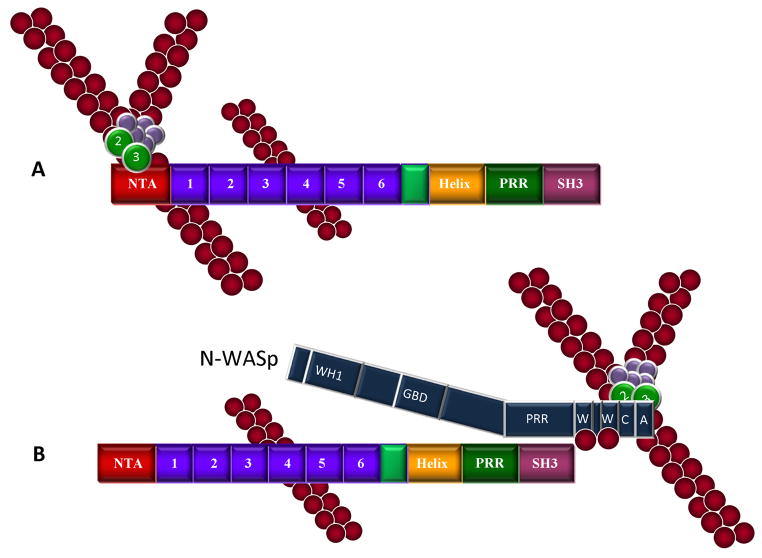
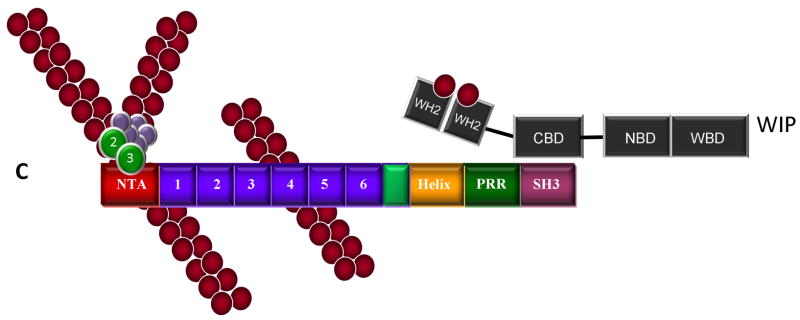
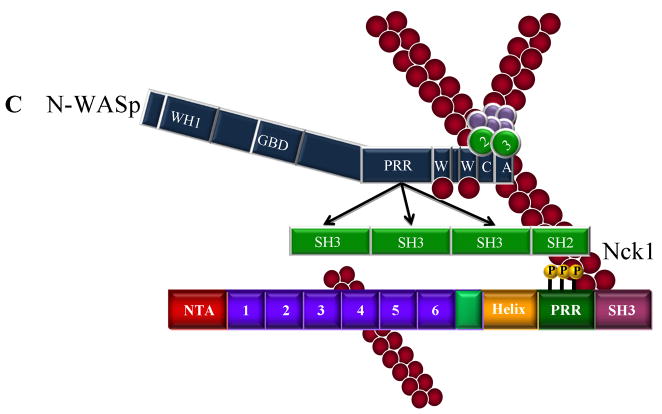
Figure 2.
Cortactin-mediated activation of Arp2/3 actin nucleation and phosphorylation-independent interactions with N-WASp and WIP. A. Cortactin functions as an NPF for Arp2/3 complex. Cortactin binds the Arp2/3 through its NTA domain and F-actin through the fourth tandem repeat, resulting in direct activation Arp2/3 actin nucleation activity. B. Cortactin activates N-WASp. Cortactin enhances N-WASp mediated Arp2/3 activation by disrupting N-WASp autoinhibition through binding of the cortactin SH3 domain to the PRR region of N-WASp. The CA domain of N-WASp binds to and activates Arp2/3, while the WH2 domain binds ATP-loaded G-actin. In this scenario, N-WASp appears to be the dominant activator of Arp2/3 complex. C. WIP enhances cortactin-mediated Arp2/3 activation. The cortactin SH3 domain interacts with the WIP cortactin binding domain (CBD), presumably bringing tandem WH2 domains with bound G-actin in close proximity with Arp2/3 complex activated by the cortactin NTA, serving to enhance cortactin-mediated Arp2/3 activation.
Similar to cortactin, N-WASp and WAVE/Scar proteins bind to F-actin, though this interaction occurs through a basic region and not an ABR (Suetsugu et al. 2001; Kelly et al. 2006). These proteins can bind G-actin monomers and the Arp2/3 complex by virtue of a Wasp Homology-2 (WH2)-central-acidic (WCA) region ((Marchand et al. 2001), reviewed in (Takenawa and Suetsugu 2007)). While F-actin binding is important for WASp-family protein function, the binding of G-actin to the WCA region results in dramatic enhancement of NPF activity by placing actin monomers in close proximity to activated Arp2/3 (Machesky et al. 1999).
Although cortactin-mediated Arp2/3 activation is weaker than that of N-WASp, cortactin synergizes with N-WASp in activating Arp2/3, enhancing N-WASp mediated actin nucleation (Weaver et al. 2001). How N-WASp and cortactin cooperate to initiate Arp2/3 activation is somewhat controversial. The interaction of N-WASp with the side of an actin filament enhances recruitment of and activates Arp2/3, through binding to Arp2, Arp3, and ARPC1 subunits (Suetsugu et al. 2001; Pollard 2007). The binding of the cortactin NTA domain displaces the N-WASp WCA domain from the Arp3 subunit, but not Arp2 or ARPC1 (Weaver et al. 2002). In this manner, N-WASp and cortactin may both bind to and activate Arp2/3 complex concomitantly, forming a ternary complex of Arp2/3/WCA/NTA that results in enhanced NPF activity. Alternately, evidence exists that the activation of Arp2/3 by N-WASP and cortactin occurs in a sequential manner. Cortactin has a higher binding affinity for activated Arp2/3 complex at branch points compared to non-activated, non-actin associated Arp2/3 (Uruno et al. 2003). After Arp2/3 is activated by N-WASp, cortactin effectively binds and displaces N-WASp from the Arp2/3, presumably stabilizing the Arp2/3/cortactin/F-actin complex (Uruno et al., 2003). Whether one or both of these mechanisms are employed at the cellular level to drive Arp2/3 activity remains to be determined.
In addition to directly interacting with Arp2/3 via its NTA, cortactin also promotes actin-nucleation activity indirectly through the SH3 domain. The SH3 domain of cortactin binds directly to a proline-rich region on N-WASp, liberating N-WASp from its auto-inhibited state, resulting in N-WASp-mediated Arp2/3 nucleation activity (Mizutani et al. 2002; Martinez-Quiles et al. 2004) (Fig. 2B). The interaction of cortactin and N-WASp promotes the localization of N-WASp to sites of actin polymerization within invadopodia/podosomes (Mizutani et al. 2002) and enhances cell movement (Kowalski et al. 2005). Collectively these studies suggest cortactin and N-WASp are intimately intertwined in regulating Arp2/3 activity responsible for cortical actin assembly.
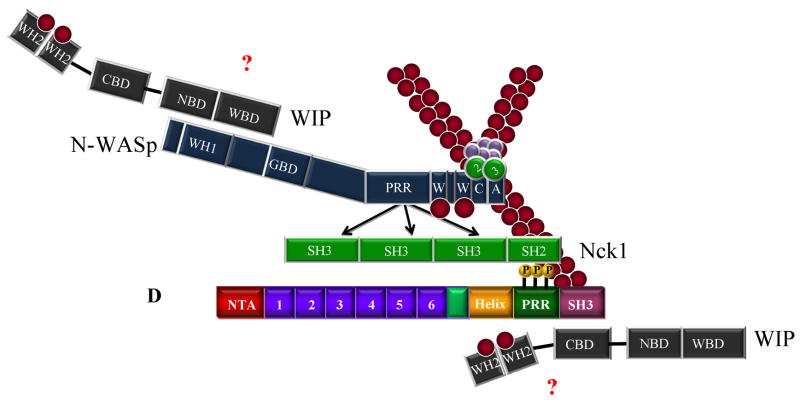
WASP-interacting protein (WIP) also binds to the SH3 domain of cortactin and enhances cortactin-mediated Arp2/3 activation (Kinley et al. 2003) (Fig. 2C, D). Similar to other SH3 binding partners, localization of WIP to the cell cortex area is dependent upon the cortactin binding domain (Kinley et al. 2003). Maximal Arp2/3 activity is achieved when the cortactin-WIP complex is associated with actin filaments, suggesting a novel role for cortactin in linking WIP to pre-existing filaments. WIP also inhibits the depolymerization of actin filaments (Martinez-Quiles et al. 2001), resulting in the stabilization of actin branch points and inhibition of filament depolymerization.
Characterization of cortactin binding to F-actin
Cortactin was initially identified as an F-actin binding protein in cosedimentation assays using recombinant truncation mutants (Wu and Parsons 1993). Topical mapping analysis determined that the repeat region was responsible for F-actin binding, with a binding stoichiometry of one cortactin molecule per 14 actin monomer subunits. Deletion mapping of the cortactin repeats region indicates that the presence of the fourth repeat is required for optimal F-actin binding (Weed et al. 2000). This finding is supported by F-actin binding studies with different naturally-occurring cortactin splice variants isolated from human squamous cell carcinoma cell lines (van Rossum et al. 2003). Alternate splicing of the cortactin transcript results in the loss of the 6th ABR (termed splice variant 1; SV1-cortactin) or the loss of the 5th and 6th ABR (SV2-cortactin). Both splice variants retain the fourth repeat and display similar F-actin binding activities as compared to wild-type cortactin. In addition, these splice variants retain the ability to activate Arp2/3-mediated actin nucleation, although the SV2-cortactin variant is not as effective as SV1-cortactin. Collectively these studies support the importance of the fourth cortactin repeat in maintaining the actin binding activity of the protein. While other studies have shown cortactin splice variants lacking the fourth repeat are capable of binding F-actin, these isoforms are not as abundant as wild type, SV-1 or SV-2 cortactin (Katsube et al. 2004).
To determine which residues within actin filaments interact with cortactin, Pant et al. (Pant et al. 2006) performed a 3D reconstruction analysis from electron micrographs of actin filaments decorated with a recombinant protein encompassing the cortactin repeats region. Their results indicate that cortactin likely binds to the actin filament near amino acids 338–348 in an individual actin subunit, a known binding region for the F-actin associated proteins gelsolin (McLaughlin et al. 1993), vitamin D binding protein (DBP) (Otterbein et al. 2001), and ADF/cofilin (reviewed in (Dominguez 2004)). Binding of the cortactin repeats region to this area deepens the cleft between adjacent actin monomers. The authors hypothesize that this leads to a transient instability within the helix, allowing for the binding of other proteins to the filament, (i.e.; Arp2/3 complex). In addition, cortactin preferentially binds to F-actin filaments containing ATP or ATP/ADP-Pi, demonstrating a higher affinity for newly polymerized actin filaments as opposed to older ADP-loaded filaments (Bryce et al. 2005). This is in agreement with the requirement for cortactin for maximal incorporation of fluorescently-labeled G-actin monomers within extending lamellipodia (Bryce et al. 2005).
Detailed conformational information on cortactin has not been available due in part to the lack of a published crystal structure. Recently, cortactin was analyzed by a combined approach utilizing chemical cross-linking, circular dichroism and small angle X-ray scattering (Cowieson et al. 2008). This study revealed that recombinant cortactin assumes a globular “lollipop” shape in solution, with the helical domain residing next to the actin-binding region. This conformation results in a folding back of the C-terminal region, placing the second actin repeat in close proximity to the fifth repeat, and SH3 domain near the helical domain and the second repeat. The conformation deduced from these studies has important ramifications regarding how cortactin can interact with F-actin, since binding to F-actin does not induce changes in cortactin secondary structure (Cowieson et al. 2008). Actin binding studies at low temperature reveal that cortactin can assemble F-actin into sheets ~ one filament width wide, thereby bundling F-actin into parallel arrays. Whether or not this activity takes place in a cellular context remains to be evaluated.
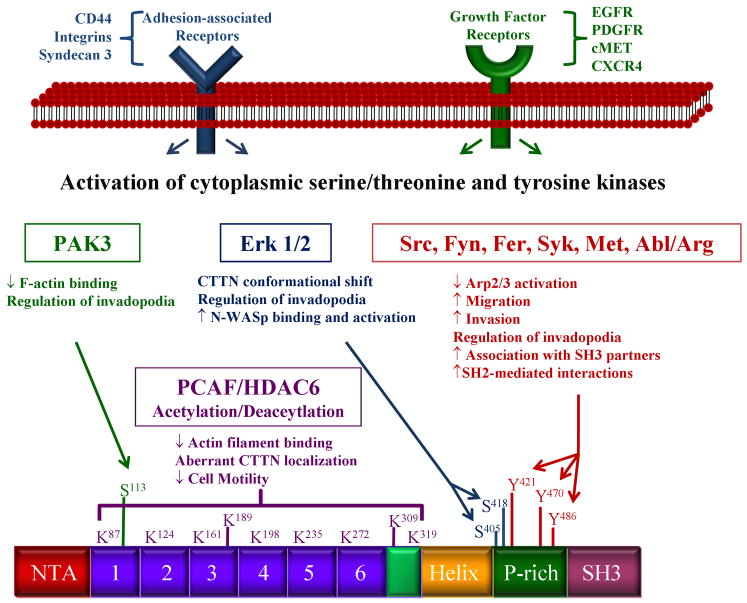
One regulatory mechanism recently identified that governs cortactin binding to F-actin involves the histone deacetylase HDAC6 (Zhang et al. 2007). Cortactin is a substrate for the histone acetyltransferase PCAF, which acetylates eleven lysine residues within the protein. It is not clear if PCAF is the only histone aceytltransferase that acetylates cortactin, as there is no significant difference in the acetylation status of cortactin from PCAF−/− fibroblasts as compared to controls (Zhang et al. 2007). Eight of the targeted lysines are located within the ABR of cortactin. These eight lysine residues are postulated to form two positively “charged patches” that facilitate the favorable interaction of cortactin with F-actin. Acetylation by PCAF neutralizes the lysine patches, inhibiting binding of the ABR to F-actin. Deacetylation by HDAC6 reverses this process and restores the ability to bind F-actin. The acetylation/deacetylation status of cortactin also has physiological consequences, as acetylated cortactin is unable to translocate to the cell periphery in a Rac-dependent manner, while deacetylated cortactin retains this ability. Cells containing acetylated cortactin also showed decreased cell migration, likely due to impaired cortactin binding to F-actin. The reversible acetylation status of cortactin provides a unique signaling mechanism that regulates F-actin binding activity.
The effects of cortactin phosphorylation on the actin cytoskeleton
The phosphorylation of cortactin and resulting functional consequences have been an intense area of study for many years, with several recent reports providing a better understanding with regard to actin dynamics and cellular behavior. In the case of tyrosine phosphorylation, initial mapping studies identified the major Src phosphorylation sites on murine cortactin as Y421, Y466 and Y482, with Y486 phosphorylated to a lesser extent (Huang et al. 1997a; Huang et al. 1998). Tyrosine 475 has also been identified as a phosphorylation site by mass spectroscopy, although the responsible kinase(s) have not been identified (Martin et al. 2006). Phosphorylation of Y421, Y466 and Y482 has known biochemical and cellular consequences with regards to actin dynamics. Tyrosine phosphorylation of cortactin at these sites is hypothesized to lead to a conformational change in the protein, resulting in more efficient cleavage by calpain proteases (Huang et al. 1997b; Perrin et al. 2006), possibly affecting the ability of cortactin to bind to and cross-link actin filaments. The majority of reports indicate that high levels of tyrosine phosphorylation correlate with elevated cell migration and cancer metastasis (Huang et al. 1998; Liu et al. 1999; Bourguignon et al. 2001; Li et al. 2001; Huang et al. 2003), although recent evidence indicates that cortactin tyrosine phosphorylation is inhibits motility in some tumor types (Jia et al. 2008). Cell type or context differences may play a role in explaining these discrepancies. Tyrosine phosphorylation can also regulate cortactin function by enhancing the binding of select SH3-ligands, including MLCK, CD2AP, and dynamin2 (Dudek et al. 2002; Lynch et al. 2003; Zhu et al. 2007). Cortactin was recently identified as a specific substrate for the tyrosine phosphatase PTP1B (Stuible et al. 2008). PTP1B binds to cortactin primarily at phosphorylated Y446, although surrounding residues may indirectly affect this interaction. EGFR activation leads to phosphorylation of Y446, as does hyperosmolarity, which activates Fyn and Fer (Kapus et al. 2000) resulting in phosphorylation of the Src-targeted sites Y421, Y470, Y486 in addition to Y446. Expression of PTP1B reduces Y421 and Y446 phosphorylation, and a Y-F mutation at codon 446 results in increased hyperosmotic apoptotic signaling, indicating that phosphorylation of Y446 is may be important for regulating apoptotic responses based on its phosphorylation status.
Several recent studies have examined the downstream cellular effects of cortactin tyrosine phosphorylation. Tyrosine phosphorylation of cortactin in osteoclasts is essential for the formation and turnover of podosomes and invadopodia, structures associated with invasive potential (Luxenburg et al. 2006; Tehrani et al. 2006; Ayala et al. 2008). Cortactin tyrosine phosphorylation is also required for efficient metastasis of cancer cells, where breast cancer cell lines expressing a tyrosine phosphorylation-deficient mutant of cortactin demonstrate a marked reduction of osteolytic metastases compared to control cells (Li et al. 2001). The phosphorylation status of cortactin also plays a role in centrosome separation at the G2-M phase in the cell cycle though its ability to link the actin cytoskeleton to the centrosome (Wang et al. 2008). Suprastimlation of pancreatic acini, used to mimic the disease state of pancreatitis, leads to an increase in cortactin tyrosine phosphorylation and the reorganization of the actin cytoskeleton, resulting in the formation of cortactin-rich membrane blebs (Singh and McNiven 2008). This process is dependent on cortactin tyrosine phosphorylation since expression of a phosphorylation-null mutant decreases actin reorganization and diminishes membrane bleb formation. Future work will likely uncover new roles for cortactin tyrosine phosphorylation in normal and pathogenic cellular processes.
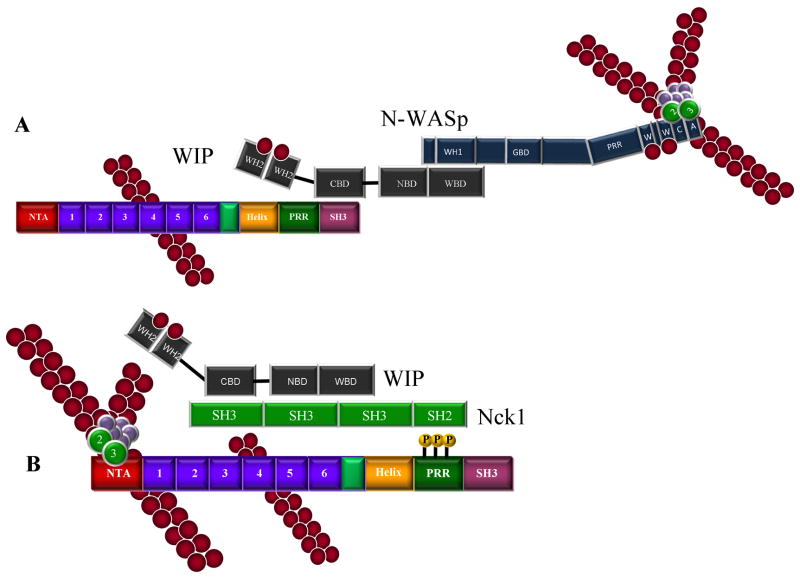
While cortactin phosphorylation plays a role in several discrete cellular functions, a link between tyrosine phosphorylation and Arp2/3 activation was initially dismissed since cortactin fragments lacking the α-helical, PRR and SH3 domains activate Arp2/3 in vitro nearly as effectively as the full-length protein (Weaver et al. 2001). However, additional work now points to a positive effect for Src-mediated cortactin phosphorylation on Arp2/3 mediated actin polymerization through Nck and N-WASp (Tehrani et al. 2007) (Fig. 2E–G). Nck1 binds to Src-phosphorylated cortactin through its SH2 domain, with the Nck1/phosphocortactin complex in turn interacting with either WIP (Fig. 2E) or N-WASp (Fig. 2F) through its SH3 domain. Trimeric cortactin/Nck1/N-WASp or /WIP complexes enhance Arp2/3 nucleation activity. Interestingly, when cortactin is the only NPF present (i.e., in conditions lacking N-WASp but retaining NCK and WIP), the Arp2/3-binding DDW motif within the cortactin NTA domain is essential for stimulating Arp2/3-mediated actin nucleation. Additionally, the SH3 domain of Src-phosphorylated cortactin is not required for Arp2/3 activation when Nck1 and N-WASP or WIP are present, suggesting that the Nck1 SH3 domain can perform in a surrogate manner to bring N-WASp or WIP to cortactin, resulting in Arp2/3 complex activation. The binding of Nck1 to phosphorylated cortactin therefore provides an indirect link to Arp2/3 complex regulation, as well as involving cortactin in an additional layer of complexity in regulating actin nucleation (Fig. 2G).
In addition to tyrosine phosphorylation, cortactin is also a substrate for several serine/ threonine kinases. Serine residues 405 and 418 were first identified to be phosphorylated downstream of MEK and can be directly phosphorylated by Erk1/2 (Campbell et al. 1999; Webb et al. 2006). Phosphorylation of S405 and 418 is associated with an 80 to 85kDa shift in Mr on SDS-PAGE, potentially due to the disruption of intramolecular interactions between the cortactin SH3 domain and amino-terminal residues (Campbell et al. 1999; Martinez-Quiles et al. 2004; Stuible et al. 2008). In support of this, phosphorylation of S405 and S418 corresponds with increased N-WASp binding and activation of Arp2/3 actin nucleation (Martinez-Quiles et al. 2004). Interestingly, Src-mediated tyrosine phosphorylation inhibits the binding between cortactin and N-WASp in this setting, resulting in decreased Arp2/3 nucleation (reviewed in (Lua and Low 2005)). This may be due to the absence of Nck in these assays. Phosphorylation of S405 and S418 is also required for efficient invadopodia formation and extra cellular matrix degradation (Ayala et al. 2008).
In addition to chemical ligand stimulation, cortactin phosphorylation and translocation can also be affected by mechanical or osmotic stress. Endothelial cell monolayers exposed to shear stress demonstrate enhanced stress fiber formation and Rac-dependent cortactin translocation to the cortical actin network independent of cortactin tyrosine phosphorylation (Birukov et al. 2002). However, monolayer exposure to sphingosine 1-phosphate (S1P) increases barrier function in a cortactin-dependent fashion, as cortactin knock-down significantly inhibits S1P-mediated barrier enhancement (Dudek et al. 2004). Translocation and tyrosine phosphorylation of cortactin is also increased in response to S1P treatment, and is critical to regulating barrier function. In addition, the interaction between MLCK and cortactin also affects barrier function, as cortactin blocking peptides inhibit S1P-mediated effects on MLC phosphorylation and barrier enhancement (Dudek et al. 2004). In the case of osmotic stress, hyperosmolarity results in cell shrinkage, triggering the tyrosine phosphorylation of cortactin independent of changes in osmolarity or intracellular pH (Kapus et al. 1999). Cortactin phosphorylation under these conditions is mediated by the Src family member Fyn. Rac-dependent cortactin translocation in response to hypertonicity is independent of shrinkage-induced phosphorylation, localizing cortactin and Arp2/3 to areas of active cortical actin reorganization known to occur in response to osmotic stress (Di Ciano et al. 2002).
Separate from exogenous stimuli, cortactin phosphorylation can also be effected by the polymerization status of actin (Fan et al. 2004). Depolymerization of actin by latrunculin B leads to enhanced tyrosine phosphorylation of cortactin mediated by Fer. Alternately, filament stabilization by jasplakinolide blocks cortactin phosphorylation induced by either hypertonic stress or treatment with latrunculin B. Stabilization results in the redistribution of cortactin to F-actin rich patches. In this way, the polymerization status of the F-actin network may regulate the actions of cortactin by altering its phosphorylation status, resulting in a feedback loop alternating between network expansion and reorganization (Gunst 2004).
Recently, the Cell Migration Consortium reported a comprehensive mass spectroscopy analysis in an attempt to identify all cortactin phosphorylation sites (Martin, et al 2006). In addition to the sites discussed above, sixteen additional phosphorylation sites were identified, twelve occurring on serine and four on threonine. Of these sites, serine 113 was independently identified as a PAK phosphorylation site, serving to downregulate F-actin binding to cortactin (Webb, et al 2006). Another intreuging phosphorylation site identified from this analysis is threonine 24, which is directly adjacent to the DDW motif in the NTA region (Martin, et al 2006). This raises the possibility that phosphorylation of this site may play a role in the regulation of Arp2/3 binding to the cortactin NTA. Future work will likely address the role of T24 phosphorylation and associated kinase(s) in cortactin function, as well as how other cortactin phosphorylation sites regulate cortactin activity.
Additional cortactin binding proteins and their role in actin organization
Given its involvement in multiple actin-based cellular processes, cortactin is capable of exerting differential effects on cortical actin cytoskeletal organization and assembly. In addition to the proteins discussed above, the ability of cortactin to alternatively regulate cortical actin dynamics stems from its direct association with additional actin-associated proteins. These interactions provide further insights into how cortactin regulates actin polymerization and organization.
Missing in metastasis (MIM)
MIM is a putative metastatic suppressor protein that is expressed at low levels in tumor cell lines (Lee et al. 2002). MIM contains a proline-rich region that mediates its binding to the cortactin SH3 domain. (Lin et al. 2005). Binding of MIM to cortactin enhances the ability of cortactin to stimulate Arp2/3 mediated actin polymerization, and decreases actin polymerization by N-WASp. However, at high concentrations MIM functions in a dominant negative fashion and inhibits the ability of cortactin to stimulate actin nucleation, possibly due to MIM sequestering G-actin through its WH2 domain (Mattila et al. 2003; Woodings et al. 2003; Woodring et al. 2003). The ability of MIM to bind to G-actin is also responsible for the inhibition of N-WASp-mediated Arp2/3 activation in the same system (Lin et al. 2005). MIM binds F-actin (Yamagishi et al. 2004), which may enhance cortactin-mediated Arp2/3 activation by placing MIM at the site of active polymerization, allowing MIM to recruit and bind to other key proteins (Lin et al. 2005). MIM proteins also contain an IRSp53 MIM domain (IMD) that shows structural similarities to the Bin-Amphiphysin-RSV (BAR) domain that plays a role in both sensing and inducing membrane curvature (Millard et al. 2005). Similar to other BAR-containing proteins, MIM is able to bind to and deform negatively-charged membranes composed of either phosphatidylinositol 4,5 bisphosphate (PIP2) or phosphatidylserine (Suetsugu et al. 2006; Mattila et al. 2007). The IMD of MIM may function to link the actin cytoskeleton and the plasma membrane by inducing outward protrusions of the plasma membrane similar to filopodia (Machesky and Johnston 2007). However, MIM proteins appear to play a role in lamellipodia, not filopodia formation (Bompard et al. 2005; Machesky and Johnston 2007). While it has not been identified as a component in invadopodia to date, the binding of MIM to cortactin may position MIM to play a role in membrane protrusion associated with invadopodia.
Dynamin-2
Dynamin-2 is a large GTPase that performs fundamental roles in vesicle formation, trafficking, and invadopodia formation (Urrutia et al. 1997; McNiven 1998; van der Bliek 1999; McNiven et al. 2000; Weaver 2006). The proline-rich region of dynamin-2 binds to the cortactin SH3 domain, and both proteins colocalize at the lamellipodial edge and within invadopodia (McNiven et al. 2000; McNiven et al. 2004; Zhu et al. 2005). Dynamin-2 binding to cortactin enhances Arp2/3-mediated actin polymerization to a moderate degree, and influences the architecture of resultant Arp2/3-F-actin networks (Schafer et al. 2002). The interaction between cortactin and dynamin is also essential for CDR and invadopodia formation (Baldassarre et al. 2003; Krueger et al. 2003). Cortactin and dynamin play a central role in receptor-mediated endocytosis, with efficient clathrin-dependent endocytosis requiring the concerted activities of the Arp2/3 complex, dynamin, and cortactin (McNiven et al. 2000; Cao et al. 2003; Zhu et al. 2005). Expression of a cortactin mutant deficient in Arp2/3 binding decreases the interaction between cortactin and dynamin as well as blocking transferrin uptake (Zhu et al. 2005). The cortactin/dynamin complex is essential for vesicle formation at the plasma membrane (Cao et al. 2003), and binding is enhanced by the Src-mediated tyrosine phosphorylation of cortactin (Zhu et al. 2007). These studies indicate that the interaction between dynamin-2 and cortactin plays an important role in multiple actin-based processes that involve movement and remodeling of cellular membranes.
p120 Catenin
p120 catenin (Ctn) is an armadillo-family protein and a Src substrate involved in cell-cell junction maintenance, transcriptional regulation and cell migration (Sarrio et al. 2004; Shibata et al. 2004; Bellovin et al. 2005; Yanagisawa and Anastasiadis 2006; Daniel 2007; Reynolds 2007). p120 Ctn binds to the amino-terminal portion of cortactin encompassing the NTA and cortactin repeats region (Boguslavsky et al. 2007). The binding of p120 Ctn to cortactin regulates lamellipodia persistence, as MCF-7 cells with reduced p120 Ctn expression display defective persistence that is phenotypically similar to cells with reduced cortactin expression (Bryce et al. 2005). While the loss of p120 Ctn expression leads to a reduction in the amount of cortactin and Arp2/3 from the leading edge, loss of cortactin expression does not affect the localization pattern of p120 Ctn (Boguslavsky et al. 2007). These data indicate that p120 Ctn may serve to sequester and stabilize cortactin within lamellipodia, allowing for subsequent cortactin-based regulation of lamellipodia dynamics.
Alix
Alix, (AIP1 or Hp95), is a conserved F- and G-actin binding adaptor protein that is associated with regulation of fibroblast morphology (Wu et al. 2002; Pan et al. 2006). Reduced Alix expression due to siRNA silencing results in decreased cellular F-actin and fewer stress fibers. In accordance with the effects on actin organization, cells lacking Alix expression contain fewer membrane protrusions and display an altered morphology (Pan et al. 2006). Alix binds to cortactin through a central region containing tandem binding sites (Pan et al. 2006), positioning Alix to function in F-actin filament bundling and stress fiber formation. Loss of Alix expression results in decreased cortactin localization within lamellipodia and a reduced association of cortactin with F-actin (Pan et al. 2006). Although these data suggest that Alix may play a role in regulating cortactin function within lamellipodia, how the interaction between these two proteins function together to influence actin dynamics is still not clear.
Caldesmon
Caldesmon is an F-actin-binding protein well characterized in smooth muscle that is also present in non-muscle cells (reviewed in (Hodgkinson 2000; Kordowska et al. 2006)). Caldesmon binds to the cortactin NTA region, resulting in attenuated Arp2/3-mediated actin nucleation (Huang et al. 2006). However, the actual biological significance and potential downstream effects of the caldesmon/cortactin interaction remains unknown. In non-dividing rat aortic fibroblasts (RAF) cells, caldesmon binds and stabilizes actin filaments (Kordowska et al. 2006). Upon cell division, caldesmon colocalizes with cortactin at the cell periphery where it is subsequently phosphorylated. Phosphorylation of caldesmon functions to decrease the binding affinity to F-actin, facilitating efficient actin cytoskeletal remodeling required for cell division and migration. Likewise, expression of a non-phosphorylatable caldesmon mutant results in the inability of RAFs to disassemble large actin bundles when induced to migrate by phorbol 12-myristate 13-acetate (PMA) (Kordowska et al. 2006). The differential binding of caldesmon to actin upon phosphorylation, coupled with its interaction with cortactin, positions these proteins to play an essential role in regulating actin cytoskeletal remodeling in response to mitotic signals.
Hip1R
Hip1R is an F-actin and clathrin binding protein that functions in receptor-mediated endocytosis (Engqvist-Goldstein et al. 1999; Engqvist-Goldstein et al. 2001; Cao et al. 2003; Engqvist-Goldstein et al. 2004; Veiga and Cossart 2005). Hip1R contains a conserved talin-HIP1/R/Sla2p actin-tethering C-terminal homology (THATCH) domain that mediates binding to F- or G-actin. Adjacent to the THATCH domain is a proline-rich region (PRR) that interacts with the SH3 domain of cortactin, positioning Hip1R to regulate actin polymerization dynamics during endocytic events (Brett et al. 2006; Le Clainche et al. 2007). In accordance with this, recent findings demonstrate that Hip1R expression negatively regulates actin assembly (Kaksonen et al. 2003; Engqvist-Goldstein et al. 2004), while a loss of Hip1R promotes cortactin-mediated actin polymerization at sites of clathrin coated pit formation (Le Clainche et al. 2007). Expression of a Hip1R construct defective in cortactin binding results in the formation of abnormal F-actin structures. In vitro actin assembly assays lacking Arp2/3 indicate that Hip1R and cortactin in combination inhibit actin assembly, while either protein alone has no effect. The inhibition of actin assembly is attributed to blocked barbed end filament elongation, similar to the effects of capping protein. These studies suggest that binding of Hip1R to cortactin may function as a filament barbed end cap at sites of endocytic vesicle formation, limiting filament elongation and facilitating endocytic vesicle internalization (Le Clainche et al. 2007).
Cortactin function in the formation of actin-based protrusive structures
The localization of cortactin with the cortical actin cytoskeleton combined with its role in regulating actin polymerization positions it to play a governing role in a variety of cellular events involving actin-based protrusion of the cell membrane. Consequently, the contribution of cortactin in the formation and function of lamellipodia, invadopodia and CDRs has been the subject of much recent investigation, leading to a better understanding about the molecular events and signaling pathways that regulate these structures.
Lamellipodia formation and adhesion
Regulation of the actin network during lamellipodia extension is one of the best characterized cellular actin-based processes. Lamellipodia are thin, flat membrane extensions containing a polarized array of actin filaments that form an orthogonal cross-linked actin network due to the distinctive nucleation signature of the Arp2/3-complex activity (Small 1988; Ponti et al. 2004). Directly behind the lamellipodium is a thicker, actin- and myosin II-rich region termed the lamella (Ponti et al. 2004; Gupton et al. 2005). This region separates the leading edge from the cell body, and is the site where integrin-rich cell-substrate adhesions first couple to the actomyosin contractile network to transmit translocating force (Ponti et al. 2004). The orthogonal arrangement of actin filaments allows filaments to move laterally across the cell front as actin polymerization progresses (Small 1994; Small and Resch 2005). The lamellipodium also initiates new sites of adhesion with the substratum, providing points of traction required for forward motility to occur (Small et al. 2002). Several proteins regulate the formation and maturation of lamellipodia; most notable are Rho, Rac, and Cdc42, members of the Rho family of GTPases (Ridley and Hall 1992; Hall 1998; Hall 2005). Rho regulates the formation of actin-myosin filaments and stress fiber assembly, while Rac and Cdc42 regulate the formation of lamellipodia and filopodia, respectively. All three GTPases contribute to the development of integrin-based focal contact formation, a process that is essential for migration (reviewed in (Raftopoulou and Hall 2004)).
Lamellipodia protrusion is initiated by actin filament elongation, a process which requires the production of free barbed ends. Free barbed ends within lamellipodia are generated by cofilin-mediated filament severing, uncapping of existing filament ends, or de novo nucleation by Arp2/3 (Condeelis 2001; Zigmond et al. 2003). Actin monomer addition to free barbed ends generates the protrusive force that pushes the membrane forward. Following extension, lamellipodia must adhere to the substratum in order to be functional in initiating and maintaining motility. Newly formed adhesions within the lamellipodia induced by Rac are termed focal “contacts”, while the more stable focal adhesions in the lamella and cell body are controlled by Rho activity (Ridley et al. 2003). Linkage of the lamellipodia to the substrate through focal contacts allows the cell to generate the traction force necessary for cell translocation. In the absence of adhesion, lamellipodia can lift and fold back towards the cell body, generating what have been classically termed “membrane ruffles“.
Recently, it has been shown that cortactin plays a role in regulating both actin dynamics and the formation of adhesive structures within the lamellipodia. Despite the ability of cortactin to influence Arp2/3 activity and actin network stability, cortactin does not appear to play a role in the initial protrusion of lamellipodia, since cortactin depletion from cells by siRNA does not affect the overall rate of protrusion (Bryce et al. 2005; Kempiak et al. 2005; Yamaguchi and Condeelis 2007). Cortactin is involved in actin network assembly and formation of lamellae-associated adhesion structures subsequent to initial lamellipod extension. Suppression of cortactin expression by siRNA in fibrosarcoma cells also reduces the rate of new adhesion formation in lamellipodia, as measured by altered paxillin dynamics in nascent focal contacts, leading to decreased lamellipodia persistence (Bryce et al. 2005) Similarly, Boguslavsky et al. (Boguslavsky et al. 2007) reported that siRNA-mediated reduction of cortactin expression in MCF-7 cells reduces lamellipodial persistence, mirroring the phenotype of cells lacking p120 catenin expression. siRNA-mediated depletion of cortactin also appears to impair cell spreading on fibronectin (Illes et al. 2006).
In contrast to these studies, several other groups have determined that cortactin depletion by siRNA has opposing effects on lamellipodia behavior. In one study, reduced cortactin expression in MTLn3 rat adenocarcinoma cells results in enhanced lamellipodia formation in MTLn3 cells exposed to EGF-coated beads (Kempiak et al. 2005). Stable down-regulation of cortactin expression in breast epithelial cells results in decreased cell migration and invasion, enhanced cell spreading, and elevated cell-cell adhesion (van Rossum et al. 2006). Finally, we have found that cortactin depletion in MTLn3 and 1483 head and neck squamous cell carcinoma cells results in impaired lamellipodia retraction following stimulation with soluble EGF and alters the lamellipodia F-actin architecture1. The apparent differences in observed effects between these studies may be due to a variety of factors, including cell type, matrix involved, and chemotatic stimulus. While potentially conflicting, these studies do highlight a putative functional role for cortactin in matrix adhesion and lamellipodial stability. Expansion of these studies with further investigation will help delineate a more precise role for cortactin function in regulating lamellipodia dynamics.
Additional evidence for cortactin in lamellipodia adhesion can be inferred from work demonstrating direct binding of Arp2/3 complex to vinculin, a protein found in focal contacts that targets Arp2/3 to sites of matrix adhesion (DeMali et al. 2002). The interaction between Arp2/3 and vinculin raises the possibility that a cortactin/Arp2/3/vinculin ternary complex may be present within newly forming focal contacts, providing a simultaneous means to regulate Arp2/3 activation and actin network stability coupled to nascent adhesion formation. Additionally, FAK binds directly to Arp3, and this interaction enhances Arp2/3-mediated actin polymerization required for the formation of nacent lamellipodia and subsequent cell spreading (Serrels et al. 2007). The release of Arp3 from FAK at the periphery of the cell places the Arp2/3 complex away from more mature adhesion structures and is associated with the formation of lamellipodia from initial transient adhesion structures (Serrels et al. 2007). FAK can also bind to and phosphorylate N-WASP (Wu et al. 2004), suggesting that FAK may be responsible for the correct positioning of multiple components of the actin polymerization machinery required for efficient lamellipodia formation.
In addition to a potential role in adhesion, other work has shown that cortactin binds directly to endothelial cell (EC) MLCK, inhibiting cortactin-mediated Arp2/3 actin polymerizing activity and reducing the affinity of MLCK for F-actin (Dudek et al. 2002). MLCK and cortactin undergo retrograde flow from the cell periphery towards the lamellar region (Kaksonen et al. 2000; Giannone et al. 2004), where myosin-dependent contractile complexes are required for lamellipodia formation and adhesion (reviewed in (Small and Resch 2005)). Therefore, an additional function for cortactin is that it may serve to chaperone MLCK to distinct areas within the lamella, aiding to promote MLCK activation required for the myosin-dependent contractile force utilized in loco-regional focal contact assembly. While experimental validation for this is needed in a cellular context, evidence to date suggests that cortactin has the capability to link actin polymerization and lamellipodia formation to adhesion and force generation, thereby influencing critical actin-based processes essential for leading edge protrusion and directional cell migration.
Formation and function of invadopodia
While the precise role(s) of cortactin in lamellipodia and adhesion formation remains somewhat unclear, it is well established that cortactin is critical for the formation and function of invadopodia. Invadopodia are ventral membrane protrusions that extend into the extracellular matrix and facilitate cellular invasion by focally sequestering membrane-bound matrix metalloproteases. Invadopodia also function as sites of exocytosis for secreted matrix metalloproteases, thus serving a dual role in directing enzymatic digestion of the ECM (reviewed in (Weaver 2006)). Invadopodia and similar structures (alternatively termed podosomes, invadosomes, invadopods or ipods) are present in several cell types, including osteoclasts, macrophages, vascular smooth muscle cells and metastatic tumor cells (Tarone et al. 1985; Linder et al. 1999; Fultz et al. 2000; Hai et al. 2002; Gimona et al. 2003; Linder and Aepfelbacher 2003; Buccione et al. 2004; Gimona and Buccione 2006; Clark et al. 2007). Invadopodia are characterized by several distinctive properties that separate them from other actin-based protrusive structures. These include a reliance on Src kinase activity for their formation, the assembly of focal adhesion proteins in close proximity to actin branching machinery, and the localized degradation of ECM at the site of development (reviewed in (Weaver 2006)). siRNA-mediated knockdown of cortactin in Src-transformed fibroblasts, breast, head and neck and melanoma cancer cells prevents the formation of invadopodia and subsequent ECM degradation (Artym et al. 2006; Clark et al. 2007; Webb et al. 2007; Ayala et al. 2008). Other actin-regulatory proteins required for invadopodia formation and function reported to date include Arp2/3 complex (Yamaguchi et al. 2005), N-WASp (Mizutani et al. 2002; Yamaguchi et al. 2005), cofilin (Yamaguchi et al. 2005) and dynamin2 (McNiven et al. 2004).
Several recent studies have analyzed the molecular events required for invadopodia formation and activity. A step-wise model explaining the sequence of events involved in invadopodia formation in breast carcinoma cells derived from live cell imaging has been reported (Artym et al. 2006). Based on this work, actin and cortactin initially accumulate at areas where the cell membrane is in contact with the ECM, with cortactin potentially serving as an adaptor to recruit other essential proteins to sites of formation. After this initiation step, recruitment of the membrane-bound matrix metalloproteinase MT1-MMP to the initiation sites determines a “preinvadopodia” stage. Subsequent degradation of the ECM by MT1-MMP and other proteases marks the onset of invadopodia maturation, with mature invadopodia enriched in cortactin, actin, and MT1-MMP. A final late stage of mature invadopodia is marked by the dissolution of cortactin and F-actin from mature invadopodia. These late stage invadopodia still retain MT1-MMP and continue to degrade matrix. Additional studies examining the role of cortactin and matrix metalloprotease secretion from invadopodia (Clark et al. 2007) show a link between the expression level of cortactin and the amount of MMP-2 and MMP-9 release in HNSCC cells. Suppressed cortactin expression in these cells affects ECM degradation to a larger extent than can be reconciled to the focal action of invadopodia alone, with matrix degradation occurring at regions not intimately associated with the invadopodia interface. Overexpression of cortactin in these cells results in increased secretion of MMP-2 and MMP-9, while reduced cortactin expression correlates with decreased metalloprotease secretion. Cortactin protein levels also regulate the secretion of non-invadopodial protein ApoA1, suggesting that cortactin is involved in governing additional secretory pathways apart from metalloprotease delivery (Clark et al. 2007).
Recent work in v-Src transformed fibroblasts examined the specific cortactin domains responsible for invadopodia formation (Webb et al. 2007). Using deletion mutants of cortactin in transient transfection assays, the actin-binding region of cortactin is the sole determinant required for v-Src induced invadopodia formation and matrix degradation in cells where endogenous cortactin expression is suppressed by siRNA. However, Src-mediated cortactin phosphorylation is also involved, since expression of a construct containing tyrosine-phenylalanine mutations codons 421, 466 and 482 partially restores the invasive phenotype. Similar experiments in A375MM melanoma cells indicate that the cortactin NTA and SH3 domain are required for invadopodia formation (Ayala et al. 2008). This same study also demonstrated that multiple different phosphorylation sites on cortactin are responsible for optimal invadopodia matrix degradation activity, including tyrosine 421, 466 and 482 and serines 113, 405 and 418 (Ayala et al. 2008). These data indicate that cortactin function in invadopodia is highly regulated by multiple signaling pathways. A role for tyrosine phosphorylation of cortactin in invadopodia function is further supported by the correlation of tyrosine phosphorylated cortactin with matrix degradation (Bowden et al. 2006), possible reflecting a role in regulating podosome turnover similar to that reported in osteoclasts (Luxenburg et al. 2006).
Role in Circular Dorsal Ruffles
Circular dorsal ruffles (CDRs, or “dorsal waves”) are another protrusive membranous structure dependent on cortactin activity for proper formation. CDRs are dynamic actin-rich structures that consist of a ridge of extended dorsal plasma membrane and form along peripheral cell edges of migrating cells in response to growth factor stimulation ((Mellstroom et al. 1983; Mellstrom et al. 1988; Dowrick et al. 1993); reviewed in(Buccione et al. 2004)). Unlike peripheral lamellipodia-associated ruffles that typically exhibit continuous rapid membrane dynamics, CDRs form only once in resting cells several minutes after stimulation and persist for 5–20 minutes (Dowrick et al. 1993; Dharmawardhane et al. 1997). Functionally, CDRs are hypothesized to participate in cytoplasmic remodeling (Krueger et al. 2003), establishment of polarity in migrating cells, and internalization of activated receptor tyrosine kinases (Orth and McNiven 2003). CDRs also facilitate the development and formation of lamellipodia, as there is a high correlation between the development of CDRs and lamellipodia production following growth factor stimulation (Krueger et al. 2003). Localization of MMP-2 at the tips of CDRs suggests a potential additional role in matrix degradation during three-dimensional cell translocation (Suetsugu et al. 2003).
CDRs contain several of the same signaling and structural proteins that are found in lamellipodia and invadopodia. The main structural element of CDRs is F-actin, as treatment of cells with cytochalasin D or latrunculin A to disrupt assembly dynamics prevents CDR formation (Hedberg et al. 1993; Warn et al. 1993; Westphal et al. 2000). CDRs also contain Arp2/3 complex, indicating that nucleation of branched actin networks are involved in their formation (Krueger et al. 2003). N-WASp, WAVE-1 and -2, WIP, dynamin, Abl/Arg tyrosine kinases, and cortactin are also found within CDRs, providing an assemblage of signaling and actin regulatory proteins needed for actin-based CDR regulation (Westphal et al. 2000; Anton et al. 2003; Krueger et al. 2003; Suetsugu et al. 2003; Kovacs et al. 2006).
Cortactin plays an essential role in CDR formation. Disruption of the interaction between cortactin and dynamin using either an inhibitory monoclonal antibody against the Dyn2 PRR or a dominant-negative construct of Dyn2 that is missing the PRR domain alters the formation of CDRs by affecting cortactin-mediated activation of Arp2/3 (Schafer et al. 2002; Krueger et al. 2003). Cortactin is also able to influence CDR formation through phosphorylation by and binding to Abl and Arg, cytoplasmic tyrosine kinases that play an active role in actin regulation and CDR formation (McWhirter and Wang 1991; Van Etten et al. 1994; Woodring et al. 2003; Boyle et al. 2007). Abl/Arg phosphorylates cortactin at the Src-targeted Y421/Y466/Y482 sites, and mutation of these residues inhibits PDGF-induced CDR formation (Boyle et al. 2007). This requirement for cortactin tyrosine phosphorylation, coupled with the dependence for dynamic actin assembly in CDR formation, may suggest that phospho-tyrosine-mediated binding of NCK1/N-WASp or NCK1/WIP complexes to cortactin may serve as a fundamental step in the genesis of CDRs.
Concluding Remarks
Cortactin has emerged as an essential protein in regulation of the cortical actin cytoskeleton through its ability to influence cytoskeletal reorganization in response to extra- and intracellular stimuli. By interacting with a variety of binding partners, cortactin bridges the actin cytoskeleton to many effector proteins that control actin-based cellular processes. Although much of the recent focus on cortactin function has biochemically centered on its role in regulating Arp2/3 complex activity and F-actin interactions, how these functions tie into regulating individual or multicellular behavior remains to be comprehensively discerned. Several important questions, including roles for unstudied phosphorylation sites, additional post-translational modifications and potential participation in other actin-based events remain unevaluated. How cortactin impacts actin dynamics during vertebrate development is also unknown, hampered in part by the lack of transgenic knock-out rodent models. Precise roles of cortactin in tumor progression and other disease types are becoming rapidly expanded or just beginning to be identified. The field remains replete with opportunities for further exciting and important discoveries.
Figure 3.
Increased complexity and role of tyrosine phosphorylation in cortactin-mediated Arp2/3 complex activation. A. Activation of Arp2/3 complex by a cortactin/WIP/N-WASp complex. Binding of the cortactin SH3 domain to the CBD of WIP, coupled with the interaction of the WIP WASp binding domain (WBD) to the WH1 domain of N-WASp creates a trimeric complex capable of activating Arp2/3 actin nucleation activity. B. Tyrosine phosphorylation of cortactin influences Arp2/3 activation. The adaptor NCK binds cortactin through an SH2-phosphotyrosine mediated interaction. An SH3 domain of Nck1 in turn interacts with proline-rich regions on WIP, thereby positioning WIP to enhance Arp2/3 activation in an analogous manner to the positioning of WIP in Fig. 2C. C. N-WASP induced Arp2/3 activation mediated by an Nck1/cortactin complex. Nck1 binding to tyrosine phosphorylated cortactin as in B can also interact with and activate N-WASp through binding of one or more Nck1 SH3 domains binding to the proline rich region (PRR) of N-WASp, resulting in Arp2/3 activation. D. Multiprotein complexes of tyrosine phosphorylated cortactin, Nck1, N-WASp and WIP incorporating aspects of Fig 2 and 3 are conceivable, with the possible binding of WIP to the cortactin SH3 domain and/or N-WASp in addition to the Nck1/N-WASp complex described in C. In instances where N-WASp is present, cortactin appears to function as an adaptor rather than an NPF. Abbreviations: N-WASp-WH1: WASP-homology 1; GBD: GTPase-binding domain; PRR: proline-rich region; W: WASP-homology 2 domain (WH2); CA: Arp2/3 binding connector and acidic domain. WIP – CBD: cortactin binding domain; NBD: Nck binding domain; WBD: N-WASp binding domain. Nck1 – SH3: Src-homology 3 domain; SH2: Src-homology 2 domain. Cortactin – NTA: N-terminal acidic domain; Helix: alpha-helical region; P-rich: proline-rich region. For simplicity, actin filaments are shown where the Arp2/3 complex is located to demonstrate nucleation activity.
Figure 4.
Schematic illustration of membrane- and cytoplasmic-based signaling molecules and their post-translational modifications on cortactin function. Kinase-based signaling cascades initiated by adhesion or growth factor receptors are mediated by select serine/threonine and tyrosine kinases that serve to modify cortactin at the indicated amino acids. Cortactin function is also regulated by a cycle of acetylation/deacetylation. Functional consequences of each specific modification are indicated.
Table 1. Summary of cortactin binding proteins that regulate actin cytoskeletal dynamics.
Summary of cortactin-binding proteins that interact directlvy with the actin cytoskeleton, including binding characteristics to cortactin and actin, as well as functional and physiological effects.
| Protein | Cttn interaction site | F or G actin binding | Function | Resulting effects on cortactin/F-actin | References |
|---|---|---|---|---|---|
| Alix | Undetermined | F | Regulation of fibroblast morphology | Involved in localization of cortactin to lamellipodia | (Wu et al. 2002; Pan et al. 2006) |
| Arp2/3 | NTA (aa 20–22) | F | de novo actin nucleation | Activation of Arp2/3-mediated actin nucleation | (Uruno et al. 2001; Weaver et al. 2001) |
| Non-muscle - Assembly and stabilization of microfilaments | |||||
| Caldesmon | NTA (aa 4–47)) | F | Muscle - with tropomyosin - Mediating factor for Ca+2 dependent inhibition of smooth muscle contraction | Attenuates Arp2/3-mediated actin polymerization | (Sobue et al. 1981; Huang et al. 2006) |
| CD2AP | SH3 | - | Scaffolding protein involved in endocytosis | Link between endocytic machinery and the actin cytoskeleton for trafficking of receptor-containing vesicles | (Lynch et al. 2003) |
| Shank2 | SH3 | - | Scaffolding protein | Potential link between the postsynaptic NMDA receptor-PSD-95 complex and the actin cytoskeleton | (Du et al. 1998; Naisbitt et al. 1999) |
| Dynamin 2 | SH3 | - | GTPase associated with endocytosis and cell migration | Vesicle formation at the plasma membrane and the trans-Golgi network | (McNiven et al. 2000; Cao et al. 2003; Cao et al. 2005; Kruchten and McNiven 2006) |
| Fgd1 | SH3 | - | Cdc42 GEF | Enhances cortactin-mediated actin polymerization through Arp2/3 activation | (Zheng et al. 1996; Hou et al. 2003; Kim et al. 2004) |
| HDAC6 | repeat region (aa 84–330) | - | Deacetylation of target proteins | Promotes cortactin association with F-actin | (Zhang et al. 2007) |
| HIP1R | SH3 | F | Negative regulator of actin assembly in clathrin- mediated endocytosis and clathrin-coated vesicle budding from TGN | Forms a complex with cortactin that blocks actin filament barbed end elongation | (Engqvist-Goldstein et al. 1999; Kaksonen et al. 2003; Engqvist-Goldstein et al. 2004; Le Clainche et al. 2007) |
| MIM | SH3 | G | Putative metastatic suppressor | Enhances cortactin-mediated actin polymerization through Arp2/3 activation | (Lee et al. 2002; Mattila et al. 2003; Lin et al. 2005) |
| Nck | pY421, pY466 and/or pY482 | - | Adaptor protein | Enhances Arp2/3-mediated actin polymerization | (Okamura and Resh 1995; Tehrani et al. 2007) |
| p120 Ctn | N-terminal region (aa 1–330) | - | Stabilization of cell-cell junctions | Decrease in lamellipodial extention and focal adhesion formation | (Boguslavsky et al. 2007) |
| N-WASp | SH3 | G/F | Activator of Arp2/3 complex | Enhances Arp2/3-mediated actin polymerization | (Suetsugu et al. 2001; Takenawa and Itoh 2001; Pollard and Borisy 2003; Takenawa and Suetsugu 2007) |
| PTP1B | Y446 | - | Tyrosine-specific phosphatase | Dephosphorylates cortactin on Y421 and Y446 | (Stuible et al. 2008) |
| WIP | SH3 | F and G | Regulation of actin polymerization | Enhances cortactin-mediated actin polymerization through Arp2/3 activation; stimulates membrane protrusion | (Kinley et al. 2003; Aspenstrom 2004; Anton and Jones 2006) |
| ZO-1 | SH3 | - | Scaffolding protein between transmembrane and cytoplasmic proteins | Regulation of chemotactic response | (Katsube et al. 2004; Lee et al. 2006) |
Acknowledgments
The authors thank the members of the Weed laboratory for critical reading of the manuscript, Cliff Martin and Joel George for technical support.
Contract grant sponsor: National Institutes of Health grants R01 DE014578 and P20 RR16440 (SAW)
Abbreviations used
- ABR
actin binding region
- ADF
Actin depolymerization factor
- Alix
ALG-2 interacting protein X
- Arp2/3
Actin related proteins 2 and 3
- BAR
Bin-Amphiphysin-RSV domain
- CDR
circular dorsal ruffle
- CP
Capping protein
- DBP
Vitamin D binding protein
- ECM
extracellular matrix
- EGF
Epidermal growth factor
- ERK
Extracellular signal Regulated Kinase
- FGF
Fibroblast growth factor
- HDAC6
Histone deacetylase 6
- Hip1R
Huntingtin-interacting protein 1 related
- HNSCC
head and neck squamous cell carcinoma
- IMD
IRSp53 MIM domain
- IRSp53
insulin receptor tyrosine kinase substrate p53
- LIMK
LIM domain kinase
- MEK
MAP kinase or ERK kinase
- MIM
missing in metastasis
- MLCK
myosin light chain kinase
- MMP
matrix metalloprotease
- MT1-MMP
Membrane type-1 matrix metalloproteinase
- NPF
nucleation promoting factor
- NTA
N-terminal acidic region
- p120 ctn
p120 catenin
- PAK
p21 activated kinase
- PCAF
p300/CBP-associated factor
- PDGF
Platelet-derived growth factor
- PIP2
phosphatidylinositol 4,5 bisphosphate
- PMA
phorbol 12-myristate 13-acetate
- PRR
proline-rich region
- S1P
Sphingosine 1-phosphate
- Scar
Suppressor of cyclic AMP receptor
- SH
Src-homology
- THATCH
talin-HIP1/R/Sla2p actin-tethering C-terminal homology
- WASP
Wiskott-Aldrich syndrome protein
- WAVE
WASP family Verprolin-homologous protein
- WCA
Wasp Homology-2 (WH2)-central-acidic region
- WIP
WASp-interacting protein
Footnotes
A.G. Ammer and S.A. Weed, unpublished data
References
- Anton IM, Jones GE. WIP: a multifunctional protein involved in actin cytoskeleton regulation. Eur J Cell Biol. 2006;85(3–4):295–304. doi: 10.1016/j.ejcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Anton IM, Saville SP, Byrne MJ, Curcio C, Ramesh N, Hartwig JH, Geha RS. WIP participates in actin reorganization and ruffle formation induced by PDGF. J Cell Sci. 2003;116(Pt 12):2443–51. doi: 10.1242/jcs.00433. [DOI] [PubMed] [Google Scholar]
- Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66(6):3034–43. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- Aspenstrom P. The mammalian verprolin homologue WIRE participates in receptor-mediated endocytosis and regulation of the actin filament system by distinct mechanisms. Exp Cell Res. 2004;298(2):485–98. doi: 10.1016/j.yexcr.2004.04.050. [DOI] [PubMed] [Google Scholar]
- Ayala I, Baldassarre M, Caldieri G, Buccione R. Invadopodia: a guided tour. Eur J Cell Biol. 2006;85(3–4):159–64. doi: 10.1016/j.ejcb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Ayala I, Baldassarre M, Giacchetti G, Caldieri G, Tete S, Luini A, Buccione R. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J Cell Sci. 2008;121(Pt 3):369–78. doi: 10.1242/jcs.008037. [DOI] [PubMed] [Google Scholar]
- Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11(8):620–5. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- Baldassarre M, Pompeo A, Beznoussenko G, Castaldi C, Cortellino S, McNiven MA, Luini A, Buccione R. Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol Biol Cell. 2003;14(3):1074–84. doi: 10.1091/mbc.E02-05-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Baum B, Kunda P. Actin nucleation: spire - actin nucleator in a class of its own. Curr Biol. 2005;15(8):R305–8. doi: 10.1016/j.cub.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bellovin DI, Bates RC, Muzikansky A, Rimm DL, Mercurio AM. Altered localization of p120 catenin during epithelial to mesenchymal transition of colon carcinoma is prognostic for aggressive disease. Cancer Res. 2005;65(23):10938–45. doi: 10.1158/0008-5472.CAN-05-1947. [DOI] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336(2):551–65. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Birukov KG, Birukova AA, Dudek SM, Verin AD, Crow MT, Zhan X, DePaola N, Garcia JG. Shear stress-mediated cytoskeletal remodeling and cortactin translocation in pulmonary endothelial cells. Am J Respir Cell Mol Biol. 2002;26(4):453–64. doi: 10.1165/ajrcmb.26.4.4725. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000a;404(6781):1007–11. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000b;10(20):1273–82. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Boguslavsky S, Grosheva I, Landau E, Shtutman M, Cohen M, Arnold K, Feinstein E, Geiger B, Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci U S A. 2007;104(26):10882–7. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118(Pt 22):5393–403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J Biol Chem. 2001;276(10):7327–36. doi: 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, Mueller SC. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312(8):1240–53. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Boyle SN, Michaud GA, Schweitzer B, Predki PF, Koleske AJ. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr Biol. 2007;17(5):445–51. doi: 10.1016/j.cub.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Brett TJ, Legendre-Guillemin V, McPherson PS, Fremont DH. Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat Struct Mol Biol. 2006;13(2):121–30. doi: 10.1038/nsmb1043. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15(14):1276–85. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5(8):647–57. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta. 2007;1775(2):263–73. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Campbell DH, Sutherland RL, Daly RJ. Signaling pathways and structural domains required for phosphorylation of EMS1/cortactin. Cancer Res. 1999;59(20):5376–85. [PubMed] [Google Scholar]
- Cao H, Orth JD, Chen J, Weller SG, Heuser JE, McNiven MA. Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol Cell Biol. 2003;23(6):2162–70. doi: 10.1128/MCB.23.6.2162-2170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7(5):483–92. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Ressad F, Pantaloni D. Control of actin dynamics in cell motility. Role of ADF/cofilin. J Biol Chem. 1999;274(48):33827–30. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Bamburg JR. Regulating actin-filament dynamics in vivo. Trends Biochem Sci. 2000;25(1):19–23. doi: 10.1016/s0968-0004(99)01511-x. [DOI] [PubMed] [Google Scholar]
- Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007;67(9):4227–35. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- Condeelis J. How is actin polymerization nucleated in vivo? Trends Cell Biol. 2001;11(7):288–93. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]
- Cosen-Binker LI, Kapus A. Cortactin: the gray eminence of the cytoskeleton. Physiology (Bethesda) 2006;21:352–61. doi: 10.1152/physiol.00012.2006. [DOI] [PubMed] [Google Scholar]
- Cowieson NP, King G, Cookson D, Ross I, Huber T, Hume DA, Kobe B, Martin JL. Cortactin adopts a globular conformation and bundles actin into sheets. J Biol Chem. 2008 doi: 10.1074/jbc.M708917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crostella L, Lidder S, Williams R, Skouteris GG. Hepatocyte Growth Factor/scatter factor-induces phosphorylation of cortactin in A431 cells in a Src kinase-independent manner. Oncogene. 2001;20(28):3735–45. doi: 10.1038/sj.onc.1204474. [DOI] [PubMed] [Google Scholar]
- Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382(Pt 1):13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 2007;1773(1):59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159(5):881–91. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhane S, Sanders LC, Martin SS, Daniels RH, Bokoch GM. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J Cell Biol. 1997;138(6):1265–78. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ciano C, Nie Z, Szaszi K, Lewis A, Uruno T, Zhan X, Rotstein OD, Mak A, Kapus A. Osmotic stress-induced remodeling of the cortical cytoskeleton. Am J Physiol Cell Physiol. 2002;283(3):C850–65. doi: 10.1152/ajpcell.00018.2002. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29(11):572–8. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Dowrick P, Kenworthy P, McCann B, Warn R. Circular ruffle formation and closure lead to macropinocytosis in hepatocyte growth factor/scatter factor-treated cells. Eur J Cell Biol. 1993;61(1):44–53. [PubMed] [Google Scholar]
- Dudek SM, Birukov KG, Zhan X, Garcia JG. Novel interaction of cortactin with endothelial cell myosin light chain kinase. Biochem Biophys Res Commun. 2002;298(4):511–9. doi: 10.1016/s0006-291x(02)02492-0. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Jacobson JR, Chiang ET, Birukov KG, Wang P, Zhan X, Garcia JG. Pulmonary endothelial cell barrier enhancement by sphingosine 1-phosphate: roles for cortactin and myosin light chain kinase. J Biol Chem. 2004;279(23):24692–700. doi: 10.1074/jbc.M313969200. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Kessels MM, Chopra VS, Hayden MR, Drubin DG. An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J Cell Biol. 1999;147(7):1503–18. doi: 10.1083/jcb.147.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154(6):1209–23. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15(4):1666–79. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Di Ciano-Oliveira C, Weed SA, Craig AW, Greer PA, Rotstein OD, Kapus A. Actin depolymerization-induced tyrosine phosphorylation of cortactin: the role of Fer kinase. Biochem J. 2004;380(Pt 2):581–91. doi: 10.1042/BJ20040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz ME, Li C, Geng W, Wright GL. Remodeling of the actin cytoskeleton in the contracting A7r5 smooth muscle cell. J Muscle Res Cell Motil. 2000;21(8):775–87. doi: 10.1023/a:1010396429297. [DOI] [PubMed] [Google Scholar]
- Gallet C, Rosa JP, Habib A, Lebret M, Levy-Toledano S, Maclouf J. Tyrosine phosphorylation of cortactin associated with Syk accompanies thromboxane analogue-induced platelet shape change. J Biol Chem. 1999;274(33):23610–6. doi: 10.1074/jbc.274.33.23610. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116(3):431–43. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- Gimona M, Buccione R. Adhesions that mediate invasion. Int J Biochem Cell Biol. 2006;38(11):1875–92. doi: 10.1016/j.biocel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Gimona M, Kaverina I, Resch GP, Vignal E, Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol Biol Cell. 2003;14(6):2482–91. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Kruchten AE, Chen J, McNiven MA. A dynamin-3 spliced variant modulates the actin/cortactin-dependent morphogenesis of dendritic spines. J Cell Sci. 2005;118(Pt 6):1279–90. doi: 10.1242/jcs.01711. [DOI] [PubMed] [Google Scholar]
- Gunst SJ. Actions by actin: reciprocal regulation of cortactin activity by tyrosine kinases and F-actin. Biochem J. 2004;380(Pt 2):e7–8. doi: 10.1042/BJ20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, et al. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168(4):619–31. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280(1):64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–14. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33(Pt 5):891–5. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Head JA, Jiang D, Li M, Zorn LJ, Schaefer EM, Parsons JT, Weed SA. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol Biol Cell. 2003;14(8):3216–29. doi: 10.1091/mbc.E02-11-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg KM, Bengtsson T, Safiejko-Mroczka B, Bell PB, Lindroth M. PDGF and neomycin induce similar changes in the actin cytoskeleton in human fibroblasts. Cell Motil Cytoskeleton. 1993;24(2):139–49. doi: 10.1002/cm.970240207. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem. 2001;70:649–76. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- Hodgkinson JL. Actin and the smooth muscle regulatory proteins: a structural perspective. J Muscle Res Cell Motil. 2000;21(2):115–30. doi: 10.1023/a:1005697301043. [DOI] [PubMed] [Google Scholar]
- Hou P, Estrada L, Kinley AW, Parsons JT, Vojtek AB, Gorski JL. Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum Mol Genet. 2003;12(16):1981–93. doi: 10.1093/hmg/ddg209. [DOI] [PubMed] [Google Scholar]
- Huang C, Liu J, Haudenschild CC, Zhan X. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J Biol Chem. 1998;273(40):25770–6. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997a;272(21):13911–5. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Huang C, Tandon NN, Greco NJ, Ni Y, Wang T, Zhan X. Proteolysis of platelet cortactin by calpain. J Biol Chem. 1997b;272(31):19248–52. doi: 10.1074/jbc.272.31.19248. [DOI] [PubMed] [Google Scholar]
- Huang J, Asawa T, Takato T, Sakai R. Cooperative roles of Fyn and cortactin in cell migration of metastatic murine melanoma. J Biol Chem. 2003;278(48):48367–76. doi: 10.1074/jbc.M308213200. [DOI] [PubMed] [Google Scholar]
- Huang R, Cao GJ, Guo H, Kordowska J, Albert Wang CL. Direct interaction between caldesmon and cortactin. Arch Biochem Biophys. 2006;456(2):175–82. doi: 10.1016/j.abb.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illes A, Enyedi B, Tamas P, Balazs A, Bogel G, Melinda Lukacs, Buday L. Cortactin is required for integrin-mediated cell spreading. Immunol Lett. 2006;104(1–2):124–30. doi: 10.1016/j.imlet.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Jia L, Uekita T, Sakai R. Hyperphosphorylated cortactin in cancer cells plays an inhibitory role in cell motility. Mol Cancer Res. 2008;6(4):654–62. doi: 10.1158/1541-7786.MCR-07-0220. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci 113 Pt. 2000;24:4421–6. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115(4):475–87. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kanner SB, Reynolds AB, Vines RR, Parsons JT. Monoclonal antibodies to individual tyrosine-phosphorylated protein substrates of oncogene-encoded tyrosine kinases. Proc Natl Acad Sci U S A. 1990;87(9):3328–32. doi: 10.1073/pnas.87.9.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapus A, Di Ciano C, Sun J, Zhan X, Kim L, Wong TW, Rotstein OD. Cell volume-dependent phosphorylation of proteins of the cortical cytoskeleton and cell-cell contact sites. The role of Fyn and FER kinases. J Biol Chem. 2000;275(41):32289–98. doi: 10.1074/jbc.M003172200. [DOI] [PubMed] [Google Scholar]
- Kapus A, Szaszi K, Sun J, Rizoli S, Rotstein OD. Cell shrinkage regulates Src kinases and induces tyrosine phosphorylation of cortactin, independent of the osmotic regulation of Na+/H+ exchangers. J Biol Chem. 1999;274(12):8093–102. doi: 10.1074/jbc.274.12.8093. [DOI] [PubMed] [Google Scholar]
- Katsube T, Togashi S, Hashimoto N, Ogiu T, Tsuji H. Filamentous actin binding ability of cortactin isoforms is responsible for their cell-cell junctional localization in epithelial cells. Arch Biochem Biophys. 2004;427(1):79–90. doi: 10.1016/j.abb.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281(15):10589–97. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempiak SJ, Yamaguchi H, Sarmiento C, Sidani M, Ghosh M, Eddy RJ, Desmarais V, Way M, Condeelis J, Segall JE. A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J Biol Chem. 2005;280(7):5836–42. doi: 10.1074/jbc.M410713200. [DOI] [PubMed] [Google Scholar]
- Kim K, Hou P, Gorski JL, Cooper JA. Effect of Fgd1 on cortactin in Arp2/3 complex-mediated actin assembly. Biochemistry. 2004;43(9):2422–7. doi: 10.1021/bi036173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinley AW, Weed SA, Weaver AM, Karginov AV, Bissonette E, Cooper JA, Parsons JT. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr Biol. 2003;13(5):384–93. doi: 10.1016/s0960-9822(03)00107-6. [DOI] [PubMed] [Google Scholar]
- Knoll B, Drescher U. Src family kinases are involved in EphA receptor-mediated retinal axon guidance. J Neurosci. 2004;24(28):6248–57. doi: 10.1523/JNEUROSCI.0985-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordowska J, Hetrick T, Adam LP, Wang CL. Phosphorylated l-caldesmon is involved in disassembly of actin stress fibers and postmitotic spreading. Exp Cell Res. 2006;312(2):95–110. doi: 10.1016/j.yexcr.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Kovacs EM, Makar RS, Gertler FB. Tuba stimulates intracellular N-WASP-dependent actin assembly. J Cell Sci. 2006;119(Pt 13):2715–26. doi: 10.1242/jcs.03005. [DOI] [PubMed] [Google Scholar]
- Kowalski JR, Egile C, Gil S, Snapper SB, Li R, Thomas SM. Cortactin regulates cell migration through activation of N-WASP. J Cell Sci. 2005;118(Pt 1):79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- Kruchten AE, McNiven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119(Pt 9):1683–90. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- Krueger EW, Orth JD, Cao H, McNiven MA. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol Biol Cell. 2003;14(3):1085–96. doi: 10.1091/mbc.E02-08-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388(6637):78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Pauly BS, Zhang CX, Engqvist-Goldstein AE, Cunningham K, Drubin DG. A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis. EMBO J. 2007;26(5):1199–210. doi: 10.1038/sj.emboj.7601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia. 2002;4(4):291–4. doi: 10.1038/sj.neo.7900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tondravi M, Liu J, Smith E, Haudenschild CC, Kaczmarek M, Zhan X. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 2001;61(18):6906–11. [PubMed] [Google Scholar]
- Lin J, Liu J, Wang Y, Zhu J, Zhou K, Smith N, Zhan X. Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24(12):2059–66. doi: 10.1038/sj.onc.1208412. [DOI] [PubMed] [Google Scholar]
- Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13(7):376–85. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci U S A. 1999;96(17):9648–53. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Huang C, Zhan X. Src is required for cell migration and shape changes induced by fibroblast growth factor 1. Oncogene. 1999;18(48):6700–6. doi: 10.1038/sj.onc.1203050. [DOI] [PubMed] [Google Scholar]
- Lua BL, Low BC. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 2005;579(3):577–85. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Parsons JT, Addadi L, Geiger B. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J Cell Sci. 2006;119(Pt 23):4878–88. doi: 10.1242/jcs.03271. [DOI] [PubMed] [Google Scholar]
- Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;278(24):21805–13. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Johnston SA. MIM: a multifunctional scaffold protein. J Mol Med. 2007;85(6):569–76. doi: 10.1007/s00109-007-0207-0. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96(7):3739–44. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3(1):76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Martin KH, Jeffery ED, Grigera PR, Shabanowitz J, Hunt DF, Parsons JT. Cortactin phosphorylation sites mapped by mass spectrometry. J Cell Sci. 2006;119(Pt 14):2851–3. doi: 10.1242/jcs.03034. [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24(12):5269–80. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Quiles N, Rohatgi R, Anton IM, Medina M, Saville SP, Miki H, Yamaguchi H, Takenawa T, Hartwig JH, Geha RS, et al. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat Cell Biol. 2001;3(5):484–91. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- Martinez MC, Ochiishi T, Majewski M, Kosik KS. Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J Cell Biol. 2003;162(1):99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176(7):953–64. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J Biol Chem. 2003;278(10):8452–9. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364(6439):685–92. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- McNiven MA. Dynamin: a molecular motor with pinchase action. Cell. 1998;94(2):151–4. doi: 10.1016/s0092-8674(00)81414-2. [DOI] [PubMed] [Google Scholar]
- McNiven MA, Baldassarre M, Buccione R. The role of dynamin in the assembly and function of podosomes and invadopodia. Front Biosci. 2004;9:1944–53. doi: 10.2741/1348. [DOI] [PubMed] [Google Scholar]
- McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol. 2000;151(1):187–98. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter JR, Wang JY. Activation of tyrosinase kinase and microfilament-binding functions of c-abl by bcr sequences in bcr/abl fusion proteins. Mol Cell Biol. 1991;11(3):1553–65. doi: 10.1128/mcb.11.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellstrom K, Heldin CH, Westermark B. Induction of circular membrane ruffling on human fibroblasts by platelet-derived growth factor. Exp Cell Res. 1988;177(2):347–59. doi: 10.1016/0014-4827(88)90468-5. [DOI] [PubMed] [Google Scholar]
- Mellstroom K, Hoglund AS, Nister M, Heldin CH, Westermark B, Lindberg U. The effect of platelet-derived growth factor on morphology and motility of human glial cells. J Muscle Res Cell Motil. 1983;4(5):589–609. doi: 10.1007/BF00712117. [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24(2):240–50. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Miki H, He H, Maruta H, Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62(3):669–74. [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95(11):6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108(2):233–46. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Okamura H, Resh MD. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270(44):26613–8. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15(1):31–9. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- Orth JD, McNiven MA. Get off my back! Rapid receptor internalization through circular dorsal ruffles. Cancer Res. 2006;66(23):11094–6. doi: 10.1158/0008-5472.CAN-06-3397. [DOI] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293(5530):708–11. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281(45):34640–50. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- Pant K, Chereau D, Hatch V, Dominguez R, Lehman W. Cortactin binding to F-actin revealed by electron microscopy and 3D reconstruction. J Mol Biol. 2006;359(4):840–7. doi: 10.1016/j.jmb.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Perrin BJ, Amann KJ, Huttenlocher A. Proteolysis of cortactin by calpain regulates membrane protrusion during cell migration. Mol Biol Cell. 2006;17(1):239–50. doi: 10.1091/mbc.E05-06-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–77. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305(5691):1782–6. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265(1):23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Reynolds AB. p120-catenin: Past and present. Biochim Biophys Acta. 2007;1773(1):2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16(10):522–9. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb Symp Quant Biol. 1992;57:661–71. doi: 10.1101/sqb.1992.057.01.072. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Sarrio D, Perez-Mies B, Hardisson D, Moreno-Bueno G, Suarez A, Cano A, Martin-Perez J, Gamallo C, Palacios J. Cytoplasmic localization of p120ctn and E-cadherin loss characterize lobular breast carcinoma from preinvasive to metastatic lesions. Oncogene. 2004;23(19):3272–83. doi: 10.1038/sj.onc.1207439. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12(21):1852–7. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuuring E, Verhoeven E, Litvinov S, Michalides RJ. The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol. 1993;13(5):2891–98. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrels B, Serrels A, Brunton VG, Holt M, McLean GW, Gray CH, Jones GE, Frame MC. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol. 2007;9(9):1046–56. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Sekine S, Kanai Y, Hirohashi S. Cytoplasmic p120ctn regulates the invasive phenotypes of E-cadherin-deficient breast cancer. Am J Pathol. 2004;164(6):2269–78. doi: 10.1016/S0002-9440(10)63783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VP, McNiven MA. Src-mediated Cortactin Phosphorylation Regulates Actin Localization and Injurious Blebbing in Acinar Cells. Mol Biol Cell. 2008;19(5):2339–47. doi: 10.1091/mbc.E07-11-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV. The actin cytoskeleton. Electron Microsc Rev. 1988;1(1):155–74. doi: 10.1016/s0892-0354(98)90010-7. [DOI] [PubMed] [Google Scholar]
- Small JV. Lamellipodia architecture: actin filament turnover and the lateral flow of actin filaments during motility. Semin Cell Biol. 1994;5(3):157–63. doi: 10.1006/scel.1994.1020. [DOI] [PubMed] [Google Scholar]
- Small JV, Resch GP. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr Opin Cell Biol. 2005;17(5):517–23. doi: 10.1016/j.ceb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12(3):112–20. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- Sobue K, Muramoto Y, Fujita M, Kakiuchi S. Purification of a calmodulin-binding protein from chicken gizzard that interacts with F-actin. Proc Natl Acad Sci U S A. 1981;78(9):5652–5. doi: 10.1073/pnas.78.9.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuible M, Dube N, Tremblay ML. PTP1B regulates cortactin tyrosine phosphorylation by targeting Y446. J Biol Chem. 2008 doi: 10.1074/jbc.M710534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Yamaguchi H, Obinata T, Takenawa T. Enhancement of branching efficiency by the actin filament-binding activity of N-WASP/WAVE2. J Cell Sci. 2001;114(Pt 24):4533–42. doi: 10.1242/jcs.114.24.4533. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281(46):35347–58. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5(4):595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145(5):1009–26. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Itoh T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim Biophys Acta. 2001;1533(3):190–206. doi: 10.1016/s1388-1981(01)00165-2. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8(1):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tarone G, Cirillo D, Giancotti FG, Comoglio PM, Marchisio PC. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp Cell Res. 1985;159(1):141–57. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Cortactin has an essential and specific role in osteoclast actin assembly. Mol Biol Cell. 2006;17(7):2882–95. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Src phosphorylation of cortactin enhances actin assembly. Proc Natl Acad Sci U S A. 2007;104(29):11933–8. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA. Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J Cell Biol. 1997;136(6):1165–8. doi: 10.1083/jcb.136.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci U S A. 1997;94(2):377–84. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T, Liu J, Li Y, Smith N, Zhan X. Sequential interaction of actin-related proteins 2 and 3 (Arp2/3) complex with neural Wiscott-Aldrich syndrome protein (N-WASP) and cortactin during branched actin filament network formation. J Biol Chem. 2003;278(28):26086–93. doi: 10.1074/jbc.M301997200. [DOI] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3(3):259–66. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9(3):96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- Van Etten RA, Jackson PK, Baltimore D, Sanders MC, Matsudaira PT, Janmey PA. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J Cell Biol. 1994;124(3):325–40. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum AG, de Graaf JH, Schuuring-Scholtes E, Kluin PM, Fan YX, Zhan X, Moolenaar WH, Schuuring E. Alternative splicing of the actin binding domain of human cortactin affects cell migration. J Biol Chem. 2003;278(46):45672–9. doi: 10.1074/jbc.M306688200. [DOI] [PubMed] [Google Scholar]
- van Rossum AG, Moolenaar WH, Schuuring E. Cortactin affects cell migration by regulating intercellular adhesion and cell spreading. Exp Cell Res. 2006;312(9):1658–70. doi: 10.1016/j.yexcr.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Veiga E, Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat Cell Biol. 2005;7(9):894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen L, Ding Y, Jin J, Liao K. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J Cell Sci. 2008;121(Pt 8):1334–43. doi: 10.1242/jcs.018176. [DOI] [PubMed] [Google Scholar]
- Warn R, Brown D, Dowrick P, Prescott A, Warn A. Cytoskeletal changes associated with cell motility. Symp Soc Exp Biol. 1993;47:325–38. [PubMed] [Google Scholar]
- Watanabe N, Higashida C. Formins: processive cappers of growing actin filaments. Exp Cell Res. 2004;301(1):16–22. doi: 10.1016/j.yexcr.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis. 2006;23(2):97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Cortactin in tumor invasiveness. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12(15):1270–8. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11(5):370–4. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Webb BA, Jia L, Eves R, Mak AS. Dissecting the functional domain requirements of cortactin in invadopodia formation. Eur J Cell Biol. 2007;86(4):189–206. doi: 10.1016/j.ejcb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Webb BA, Zhou S, Eves R, Shen L, Jia L, Mak AS. Phosphorylation of cortactin by p21-activated kinase. Arch Biochem Biophys. 2006;456(2):183–93. doi: 10.1016/j.abb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151(1):29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–88. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19(17):4589–600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B, Schafer DA. Cordon-bleu: a new taste in actin nucleation. Cell. 2007;131(2):236–8. doi: 10.1016/j.cell.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Woodings JA, Sharp SJ, Machesky LM. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem J. 2003;371(Pt 2):463–71. doi: 10.1042/BJ20021962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring PJ, Hunter T, Wang JY. Regulation of F-actin-dependent processes by the Abl family of tyrosine kinases. J Cell Sci. 2003;116(Pt 13):2613–26. doi: 10.1242/jcs.00622. [DOI] [PubMed] [Google Scholar]
- Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120(6):1417–26. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Reynolds AB, Kanner SB, Vines RR, Parsons JT. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11(10):5113–24. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL. Focal adhesion kinase regulation of N-WASP subcellular localization and function. J Biol Chem. 2004;279(10):9565–76. doi: 10.1074/jbc.M310739200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Pan S, Luo W, Lin SH, Kuang J. Hp95 promotes anoikis and inhibits tumorigenicity of HeLa cells. Oncogene. 2002;21(44):6801–8. doi: 10.1038/sj.onc.1205849. [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem. 2004;279(15):14929–36. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773(5):642–52. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168(3):441–52. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Anastasiadis PZ. p120 catenin is essential for mesenchymal cadherin-mediated regulation of cell motility and invasiveness. J Cell Biol. 2006;174(7):1087–96. doi: 10.1083/jcb.200605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Hu X, Hampton B, Burgess WH, Friesel R, Maciag T. Murine cortactin is phosphorylated in response to fibroblast growth factor-1 on tyrosine residues late in the G1 phase of the BALB/c 3T3 cell cycle. J Biol Chem. 1993;268(32):24427–31. [PubMed] [Google Scholar]
- Zhan X, Plourde C, Hu X, Friesel R, Maciag T. Association of fibroblast growth factor receptor-1 with c-Src correlates with association between c-Src and cortactin. J Biol Chem. 1994;269(32):20221–4. [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Fischer DJ, Santos MF, Tigyi G, Pasteris NG, Gorski JL, Xu Y. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271(52):33169–72. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yu D, Zeng XC, Zhou K, Zhan X. Receptor-mediated endocytosis involves tyrosine phosphorylation of cortactin. J Biol Chem. 2007;282(22):16086–94. doi: 10.1074/jbc.M701997200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhou K, Hao JJ, Liu J, Smith N, Zhan X. Regulation of cortactin/dynamin interaction by actin polymerization during the fission of clathrin-coated pits. J Cell Sci. 2005;118(Pt 4):807–17. doi: 10.1242/jcs.01668. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13(20):1820–3. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]