Abstract
The Trp53 gene family member Trp73 encodes two major groups of protein isoforms, TAp73 and ΔNp73, with opposing pro- and anti-apoptotic functions; consequently, their relative ratio regulates cell fate. However, the precise roles of p73 isoforms in cellular events such as tumor initiation, embryonic development, and cell death remain unclear. To determine which aspects of p73 function are attributable to the TAp73 isoforms, we generated and characterized mice in which exons encoding the TAp73 isoforms were specifically deleted to create a TAp73-deficient (TAp73−/−) mouse. Here we show that mice specifically lacking in TAp73 isoforms develop a phenotype intermediate between the phenotypes of Trp73−/− and Trp53−/− mice with respect to incidence of spontaneous and carcinogen-induced tumors, infertility, and aging, as well as hippocampal dysgenesis. In addition, cells from TAp73−/− mice exhibit genomic instability associated with enhanced aneuploidy, which may account for the increased incidence of spontaneous tumors observed in these mutants. Hence, TAp73 isoforms exert tumor-suppressive functions and indicate an emerging role for Trp73 in the maintenance of genomic stability.
Keywords: p73, tumor-prone phenotype, meiosis, infertility, genomic instability
p73 belongs to a small but important family of p53-related proteins (p53, p63, p73). These proteins, which are encoded by the Trp53, Trp63, and Trp73 genes, respectively, are all transcription factors involved in the regulation of development, cell death, proliferation, stem cell renewal, and cell fate commitment, as well as tumorigenesis (Yang et al. 2002; Vousden and Lane 2007). Like p53, several different protein isoforms of p63 and p73 have been reported, whose functions may compete with, synergize with, or be unrelated to those of p53 (Bourdon et al. 2005; Li and Prives 2007).
The Trp73 gene, discovered in 1997 (Kaghad et al. 1997), contains two promoters that drive the expression of two major groups of p73 isoforms with opposing cellular actions: The TAp73 isoforms contain the p73 transactivation domain (TA) and exhibit proapoptotic activities (Müller et al. 2005; Wang et al. 2007), whereas the ΔNp73 isoforms lacking the N-terminal TA domain are anti-apoptotic (Grob et al. 2001). Consequently, due to the complexity of the cross-talk between all p53 family members and the opposing functions of p73 isoforms harbored in the same gene, the specific role of Trp73 in various biological processes is still debated (McKeon and Melino 2007; Stiewe 2007).
Mouse gene targeting studies have revealed that both Trp63 and Trp73 are required for normal embryogenesis (Mills et al. 1999; Yang et al. 1999, 2000). In contrast, the majority of mice deficient in Trp53 are viable and appear normal, although ∼30% of p53-deficient females die in utero with neuronal defects (Donehower et al. 1992). In addition, Trp53−/− mice of both sexes are more susceptible than the wild type to the development of spontaneous tumors (Donehower et al. 1992). In comparison, studies of mice that are homozygous or heterozygous for a complete deletion of Trp73 have produced conflicting results. Indeed, while inactivation of all p73 isoforms in mice does not enhance susceptibility to spontaneous tumors (Yang et al. 2000), Trp73 haploinsufficiency contributes to an increased incidence of spontaneous tumors such as lung adenoma, lung adenocarcinoma, thymic lymphoma, squamous cell hyperplasia, or hemangiosarcoma, particularly when combined with Trp53 heterozygosity (Flores et al. 2005). However, this gene interaction appears to be tissue-specific as Trp73 does not contribute to Trp53 tumor suppressor activity during lymphomagenesis (Perez-Losada et al. 2005). It is likely that Trp73−/− mouse models do not show enhanced spontaneous tumor formation be due to shortened life span. Despite the fact that studies involving Trp73+/− revealed enhanced tumor formation, the Trp73−/− mouse model does not seem to fully mirror the molecular changes seen in human cancers, where an altered ratio of TAp73/ΔNp73 isoforms is often present rather than a total loss or mutation of p73 isoforms (Coates 2006). Thus, despite the usefulness of Trp73−/− mice, isoform-selective knockouts of Trp73 are needed to truly evaluate p73’s impact on tumorigenesis and other cellular processes.
The complexity of the overlapping, combined, and opposing functions of Trp53 family members and their protein isoforms suggests the existence of an intricate functional network (Deyoung and Ellisen 2007). Over the last few years, several studies have suggested specific roles for particular p73 isoforms (De Laurenzi et al. 1998; Murray-Zmijewski et al. 2006; Klanrit et al. 2008). Thus, these data indicate that the ratio of various p73 isoforms present in an individual cell determines the outcome of p73 function, rather than the sum of the activities of each isolated p73 isoform. To investigate this hypothesis and elucidate the biological role of TAp73 isoforms, we generated mice that are missing exons encoding the TA domain of p73 (TAp73−/− mice). These mutants are deficient only in TAp73 isoforms and remain capable of producing ΔNp73 isoforms causing a great imbalance in the TAp73/ΔNp73 ratio. We clearly demonstrate that TAp73 isoforms are tumor-suppressive, which may be a consequence of TAp73’s apparent role in the maintenance of genomic stability. In addition, we demonstrate that TAp73 is a novel maternal-lethal gene whose deficiency in mice causes infertility, and its loss mimics Trp73 deficiency causing hypocampal dysgenesis.
Results
Characterization of TAp73 deletion and its effects on the expression and activities of Trp53 family members
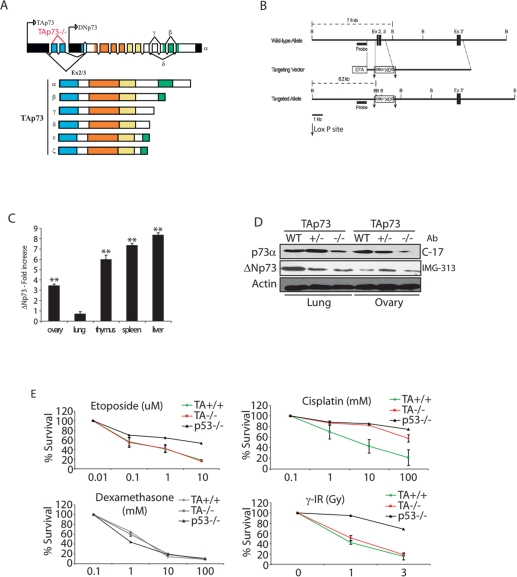
To determine the specific role of the TAp73 isoforms of p73 (Fig. 1A), we generated TAp73-deficient mice in which deletion of exons 2 and 3 of the Trp73 gene (Fig. 1B) selectively abolished expression of all TAp73 isoforms (Supplemental Fig. S1A–C). Firstly, we confirmed the specific deletion of TAp73, and we analyzed the expression patterns of ΔNp73, p53, and p63 in various tissues of TAp73−/− mice. Although p53 and p63 expression were not modified (Supplemental Fig. S2A,D; data not shown), ΔNp73 mRNA was up-regulated in most tissues of TAp73−/− mice (Fig. 1C; Supplemental Fig. S2B). The exception was the lung, where ΔNp73 mRNA was not significantly different from that of wild-type mice (Fig. 1C). We also observed enhanced ΔNp73 mRNA expression in TAp73−/− mouse embryonic fibroblasts (MEFs), and this increased expression was persistent even after the excision of the PGK-neo cassette (Supplemental Fig. S1D,E), indicating that the observed up-regulation of ΔNp73 mRNA is due to the absence of TAp73 proteins rather than to the insertion of the PGK-neo cassette close to the ΔNp73 promoter. However, the enhanced ΔNp73 mRNA level is not reflected in a significant increase in ΔNp73 protein level. Indeed, we analyzed ΔNp73 protein level in lung and ovaries from TAp73−/− tissues versus wild-type tissues by using two antibodies—one specific of the ΔN region (IMG-313) and one specific of the a-C-terminal region from all p73 isoforms (C-17)—and we compared the bands corresponding to the ΔNp73 isoforms (Fig. 1D). Therefore, the antibody data would suggest that the increases in ΔNp73 transcript levels are not accompanied by parallel increases in expression of ΔNp73 protein. The elevated levels of ΔNp73 mRNA in TAp73−/− mice were unexpected because TAp73 is a known transcriptional inducer of ΔNp73 (Grob et al. 2001). However, further examination showed that, in wild-type mice, ΔNp73 levels decreased more rapidly than levels of TAp73 following DNA damage (Maisse et al. 2004; Toh et al. 2004) due to the ubiquitination and proteasomal degradation of ΔNp73 by an as-yet-uncharacterized ubiquitin ligase (B.S. Sayan, C. Piro, A.L. Yang, and G. Melino, unpubl.). These data suggest that there are multiple mechanisms that operate post-transcriptionally to control the levels of various p73 isoforms, resulting in a complex inteplay between TAp73 and ΔNp73. Nonetheless, our TAp73−/− knockout seems to be a pure TAp73-deficient mouse at the protein level, reminiscent of the TAp73/ΔNp73 imbalance observed in human tumors.
Figure 1.
Generation of TAp73−/− mice and up-regulation of ΔNp73 expression. (A) Structure of the murine Trp73 gene showing the TAp73 and ΔNp73 isoform families. Domains: (blue) transactivation; (orange) DNA binding; (yellow) oligomerization; (green) sterile α motif. (B) Targeting strategy. Homologous recombination of the indicated targeting vector with the wild-type Trp73 gene resulted in the replacement of ∼1.5 kb with the neo gene and the deletion of exons 2 and 3 specific to the TAp73 isoform. LoxP sites and the probe used for Southern blotting are indicated. (C) Quantitation of ΔNp73 expression in tissues from TAp73+/+ and TAp73−/− mice. Fold increase of ΔNp73 mRNA level in TAp73−/− tissues compared with wild-type tissues by real-time PCR. (**) P < 0.01 (Student’s t-test). (D) Western blot analysis of ΔNp73 level in lung and ovary from 10-mo-old wild-type and TAp73-deficient mice. C-17 antibody is specific from α-C-terminal isoforms, and IMG-313 antibody is specific from ΔNp73 isoforms. (Ab) Antibody. (E) Thymocyte death. Thymocytes from TAp73+/+, TAp73−/−, and p53−/− mice were treated for 24 h with the indicated agents at the indicated concentrations, and percent cell death was assayed by annexinV staining and flow cytometry. Data represent the mean ± SD of two experiments.
Since ΔNp73 is a functional inhibitor of p53 and TAp73 isoforms (Grob et al. 2001; Deyoung and Ellisen 2007), and as we observed an increase in ΔNp73 mRNA level, we wanted to be sure that no ΔNp73 activity was induced, potentially interfering with p53 functions. We showed that p53 activation was comparable in wild-type and TAp73−/− MEFs (Supplemental Fig. S2C,D). Furthermore, we detected no differences between wild-type and TAp73−/− thymocytes (Fig. 1E), primary MEFs (Supplemental Fig. S2E), or E1A-transformed MEFs (Supplemental Fig. S2F) in p53-dependent cell death induced by γ-irradiation, etoposide, cisplatin, or doxorubicin. The normal sensitivity to death of TAp73−/− thymocytes is particularly interesting in view of the finding that E2F binding to the TAp73 promoter is required for the TCR-induced death of human peripheral T cells from wild-type mice as well as for the death of cancer cells (Irwin et al. 2000; Lissy et al. 2000; Stiewe and Pützer 2000). Our data suggest that the ΔNp73 mRNA increase observed in most tissues from TAp73-deficient mice is not relevant since ΔNp73 protein levels appear normal. A compensatory mechanism may have been elicited in the TAp73-deficient mice to guarantee normal ΔNp73 protein expression under normal or physiological conditions.
Loss of TAp73 leads to infertility due to defects in early embryonic development
A significant percentage of TAp73−/− mice die during embryogenesis, resulting in an abnormal Mendelian ratio at birth (+/+, 29%; +/−, 52%; −/−, 19%) either in mixed or pure background mice (Supplemental Fig. S3A). This partial embryonic lethality could theoretically be caused by the potential enhanced ΔNp73 protein not balanced by the TAp73 at specific stages of embryonic development. Indeed, strongly deregulated ΔNp73 expression in ΔNp73 transgenic mice leads to 100% embryonic lethality partially due to inhibition of p53 activities (Erster et al. 2006; Huttinger-Kirchhof et al. 2006).
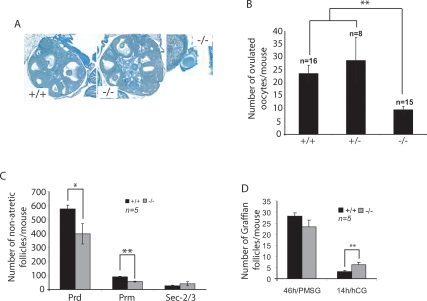
We were unable to obtain pups from mating trials involving mating a TAp73−/− male or female with wild-type mice, suggesting that both sexes are infertile (Supplemental Fig. S3B). In contrast to Trp73−/− mice, in which defects in sensory and hormonal pathways contribute to reproductive and behavioral phenotypes that lead to male and female infertility, TAp73−/− mice mate normally and the females display normal cyclicity (Supplemental Fig. S3B; data not shown). To investigate the nature of TAp73−/− female infertility, we induced superovulation in 3- to 4-wk-old TAp73−/− females and their wild-type littermate sisters using exogenous gonadotropins. In contrast to wild-type females, no oocytes were present in the fallopian tubes of TAp73−/− females 16 h after administration of hCG (human choriogonadotropin). When the ovaries and oviducts of TAp73−/− females were dissected, we found that the ovulated oocytes were trapped under the bursa and did not progress toward the fallopian tubes (Fig. 2A, right). Upon collection of these oocytes, it became clear that TAp73−/− females also ovulated fewer gametes (Fig. 2B).
Figure 2.
Characterization of reproductive defects in TAp73−/− female mice. (A) Normal TAp73−/− ovarian structure. (Left and center panels) Sections of ovaries from 4-wk-old TAp73+/+ and TAp73−/− mice stained with picric acid and methyl blue. Magnification, 5 × 10. (Right panel) While no obvious differences in gross ovarian morphology were detected, ovulated oocytes were trapped under the bursa in TAp73−/− ovary. Magnification, 40 × 10. (B) TAp73-deficient females ovulate fewer oocytes. Immature TAp73+/+, TAp73+/−, and TAp73−/− females were superovulated, and the number of oocytes per mouse was counted. The results shown are the mean ovulated oocyte number ± SE for the indicated number (n) of mice/genotype. (**) P < 0.001 (Student’s t-test). (C) Ovarian reserve is diminished by TAp73 deficiency. Serial sections of ovaries from superovulated immature TAp73+/+ and TAp73−/− mice were subjected to histomorphometric analysis. The results shown are the mean number of nonatretic follicles/mouse ± SE for 5 mice per group. (*) P < 0.01; (**) P < 0.001 (Student’s t-test). The number of both primordial (Prd) and primary (Prm) follicles is significantly decreased, but the growth rate evidenced by the number of secondary follicles with two to three layers of granulosa cells (Sec-2/3) is not altered. (D) The decreased number of ovulated oocytes is caused by impaired ovulation and not by insufficient follicular growth. The number of Graffian follicles with oocytes before (46 h/PMSG) and the number of luteinized follicles with oocytes after ovulation (14h/HCG) was determined in 3-wk-old TAp73+/+ and TAp73−/− female mice. The results shown are the mean number of Graffian follicles/mouse ± SE for n = 5 mice per group. (**) P < 0.001 (Student’s t-test).
To establish if altered follicular development was responsible for the oocyte deficit in TAp73−/− female mice, we performed morphometric analyses (based on the counting of nonatretic follicles) of ovaries obtained from adolescent TAp73+/+ and TAp73−/− females. Although significant differences were observed in the number of quiescent (primordial) or early growing (primary) follicles, numbers of early secondary follicles were comparable (Fig. 2C). To examine the later “Graffian follicle” stages, which are dependent on gonadotropins, we treated prepubertal TAp73+/+ and TAp73−/− females with a single dose of pregnant mare serum gonadotropin (PMSG) to synchronize follicular development. Ovaries collected 46 h later revealed no significant differences in the competence of TAp73−/− gonads to form large antral ovulatory follicles (Fig. 2D, left). However, when we assessed the ovulatory capacity of these follicles, we found that a greater number of oocytes remained trapped within the luteininzing granulosa cells of TAp73−/− ovaries compared with wild-type females (Fig. 2D, right). To confirm that ΔNp73 was not responsible for that phenotype, we determined that ΔNp73 mRNA expression level was even reduced in MII oocytes from TAp73−/− mice compared with wild-type MII oocytes (Supplemental Fig. S4A). These results suggest that the decreased number of ovulated oocytes in TAp73−/− mice is likely due to the retention of some germ cells within the ovary.
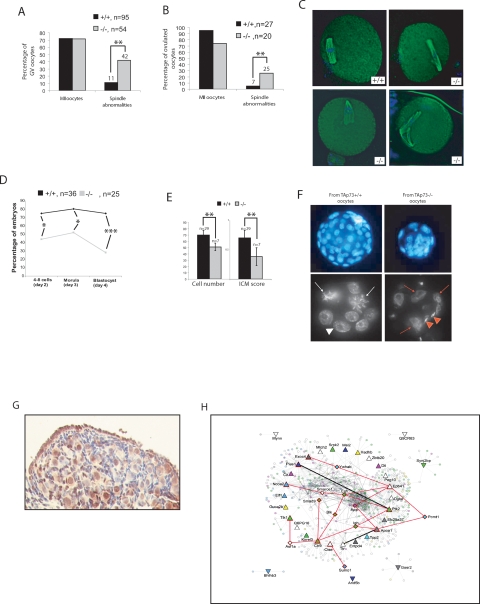
We next investigated whether oocyte quality contributed to TAp73−/− female sterility. Germinal vesicle oocytes as well as ovulated oocytes from 3- to 4-wk-old TAp73+/+ and TAp73−/− mice were induced to mature in vitro. In both cases, oocytes of both genotypes showed the same rate of arrest in metaphase II (Fig. 3A,B, left). However, TAp73−/− oocytes exhibited a striking increase in spindle abnormalities (Fig. 3A,B, right), which included multipolar spindles, spindle relaxation, and spindle scattering accompanied by varying degrees of misalignment (Fig. 3C). We next compared the developmental competence of ovulated TAp73+/+ and TAp73−/− oocytes by assessing their ability to undergo in vitro fertilization (IVF) and preimplantation development in vitro. While the fertilization rate was the same, only 28% of zygotes originating from TAp73−/− oocytes became blastocysts, compared with 75% of zygotes originating from TAp73+/+ oocytes (Fig. 3D). The majority of embryos obtained from TAp73−/− oocytes arrested during early cleavage, resulting in embryos with multinucleated blastomeres, and blastocysts of inferior quality with an abnormal cell number (Fig. 3E,F). Thus, Trp73 is a maternal-lethal effect gene, because a lack of TAp73 in developing oocytes leads to a failure of preimplantation embryonic development.
Figure 3.
TAp73 deficiency effects on oocyte quality. (A,B) Quantitation of spindle abnormalities in TAp73+/+ and TAp73−/− oocytes matured in vitro (A) or in vivo (B). While no difference was observed in ability to proceed through the meiosis and arrest in metaphase II (MII), higher numbers of spindle abnormalities were detected in TAp73 oocytes. (**) P < 0.001 (Student’s t-test). (C) Representative examples of B. Magnification, 40 × 10. (D) The IVF procedure showed that no difference exists in the fertilization rate caused by TAp73 deficiency, but progression of zygotes through preimplantation development is abnormal. The percentage of TAp73−/− oocytes that upon fertilization reached the four- to eight-cell, morula, and blastocyst stages at appropriate developmental days is greatly diminished. (*) P < 0.01; (***) P < 0.0001. Statistical analyses were done using an X2 test. (E) Poor quality of IVF-derived blastocysts obtained from TAp73−/− oocytes as assessed by total cell number (left) and inner cell mass (ICM) score (right). (**) P < 0.001 (Student’s t-test). (F) DAPI staining of blastocysts (top, magnification, 20 × 10) and eight-cell stage-arrested embryos (bottom, magnification, 40 × 10) of B. (White arrows) Mitotic cells; (white arrowhead) normal nucleus; (red arrows) multinuclear cells; (red arrowheads) sperm heads. (G) TAp73 expression in a post-natal day 4 TAp73+/+ ovary as detected by immunohistochemical staining with H79 antibody. (H) Network visualization of genes that were up-regulated in TAp73+/+ and TAp73−/− neonatal ovaries. Node color represents GO (The Gene Ontology) function, as shown in the legend. Node shape represents different targets: Triangles pointed up are targets up-regulated in TAp73−/− ovary; triangles pointing down are targets up-regulated in TAp73+/+ ovary. Diamonds and triangles outlined in red represent critical proteins within the network. Thicker lines indicate direct interactions among targets. Red lines represent critical interactions within the network. All other lines represent secondary interacting proteins.
Poor oocyte quality leading to abnormal embryonic development is also associated with maternal aging, and is one of the most important prognostic factors of human infertility. Because oocytes from young TAp73−/− mice displayed a similar spectrum of spindle abnormalities to that observed in oocytes from aged wild-type mice (Tarin et al. 2001), we evaluated TAp73 expression in the ovaries of 8- and 55-wk-old wild-type mice. We found that natural aging abolishes TAp73 expression in oocytes (Supplemental Fig. S4B,C). This observation suggests that loss of TAp73 may be responsible for the compromised developmental capacity of aged normal oocytes.
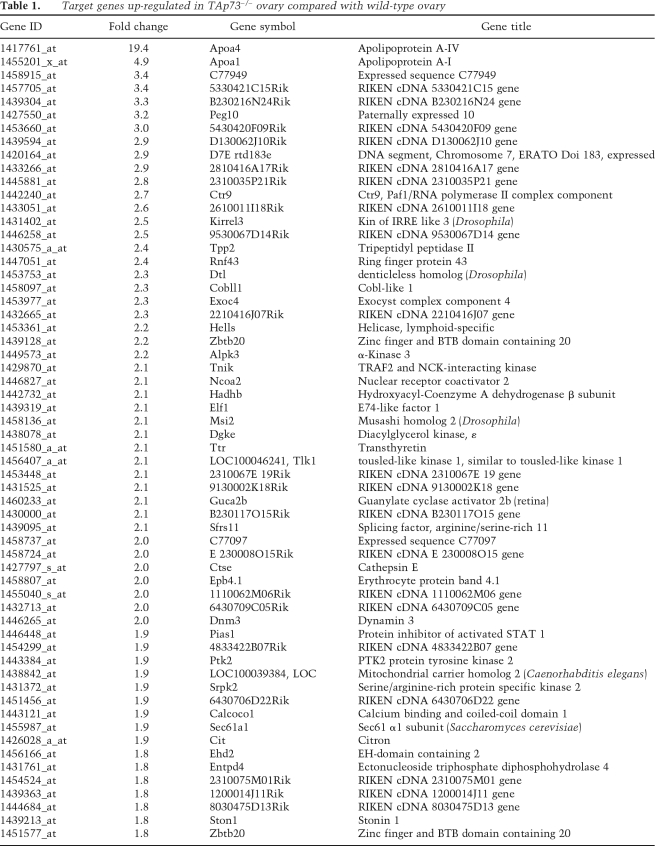
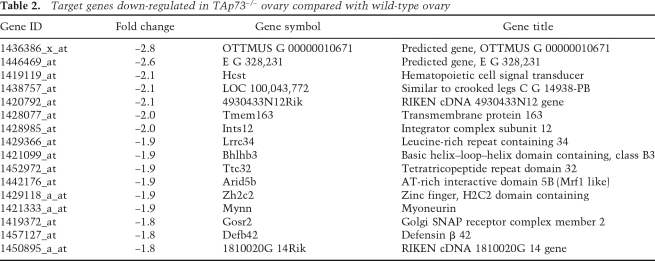
To identify TAp73 transcriptional targets potentially involved in the regulation of developmental competence, we decided to perform a gene microarray analysis of ovarian tissue. We first used immunocytochemistry to determine what stages of oogenesis were most likely to be affected by a lack of TAp73. Growing wild-type oocytes from the primary follicle stage onward displayed only cytoplasmic expression of TAp73, but the oocytes of wild-type primordial follicles often exhibited nuclear staining (Fig. 3G; Supplemental Fig. S4B). Thus, we used neonatal ovaries, which are enriched in oocytes at this developmental stage, as the target tissue for our microarray analysis. Our results showed that the expression of 79 annotated genes in neonatal ovary was altered by at least 1.8-fold by an absence of TAp73. We confirmed for some genes their modulation of expression in TAp73−/− ovaries compared with wild-type ovaries (Supplemental Fig. S5A,B; data not shown). Among these 79 genes, many were involved in the regulation of transcription and translation via control of RNA processing (see Tables 1, 2). We confirmed that most of these genes are potential or already known p53 targets (Supplemental Fig. S5C). To determine functional links among the differentially regulated targets, we mapped them to the I2D protein interaction database (Brown and Jurisica 2007). Extensive analyses identified several proteins that interacted strongly with TAp73, and also showed that all of these proteins were at points of network disconnect, which suggests that they are essential for signaling within this network (Fig. 3H). Indeed, several of these genes (as example: PEG10 and Ctr9) have been disrupted in animal models with resulting embryonic lethality or infertility (Ono et al. 2006; Akanuma et al. 2007). Taken together, these data suggest that these target genes may normally be regulated by TAp73 or are physical interactors of TAp73, as, for example, in the case of PIAS1 that regulates cell cycle checkpoint progression by sumoylation of TAp73 (Munarriz et al. 2004), and thus their dysregulation may contribute to preimplantation lethality in the absence of TAp73.
Table 1.
. %Target genes up-regulated in TAp73−/− ovary compared with wild-type ovary
Table 2.
Target genes down-regulated in TAp73−/− ovary compared with wild-type ovary
TAp73 is necessary for hippocampal development but not for neural cell maintenance
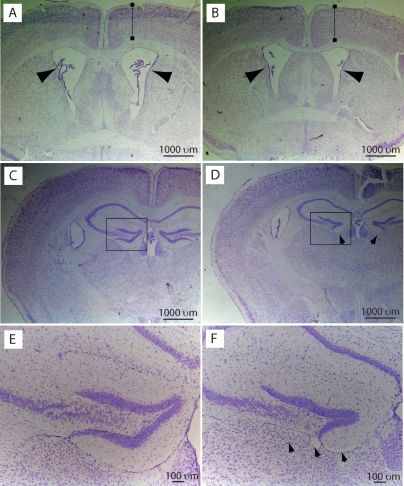
Previous analyses of Trp73−/− mice have demonstrated that p73 is critical for the development and maintenance of the nervous system (Yang et al. 2000). Indeed, Trp73−/− mice exhibit hippocampal dysgenesis with a gradual but persistent postnatal loss of neurons with greatly enlarged ventricles (hydrocephalus) and greatly reduced cortical tissue (Yang et al. 2000; Pozniak et al. 2002). To investigate whether either of these phenotypes might be due to the loss of TAp73 isoforms, we carried out coronal sectioning of the brains of 10-wk-old wild-type and TAp73−/− mice. Nissl staining of the forebrain revealed that TAp73−/− brains were similar to their wild-type counterparts with regard to the size of the lateral ventricles and the thickness of the cortex (Fig. 4A,B) but that hippocampal anatomy was abnormal. In particular, the lower blade of the dentate gyrus was either completely missing or greatly truncated, depending on the level of the section (Fig. 4C–F). This hippocampal dysgenesis was strikingly similar to that seen in Trp73−/− mice at postnatal day 14, before the occurrence of the ventricular enlargement and loss of neural tissue that further perturb hippocampal morphology in adult Trp73−/− mice. The similarity between TAp73−/− mice and p73−/− mice on this particular phenotype suggests that it is due to TAp73 loss and not to ΔNp73 loss. Thus, our data support previous work (Pozniak et al. 2000, 2002; Walsh et al. 2004) showing that TAp73 is essential for normal hippocampal development, while ΔNp73 seems to prevent neural tissue loss and ventricular enlargement in adult life.
Figure 4.
TAp73 is essential for normal hippocampal development. Representative pictures from multiple experiments using seven TAp73+/+ and seven TAp73−/− mice of coronal forebrain sections of 4-mo-old TAp73+/+ (A,C,E) and TAp73−/− (B,D,F) C57B6 background mice stained with cresyl violet. (A,B) Nissl-stained coronal sections of forebrain at the level of the anterior commissure. Arrows indicate the lateral ventricles. Cortical thickness (brackets) was similar in TAp73+/+ and TAp73−/− brains. (C–F) Nissl-stained coronal sections of the forebrain at the level of the rostral hippocampus. The area denoted by the boxes in C and D is shown at higher magnification in E and F, respectively. Arrows in D and F denote the site of the lower blade of the dentate gyrus, which is missing in the TAp73−/− brain.
TAp73−/− mice are tumor-prone and sensitive to carcinogens
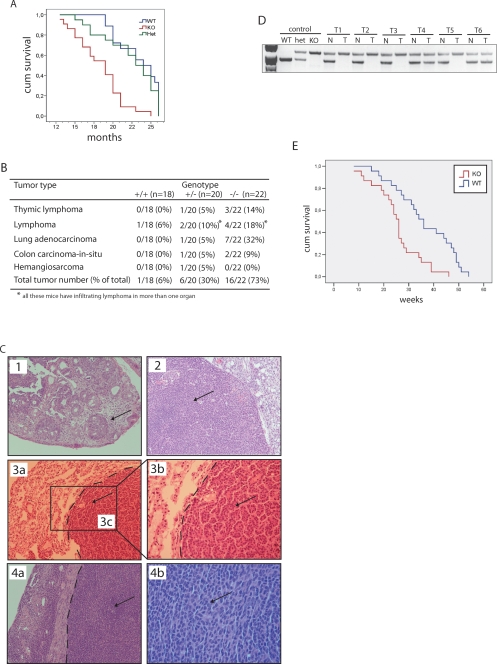
While infertility and neural abnormalities detected in the TAp73−/− mice were not identical but reminiscent of those described for Trp73−/− mice (Yang et al. 2000), other aspects of the phenotype were markedly different. Indeed, although the life span of TAp73−/− mice was significantly shorter than that of wild-type mice (Fig. 5A), it still exceeded that of Trp73−/− mice (Yang et al. 2000). Furthermore, whereas untreated Trp73−/− mice do not develop tumors, 30% of TAp73+/− and 73% of TAp73−/− mice of a mixed genetic background spontaneously developed malignancies (Fig. 5B,C). Lung adenocarcinoma was the most frequent cancer that developed in TAp73−/− mice, being observed in 32% of all TAp73−/− mice analyzed and representing 44% of all tumors excised from TAp73−/− mice. In the lung of the TAp73−/− knockout, no changes in either ΔNp73 mRNA or protein compared with wild-type were observed (see above). Importantly, no correlation was found between tumor onset and p53, p63, or ΔNp73 protein expression in any tumor analyzed (data not shown), suggesting that the appearance of tumors in TAp73+/− and TAp73−/− mice was specifically due to TAp73 deletion rather than to ΔNp73, p63, or p53 deregulation. Consistent with a previous report (Roman-Gomez et al. 2006), loss-of-heterozygosity (LOH) of p73 had occurred in 66% of tumors developing in TAp73+/− mice (Fig. 5D).
Figure 5.
TAp73−/− mice are tumor-prone and sensitive to carcinogens. (A) Kaplan-Meyer survival curves of TAp73+/+ (n = 18), TAp73+/− (n = 20), and TAp73−/− (n = 22) mice. P < 0.001 (Logrank test). (B) Spontaneous tumor spectrum in TAp73+/+, TAp73+/−, and TAp73−/− mice. Spontaneous tumor development in TAp73−/− mice is significative with a P-value of P < 0.005 (X2 Test). (C, arrows) H&E-stained tumors in tissues of TAp73−/− mice. (1) Colon carcinoma in situ, 10× magnification. (2) Invasive lymphoma in ovary, 10× magnification. (3a–3c) Lung adenocarcinoma, 10× magnification, with 3b being a higher magnification of 3c, 20× magnification. (4a–4b) Invasive lymphoma in uterus, 10× magnification, with 4b being a higher magnification of 4a, 40× magnification. Dotted lines denote the border between healthy and tumoral tissue. (D) Representative LOH analysis of six TAp73+/− tumor-bearing mice (T1-6); (T) tumor; (N) normal tissue. (E) Kaplan-Meyer survival curves of DMBA-treated TAp73+/+ (n = 23) and TAp73−/− (n = 23) mice in weeks after treatment. P < 0.005 (Logrank test).
To confirm the tumor-prone phenotype of TAp73−/− mice, we induced carcinogenesis by intraperitoneal injection of DMBA into TAp73+/+ and TAp73−/− female littermates. The mean latency time for tumor development in TAp73−/− mice was 18.7 ± 7.1 wk, significantly shorter than the 32 ± 3.4-wk mean latency observed in DMBA-treated TAp73+/+ mice (Fig. 5E). However, there was no difference between DMBA-treated TAp73+/+ and TAp73−/− mice in sites of tumor development, which included the colon, intestine, liver, and stomach (data not shown). These results indicate that TAp73 isoforms mediate the tumor-suppressive function of Trp73, and that loss of TAp73 alone is sufficient to promote oncogenic transformation.
Taking into account the observation that TAp73−/− mice develop predominantly lung adenocarcinoma, we decided to investigate the expression of p73 isoforms in human lung cancer. Examination of the expression of TAp73 and ΔNp73 in matched normal and tumoral lung tissue samples from 18 lung cancer patients revealed that most tumoral lung samples exhibited a down-regulation of TAp73 and an up-regulation of ΔNp73, with a consequent decrease in the TAp73/ΔNp73 ratio (Supplemental Fig. S6). Notably, in the two samples (patients 133 and 177) in which TAp73 expression was up-regulated, greater amounts of ΔNp73 were also expressed. These results are consistent with the postulated inactivation of p53 in human lung cancer (Haruki et al. 2001).
TAp73 participates in maintenance of genomic stability
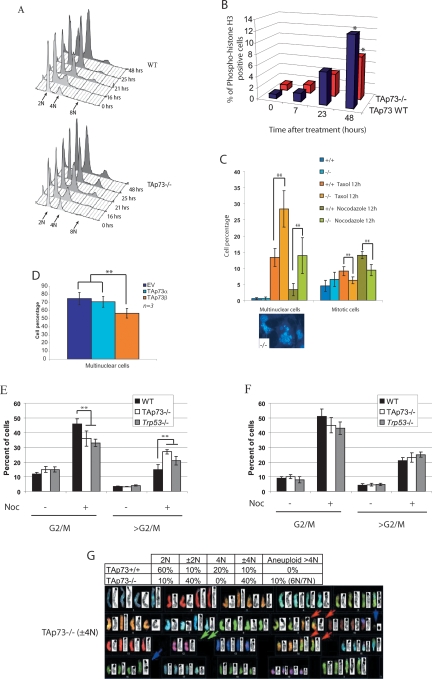
Since we observed an increased incidence of spontaneous tumors and also an abnormal spindle formation in the oocytes of TAp73−/− mice, we next investigated the possible impact of TAp73 deficiency on the maintenance of genomic stability. Accurate chromosome segregation during meiosis and mitosis is critical to the preservation of euploidy in eukaryotic cells (Yuen et al. 2005), and errors in the molecular mechanisms regulating segregation result in aneuploidy (Hassold and Hunt 2001; Thomas et al. 2001). We analyzed the ability of TAp73-deficient cells to undergo mitotic arrest and observed that TAp73−/− MEFs showed a significant decrease in nocodozole-induced mitotic arrest. Indeed, at 25 h post-nocodozole, the percentages of cells in G1 and >G2/M were increased in TAp73−/− MEFs compared with TAp73+/+ MEFs (Fig. 6A; Supplemental Fig. S7A). Immunoblotting of mitotic phase marker proteins such as cyclin B1 and securin revealed that mitotic “slippage” had occurred in TAp73−/− MEFs (data not shown). Premature mitotic exit in TAp73−/− MEFs was confirmed by immunostaining to detect phospho-histone H3-positive cells (Fig. 6B) and by counting mitotic figures (Fig. 6C, right). Thus, in the presence of nocodazole, TAp73 deficiency results in the abnormal mitotic progression of cells that otherwise would not be permitted to pass into the G1 phase. We observed a similar premature mitotic exit in p73−/− MEF cells treated with nocodazole, strongly suggesting that TAp73 isoforms are the functional isoforms in mitotic regulation rather than ΔNp73 isoforms (Supplemental Fig. S7B,C).
Figure 6.
Defective maintenance of genomic stability in the absence of TAp73. (A) Cell cycle analysis by flow cytometry of TAp73+/+ (top) and TAp73−/− (bottom) MEFs treated with nocodazole for the indicated number of hours. The graph was created using FlowJo 7.2.2 software. (B) Percentage of phospho-histone H3+ cells in cultures of TAp73+/+ (TAp73 wild type) and TAp73−/− MEFs treated with nocodazole for indicated times. (*) P < 0.01 (Student’s t-test). (C) Percentages of multinuclear (left) and mitotic (right) cells in TAp73+/+ and TAp73−/− MEFs treated with taxol or nocodazole as indicated. (Bottom) DAPI-stained multinuclear TAp73−/− MEFs; 40× magnification. The data shown represent the mean ± SE of three different experiments. (**) P < 0.001 (Student’s t-test). (D) Percentage of multinuclear cells in H1299 cultures overexpressing empty vector (EV) or vector containing the indicated TAp73 isoforms and treated with taxol for 24 h. (**) P < 0.001 (Student’s t-test). (E,F) Percentages of cells in G2/M phase or in >G2/M phase (aneuploid cells). Cells were extracted from the lung (E) or thymus (F) of wild-type, TAp73−/−, and Trp53−/− mice and treated for 12 h with nocodazole. The results shown are the mean percentage of cells ± SE of three independent trials. (**) P < 0.001 (Student’s t-test). (G) Increased aneuploidy. (Top) Percentages of cells in TAp73+/+ and TAp73−/− 3T3 cultures showing diploid (2N), near-diploid (±2N), tetraploid (4N), near-tetraploid (±4N), and aneuploid (>4N) DNA content. (Bottom) Representative karyotype of a ±4N TAp73−/− 3T3 cell.
After 25 h of nocodazole treatment, an 8N population of polyploid cells appeared frequently among TAp73−/− MEFs when compared with TAp73+/+ MEFs (Fig. 6A). Indeed, ∼14% of TAp73−/− MEFs showed aneuploidy compared with 5% of cells in TAp73+/+ MEF cultures. The percentage of multinuclear cells was also increased in TAp73−/− MEFs treated with taxol or nocodozole (Fig. 6C, left). When H1299 cells were engineered to overexpress the TAp73β isoform, they were protected from taxol-induced formation of multinuclear cells (Fig. 6D). In contrast, overexpression of ΔNp73α or ΔNp73β in Trp73−/− MEFs did not reduce the enhanced formation of multinuclear cells (data not shown).
We next examined lung and thymic cell populations from TAp73−/−, Trp53−/−, and wild-type mice that were treated or not with nocodazole for 12 h. TAp73−/− lung fibroblasts showed a higher frequency of >G2/M cells, demonstrating a loss of euploidy, which are likely aneuploid, than did Trp53−/− or wild-type lung fibroblasts, whereas no differences were observed among TAp73−/−, Trp53−/−, and wild-type thymic cells (Fig. 6E,F). Thus, the effect of TAp73 on aneuploidy appears to be tissue-specific, perhaps explaining why TAp73−/− mice preferentially develop lung adenocarcinomas. Our results indicate that, at least in lung tissue, TAp73 loss is associated with decreased genomic stability.
To confirm more directly that TAp73 deficiency is associated with an altered DNA content, we karyotyped TAp73−/− and TAp73+/+ 3T3 cells and observed that the mutant cultures contained a significantly higher percentage of cells with an abnormal karyotype compared with wild-type controls (Fig. 6G). The affected cells showed a loss or gain of a single chromosome, indicating aneuploidy. These data further support a role for TAp73 in maintaining genomic stability.
Discussion
As mentioned above, mouse models featuring deletion of all p73 isoforms (Yang et al. 2000), or p73 deletion in combination with Trp53 deficiency (Flores et al. 2005; Perez-Losada et al. 2005), or overexpression of specific p73 isoforms (Huttinger-Kirchhof et al. 2006), have all been helpful in dissecting the role of Trp73 in various biological processes. However, these mouse models cannot distinguish between the effects of the TAp73 and ΔNp73 isoforms, so that the relative importance of these isoforms in specific biological processes remains unclear or even conflicting. In our study, we generated and analyzed the effects of a selective deficiency of TAp73 isoforms. Our comparison of the phenotypes of Trp73−/− and TAp73−/− mice has clearly identified several functions that require TAp73 as opposed to ΔNp73. However, single isoform deletion models, the complete deletion of all isoforms, and even partial deletions and hypomorphic models will all be needed to completely understand the complex functional network mediated by p53 family members.
Both Trp73−/− and TAp73−/− mice are infertile, but the root causes of this infertility differ. Trp73−/− mice have sensory defects that prevent them from mating normally (Yang et al. 2000). In contrast, TAp73−/− female mice mate normally but have oocytes of reduced developmental competence that result in impaired early embryogenesis. The increased frequency of spindle defects in TAp73−/− ovulated oocytes may also contribute to the sterility of TAp73−/− females, in line with the strong correlation of this parameter with human infertility (Baird et al. 2005). Importantly, our work has identified Trp73 as one of a handful of genes whose mutation generates a maternal lethal effect phenotype. Genes belonging to this group are required for the transition from maternal to embryonic control over embryogenesis. Accordingly, the spectrum of cellular defects observed in embryos originating from TAp73−/− oocytes is remarkably similar to the range of embryonic defects observed in infertile patients undergoing IVF, particularly older patients (Hardarson et al. 2001; Hardy et al. 2001). These data are also consistent with the down-regulation of p73 transcripts that occurs in the oocytes of older patients (Steuerwald et al. 2007). The p53-family members are pivotal in the control of aging, as shown for p63 (Keyes et al. 2005; Suh et al. 2006), p53 mutant (Tyner et al. 2002), or deficient mice (Ohkusu-Tsukada et al. 1999). Regarding our results, it would be of interest that TAp73 down-regulation, due to aging or loss-of-function mutations, may be a potential prognostic factor in cases of human female infertility.
In addition to infertility, TAp73−/− mice exhibited a mild defect in brain morphology. Hippocampal dysgenesis was observed in TAp73−/− mice, but there were no signs of neural tissue loss or ventricular enlargement as occur in Trp73−/− mice. Based on our studies and those of Trp73-deficient mice, we suggest that a lack of the ΔNp73 isoform rather than TAp73 is responsible for neural tissue loss and ventricular enlargement in the brain, whereas hippocampal dysgenesis with loss or severe truncation of the dentate gyrus is specifically linked to TAp73 deficiency. The generation and characterization of a mouse with a selective deficiency of ΔNp73 is needed to completely resolve this question.
Our study is the first clear demonstration that TAp73 isoforms possess the tumor-suppressive ability of Trp73, and thus resolves previous conflicting results (McKeon and Melino 2007). It has been proposed previously that a loss of p73 function might lead to tumorigenesis (Flores et al. 2002, 2005), and that TAp73 down-regulation may be coupled with ΔNp73 up-regulation in some tumors (Zaika et al. 2002). We found that mice with a selective deficiency of TAp73 develop spontaneous tumors, particularly lung adenocarcinomas, and are more sensitive to chemical carcinogens. The development of spontaneous tumors in TAp73−/− mice but not in Trp73−/− mice illustrates the complex relationship between Trp53 family members and cancer.
Intriguingly, the types of tumors detected in TAp73−/− mice, as well as those detected in Trp63+/− and Trp73+/− mice, are quite different from the range of malignancies displayed by Trp53−/− mice (even taking strain differences into account) (Flores et al. 2002, 2005). Furthermore, although p63 and p73 do not contribute to p53-mediated suppression of murine lymphomagenesis in vivo (Perez-Losada et al. 2005), allelic loss and genetic instability of Trp73 have been linked to non-Hodgkin’s lymphomas (Martinez-Delgado et al. 2002). These potentially conflicting data and the diversity in tumor spectra in these various models are likely due to the intricate pattern of expression of multiple p53, p63, and p73 isoforms. Human cancer cells can express peculiarly spliced isoforms of p73, such as Δ2p73 and Δ2,3p73 (Pützer et al. 2003; Concin et al. 2004), that involve selective deletions of Trp73 exons 2 and 3. These cancer cells show both elimination of the TAp73 isoform and up-regulation of a ΔNp73 variant, mimicking our TAp73−/− mice, and developing a strikingly similar phenotype. Our mouse model therefore highlights the complex inter-regulation of TAp73 and ΔNp73 isoforms and closely reflects the aberrant pattern of p73 expression in many human cancers.
More and more evidence implicates p53 and its family members in the prevention of genomic instability, aneuploidy, and cancer, although the details of the molecular links remain unclear (Duensing and Duensing 2005). Inactivation of p53 family members may lead to a chromosomal instability phenotype that is a hallmark of cancers in which specific mitotic proteins are dysregulated (Carter et al. 2006; for review, see Tomasini et al. 2008). A recent report has shown that the combined loss of Trp73 and Trp53 induces severe genomic instability and a rapid increase in polyploidy and aneuploidy that markedly exceeds that induced by loss of Trp53 alone (Talos et al. 2007). In addition, Trp73 seems necessary for the death of cells undergoing abnormal mitosis (Niikura et al. 2007). These results are in line with the abnormal survival described in our TAp73−/− cells with elevated (more than 4N) DNA content. The persistence of these genetically unstable cells could then set the stage for tumorigenesis.
Our data demonstrate that TAp73 isoforms are bona fide tumor suppressors, contributing to the growing body of evidence linking genomic instability and aneuploidy with tumorigenesis (Pérez de Castro et al. 2007; Weaver and Cleveland 2007). Specifically, our findings point to a critical emerging role for the TAp73 isoform in the maintenance of genomic stability that prevents both tumor formation and infertility. Indeed, like p53 (Hu et al. 2007) and p63 (Suh et al. 2006), the entire p53 family seems to act as “the guardian of the female germline.”
Materials and methods
Generation of TAp73−/− mice and LOH analysis
Mutant mice deficient for Trp73 exons 2 and 3, which are specific for the TAp73 isoforms, were generated by conventional gene targeting procedures in Sv129Ola embryonic stem cells. Blastocysts were transferred to pseudopregnant C57BL/6J female mice. TAp73 wild-type (wild type, +/+; TAp73+/+), heterozygous (+/−; TAp73+/−), and homozygous (−/−; TAp73−/−) littermate mice (Sv129Ola × C57BL/6J) were created by intercrossing TAp73+/− mice. Offspring were genotyped by PCR analysis using specific primer pairs to detect the wild-type Trp73 and Trp73ΔTA alleles. Sense and antisense primers for the wild-type Trp73 allele were 5′-CTGGTCCAGGAGGTGAGACTGAG GC-3′ and 5′-CTGGCCCTCTCAGCTTGTGCCACTTC-3′, respectively. Sense and antisense primers for the gene-targeted Trp73ΔTA allele were 5′-GTGGGGGTGGGATTAGATAAAT GCCTG-3′ and 5′-CTGGCCCTCTCAGCTTGTGCCACTTC3′, respectively. Predicted PCR product sizes were 1.0 kb and 1.2 kb for the wild-type Trp73 and Trp73ΔTA alleles, respectively. F4 mixed background mice were used for all experiments except the analysis of the Mendelian ratio, for which we also used animals of the fifth-generation backcross in a C57B6 background. All animals were treated in accordance with the NIH Guide for Care and Use of Laboratory Animals as approved by the Ontario Cancer Institute Animal Care Committee (Toronto, ON, Canada). LOH analysis was performed using genotyping primers and PCR amplification of Trp73 exons 2 and 3 in genomic DNA from tumor and normal (tail) tissues of tumor-bearing TAp73+/− mice.
Analyses of spontaneous and induced tumors
Life span and spontaneous tumor incidence were determined in untreated TAp73+/+ and TAp73−/− mice. Mice with visible tumors and moribund mice showing weight loss or difficulties in moving were sacrificed upon detection. The remaining surviving mice were sacrificed at 26 mo of age. For induced tumorigenesis, TAp73+/+ and TAp73−/− mice (23 mice/group) were injected i.p. with 1 mg/kg dimethylbenz[a]anthracene (DMBA; Sigma-Aldrich) starting at 63 d post-partum and continuing once a week for 6 wk. This dose was sufficient to induce 100% tumor incidence in controls. Tumors and tissues of control and mutant mice were collected and processed for histopathology and RNA/DNA isolation by standard procedures.
Oocyte collection, in vitro maturation, and in vitro fertilization
Female mice (3–7 wk old) were superovulated, and mature oocytes were collected as previously described (Perez et al. 2005). For in vitro maturation, germinal vesicle stage oocytes from 3-wk-old females were induced to undergo spontaneous in vitro maturation (Yao et al. 2004), and in vitro fertilization was carried out as described previously (Acton et al. 2004). Embryonic developmental competence was assessed daily. On day 4, embryos were fixed and cell number, cell death, and mitotic rates were determined as described previously (Jurisicova et al. 1998). Statistics were obtained using the χ2 or Student’s t-test.
Microarray studies
Microarray analysis was performed on day 4 ovarian tissue from two sibling pairs of TAp73+/+ and TAp73−/− females by the Center for Applied Genomics (Hospital for Sick Children, Toronto) using the Affymetrix platform (Mouse MOE 430 2.0) with two cycle amplification steps. Data were preprocessed using RMAExpress 1.0 beta 3 (Bolstad et al. 2003), log2 transformed, and then analyzed using SAM version 3.0 (Tusher et al. 2001). Using FDR (False Discovery Rate) = 0.603%, we identified 2538 genes that were differentially expressed (1178 up-regulated and 1360 down-regulated in TAp73+/+ ovaries). To further diminish false positives, we prioritized the genes by considering only fold change >1.8, which resulted in 16 genes up-regulated and 60 genes down-regulated in wild-type ovaries.
To determine functional links among identified target genes, we mapped them to the Interologous Interaction Database I2D ver. 1.71 (http://ophid.utoronto.ca/i2d). A network was formed containing 531 proteins and 977 interactions on seven targets up-regulated in TAp73+/+ ovaries and 26 targets up-regulated in TAp73−/− ovaries. Further visualization and analysis of the resulting protein interaction network were performed using NAViGaTOR ver 2.0 (http://ophid.utoronto.ca/navigator). Shortest path calculations were used to determine critical proteins and interactions. Briefly, the shortest path among all nodes within the network was computed, and the frequency of inclusion of individual proteins and interactions was counted. The most frequent nodes and lines form the critical subgraph within the network. We applied blending to reduce network complexity (NAViGaTOR version 2.0).
Immunohistochemical analysis
For histological analysis of mouse brains, sacrificed animals were perfused with 4% paraformaldehyde, and their brains were cryoprotected, sectioned, and Nissl-stained as described previously (Pozniak et al. 2002). For histological analysis of oocytes, oocyte fixation and immunocytochemistry were performed as described previously (Yao et al. 2004). Samples were viewed on a deconvolution fluorescence microscope. Follicular histomorphology was performed on fixed ovarian tissue obtained from 3-wk-old females (Canning et al. 2003), and immunocytochemistry on ovarian sections was performed as described previously (Matikainen et al. 2001). The antibodies used for analyses of oocytes and/or ovarian sections were rat monoclonal anti-tubulin (YL1/2; Abcam), rabbit polyclonal anti-p73 (H79; Santa Cruz Biotechnologies), and goat anti-p73α (C17; Santa Cruz Biotechnologies). For spindle analyses, secondary antibody was conjugated to Oregon green (Molecular Probes). For ovarian sections, visualization was performed using a biotinylated anti-rabbit antibody detection kit (DAKO).
Cell lines and overexpression experiments
MEFs obtained from embryonic day 13.5 (E13.5) TAp73+/+, TAp73−/−, and p73−/− embryos, or 3T3 cells, were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS without antibiotics. For overexpression experiments, the human lung carcinoma cell line H1299 was plated at 60% density the day before transfections. Transient transfection of plasmids was carried out using the FuGENE reagent (Roche Applied Science) according to the manufacturer’s protocol. Control vector pcDNA3-HA was from Clontech. DNA sequences encoding the TAp73α and TAp73β isoforms were all cloned in pcDNA3-HA.
Flow cytometric analyses
TAp73−/− MEFs were seeded at 3 × 105 cells per 100-mm plate. Cultures were then either left untreated or treated with nocodazole (200 ng/mL; Sigma-Aldrich). Immediately (control) or at 16, 21, 25, or 48 h after nocodazole treatment, cells were harvested and fixed for at least 1 h at 4°C in PBS/70% ethanol. Fixed cells were resuspended in PBS containing 100 μg/mL RNase A and 50 μg/mL propidium iodide (Sigma-Aldrich) and immediately analyzed using a FACSCalibur (Becton Dickinson Biosciences). Data were collected and analyzed with CellQuest Pro (BD Biosciences) and FlowJo 7.2.2 Data Analysis Software. Phospho-histone H3 staining was performed using an anti-phospho-histone H3 antibody (#9708; Cell Signaling) according to the manufacturer’s protocol. Cell cycle analysis of total cell populations from fresh TAp73+/+, TAp73−/−, and p53−/− thymus or lung tissues was performed as previously described (Yamamoto et al. 2003).
Cell death analysis was performed by flow cytometry using propidium iodide staining together with AnnexinV-FITC as described previously.
Western blotting
Protein extracts (100 μg) were fractionated by SDS-PAGE and subjected to Western blotting by standard procedures. Western blots were quantified by band densitometry using an Odyssey Infrared Imaging System (LI-COR Biosciences). For immunoprecipitations, protein extracts from MEFs were analyzed according to standard protocols. The antibodies used for Western blot were mouse anti-ΔNp73 (IMG-313; Imgenex), goat anti-p73 (C-20; Santa Cruz), rabbit anti-phospho-p53 ser15 (#9284L; Cell Signaling), and rabbit anti-p53 total (CM5; Vector Laboratories).
Acknowledgments
We gratefully acknowledge H. Okada and R.A. Knight for technical advice and helpful discussions, P. Berthezene for statistical analysis, S.K. Lau for technical help with human real-time PCR analysis, and M.E. Saunders for scientific editing. This work was supported by grants from AIRC, EU (Active-p53, Epistem), FIRB, MIUR, and MinSan to G.M. R.T. was supported by l’Association pour la recherche contre le cancer. R.T, K.T., and A.J. were supported by the Canadian Institutes of Health Research. R.T., K.T., M.W., M.F., A.R., C.C.C., F.K., A.I.-Y., A.W., P.B., and A.J. conducted research for the paper. R.T., G.M., A.J., and T.W.M. wrote the paper. We all discussed the results and read the paper.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1695308.
References
- Acton B.M., Jurisicova A., Jurisica I., Casper R.F. Alterations in mitochondrial membrane potential during preimplantation stages of mouse and human embryo development. Mol. Hum. Reprod. 2004;10:23–32. doi: 10.1093/molehr/gah004. [DOI] [PubMed] [Google Scholar]
- Akanuma T., Koshida S., Kawamura A., Kishimoto Y., Takada S. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 2007;8:858–863. doi: 10.1038/sj.embor.7401045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D.T., Collins J., Egozcue J., Evers L.H., Gianaroli L., Leridon H., Sunde A., Templeton A., Van Steirteghem A., Cohen J., et al. Fertility and ageing. Hum. Reprod. Update. 2005;11:261–276. doi: 10.1093/humupd/dmi006. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bourdon J.C., Fernandez K., Murray-Zmijewski K., Liu G., Diot A., Xirodimas D.P., Saville M.K., Lane D.P. p53 isoforms can regulate p53 transcriptional activity. Genes & Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning J., Takai Y., Tilly J.L. Evidence for genetic modifiers of ovarian follicular endowment and development from studies of five inbred mouse strains. Endocrinology. 2003;144:9–12. doi: 10.1210/en.2002-220988. [DOI] [PubMed] [Google Scholar]
- Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Coates P.J. Regulating p73 isoforms in human tumours. J. Pathol. 2006;210:385–389. doi: 10.1002/path.2080. [DOI] [PubMed] [Google Scholar]
- Concin N., Becker K., Slade N., Erster S., Mueller-Holzner E., Ulmer H., Daxenbichler G., Zeimet A., Zeillinger R., Marth C., et al. Transdominant ΔTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V., Costanzo A., Barcaroli D., Terrinoni A., Falco M., Annicchiarico-Petruzzelli M., Levrero M., Melino G. Two new p73 splice variants, γ and δ, with different transcriptional activity. J. Exp. Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung M.P., Ellisen L.W. p63 and p73 in human cancer: Defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr., Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Duensing A., Duensing S. Guilt by association? p53 and the development of aneuploidy in cancer. Biochem. Biophys. Res. Commun. 2005;331:694–700. doi: 10.1016/j.bbrc.2005.03.157. [DOI] [PubMed] [Google Scholar]
- Erster S., Palacios G., Rosenquist T., Chang C., Moll U.M. Deregulated expression of ΔNp73α causes early embryonic lethality. Cell Death Differ. 2006;13:170–173. doi: 10.1038/sj.cdd.4401743. [DOI] [PubMed] [Google Scholar]
- Flores E.R., Tsai K.Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Flores E.R., Sengupta S., Miller J.B., Newman J.J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Grob T.J., Novak U., Maisse C., Barcaroli D., Lüthi A.U., Pirnia F., Hügli B., Graber H.U., De Laurenzi V., Fey M.F., et al. Human ΔNp73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- Hardarson T., Hanson C., Sjogren A., Lundin K. Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: Indications for aneuploidy and multinucleation. Hum. Reprod. 2001;16:313–318. doi: 10.1093/humrep/16.2.313. [DOI] [PubMed] [Google Scholar]
- Hardy K., Spanos S., Becker D., Iannelli P., Winston R.M., Stark J. From cell death to embryo arrest: Mathematical models of human preimplantation embryo development. Proc. Natl. Acad. Sci. 2001;98:1655–1660. doi: 10.1073/pnas.98.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki N., Harano T., Masuda A., Kiyono T., Takahashi T., Tatematsu Y., Shimuzu S., Mitsudomi T., Konishi H., Osada H., et al. Persistent increase in chromosome instability in lung cancer: Possible indirect involvement of p53 inactivation. Am. J. Pathol. 2001;159:1345–1352. doi: 10.1016/S0002-9440(10)62521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hu W., Feng Z., Teresky A.K., Levine A.J. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- Huttinger-Kirchhof N., Cam H., Griesmann H., Hofmann L., Beitzinger M., Stiewe T. The p53 family inhibitor ΔNp73 interferes with multiple developmental programs. Cell Death Differ. 2006;13:174–177. doi: 10.1038/sj.cdd.4401809. [DOI] [PubMed] [Google Scholar]
- Irwin M., Marin M.C., Phillips A.C., Seelan R.S., Smith D.I., Liu W., Flores E.R., Tsai K.Y., Jacks T., Vousden K.H., et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Jurisicova A., Rogers I., Fasciani A., Casper R.F., Varmuza S. Effect of maternal age and conditions of fertilization on programmed cell death during murine preimplantation embryo development. Mol. Hum. Reprod. 1998;4:139–145. doi: 10.1093/molehr/4.2.139. [DOI] [PubMed] [Google Scholar]
- Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J.C., Valent A., Minty A., Chalon P., Lelias J.M., Dumont X., et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Keyes W.M., Wu Y., Vogel H., Guo X., Lowe S.W., Mills A.A. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes & Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klanrit P., Flinterman M.B., Odell E.W., Melino G., Killick R., Norris J.S., Tavassoli M. Specific isoforms of p73 control the induction of cell death induced by the viral proteins, E1A or apoptin. Cell Cycle. 2008;7:207–215. doi: 10.4161/cc.7.2.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Lissy N.A., Davis P.K., Irwin M., Kaelin W.G., Dowdy S.F. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;5:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- Maisse C., Munarriz E., Barcaroli D., Melino G., De Laurenzi V. DNA damage induces the rapid and selective degradation of the ΔNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- Martinez-Delgado B., Melendez B., Cuadros M., Garcia M.J., Nomdedeu J., Rivas C., Fernandez-Piqueras J., Benitez J. Frequent inactivation of the p73 gene by abnormal methylation or LOH in non-Hodgkin’s lymphomas. Int. J. Cancer. 2002;102:15–19. doi: 10.1002/ijc.10618. [DOI] [PubMed] [Google Scholar]
- Matikainen T., Perez G.I., Jurisicova A., Pru J.K., Schlezinger J.J., Ryu H.-Y., Laine J., Sakai T., Korsmeyer S.J., Casper R.F., et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat. Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- McKeon F., Melino G. Fog of war: The emerging p53 family. Cell Cycle. 2007;6:229–232. doi: 10.4161/cc.6.3.3876. [DOI] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Müller M., Schilling T., Sayan A.E., Kairat A., Lorenz K., Schulze-Bergkamen H., Oren M., Koch A., Tannapfel A., Stremmel W. TAp73/ΔNp73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–1577. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- Munarriz E., Barcaroli D., Stephanou A., Townsend P.A., Maisse C., Terrinoni A., Neale M.H., Martin S.J., Latchman D.S., Knight R.A., et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol. Cell. Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Lane D.P., Bourdon J.C. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Niikura Y., Dixit A., Scott R., Perkins G., Kitagawa K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J. Cell Biol. 2007;178:283–296. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkusu-Tsukada K., Tsukada T., Isobe K. Accelerated development and aging of the immune system in p53-deficient mice. J. Immunol. 1999;163:1966–1972. [PubMed] [Google Scholar]
- Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- Perez G.I., Jurisicova A., Matikainen T., Moriyama T., Kim M., Takai Y., Pru J.K., Kolesnick R.N., Tilly J.L. A central role for ceramide in the aging-related acceleration of apoptosis in the female germ line. FASEB J. 2005;19:860–872. doi: 10.1096/fj.04-2903fje. [DOI] [PubMed] [Google Scholar]
- de Pérez Castro I., de Cárcer G., Malumbres M. A census of mitotic cancer genes: New insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J., Wu D., Delrosario R., Balmain A., Mao J.H. p63 and p73 do not contribute to p53-mediated lymphoma suppressor activity in vivo. Oncogene. 2005;24:5521–5524. doi: 10.1038/sj.onc.1208799. [DOI] [PubMed] [Google Scholar]
- Pozniak C.D., Radinovic S., Yang A., McKeon F., Kaplan D.R., Miller F.D. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- Pozniak C.D., Barnabe-Heider F., Rymar V.V., Lee A.F., Sadikot A.F., Miller F.D. p73 is required for survival and maintenance of CNS neurons. J. Neurosci. 2002;22:9800–9809. doi: 10.1523/JNEUROSCI.22-22-09800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pützer B.M., Tuve S., Tannapfel A., Stiewe T. Increased ΔN-p73 expression in tumors by upregulation of the E2F1-regulated, TA-promoter-derived ΔN′-p73 transcript. Cell Death Differ. 2003;10:612–614. doi: 10.1038/sj.cdd.4401205. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J., Gimenez-Velasco A., Agirre X., Castillejo J.A., Navarro G., Calasanz M.J., Garate L., San jose-Eneriz E., Cordeu L., Prosper F., et al. CpG island methylator phenotype redefines the prognostic effect of t(12;21) in childhood acute lymphoblastic leukemia. Clin. Cancer Res. 2006;12:4845–4850. doi: 10.1158/1078-0432.CCR-05-2592. [DOI] [PubMed] [Google Scholar]
- Steuerwald N.M., Bermúdez M.G., Wells D., Munné S., Cohen J. Maternal age-related differential global expression profiles observed in human oocytes. Reprod. Biomed. Online. 2007;14:700–708. doi: 10.1016/s1472-6483(10)60671-2. [DOI] [PubMed] [Google Scholar]
- Stiewe T. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- Stiewe T., Pützer B.M. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- Suh E.K., Yang A., Kettenbach A., Bamberger C., Michaelis A.H., Zhu Z., Elvin J.A., Bronson R.T., Crum C.P., McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Talos F., Nemajerova A., Flores E.R., Petrenko O., Moll U.M. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol. Cell. 2007;27:647–659. doi: 10.1016/j.molcel.2007.06.036. [DOI] [PubMed] [Google Scholar]
- Tarin J.J., Perez-Albala S., Cano A. Cellular and morphological traits of oocytes retrieved from aging mice after exogenous ovarian stimulation. Biol. Reprod. 2001;65:141–150. doi: 10.1095/biolreprod65.1.141. [DOI] [PubMed] [Google Scholar]
- Thomas N.S., Ennis S., Sharp A.J., Durkie M., Hassold T.J., Collins A.R., Jacobs P.A. Maternal sex chromosome non-disjunction: Evidence for X chromosome-specific risk factors. Hum. Mol. Genet. 2001;10:243–250. doi: 10.1093/hmg/10.3.243. [DOI] [PubMed] [Google Scholar]
- Toh W.H., Siddique M.M., Boominathan L., Lin K.W., Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J. Biol. Chem. 2004;279:44713–44722. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- Tomasini R., Mak T.W., Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner S.D., Ventakachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Lane D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Walsh G.S., Orike N., Kaplan D.R., Miller F.D. The invulnerability of adult neurons: A critical role for p73. J. Neurosci. 2004;24:9638–9647. doi: 10.1523/JNEUROSCI.1299-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu Y.X., Hande M.P., Wong A.C., Jin Y.J., Yin Y. TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J. Biol. Chem. 2007;282:29152–29162. doi: 10.1074/jbc.M703408200. [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Cleveland D.W. Aneuploidy: Instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takuechi O., Takeda K., Akira S. TRAM is specifically involved in the toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A., Walker N., Bronson N., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A., et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Caput D., McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- Yao L.J., Zhong Z.S., Zhang L.S., Chen D.Y., Schatten H., Sun Q.Y. Aurora-A is a critical regulator of microtubule assembly and nuclear activity in mouse oocytes, fertilized eggs, and early embryos. Biol. Reprod. 2004;70:1392–1399. doi: 10.1095/biolreprod.103.025155. [DOI] [PubMed] [Google Scholar]
- Yuen K.W., Montpetit B., Hieter P. The kinetochore and cancer: What’s the connection? Curr. Opin. Cell Biol. 2005;17:576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Zaika A.I., Slade N., Erster S.H., Sansome C., Joseph T.W., Pearl M., Chalas E., Moll U.M. ΔNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]