Abstract
During mitosis in metazoans, segregated chromosomes become enclosed by the nuclear envelope (NE), a double membrane that is continuous with the endoplasmic reticulum (ER). Recent in vitro data suggest that NE formation occurs by chromatin-mediated reorganization of the tubular ER; however, the basic principles of such a membrane-reshaping process remain uncharacterized. Here, we present a quantitative analysis of nuclear membrane assembly in mammalian cells using time-lapse microscopy. From the initial recruitment of ER tubules to chromatin, the formation of a membrane-enclosed, transport-competent nucleus occurs within ∼12 min. Overexpression of the ER tubule-forming proteins reticulon 3, reticulon 4, and DP1 inhibits NE formation and nuclear expansion, whereas their knockdown accelerates nuclear assembly. This suggests that the transition from membrane tubules to sheets is rate-limiting for nuclear assembly. Our results provide evidence that ER-shaping proteins are directly involved in the reconstruction of the nuclear compartment and that morphological restructuring of the ER is the principal mechanism of NE formation in vivo.
Introduction
A hallmark of eukaryotic cellular organization is the compartmentalization of distinct metabolic functions into membrane-enclosed organelles. The inheritance of these organelles is an essential homeostatic process in dividing cells. One of the most morphologically elaborate organelles is the ER, which is organized into membrane tubules and sheets (Shibata et al., 2006). The relative distribution of these ER domains changes continuously as cells move through the cell cycle. Dynamic processes involving fission and homotypic fusion of membranes maintain the overall architecture of the ER (Powell and Latterich, 2000; Fagarasanu et al., 2007).
Connected to the ER is the nuclear envelope (NE), an extensive spheroid membrane sheet that is composed of two lipid bilayers that enclose the nuclear genome. The outer and inner nuclear membranes are fused at sites where nuclear pore complexes (NPC) are inserted. NPCs are large multiprotein channels that mediate nucleocytoplasmic trafficking (Fahrenkrog et al., 2004). Despite the physical connection between NE and ER, the nuclear membrane contains a unique set of proteins that are not present in the ER. One class of proteins is localized in the outer NE and provides contacts to the cytoskeleton (Worman and Gundersen, 2006). A second class of proteins preferentially associates with the inner NE and has been shown to interact with the nuclear lamina and chromatin (for reviews see D'Angelo and Hetzer, 2006; Stewart et al., 2007).
Only recently have proteins been identified that exclusively localize to ER tubules and are excluded from flat membrane sheets such as the NE (Voeltz et al., 2006). This class of ER membrane proteins contains the reticulons and the functionally related protein DP1/YOP1, both of which are expressed across all eukaryotic species (Yang and Strittmatter, 2007). These proteins contain a domain of ∼200 amino acids, including two hydrophobic segments, which are thought to form a hairpin in the membrane. The ability of reticulons to oligomerize appears to allow these proteins to generate and/or stabilize tubules in vitro (Shibata et al., 2008). Although the mechanism of reticulon function is still being characterized, it is known that the levels of these proteins can shift the balance between the tubular network and the flat membrane sheets (Voeltz et al., 2006).
In addition to differences in protein composition, the NE and the ER exhibit a striking difference in their dynamic behavior during mitosis. Although it is well documented that the NE completely breaks down in prophase of metazoa (for reviews see Lenart and Ellenberg, 2003; Margalit et al., 2005), it has become clear that the ER network remains intact during cell division (Ellenberg et al., 1997). EM data have recently demonstrated that the mitotic ER is a highly interconnected network of tubules during metaphase (Puhka et al., 2007). Considering the dramatic difference in the fates of NE and ER during mitosis, questions arise as to how the disassembled NE components are dispersed, and consequently, how the nuclear membrane is reformed around the segregated chromosomes. Historically, data obtained from biochemical fractionation of ER membranes supported a model where the NE is fragmented into vesicles that remain distinct from the ER (Collas and Poccia, 2000). In contrast, live imaging has revealed that GFP-tagged NE components reside in the mitotic ER, which suggests that the lipids and membrane proteins of the NE retract into the tubular ER network (Ellenberg and Lippincott-Schwartz, 1999; Daigle et al., 2001; Anderson and Hetzer, 2007). The idea of mitotic NE retraction implies that at the end of mitosis, the nuclear membrane reemerges from the ER and does not form by fusion of a separate pool of vesicles (Collas and Poccia, 2000). In support of such a mechanism, in vitro analyses suggest that the intact tubular ER is required for NE formation (Anderson and Hetzer, 2007; Puhka et al., 2007). Using live imaging, we were able to provide new insights into the membrane dynamics of NE formation. We showed that the ER is targeted to chromosomes via tubule-end binding and subsequently immobilized on the chromatin surface. This chromatin-bound network then flattens and seals into a closed NE (Anderson and Hetzer, 2007). In vitro data also demonstrate that the targeting of membranes to chromatin is at least partially regulated by NE-specific transmembrane proteins binding to DNA and/or chromatin (Pyrpasopoulou et al., 1996; Ulbert et al., 2006; Anderson and Hetzer, 2007). The general mechanism of membrane reshaping from tubules to the double lipid-bilayer sheets of the NE is unclear, and in vivo evidence for such a process remains sparse.
Here, we set out to determine if changes in ER morphology play a role in NE formation in vivo. A novel assay to study the dynamic relationship between the organization of the ER and forming NE in dividing cells was developed. This assay and other live-cell imaging approaches provide direct evidence that NE formation occurs by the reorganization of the ER on chromatin. Furthermore, our studies revealed that reticulons and DP1 act to regulate NE formation. In addition, we show that NE expansion is mechanistically coupled to the ER organization.
Results
Displacement of reticulons from the reforming NE
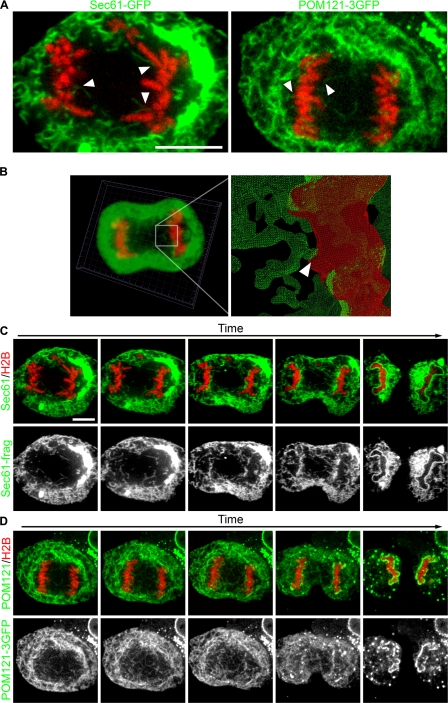
To determine if chromatin-mediated reshaping of ER tubules is the principal mechanism of NE formation in vivo, we compared ER membrane dynamics during mitosis in different tissue culture cells. We selected U2OS cells, an osteosarcoma cell line, because they remain relatively flat during mitosis, making them suitable for high-resolution time-lapse microscopy. To directly test if ER tubules are recruited to chromatin, we cotransfected the ER marker Sec61β fused to GFP (Sec61-GFP) as well as the histone subunit H2B fused to tdTomato (H2B-tdTomato) to visualize chromatin (Shaner et al., 2004). In interphase, Sec61-GFP stained the NE and the ER, highlighting the continuity of the two membrane systems (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200805140/DC1). In metaphase, chromatin was essentially free of membranes, and, as expected from recent observations, the ER remained intact during mitosis. As cells progressed to telophase, the cell cycle phase when NE reformation occurs, ER membranes contacted chromatin via tubule-end binding (Fig. 1 A). Once a few tubules were immobilized, additional tubules sliding along each other were captured on chromatin and eventually coated the entire chromatin surface (Fig. 1 C and Video 1), confirming that ER recruitment to chromatin is an early event in nuclear assembly.
Figure 1.
ER tubules bind and collapse on chromatin in vivo. U2OS cells were transfected with GFP fusion proteins and H2B-tdTomato, then imaged through mitosis with real-time spinning-disk confocal microscopy. (A) Large images of the initial interactions of Sec61-GFP and POM121-3GFP are shown. (B) z stacks were acquired every 30 s and initial membrane–chromatin contact points were characterized by 3D reconstruction. Arrowheads indicate initial membrane–chromatin contact. (C) Sec61-GFP was imaged every 5 s to monitor ER dynamics. (D) POM121-3GFP was imaged every 10 s to monitor the redistribution from the ER to the forming NE. Bars, 10 μm.
To confirm that the linear structures seen in the two-dimensional images of membrane–chromatin contact points were tubules, confocal z stacks of cells expressing Sec61-GFP and H2B-tdTomato were acquired every 30 s. After 3D reconstruction, membrane tubules contacting the surface of chromatin were clearly resolved (Fig. 1 B), which confirms that the linear structures represent membrane tubules.
We have recently shown that transmembrane components of the NPC such as POM121 are dispersed throughout the ER network during NE breakdown (Anderson and Hetzer, 2007). Therefore, we expected that POM121 should be incorporated into the reforming NE via ER tubules connected to chromatin. To test this, we expressed the NE marker POM121-GFP and found that it was indeed present in tubules that initially contacted chromatin (Fig. 1, A and D; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200805140/DC1). As NE formation proceeded, POM121-3GFP was cleared from surrounding tubules and accumulated on the chromatin surface, which supports the idea that NE proteins are recruited to chromatin from the ER network. Collectively, these results suggest that the ER serves as the precursor membrane for the NE.
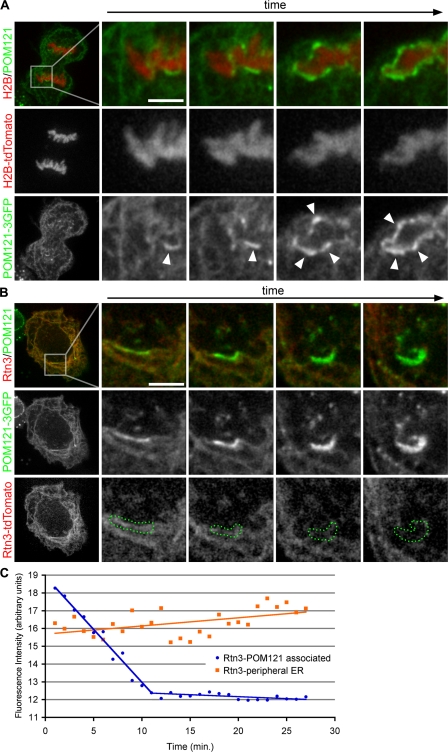
In combination with previous data, these results imply a two-step process of NE formation in which ER tubules first become immobilized on chromatin and then reorganize into flat membrane sheets. As a consequence of such a stepwise membrane reorganization process, chromatin-bound ER tubules should initially contain tubulating proteins like reticulons. However, as the transition into sheets progresses, these proteins should be displaced because they are ultimately excluded from flat membranes (Voeltz et al., 2006). To directly test for this possibility, we coexpressed POM121-3GFP and the tubular ER protein reticulon 3 (Rtn3) fused to tdTomato, and imaged their dynamic behavior by fluorescence microscopy in real time. As expected, POM121-3GFP accumulated at the forming NE and therefore served as a reporter for membrane–chromatin contact sites (Fig. 2 A). Similar to POM121-3GFP, Rtn3-dtTomato was detectable on the surface of chromatin (Fig. 2 B), but as cells moved through telophase, POM121-3GFP accumulated in the reforming NE, whereas the fluorescence intensity of Rtn3-tdTomato at the forming NE gradually decreased to background levels once the NE was formed (Fig. 2 B). The rate of reticulon decrease at the forming NE was steady for ∼10 min from the start of POM121 accumulation until the fluorescence signal reached background levels, as measured by threshold selection of regions of bright POM121-3GFP (Fig. 1 C). Strikingly, during NE formation, the intensity of Rtn3-tdTomato in the surrounding ER increased. This suggests that in order to generate an extensive membrane surface around chromosomes, a large number of tubules were flattened, and a considerable amount of reticulons were displaced from chromatin-bound membranes to surrounding ER tubules (Fig. 1 C). These results uncovered a reciprocal behavior in protein dynamics in the forming nuclear membrane, with reticulons being displaced and proteins that accumulate in NE sheets being recruited. This supports the idea that NE formation occurs by the gradual reorganization of ER tubules into flat membrane sheets on the surface of chromatin. We cannot exclude the possibility that ER patches contribute to NE formation; however, such a contribution would likely be minor, as only tubules are detected in the mitotic ER (Puhka et al., 2007).
Figure 2.
Rtn3 is removed from NE-forming tubules. (A) U2OS cells were transfected with POM121-3GFP and H2B-tdTomato to monitor the accumulation of POM121 at the forming NE. Arrowheads indicate forming NE. (B) U2OS cells were transfected with POM121-3GFP and Rtn3-tdTomato to monitor the dynamic localization of these proteins at the forming NE. Bars, 5 μm. (C) Intensity threshold selections of accumulating POM121 were used to define the regions of NE formation (dotted green regions in B). Fluorescence intensity of Rtn3-tdTomato was measured in these regions through NE formation (blue circles); intensity was also measured in the peripheral ER (orange squares). Cells were imaged every 30 s.
Kinetics of NE formation
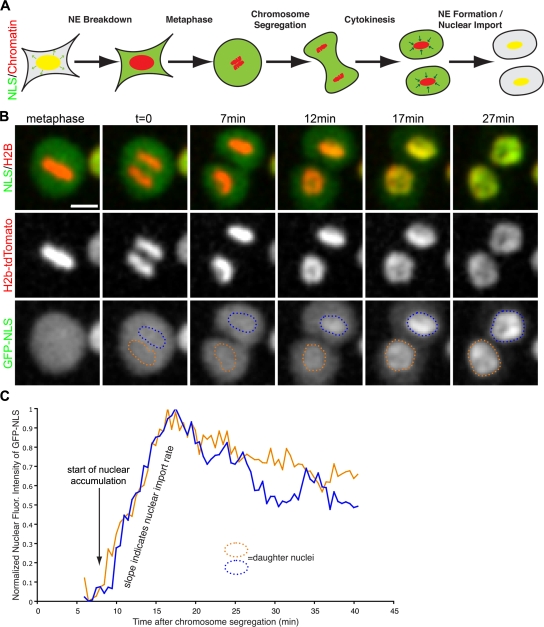
Because the observed ER membrane reorganization events occur on the highly condensed surface of chromatin (Mora-Bermudez et al., 2007), visualization of individual tubules and their transition into sheets by light microscopy in living cells was not feasible. Therefore, we developed an assay to study the kinetics and molecular mechanism of NE formation in real time in proliferating cells. The basic principles of the assay design are outlined in Fig. 3 A. To monitor mitotic progression and provide spatial reference for NE formation, we expressed histone H2B-tdTomato. To determine the exact time when an intact NE has formed, we cotransfected a triple GFP fused to the SV40 large T antigen NLS (GFP-NLS). The rationale for using this approach was that the nuclear accumulation of GFP-NLS would require an intact NE. This assumption was made because the rate of diffusion of a protein the size of our GFP-NLS reporter through a small hole the size of an NPC is approximately seven times greater than the rate of facilitated nucleocytoplasmic transport (Ribbeck and Gorlich, 2001). Thus, by cotransfecting GFP-NLS and H2B-tdTomato, we were able to determine the time from metaphase-to-anaphase transition to nuclear accumulation of GFP-NLS, encompassing the onset of NE formation to its completion.
Figure 3.
Assay developed to measure NE-formation kinetics. (A) Schematic of the dynamic localization of the reporter genes for nuclear localization (NLS) and chromatin during mitosis. Yellow represents the colocalization of NLS (green) and chromatin (red) in the intact nucleus. (B) U2OS were transfected with NLS fused to 3GFPs (GFP-NLS) and H2B-tdTomato, then imaged every 30 s through mitosis with live-cell confocal microscopy. Bar, 20 μm. (C) After chromosome segregation, regions over chromatin were selected, and GFL-NLS intensity was measured over time. Time = 0 corresponds to when the chromatid clusters have completely separated, and blue and orange traces correspond to the nuclear influx of GFP-NLS in each daughter nucleus.
We first used this assay to determine the exact time of NE formation in vivo, and the metaphase-to-anaphase transition, which was detectable by the separation of the sister chromatid clusters, was defined as time zero (Fig. 3 B). In metaphase, GFP-NLS localized diffusely in the cytoplasm (Fig. 3 B, left). During cytokinesis, ∼5 min after the onset of chromosome segregation, GFP-NLS was cytoplasmic, but accumulation of GFP-NLS into the daughter nuclei was observed after 10.2 min, followed by nuclear expansion (Fig. 3 B). To accurately determine the onset of nuclear accumulation of GFP-NLS, the fluorescence intensity of GFP was measured over the areas occupied by the daughter chromatin clusters from anaphase into G1, and then GFP-NLS accumulation measurements were synchronized starting from chromosome segregation (Fig. 3 C). In summary, our live-imaging assay allowed us to determine that NE formation occurs within ∼10 min in U2OS cells. Similar results were obtained in HeLa cells (unpublished data).
To validate the assumption that accumulation of GFP-NLS is an accurate marker for NE closure, the timing of NE formation was determined by the visual appearance of a smooth rim around chromatin, an unequivocal sign of a closed NE (Fig. 1 C). Chromosomes were completely enclosed by a smooth membrane rim within a mean 11.9 ± 2.3 min after chromosome segregation (Table I). Because the accurate assessment of nuclear rim formation requires higher resolution than the measurement of GFP-NLS accumulation, membrane dynamics were imaged at 63× magnification, whereas GFP-NLS accumulation was imaged at 20×. The increased exposure to laser light with higher magnification imaging likely explains the time discrepancy in timing of NE formation of these two methods. However, it is clear that NE formation occurs within 12 min in U2OS cells.
Table I. Kinetic events of NE formation.
| Nuclear accumulation
|
Rim formation
|
|||||||
|---|---|---|---|---|---|---|---|---|
| From chromosome segregation |
Standard deviation | n | P-value | From chromosome segregation |
Standard deviation |
n | P-value | |
| Wild type | 10.2 min | 0.2 min | 85 | n/a | 11.9 min | 2.3 min | 28 | n/a |
| V5-Rtn3 | 21.9 min | 2.0 min | 49 | <0.001 | nd | |||
| V5-Rtn4 | 26.3 min | 2.4 min | 51 | <0.001 | 25.4 min | 13.8 min | 29 | <0.001 |
| V5-DP1 | 25.2 min | 3.8 min | 34 | <0.001 | nd | |||
| Scramble RNA | 10.6 min | 0.4 min | 19 | 0.38 | 12.9 min | 2.7 min | 20 | 0.2 |
| Reticulon siRNA | 7.8 min | 0.2 min | 57 | <0.001 | 9.7 min | 2.2 min | 17 | 0.003 |
Time from chromosome segregation to nuclear accumulation and NE rim formation are represented as mean values in minutes. Standard deviation is provided alone with number of cells measured (n) and t test–calculated p-values. n/a, not applicable; nd, not determined.
Overexpression of reticulons delay NE formation
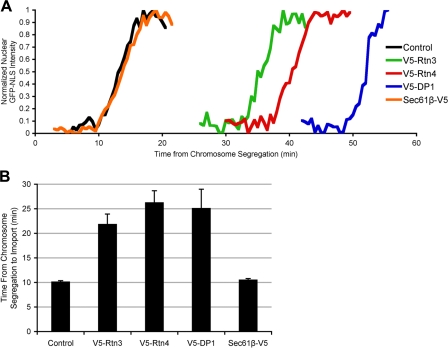
Having established a kinetic framework for NE assembly, we next wanted to study the molecular mechanisms involved in this pivotal membrane-remodeling event. The release of reticulons from the forming NE (Fig. 2; Anderson and Hetzer, 2007) suggested that these ER tubule-forming proteins might act as antagonists of membrane flattening. To directly test this possibility, human Rtn3 or Rtn4, each fused to a V5 tag at their N termini, were transfected into U2OS cells. Both V5-Rtn3 and V5-Rtn4 were expressed at the expected size and specifically localized to ER tubules, which is similar to the immunostaining pattern of endogenous Rtn4 (Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200805140/DC1). Because the analysis of reticulon overexpression using the NE kinetic assay required transfection of several genes at once, it was important to determine the cotransfection efficiency. Although GFP-NLS and H2B-tdTomato were visualized using their intrinsic fluorescence, V5-tagged reticulon proteins were detected by indirect immunofluorescence using antibodies against the V5 epitope (Fig. S1 C). More than 94% of cells expressing detectable levels of both fluorescent reporter proteins also expressed detectable levels of the V5-tagged proteins (Fig. S1 D). These data demonstrate that triple transfection efficiency into U2OS cells was robust, thus allowing us to analyze the effect of protein overexpression on NE formation kinetics.
Using this approach, we found that elevated reticulon levels significantly delayed NE formation when compared with control cells expressing only the recombinant GPF-NLS and H2B-tdTomato proteins (Fig. 4, A and B). This effect was specific for reticulons because the overexpression of full-length V5-Sec61β, a transmembrane component of the ER protein translocator, did not delay NE formation (Fig. 4, A and B). On average, in control cells, NEs were closed after 10.2 min, whereas the overexpression of Rtn3 caused a significant delay to 21.9 min and the overexpression of Rtn4 caused a delay to 26.3 min (Table I). These data suggest that overtubulation counteracts NE membrane flattening and that ER organization by reticulons and NE formation are tightly linked processes. It is important to note that the observed delay in the onset of nuclear accumulation is not caused by a defect in NPC assembly because the rate of nuclear accumulation in the delayed cells was comparable to that of control cells (Fig. 4 A).
Figure 4.
ER tubule-shaping proteins delay NE formation. U2OS cells were transfected with GFP-NLS and H2B-tdTomato (control), with V5-tagged ER-shaping protein (Rtn3, Rtn4, or DP1), or with the V5-tagged ER protein Sec61β, then NE formation was analyzed. (A) Representative traces of nuclear GFP-NLS intensity during NE formation are shown. (B) Mean times from chromosome segregation to the onset of nuclear accumulation are plotted with standard error bars.
Similar to elevated levels of reticulons, overexpression of V5-tagged DP1 (Fig. S1, A–C), a reticulon-like protein, delayed NE formation (Fig. 4, A and B). Nuclear accumulation of GFP-NLS was significantly delayed when compared with control cells and only occurs after 25.2 min as measured from chromosome segregation (Table I). DP1 is composed of the two membrane-inserting, hydrophobic domains but lacks the N-terminal extension found in most reticulons. Importantly, DP1 can be considered a functional homologue to the reticulon in the context of membrane shaping because it localizes to the tubular ER in vivo (Voeltz et al., 2006) and it's yeast homologue, yop1, has been demonstrated to directly tubulate membranes in vitro (Hu et al., 2008).
These data suggest that increased levels of DP1 caused a delay in NE formation, possibly due to the induction of membrane curvature and tubulation. They also provide strong support for the general idea that NE formation in vivo occurs by the reshaping of ER tubules.
NE protein recruitment is unaffected by Rtn4 overexpression
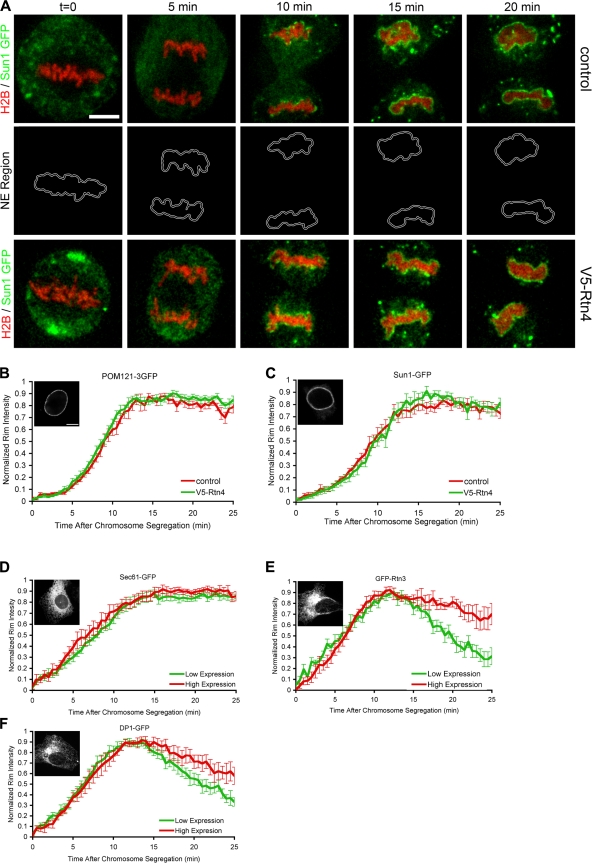
Although we did not detect any differences in ER dynamics and protein diffusion through the membrane network as determined by fluorescence recovery after photobleaching studies (unpublished data), in principal it was possible that Rtn4 overexpression inhibited the targeting of ER membranes and the NE-specific membrane proteins to chromatin. To examine this possibility, an assay was developed that allowed us to monitor the recruitment of GFP-tagged ER membrane proteins around H2B-tdTomato–labeled chromatin (Fig. 5 A). Because the NE forms tightly around the segregated chromatin clusters, a thin border around the edge of the chromatin clusters was selected and the GFP fluorescence intensity was measured from the metaphase-to-anaphase transition to G1 (Fig. 5 A).
Figure 5.
NE protein recruitment is not delayed by Rtn4 overexpression. A real-time microscopic assay was established to measure the recruitment of GFP fusion proteins to the forming NE. (A) U2OS cells were transfected with H2B-tdTomato and Sun1-GFP with or without the overexpression of V5-Rtn4 and imaged every 30 s through mitosis. A thin region surrounding the chromatin (NE region) was selected to measure the intensity of Sun1-GFP at the forming NE. (B) Fluorescence intensity of POM121-3GFP was measured at the NE region in control cells (red) or cells expressing V5-Rtn4 (green) starting at the onset of chromosome segregation, then normalized, averaged, and plotted with standard error bars. (C) Cells expressing Sun1-GFP were analyzed as in B. (D) U2OS cells were grouped for high (red) and low (green) expression of GFP-Sec61, and fluorescence intensity was measured at the forming NE as in A. (E) Cells expressing GFP-Rtn3 were analyzed as in D. (F) Cells expressing DP1-GFP were analyzed as in D. Interphase localization of each GFP fusion protein is shown in the insets in B–F. Bars: (A) 10 μm; (B) 5 μm.
Using this assay, we found that Rtn4 overexpression had no effect on the recruitment of the transmembrane nucleoporin, POM121 (Fig. 5 B), or Sun1 (Fig. 5 C), an inner nuclear membrane protein that has been implicated in the maintenance of nuclear membrane spacing (Crisp et al., 2006). These data suggest that the inhibition in NE formation caused by the overexpression of reticulons is caused by a defect in membrane flattening and not membrane targeting to chromatin. These results also suggest that NE formation in vivo, similar to the situation in cell-free assembly systems, is a two-step process where the rapid first step of membrane targeting to chromatin can be distinguished from the slower second step of enclosing chromatin by a sealed nuclear membrane.
Rtn3 and DP1 removal from the NE is concentration dependent
Because protein diffusion was not detectably affected by the overexpression of V5-Rtn4, we predicted that reticulon/DP1 removal from chromatin-bound membranes in these overexpressing cells might be causing a delay in NE formation. To test this possibility, the removal of fluorescently tagged Rtn3 and DP1 was imaged. Cells were grouped based on their expression level, and NE removal rates were compared.
Both Rtn3 and DP1 were fused to GFP, Rtn3 at the N terminus, and DP1 at the C terminus. These constructs localized to the tubule ER (Fig. 5, E and F, insets) and exhibited limited mobility when tested with fluorescence recover after photobleaching (not depicted), which is consistent with published observations (Shibata et al., 2008) and suggests that GFP-Rtn3 and DP1-GFP are functional. U2OS cells were transfected with H2B-tdTomato along with Sec61-GFP, GFP-Rtn3, or DP1-GFP, and imaged through mitosis. GFP intensity at the forming NE was measured as in Fig. 5 A, and cells were divided into low and high GFP expression. Localization of Sec61-GFP at the forming NE was similar with low or high expression levels (Fig. 5 D). However, high expression of GFP-Rtn3 or DP1-GFP caused a delay in the release of these proteins from the forming NE (Fig. 5, E and F). These data suggest that the delay in NE formation observed in cells overexpressing reticulon/DP1 is caused by the reduced release of these proteins from the chromatin-bound membrane, which may cause a delay in tubule to sheet transition.
Membrane flattening is blocked by the overexpression of Rtn4
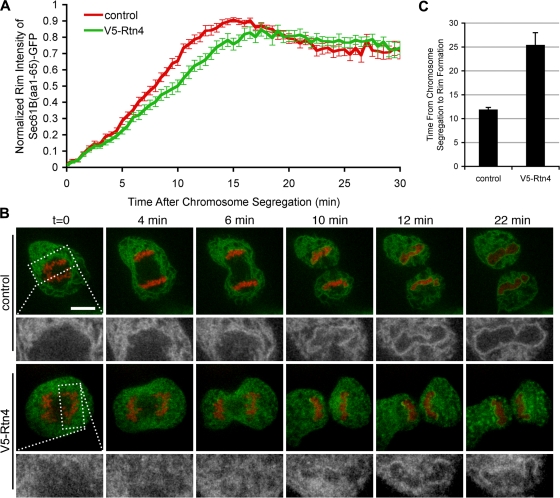
Collectively, findings thus far suggest a tug-of-war–like mechanism of NE formation in which the intrinsic propensity of the ER to move from tubules to flat sheets is shifted toward sheet formation by membrane immobilization on chromatin. To further explore this hypothesis, cells were transfected with Sec61-GFP and H2B-tdTomato, and imaged through mitosis with or without the overexpression of V5-Rtn4. The intensity of Sec61-GFP on chromatin was comparable under both conditions for the first 3 min; however, Sec61-GFP recruitment was delayed at later time points by the overexpression of V5-Rtn4 (Fig. 6 A). As described, a smooth rim, an unequivocal sign of an assembled NE, formed in control cells in 11.9 ± 2.3 min (Fig. 6, B and C; and Table I). When V5-Rtn4 was overexpressed, membranes rapidly targeted to chromatin (Fig. 5 A); however, the formation of a smooth rim was significantly inhibited, taking 25.4 ± 13.8 min (Fig. 6, B and C; and Table I). These data further suggest that overexpression of Rtn4 specifically antagonizes the second step of NE formation where, after ER tubules are targeted to chromatin, they are reshaped into flat membrane sheets.
Figure 6.
NE rim formation is delayed by the overexpression of Rtn4. U2OS cells were transfected with Sec61-GFP and H2B-tdTomato, and imaged every 30 s in real time with confocal microscopy to quantify the time required for membrane rim formation. (A) Recruitment assay described in Fig. 6 was used to measure the recruitment of Sec61-GFP in control cells (red) and cells overexpressing Rtn4 (green). (B) Control cells and cells expressing V5-Rtn4 were imaged to focus on the membrane dynamics at the forming NE (high-magnification view in the bottom rows). (C) Time from chromosome segregation to nuclear rim formation was measured and averaged. Error bars represent standard error.
Lowering the levels of endogenous reticulons accelerates NE formation
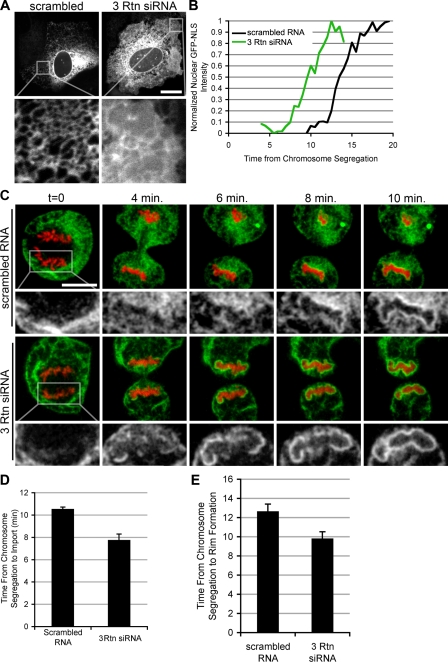
The observation that increased levels of reticulons inhibited NE formation raised the interesting possibility that displacement of reticulons might be a crucial step in NE formation. If this were the case, the rate of NE formation should be inversely proportional to the levels of reticulon proteins in the ER. To test this possibility, we measured the time of NE formation in control cells versus cells in which the overall concentrations of reticulons had been reduced by siRNA-mediated knock down. Because studies in yeast have shown that the double knockout of both reticulons, rtn1 and rtn2, was necessary to cause a transformation of the tubular ER into membrane sheets (Voeltz et al., 2006), we expected that a similar redundancy would exist in mammalian cells where four reticulon isoforms have been found. To test for a potential redundancy in reticulon function, we knocked down Rtn1, Rtn3, and Rtn4 individually or in combination. Reduction in reticulon mRNA levels was confirmed by quantitative PCR. Transfection of siRNA oligos against Rtn1, Rtn3, and Rtn4 simultaneously caused a 94, 74, and 94% reduction mRNA levels of Rtn1, 3, and 4, respectively, in comparison to scrambled RNA. As expected, when U2OS cells were transfected with Sec61-GFP and scrambled RNA, the ER contained an extensive tubular network (Fig. 7 A) in 83% of the cells imaged. In striking contrast, in cells in which Rtn1, Rtn3, and Rtn4 levels were reduced, only 34% of the cells contained a visible tubular network; the other 66% of the cells exhibited an altered ER morphology, which was almost entirely composed of membrane sheets (Fig. 7 A). The knockdown of each single reticulon had no detectable change in ER morphology (unpublished data).
Figure 7.
Reticulon knockdown accelerates NE formation. (A) U2OS cells were transfected with Sec61-GFP and scrambled RNA oligos or siRNA oligos directed against Rtn1, Rtn3, and Rtn4 (3 Rtn siRNA). ER morphology was then analyzed with confocal microscopy. Bar, 20 μm. (B) Cells transfected with scrambled RNA or 3 Rtn siRNA were analyzed using the NE formation kinetics assay as in Fig. 5. (C) Nuclear rim was analyzed as in Fig. 6, with cells transfected with either scrambled RNA or 3 Rtn siRNA. (D) The time of NE formation was averaged for cells transfected with scrambled RNA and 3 Rtn siRNA. Error bars represent standard error. (E) Time from chromosome segregation to rim formation was averaged. Error bars represent standard error.
Having established conditions to knock down reticulons to levels that alter the overall morphology of the ER, we performed our kinetic analysis in control and triple-siRNA knockdown cells. Strikingly, NE formation was accelerated in cells with reduced levels of reticulons (Fig. 7 B). Although NEs formed on average after 10.6 ± 0.4 min from chromosome segregation in cells transfected with scrambled RNA, reticulon knockdown resulted in an acceleration of nuclear assembly to 7.8 ± 0.2 min (Fig. 7 D and Table I), providing the first evidence that the flattening of the ER might be rate limiting for NE formation.
Consistent with increased rates of membrane flattening, the appearance of a smooth membrane rim around chromatin, an unequivocal sign of a closed NE, was detected earlier in cells treated with siRNAs against reticulons when compared with control cells (Fig. 7 C). Rim formation occurred in a mean of 12.9 ± 2.7 min from chromosome segregation in cells transfected with scrambled RNA, and was accelerated to 9.7 ± 2.2 min with the knockdown of Rtn1, Rtn3, and Rtn4 (Fig. 7 E and Table I). Collectively, these results suggest that the concentration of reticulons is rate-limiting for NE formation. It is possible that the reduction of reticulons partially converts the mitotic ER into membrane sheets, which more efficiently contribute to nuclear membrane formation. Alternatively, the transition from tubules to sheets of membrane bound to chromatin may be accelerated by the decrease in reticulon concentration.
Rtn4 overexpression inhibits nuclear expansion
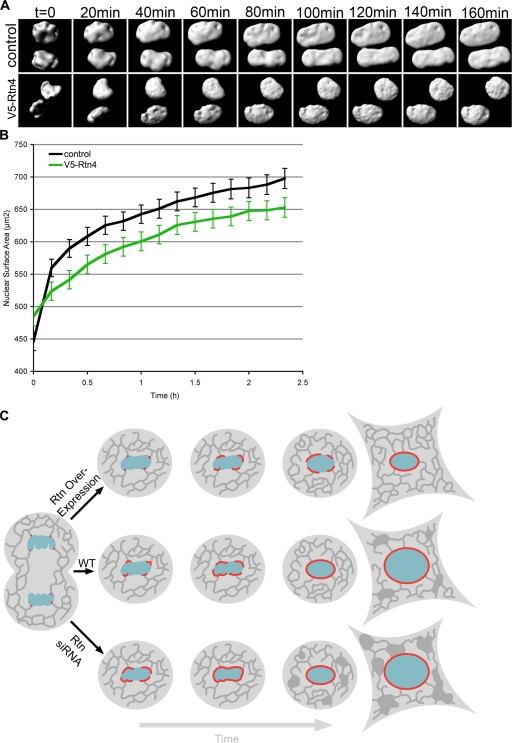
Our data revealed a tight link between the organization of the ER and NE formation. Because the closed NE is continuous with the surrounding ER network, and nuclei grow in size as they move through interphase (Maul et al., 1977), we wanted to investigate if reticulons might be involved in NE expansion. We have recently shown that nuclear expansion in vitro requires connection with the peripheral ER (Anderson and Hetzer, 2007), which suggests that membranes feed into the expanding NE through connections with ER tubules. To determine if hypertubulation inhibits nuclear expansion, the effect of Rtn4 overexpression during G1 was analyzed. Because GFP-NLS targets diffusely in the nucleus, it provides a close approximation of NE surface area. Using 3D reconstructions of confocal z stacks from nuclei stained with GFP-NLS, nuclear surface area can be accurately measured (D'Angelo et al., 2006). We found that asynchronously growing cells have a mean nuclear surface area of 1,063.3 ± 270.1 μm2. Nuclear growth was then monitored starting when the NE was fully formed, as judged by the nuclear accumulation of GFP-NLS. NE expansion was significantly impaired in cells overexpressing V5-Rtn4 compared with control cells (Fig. 8, A and B). Interestingly, nuclear growth was not accelerated by the triple knockdown of reticulons, which suggests that there is an additional rate-limiting step in nuclear expansion (unpublished data). These data suggest that the membrane-shaping proteins control nuclear size and that the propagation of the NE–ER membrane continuum during cell division is mechanistically linked.
Figure 8.
Nuclear expansion is inhibited by Rtn4. U2OS cells were transfected with GFP-NLS and imaged every 10 min in 3D from mitosis through G1 by acquiring z stacks using confocal microscopy. (A) Nuclear surfaces were reconstructed from z stacks for control cells and cells overexpressing V5-Rtn4. (B) The surface areas of expanding nuclei were synchronized starting after nuclear accumulation of GFP-NLS and then averaged. Error bars indicate standard error. (C) Schematic shows the affects of ER-shaping protein concentration on NE formation and expansion, where the ER is represented in dark gray, chromatin is represented in blue, and red represents the forming NE.
Discussion
At the end of mitosis, nuclear assembly is an essential process in establishing eukaryotic cell compartmentalization. Our data provide the first detailed kinetic analysis of NE formation in dividing cells, and uncover novel molecular regulators of this membrane-shaping process. A major conclusion from these studies is that the NE is the result of a massive reorganization of the tubular ER network on chromatin. Surprisingly, the displacement of reticulons is rate-limiting for nuclear assembly, which suggests a bottleneck at the tubule-to-sheet transition. Furthermore, the levels of reticulons, which directly affect the balance between tubules and sheets in the ER, are linked to NE expansion. Therefore, our data suggest that the intrinsic ability of the ER to move between membrane sheets and tubules is used to form the NE.
The state of the mitotic NE
Historically, NE formation has been discussed as an example of a cell cycle–specific membrane fusion event similar to the reassembly of the Golgi apparatus. This hypothesis was based on nuclear reconstitution experiments using Xenopus laevis eggs. These experiments have revealed distinct vesicle populations that contribute to NE formation, which suggests that these vesicles were not part of a continuous membrane network before extract preparation (Vigers and Lohka, 1991). When membrane vesicles were mixed with chromatin and cytosol, nuclear assembly was found to depend on the presence of ATP and GTP, which suggests that there is membrane fusion machinery involved in the creation of the NE (Boman et al., 1992; Hetzer et al., 2001). These data further suggest that the NE is formed by the fusion of NE-specific mitotic vesicles.
However, a growing line of evidence supports an alternative model where the NE resides in the ER during open mitosis. Proteins of the NE have been visualized in the continuous mitotic ER in a growing number of studies, which implies that these NE components do not reside in distinct vesicles (Ellenberg and Lippincott-Schwartz, 1999; Daigle et al., 2001; Anderson and Hetzer, 2007). In addition, a recent study used EM to detect regions of the mitotic ER with high concentration of NE components and areas where these components are undetected (Puhka et al., 2007). Combined, these data are evidence that the NE resides in certain regions of the mitotic ER, and when these membranes are purified, heterogeneous vesicles are formed (Mattaj, 2004).
In addition, Puhka et al. (2007) used 3D reconstruction methods to demonstrate that membrane tubules coated chromatin during anaphase, and these tubules were connected to the mitotic ER. In our previous study, a modified in vitro nuclear formation method, where membranes are first allowed to reform an intact ER network and then used in nuclear assembly, we found that the NE formation from an intact ER can occur in the presence of fusion inhibitors (Anderson and Hetzer, 2007). These data suggest that the fusion machinery previously thought to be required for NE formation is likely only required for ER homotypic fusion.
In this paper, we provide direct in vivo evidence to support the model that the NE reemerges from the mitotic ER. Reticulons, proteins found exclusively on ER tubules, are found on membranes of the forming NE, which demonstrates that the ER contributes directly to NE formation. In addition, our data show that elevated levels of ER-shaping proteins, reticulons and DP1, inhibit NE formation. In contrast, the reduction of reticulons by siRNA accelerates NE formation. Collectively, these data provide compelling in vivo evidence in favor of the model where the ER contributes directly to NE formation.
It is tempting to speculate that reticulons might also play a role in NE breakdown by retubulating the NE into the ER. Consistent with this idea, the knockdown of reticulon 1 and YOP1/DP1 in Caenorhabditis elegans causes a defect in NE breakdown (Audhya et al., 2007). Retraction into the ER provides a simple and direct mechanism for the disappearance and appearance of the NE during mitosis that only requires the regulation of chromatin targeting of NE proteins and subsequent coordinated ER reorganization.
Membrane reshaping
The nascent NE is a massive membrane sheet with a surface area of ∼450 μm2 (Fig. 8 B). Given that the diameter of an ER tubule is ∼60 nm, the formation of the NE would require the conversion of ER tubules measuring >4.5 mm in length. This quantity of membrane likely represents a substantial fraction of the entire ER network. How these ER membranes are targeted to chromatin specifically during anaphase remains to be determined but likely involves the coordinated effort of numerous proteins (Anderson and Hetzer, 2008). The simplest explanation is that inner nuclear membrane proteins, which have the capacity to bind DNA directly or via chromatin, mediate the targeting of membranes to the chromatin surface. How is this massive amount of tubules transformed into sheets? The simplest interpretation of our results is that some type of “tug of war” between sheet and tubule formation occurs on the surface of chromatin. Overexpression of the reticulon isoform Rtn4a leads to the proliferation of bundled ER tubules, and, therefore, NE sheet formation is reduced. In a reciprocal fashion, when the reticulons or DP1 were deleted, the tubulating activity was diminished and NE sheet formation predominated. We speculate that displacement of reticulons results in a local loss of tubule-stabilizing activity and thereby increases tubule diameter, promoting the flattening of tubules into sheets.
How reticulons are displaced from the forming NE is unclear because very little is known about the biochemical properties of these proteins. One possibility is that reticulons are pushed aside by incoming NE proteins as they become concentrated in the chromatin-bound tubules. The reciprocal behavior of Rtn3 and POM121 is consistent with this idea. Alternatively, although not mutually exclusive, reticulons are phosphorylated (Olsen et al., 2006), which may regulate their ability to oligomerize and therefore modulate their membrane-bending capacities.
Considering the regularity of the spacing between the outer and inner nuclear membranes, it is likely that structural elements are needed to stabilize the NE. It remains to be determined whether luminal domains of membrane proteins might contribute to membrane flattening by bridging the gap between the two membranes and thereby forming a stable connection to maintain a constant separation. A kinetic analysis using our NE formation assay in cells with modulated concentrations of these inner NE proteins should be useful in answering these questions.
Nuclear expansion
Nuclear expansion is an important mechanism of growth in dividing cells; however, the mechanism of regulation of this growth is currently unknown. Our data provide a novel link between the levels of membrane-deforming proteins of the ER and nuclear size. Because the NE can be viewed as a single continuous ER sheet, levels of lipid synthesis and expression of reticulons might be one determinant of the overall size of nuclei. It will be interesting to test if the total size of the ER plays a role in nuclear expansion. Because reduction of reticulons does not accelerate NE growth, additional regulators are likely to exist. For example, nuclear import or lipid synthesis might limit the rate of nuclear expansion during G1. The uncovering of this interesting interaction between the ER and NE formation and expansion provides a starting point for further investigation of the interaction between these two organelles.
Materials and methods
DNA constructs
Sec61β amino acids 1–65 with GFP fused to the C terminus (Sec61frag) has been described previously (Anderson and Hetzer, 2007). POM121 fused to a triple repeat of GFP at the C terminus was a gift from the laboratory of J. Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany; Rabut et al., 2004). H2B with tdTomato fused to the C terminus was a gift from G. Pearson (Salk Institute for Biological Studies, La Jolla, CA). The NLS from SV-40 large T antigen (PPKKKRKV) was added to tandem EGFP by PCR and then inserted the N terminus containing the cycle-3 GFP vector pcDNA6.2 DEST53 using Gateway cloning (Invitrogen). Human Rtn3 was isolated from IMAGE clone no. 3873400 (available from GenBank/EMBL/DDBJ under accession no. BC011394) by PCR and inserted in pDEST R4,R3 along with a cytomegalovirus promoter and a C-terminal tdTomato using multisite Gateway cloning. Human Rtn4 was isolated from IMAGE clone no. 3505850 (accession no. BC016165), and human DP1 was isolated from IMAGE clone no. 3350749 (accession no. BC000232), and both along with Rtn3 were inserted into pcDNA6.2-nLumio, an N-terminal V5 epitope containing vector, using Gateway cloning. Human Sun1 was isolated from IMAGE clone no. 3864251 (accession no. BC013613) and cloned into pcDNA6.2 DEST47 using Gateway cloning.
Cell transfection and live-cell imaging
U2OS cells were grown and imaged in DME with 10% fetal bovine serum with 1× antibiotic-antimycotic (Invitrogen). Cells were plated on 8-well μ-slides (ibidi) and transfected with 0.6 μ of Lipofectamine 2000 (Invitrogen) and 0.3–0.6 μg of each DNA construct 2 d before live-cell imaging, as recommended by Invitrogen. For siRNA knockdown, cells were transfected 25–50 nmol of RNA 2 and 4 d before imaging. siRNA oligo sequences used to knock down reticulons were as follows: Rtn1, 5′-UAGAUGCGGAAACUGAUGGTT-3′; Rtn3, 5′-CCUUCUAAUUCUUGCUGAATT-3′; and Rtn4, 5′-GAAUCUGAAGUUGCUAUATT-3′ (Invitrogen). Live cells were imaged at 37°C maintained by an air stream incubator and enriched with CO2 (Solent Scientific). Time-lapse images were taken on a spinning disk confocal microscope (McBain Instruments) built around a DMRIE2 inverted-stage microscope (Leica). Images were captured on an EM charge-coupled device digital camera (Hamamatsu) and acquired using SimplePCI (Hamamatsu). Cells were imaged using a 63× oil objective with a 1.4 numerical aperture (Leica).
Antibody production and immunohistochemistry
Amino acids 1–117 of human Rtn4 were inserted into pDEST15 using Gateway cloning. This GST-tagged protein fragment was then expressed in BL21 Escherichia coli cells and purified using glutathione–agarose beads (Sigma-Aldrich). Guinea pigs were injected with two rounds ∼100 μg of protein and then bled 10 d after the second injection. Serum was used at a 1:1,000 dilution for indirect immunofluorescence and 1:5,000 for Western blotting. Monoclonal antibodies against the V5 epitope were used at a dilution of 1:1,000 for indirect immunofluorescence and 1:5,000 for Western blotting.
Image analysis and statistics
Regional and intensity threshold selections as well as subsequent mean pixel intensities were measured using Photoshop Extended CS3 (Adobe). Nuclear surface area was measured as described previously (D'Angelo et al., 2006). Numerical data were then analyzed and summarized graphically using Excel (Microsoft). Data for each experiment was collected from three independent experiments and combined for statistical analysis. p-values were determined using a student's t test, error bars represent standard errors, and ± values represent the standard deviations. 3D reconstructions were generated using the iso-surface function of Imaris (Bitplane).
Online supplemental material
Fig. S1 shows the localization of V5-tagged constructs and the efficiency of triple transfection. Video 1 shows the localization of ER membrane dynamics during NE formation as presented in Fig. 1 C. Video 2 shows the localization of POM121-3GFP during NE formation as presented in Fig. 1 D. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200805140/DC1.
Supplementary Material
Acknowledgments
Joshua Hsiao aided in the cloning of several constructs used in this manuscript. We thank Maximiliano D'Angelo, Maya Capelson, Christine Doucet, Sebastian Gomez, Robbie Schulte, Jessica Talamas, and Jesse Vergas for critically reading the manuscript and helpful discussion.
This work was supported by a grant from National Institutes of Health (R01 GM073994).
Abbreviations used in this paper: NE, nuclear envelope; NPC, nuclear pore complex; Rtn3 and 4, reticulon 3 and 4, respectively.
References
- Anderson, D.J., and M.W. Hetzer. 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9:1160–1166. [DOI] [PubMed] [Google Scholar]
- Anderson, D.J., and M.W. Hetzer. 2008. The life cycle of the metazoan nuclear envelope. Curr. Opin. Cell Biol. 20:386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., A. Desai, and K. Oegema. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., M.R. Delannoy, and K.L. Wilson. 1992. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J. Cell Biol. 116:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas, P., and D. Poccia. 2000. Membrane fusion events during nuclear envelope assembly. Subcell. Biochem. 34:273–302. [DOI] [PubMed] [Google Scholar]
- Crisp, M., Q. Liu, K. Roux, J.B. Rattner, C. Shanahan, B. Burke, P.D. Stahl, and D. Hodzic. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, M.A., and M.W. Hetzer. 2006. The role of the nuclear envelope in cellular organization. Cell. Mol. Life Sci. 63:316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, M.A., D.J. Anderson, E. Richard, and M.W. Hetzer. 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 312:440–443. [DOI] [PubMed] [Google Scholar]
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J., and J. Lippincott-Schwartz. 1999. Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods. 19:362–372. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., E.D. Siggia, J.E. Moreira, C.L. Smith, J.F. Presley, H.J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu, A., M. Fagarasanu, and R.A. Rachubinski. 2007. Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23:321–344. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., J. Koser, and U. Aebi. 2004. The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 29:175–182. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., H.H. Meyer, T.C. Walther, D. Bilbao-Cortes, G. Warren, and I.W. Mattaj. 2001. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3:1086–1091. [DOI] [PubMed] [Google Scholar]
- Hu, J., Y. Shibata, C. Voss, T. Shemesh, Z. Li, M. Coughlin, M.M. Kozlov, T.A. Rapoport, and W.A. Prinz. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250. [DOI] [PubMed] [Google Scholar]
- Lenart, P., and J. Ellenberg. 2003. Nuclear envelope dynamics in oocytes: from germinal vesicle breakdown to mitosis. Curr. Opin. Cell Biol. 15:88–95. [DOI] [PubMed] [Google Scholar]
- Margalit, A., S. Vlcek, Y. Gruenbaum, and R. Foisner. 2005. Breaking and making of the nuclear envelope. J. Cell. Biochem. 95:454–465. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W. 2004. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 5:65–69. [DOI] [PubMed] [Google Scholar]
- Maul, G.G. 1977. Nuclear pore complexes. Elimination and reconstruction during mitosis. J. Cell Biol. 74:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermudez, F., D. Gerlich, and J. Ellenberg. 2007. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9:822–831. [DOI] [PubMed] [Google Scholar]
- Olsen, J.V., B. Blagoev, F. Gnad, B. Macek, C. Kumar, P. Mortensen, and M. Mann. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127:635–648. [DOI] [PubMed] [Google Scholar]
- Powell, K.S., and M. Latterich. 2000. The making and breaking of the endoplasmic reticulum. Traffic. 1:689–694. [DOI] [PubMed] [Google Scholar]
- Puhka, M., H. Vihinen, M. Joensuu, and E. Jokitalo. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpasopoulou, A., J. Meier, C. Maison, G. Simos, and S.D. Georgatos. 1996. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Rabut, G., V. Doye, and J. Ellenberg. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N.C., R.E. Campbell, P.A. Steinbach, B.N. Giepmans, A.E. Palmer, and R.Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572. [DOI] [PubMed] [Google Scholar]
- Shibata, Y., G.K. Voeltz, and T.A. Rapoport. 2006. Rough sheets and smooth tubules. Cell. 126:435–439. [DOI] [PubMed] [Google Scholar]
- Shibata, Y., C. Voss, J.M. Rist, J. Hu, T.A. Rapoport, W.A. Prinz, and G.K. Voeltz. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, C.L., K.J. Roux, and B. Burke. 2007. Blurring the boundary: the nuclear envelope extends its reach. Science. 318:1408–1412. [DOI] [PubMed] [Google Scholar]
- Ulbert, S., M. Platani, S. Boue, and I.W. Mattaj. 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigers, G.P., and M.J. Lohka. 1991. A distinct vesicle population targets membranes and pore complexes to the nuclear envelope in Xenopus eggs. J. Cell Biol. 112:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G.K., W.A. Prinz, Y. Shibata, J.M. Rist, and T.A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. [DOI] [PubMed] [Google Scholar]
- Worman, H.J., and G.G. Gundersen. 2006. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 16:67–69. [DOI] [PubMed] [Google Scholar]
- Yang, Y.S., and S.M. Strittmatter. 2007. The reticulons: a family of proteins with diverse functions. Genome Biol. 8:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.