Abstract
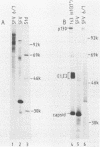
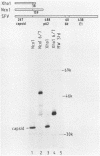
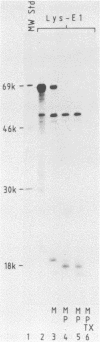
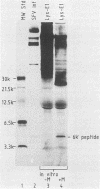
The protease activities responsible for the cotranslational processing of the Semliki Forest virus structural polyprotein were investigated by using an in vitro transcription-translation system. Three cleavages released the individual chains from the nascent polyprotein in the order capsid, p62, 6K (a nonstructural peptide), and E1. We showed directly that the protease activity responsible for the release of the capsid protein resides in the capsid itself: by progressive truncation of the cDNA used for the SP6 transcription, we showed that a precursor containing as few as 38 residues of the p62 protein left at the C terminus of the capsid was still very efficiently cleaved in vitro. We further tested the possibility that serine-219 of the capsid is involved in autoproteolysis by site-directed in vitro mutagenesis. A change in the sequence Gly-Asp-Ser(219)-Gly, a tetrapeptide conserved among several animal serine proteases, to Gly-Asp-Arg-Ser-Thr was shown to completely abolish in vitro cleavage. This supports the notion that the capsid is a serine protease. The role of the capsid protease in the processing of the 6K junctions was then investigated by translations of a hybrid polyprotein in which the capsid and most of the p62 sequences are replaced by those of the secretory protein lysozyme. The cleavages and concomitant appearance of the 6K peptide occurred efficiently and were shown to require the presence of membranes. This demonstrates that the capsid protease is not required for those cleavages and suggests that a membrane-associated host protease is responsible for the cleavage.
Full text
PDF

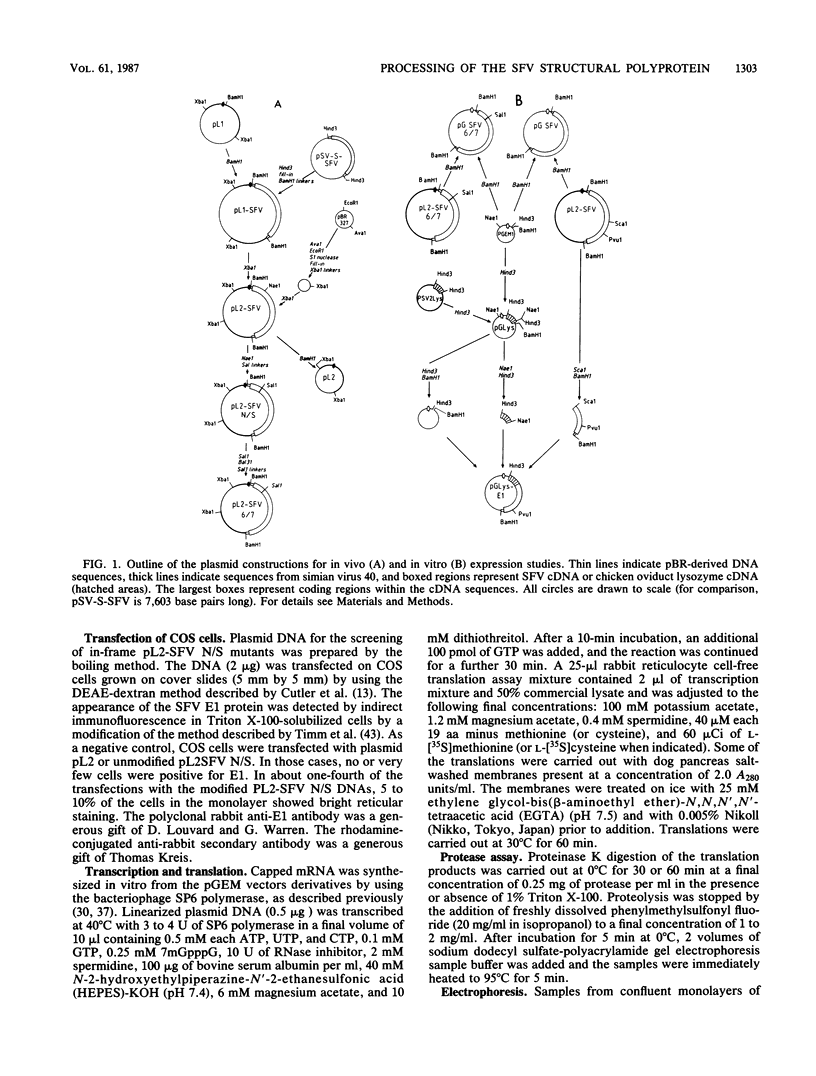


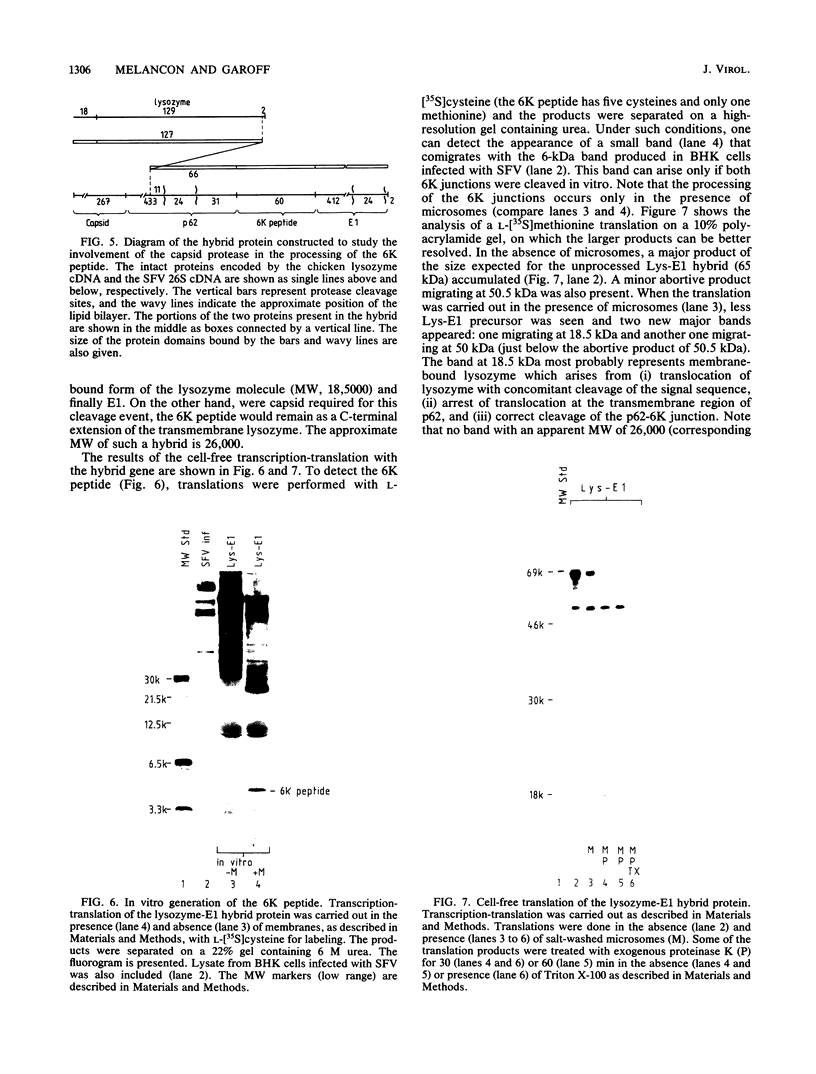
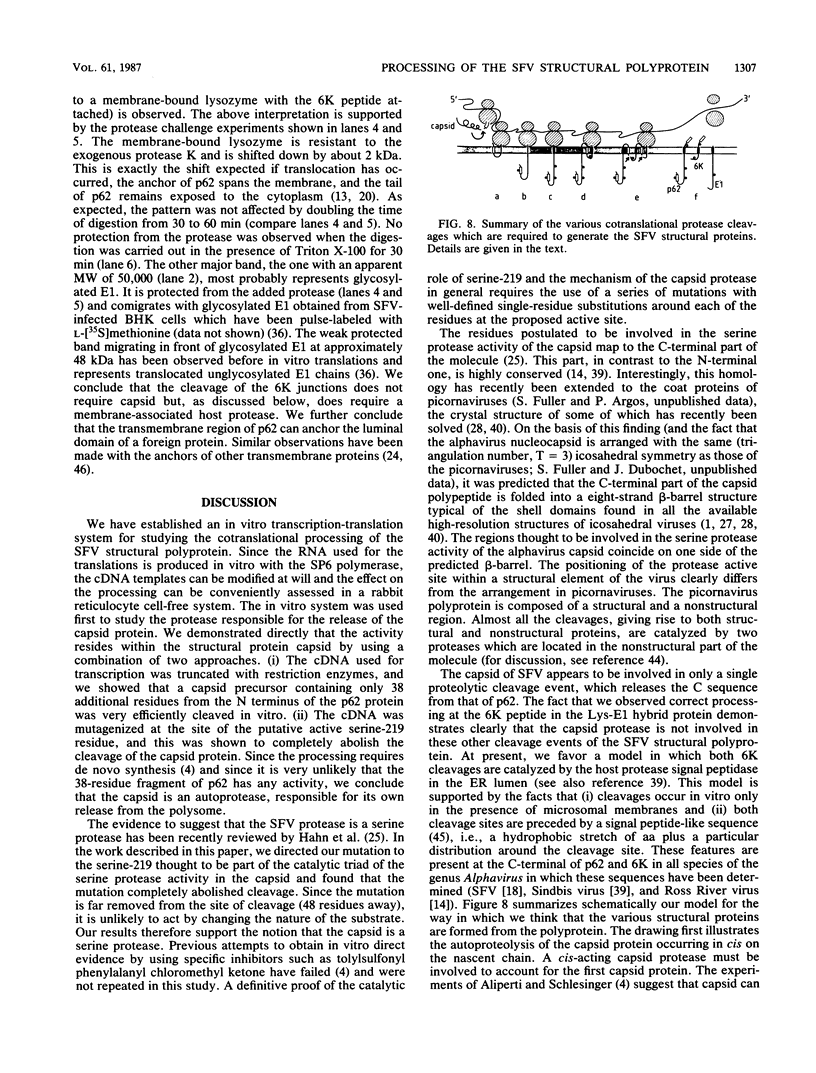


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams G. A., Rose J. K. Incorporation of a charged amino acid into the membrane-spanning domain blocks cell surface transport but not membrane anchoring of a viral glycoprotein. Mol Cell Biol. 1985 Jun;5(6):1442–1448. doi: 10.1128/mcb.5.6.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams G. A., Rose J. K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985 Jul;41(3):1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- Aliperti G., Schlesinger M. J. Evidence for an autoprotease activity of sindbis virus capsid protein. Virology. 1978 Oct 15;90(2):366–369. doi: 10.1016/0042-6822(78)90321-5. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boege U., Wengler G., Wengler G., Wittmann-Liebold B. Primary structures of the core proteins of the alphaviruses Semliki Forest virus and Sindbis virus. Virology. 1981 Aug;113(1):293–303. doi: 10.1016/0042-6822(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Bonatti S., Cancedda R., Blobel G. Membrane biogenesis. In vitro cleavage, core glycosylation, and integration into microsomal membranes of sindbis virus glycoproteins. J Cell Biol. 1979 Jan;80(1):219–224. doi: 10.1083/jcb.80.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatti S., Migliaccio G., Blobel G., Walter P. Role of signal recognition particle in the membrane assembly of Sindbis viral glycoproteins. Eur J Biochem. 1984 May 2;140(3):499–502. doi: 10.1111/j.1432-1033.1984.tb08130.x. [DOI] [PubMed] [Google Scholar]
- Cancedda R., Swanson R., Schlesinger M. J. Viral proteins formed in a cell-free rabbit reticulocyte system programmed with RNA from a temperature-sensitive mutant of Sindbis virus. J Virol. 1974 Sep;14(3):664–671. doi: 10.1128/jvi.14.3.664-671.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Clegg J. C., Kennedy S. I. In vitro synthesis of structural proteins of Semliki Forest virus directed by isolated 26 S RNA from infected cells. FEBS Lett. 1974 Jun 15;42(3):327–330. doi: 10.1016/0014-5793(74)80757-x. [DOI] [PubMed] [Google Scholar]
- Cutler D. F., Melancon P., Garoff H. Mutants of the membrane-binding region of Semliki Forest virus E2 protein. II. Topology and membrane binding. J Cell Biol. 1986 Mar;102(3):902–910. doi: 10.1083/jcb.102.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno L., Rice C. M., Strauss J. H. Ross River virus 26 s RNA: complete nucleotide sequence and deduced sequence of the encoded structural proteins. Virology. 1983 Aug;129(1):170–187. doi: 10.1016/0042-6822(83)90404-x. [DOI] [PubMed] [Google Scholar]
- Davis N. G., Boeke J. D., Model P. Fine structure of a membrane anchor domain. J Mol Biol. 1985 Jan 5;181(1):111–121. doi: 10.1016/0022-2836(85)90329-8. [DOI] [PubMed] [Google Scholar]
- Davis N. G., Model P. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell. 1985 Jun;41(2):607–614. doi: 10.1016/s0092-8674(85)80033-7. [DOI] [PubMed] [Google Scholar]
- Garoff H., Kondor-Koch C., Riedel H. Structure and assembly of alphaviruses. Curr Top Microbiol Immunol. 1982;99:1–50. doi: 10.1007/978-3-642-68528-6_1. [DOI] [PubMed] [Google Scholar]
- Garoff H., Simons K., Dobberstein B. Assembly of the Semliki Forest virus membrane glycoproteins in the membrane of the endoplasmic reticulum in vitro. J Mol Biol. 1978 Oct 5;124(4):587–600. doi: 10.1016/0022-2836(78)90173-0. [DOI] [PubMed] [Google Scholar]
- Garoff H., Söderlund H. The amphiphilic membrane glycoproteins of Semliki Forest virus are attached to the lipid bilayer by their COOH-terminal ends. J Mol Biol. 1978 Sep 25;124(3):535–549. doi: 10.1016/0022-2836(78)90186-9. [DOI] [PubMed] [Google Scholar]
- Garoff H. Using recombinant DNA techniques to study protein targeting in the eucaryotic cell. Annu Rev Cell Biol. 1985;1:403–445. doi: 10.1146/annurev.cb.01.110185.002155. [DOI] [PubMed] [Google Scholar]
- Glanville N., Morser J., Uomala P., Käri5AAINEN L. Simultaneous translation of structural and nonstructural proteins from Semliki-forest-virus RNA in two eukaryotic systems in vitro. Eur J Biochem. 1976 Apr 15;64(1):167–175. doi: 10.1111/j.1432-1033.1976.tb10285.x. [DOI] [PubMed] [Google Scholar]
- Green J., Griffiths G., Louvard D., Quinn P., Warren G. Passage of viral membrane proteins through the Golgi complex. J Mol Biol. 1981 Nov 15;152(4):663–698. doi: 10.1016/0022-2836(81)90122-4. [DOI] [PubMed] [Google Scholar]
- Guan J. L., Rose J. K. Conversion of a secretory protein into a transmembrane protein results in its transport to the Golgi complex but not to the cell surface. Cell. 1984 Jul;37(3):779–787. doi: 10.1016/0092-8674(84)90413-6. [DOI] [PubMed] [Google Scholar]
- Hahn C. S., Strauss E. G., Strauss J. H. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid protein autoprotease. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hogle J. M., Chow M., Filman D. J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985 Sep 27;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Kondor-Koch C., Burke B., Garoff H. Expression of Semliki Forest virus proteins from cloned complementary DNA. I. The fusion activity of the spike glycoprotein. J Cell Biol. 1983 Sep;97(3):644–651. doi: 10.1083/jcb.97.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P., Strachan R., Wallis E., Tabe L., Colman A. Efficient expression of cloned complementary DNAs for secretory proteins after injection into Xenopus oocytes. J Mol Biol. 1984 Dec 15;180(3):615–643. doi: 10.1016/0022-2836(84)90030-5. [DOI] [PubMed] [Google Scholar]
- Lachmi B. E., Glanville N., Keränen S., Läriäinen L. Tryptic peptide analysis on nonstructural and structural precursor proteins from Semliki Forest virus mutant-infected cells. J Virol. 1975 Dec;16(6):1615–1629. doi: 10.1128/jvi.16.6.1615-1629.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perara E., Lingappa V. R. A former amino terminal signal sequence engineered to an internal location directs translocation of both flanking protein domains. J Cell Biol. 1985 Dec;101(6):2292–2301. doi: 10.1083/jcb.101.6.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Arnold E., Erickson J. W., Frankenberger E. A., Griffith J. P., Hecht H. J., Johnson J. E., Kamer G., Luo M., Mosser A. G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985 Sep 12;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Simmons D. T., Strauss J. H. Translation of Sindbis virus 26 S RNA and 49 S RNA in lysates of rabbit reticulocytes. J Mol Biol. 1974 Jun 25;86(2):397–409. doi: 10.1016/0022-2836(74)90027-8. [DOI] [PubMed] [Google Scholar]
- Swank R. T., Munkres K. D. Molecular weight analysis of oligopeptides by electrophoresis in polyacrylamide gel with sodium dodecyl sulfate. Anal Biochem. 1971 Feb;39(2):462–477. doi: 10.1016/0003-2697(71)90436-2. [DOI] [PubMed] [Google Scholar]
- Timm B., Kondor-Koch C., Lehrach H., Riedel H., Edström J. E., Garoff H. Expression of viral membrane proteins from cloned cDNA by microinjection into eukaryotic cell nuclei. Methods Enzymol. 1983;96:496–511. doi: 10.1016/s0076-6879(83)96043-3. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Yost C. S., Hedgpeth J., Lingappa V. R. A stop transfer sequence confers predictable transmembrane orientation to a previously secreted protein in cell-free systems. Cell. 1983 Oct;34(3):759–766. doi: 10.1016/0092-8674(83)90532-9. [DOI] [PubMed] [Google Scholar]
- Zerial M., Melancon P., Schneider C., Garoff H. The transmembrane segment of the human transferrin receptor functions as a signal peptide. EMBO J. 1986 Jul;5(7):1543–1550. doi: 10.1002/j.1460-2075.1986.tb04395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]