Abstract
The Rab11-Family Interacting Protein (Rab11-FIP) group of effector proteins contain a highly conserved region in their C-termini that bind the GTPase, Rab11. Rab11 belongs to the largest family of small GTPases and is believed to regulate vesicle docking with target membranes and vesicle fusion. The amino acid sequence of the Rab11-FIP proteins predicts coiled-coil formation in the conserved C-terminal domain. In this study on Rab11-FIP2, we found experimental evidence for the coiled-coil and then defined the minimal structured core using limited proteolysis. We also showed that the Rab11-FIP2 coiled-coil domain forms a parallel homodimer in solution using cross-linking and mutagenesis and sedimentation equilibrium experiments. Various constructs representing the C-terminal domain of Rab11-FIP2 were characterized by circular dichroism and their affinity with Rab11 measured using isothermal titration calorimetry. The longest construct was both well-structured and bound Rab11. A construct truncated at the N-terminus was poorly structured, but retained the same affinity for binding to Rab11. Conformational changes were also demonstrated upon complex formation between Rab11 and Rab11-FIP2. A construct truncated at the C-terminus, which was the minimal coiled-coil domain defined by limited proteolysis, did not retain the ability to interact with Rab11 although it was as well-structured as the longer peptide. These data show that coiled-coil formation and Rab11 binding are separable functions of the C-terminal domain of Rab11-FIP2. The dissection of Rab11 binding from the formation of defined structure in a coiled-coil provides a potential mechanism for regulating Rab11-dependent endosomal trafficking.
Keywords: Rab11-FIP2, Coiled-coil domain, ITC, CD, Limited proteolysis
Endocytotic membrane trafficking requires a strictly regulated system to ensure organelle function and transport specificity. The Rab GTPases and their effectors have been implicated to be the most likely candidates for the regulatory system that insures the accurate association of targeting molecules on the surfaces of transport vesicles with their corresponding membrane acceptors (1). In particular, the Rab11 isoforms (Rab11a, Rab11b, and Rab25) have been established to play a central role in plasma membrane recycling. In nonpolarized cells, Rab11 has been shown to be necessary for transferrin receptor recycling through perinuclear recycling endosome (2); in MDCK1 cells, Rab11a can regulate the transcytosis of polymeric IgA, but not basolateral recycling of transferrin (2). Rab11 has also been reported to regulate efficient transport from early endosomes to the trans-Golgi network (3).
Rab11 exerts its functions by binding other proteins, typically in a GTP-dependent manner. Several Rab11-binding proteins have been identified (4-10). Recently, a new family of Rab11 interacting proteins (FIP) has been found that includes six members to date: Rab11-FIP1 (FIP1) (5), Rab11-FIP2 (FIP2) (5), and Rab11-FIP3 (FIP3/eferin) (5), Rip11 (4), Rab Coupling Protein (RCP) (6), and Rab11-FIP4 (FIP4) (11). Proteins from this family share similar characteristics. They all have a Rab11-interacting site within their C-terminal homology domain known as the Rab11/25-binding domain (RBD) (12) that is partially overlapping a predicted coiled-coil. Rab11-FIP proteins also localize on recycling endosomes and participate in Rab11-mediated endosomal recycling. When classified according to the presence of other structural domains, there are three types of FIPs: class I FIPs (Rip11, FIP2, and RCP) containing a C2 domain at the N terminus, class II FIPs (FIP3 and FIP4) containing EF-hand motifs, and the class III FIP (FIP1) which, except for the coiled-coil domain, has no homology to other domains (13).
The focus of this manuscript is on Rab11-FIP2 (FIP2). FIP2 has biological roles different from the other FIPs. As well as its role in early endosome trafficking through Rab11, FIP2 coordinates EGF-receptor signaling through Reps1 (14). Rab11-FIP2 contains three Asn-Pro-Phe sequences and it binds the EH domain of Reps1. Rab11-FIP2 also is a coordinator of the interaction of Rab11 with myosin Vb (15). The C-terminal region of FIP2 was found to be essential for its colocalization with Rab11 on early endosomes and enables FIP2 to form higher order oligomers (14). For recycling of the transferrin receptor, expression of the C-terminal region of FIP2 induces aberrant tubulation of the compartment containing transferrin receptors, which can not be reversed by over-expression of Rab11 implying the importance of the C-terminal region of FIP2 and not other Rab11 effectors in transferrin receptor recycling (16).
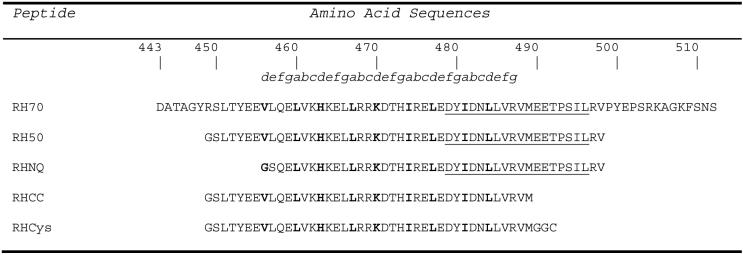
Since the identification of the importance of the C-terminal region of FIP2, efforts have been put into understanding the mechanisms of specificity between Rab11 and FIP interaction. The Rab11-binding domain of FIP2 (Figure 1) was demonstrated to be a region that codes specificity for Rab11 (7). Larger C-terminal fragments of Rip11 (amino acids 490-652), FIP2 (amino acid 378-511), RCP (amino acids 479-659) were shown to bind to the GTP-analogue bound form of Rab11 with similar affinity, while a weaker interaction was seen with the GDP-bound form of Rab11 (17). The RBD overlaps the last two heptad repeats of the predicted coiled-coil which contains 35 residues (18)(Figure 1). A 162-residue Rip11 fragment (amino acids 490-652) was shown to form a homodimer in solution (17). Experimental evidence for coiled-coil formation has not been published and the minimal, structured core of the C-terminal coiled-coil domain of Rip11 or any other FIP has not been defined. It is also not clear how oligomer formation relates to the ability of FIP proteins to bind Rab11.
Figure 1.
Peptide sequences representing regions of FIP2 containing the coiled-coil domain. The parent construct, RH70, comprises amino acid residues 443 to 512 of FIP2. The italic letters above the sequences denote the predicted heptad repeats within the coiled-coil. Residues in the a and d positions are in bold and predicted to participate in the formation of a hydrophobic interface. The residues comprising the minimal Rab-11 binding domain (7) are underlined.
In this paper, we set out to define the limits of the C-terminal homology region in FIP2 encompassing both the predicted coiled-coil and the Rab-11 binding domain. Through these studies, a 41 residue coiled-coil domain was characterized biophysically and demonstrated to be dimeric in solution. Various constructs containing different domains were assayed biochemically for their interaction with Rab11. Our studies contribute to a model in which the RBD domain without a pre-formed coiled-coil conformation is used for interaction with Rab11.
MATERIALS AND METHODS
FIP2 Cloning, Expression, and Purification
Several constructs were designed to investigate the determinants of oligomer formation and Rab11 interaction in the C-terminal region of FIP2. The initial expression construct comprised the C-terminal 70 residues of FIP2 (residues 443-512) subcloned into pGEX-4T-1 (14). New protein fragments were generated by PCR amplification followed by subcloning using BamHI and EcoRI sites into pGEX-4T-1. Clones were verified by sequencing.
Glutathione S-transferase (GST) fusion proteins were expressed in E. coli BL21(DE3) Codon Plus (RIL) cells. One liter of LB medium was inoculated with 50 ml stationary culture and grown until the O.D.600 was 0.7 before induction with 0.4 mM IPTG for 3h. Cells were harvested, and the pellet resuspended in BugBuster™ (Novagen) containing Benzonase and a cocktail of protease inhibitors, and subjected to rotary shaking for 30 min at room temperature. The supernatant after centrifugation was mixed with glutathione-Sepharose beads and subjected to rotary shaking for 2 h at 4°C. The beads were collected by centrifugation, washed three times with 20 volumes of 20 mM sodium phosphate buffer (pH 7.2) and three times with 20 volumes of 50 mM NH4HCO3. Thrombin digestion was carried out in 50 mM NH4HCO3, and the GST-free peptide was released into the supernatant. Proteins were further purified through size exclusion chromatography (Sephadex G75 superfine). The purity (>95%) of the FIP2 coiled-coil domains were verified by SDS-PAGE and mass spectrometry.
Rab11 Cloning, Expression, and Purification
A construct expressing GST- Rab11a in pGEX-5X (14) was modified using site-directed mutagenesis (Stratagene) to be cleavable by thrombin instead of Factor X because of the multiple cleavages observed with Factor X. GST-fusion proteins were expressed in E. coli BL21 (DE3) Codon Plus (RIL) cells in LB medium as described for FIP2. The cells were harvested and the pellets resuspended in a lysis buffer containing 10 mM phosphate, 100 mM NaCl, 1% Triton X-100, 2 mM MgCl2, 3 mM DTT, 100 μM AEBSF and 0.1% (v/v) Protease Inhibitor Cocktail (Sigma). The cells were lysed using an EmulsiFlex-C5 cell homogenizer (Avestin, Inc.). The clarified lysate was mixed with glutathione-Sepharose beads and subjected to rotary shaking for 2 h. The beads were washed with 10 volumes of lysis buffer and 10 volumes of lysis buffer without Triton X-100. The protein was then subjected to a nucleotide exchange protocol to replace GDP with GMP-PNP as previously described (19). The beads were washed with a buffer containing 20 mM HEPES, 150 mM NaCl, 2 mM Mg++, pH 8.0, thrombin was added, and the mixture nutated at room temperature for 2 h. The protein was eluted with 5 column volumes of a buffer containing 20 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 3 mM DTT, 100 μM AEBSF, pH 7.4, and passed through a Benzamidine Sepharose column (Amersham). Protein was further purified by gel filtration on Sephadex G-50 and assayed for concentration using absorption at 280 nm (20).
Circular Dichroism (CD) Spectroscopy and Denaturation Measurements
Far-UV CD spectra were obtained by averaging 2 scans with a step size of 0.5 nm on a JASCO model 810 spectropolarimeter. All measurements were performed in cuvettes with path lengths of 0.1 cm. Spectra were baseline-corrected by buffer subtraction and data were smoothed before curve fitting. Peptide concentrations were determined by UV absorbance at 280 nm. For thermal denaturation, ellipticity at 222 nm was measured as a function of temperature. The data were fit using a two-state model (folded dimer and unfolded monomer) as described (21-23):
where ΘN and ΘD represent the pre-transition (N) and post-transition (D) ellipticity values and fD is the fraction of dissociation of the dimer at each observed ellipticity (Θ). The equilibrium constant (K) was determined at each temperature using the fitted fd values:
where Ct represents the total concentration of the monomer.
Using the relationship ΔG = -RT ln K where ΔG is the Gibb-Helmholtz free energy of unfolding, the Tm was determined for the midpoint of the transition curve where fD = 0.5 and
where ΔCp is the apparent heat capacity of unfolding at constant pressure and ΔH(Tm) is the apparent enthalpy of unfolding at Tm.
Limited Proteolysis
The purified protein was subjected to limited protease digestion at 25 °C in 20 mM phosphate buffer containing 100 mM NaCl using trypsin at molar ratio of 1:2000 to protein. Proteolysis was stopped by boiling the sample in SDS-containing buffer prior to analysis on SDS-PAGE.
N-terminal Microsequencing of Proteolyzed Fragments
The fragments generated from proteolysis were separated by Tris-tricine SDS-PAGE and blotted onto a polyvinylidene difluoride (PVDF) membrane with electroblotting buffer (10 mM N-Cyclohexyl-3-aminopropanesulfonic acid, pH 11, 10% (v/v) methanol). Protein fragments were visualized by Coomassie brilliant blue R250 staining, excised and subjected to N-terminal sequencing.
Chemical Cross-linking Experiments
BS3 is an N-hydroxysuccinimide ester that reacts in pH 7-9 buffers with primary amino groups to form stable amide bonds. Primary amines are available from the side chains of lysine residues and the N-terminus of each polypeptide. FIP2 (40 μM) was incubated with 4 to 60 mM freshly prepared BS3 (Bis(sulfosuccinimidyl)suberate) for 30 min at room temperature. The cross-linking reaction was stopped by incubating with 0.1 M Tris, pH 7.5, for 10 min. The samples were electrophoresed, and the gels were stained with Coomassie Blue to visualize cross-linked products.
ITC Experiments
Samples were dialyzed against 20 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 2 mM TCEP overnight (24). ITC experiments were conducted at 25°C on a Microcal VP-ITC calorimeter by injecting aliquots of 100 μM FIP2 peptide into 10 μM Rab11-GMP-PNP. Data were analyzed using Origin to integrate peak volumes and calculate baselines, and baselines were then visually inspected and corrected.
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed on a BeckmanCoulter XL-I with interference optics. Ultracentrifugation samples were prepared from a concentrated protein stock solution and dialyzed against 20 mM sodium phosphate (pH 7.2) overnight. Protein was tested at four concentrations (1.4 mg/ml, 0.7 mg/ml, 0.3 mg/ml, and 0.175 mg/ml) with no apparent aggregation and at two different speeds (50,000 rpm and 35,000 rpm). The collected data were edited and analyzed using the program WINNONLIN v.1.06 (25). A global fit for each of the eight curves was performed to obtain a single species average molecular weight, M, from the following relationship:
where σ was the reduced molecular weight and obtained through curve fitting, is the partial specific volume; ρ is the solvent density; ω is the rotor speed (radians/s), R is the gas constant; and T is the temperature (K).
Equilibrium solute distributions were obtained using the equation:
where Cr is the solute concentration at radial position r and C0 is the concentration at a reference position r0; H is the constant n is the multimer ratio, Ka is the association constant, and E is the optical baseline offset (26).
RESULTS
Determination of the Boundaries of the FIP2 Coiled-coil Domain by Limited Proteolysis
A peptide comprising the last 70 residues of FIP2, RH70 (Figure 1), was incubated with trypsin at 25°C in 10 mM sodium phosphate, pH 7.0. After incubation, a smaller band appeared on a 15% Tris-tricine gel that was found to be 5976 Da by mass spectrometry. N-terminal sequencing showed the N-terminus started from Ser450 indicating the cleavage site after Arg449. The mass was consistent with a second C-terminal trypsin cleavage site after Arg497 (Figure 2).
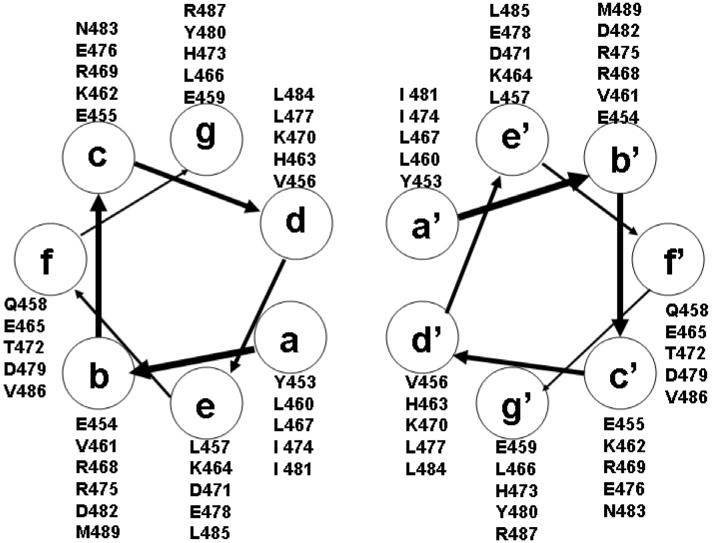
Figure 2.
Helical wheel representation of residues of the RHCC dimer (453-489) in heptad repeats of a coiled-coil model.
A new construct containing the trypsin resistant core (RH50, residues 450-497) was cloned, expressed, and purified (Figure 1). N-terminal amino acid sequencing of a degradation product that slowly appeared with time had the same N-terminus as the trypsin resistant core, indicating degradation from the C-terminus. The observed mass (5039 Da) indicated cleavage after Met489, consistent with cleavage by a contaminating enzyme with chymotrypsin-like activity.
Another construct comprising this Rab11-FIP homology region coiled-coil (RHCC, residues 450-489) was cloned, expressed, and purified (Figure 1). This construct represents the boundaries of the core FIP2 coiled-coil domain with respect to a chymotrypsin-like activity in E. coli and to trypsin. This resulting peptide was resistant to limited proteolysis with Glu-C (data not shown).
Examination of the sequence of RHCC shows a hydrophobic residue every seventh position. Assigning these hydrophobic residues at position ‘a’ of a helical wheel, results in residues at position‘d’ that are mostly hydrophobic (Figure 2). An alternative registry that places the initial group of hydrophobics at position d, results in negatively-charged Glu and Asp residues at position ‘a’ in the helical wheel, which is not seen in coiled-coil structures (27).
Oligomerization State of the FIP2 Coiled-coil Domain by Cross-linking Experiments
FIP2 and most of its family members have been reported to self-interact both in vivo and in vitro (11). While a gel filtration experiment on RCP indicates a dimer (17), no one has determined the oligomerization state of FIP2. A cross-linking experiment was performed using the reagent BS3 [Bis(sulfosuccinimidyl)suberate] (Figure 3). Non-cross-linked peptide appeared at ∼4 kDa, whereas only one cross-linked product with an apparent molecular weight of ∼8 kDa was observed, which clearly shows that the FIP2 coiled-coil domain primarily exists as dimer at a concentration of about 0.2 mg/ml.
Figure 3.
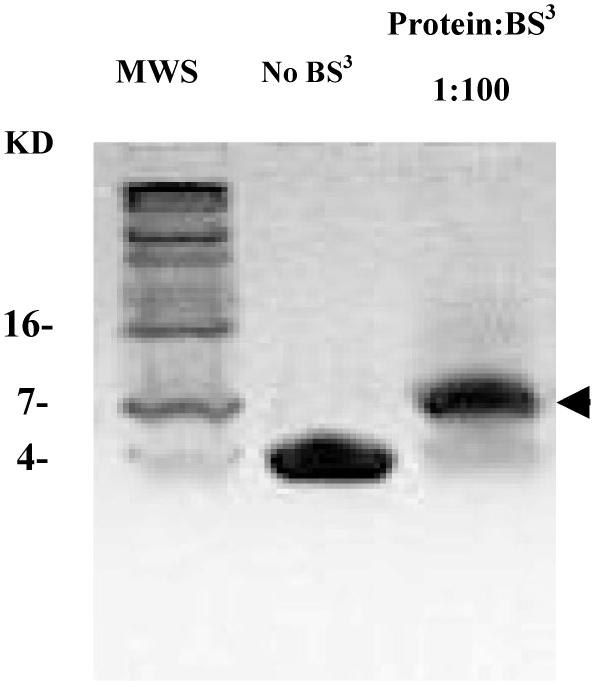
Chemical cross-linking reveals that the coiled-coil forms a dimer. Purified RHCC domain (40 μM) was incubated with BS3 cross-linker for 30 min at 25 °C and the reaction was stopped by the addition of 0.1M Tris. Cross-linked products were analyzed by a 15% Tris-tricine gel. The arrow indicates the major cross-link product at MW8000.
Sedimentation Equilibrium Studies
FIP2 was also assessed by sedimentation equilibrium to assess its oligomeric state over a range of concentrations. The equilibrium solute distributions were analyzed as indicated under “Materials and Methods” and residuals evidenced the high quality of the fit (Figure 4). At all velocities and concentrations, an estimated average molecular mass of 9957.5 Da was obtained using an ideal single species model. The mass is consistent with a dimeric model for FIP2 coiled-coil domain (calculated mass 9986 Da). We saw no monomer in the sedimentation experiments, consistent with a high association constant for RHCC dimer formation. Using an oligomeric model, data fitting indicated a mass of 9957 Da data (the dimer) in equilibrium with a second component having a multimer ratio of 3 corresponding to a mass of 29870 Da (a hexamer). Because the dimer mass of the oligomeric model agrees with that of the single-species model, the percentage of hexamer is assumed to be small (less than 5%). The association constant was not consistent among the eight data sets (lnKa varied from -5.05 to 1.49), indicating that the system was not in chemical equilibrium on the time scale of the experiment. Both chemical cross-linking experiments and analytical centrifugation experiments reflect the existence of a homodimeric FIP2 coiled-coil domain.
Figure 4.

Sedimentation equilibrium solute distribution for RHCC. Centrifugation was performed at various loading concentrations and two speeds. Data are shown for a concentration of 0.7 mg/ml collected at 35,000 rpm. The data were initially fitted with an equation for a single, ideal species but a better fit was obtained with a dimer-hexamer form of the equation. The line is the best fit through the data points (circles). The residuals are shown in the panel above the data. This fit was typical of data obtained with all sample concentrations of 1.4 mg/ml, 0.7 mg/ml, 0.35 mg/ml, and 0.175 mg/ml respectively at both centrifugation speeds (35,000 and 50,000 rpm). The global fit from all eight data sets was consistent with a dimer-hexamer two state model with a predominant dimer (>95%) with a molecular weight of 9957 Da.
Thermal Denaturation Studies
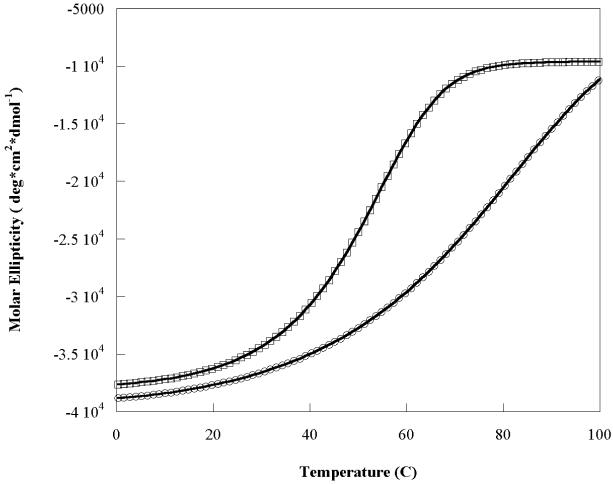
Preliminary NMR analysis (data not shown) indicated that the RHCC peptide monomers were arranged in a parallel rather than antiparallel manner. A new peptide was designed, RHcys, which had cysteine added at the C-terminus (28). If the dimer were parallel, the introduced disulfide bridge would be expected to be stabilizing. Purified RHcys peptide was dissolved in ammonium bicarbonate buffer (50 mM, pH 8.5) and stirred in an open beaker overnight to allow oxidation of cysteines to cystine. The peptide was dimeric when analyzed on a Coomassie-stained SDS-PAGE gel. In the presence of the BS3 cross-linker, the RHCys peptide showed quantitatively the same pattern as the RHCC core peptide, except the extent of monomer was slightly greater. RHCC and RHCys (and RH50) showed bands consistent with about 90% dimer and some small amount (ca. 5%) of higher order aggregates with molecular weights of approximately 12 and 16 kDa, consistent with tetramer and hexamer formation (Figure 1S). In addition, analysis of the RHCys peptide by gel filtration on a FPLC system showed little evidence of tetramer formation with predominant peaks corresponding to dimer and some monomer. In thermal denaturation experiments carried out on peptides at a monomer concentration of 20 μM, cooperative transitions were observed for both RHCC and RHcys (Figure 5). The unfolding data were fitted to a two-state model with simultaneous dissociation and unfolding processes. The fitted value of Tm was 71 °C for RHcys and 23 °C higher than RHCC (Tm= 48 °C) consistent with observations made for the disulfide cross-linking of the coiled-coil domain of GCN4 (28). The higher thermal stability for RHcys evidenced that the disulfide-bridge stabilizes the coiled-coil structure and suggests that the RHCC dimer is parallel.
Figure 5.
The thermal stability increases by introducing a disulfide-bridge into the C-terminus of the FIP2 coiled-coil domain. Data shows the temperature denaturation profile of RHCC (squares) and RHcys (circles). Molar ellipticity data were fit to obtain Tm values. The Tm was 48 °C for RHCC (at a peptide concentration of 0.11 mg/ml) and 71 °C for RHcys. Samples were prepared in 10 mM Tris buffer, pH 7.3.
Assessment of Structure by Circular Dichroism Spectroscopy
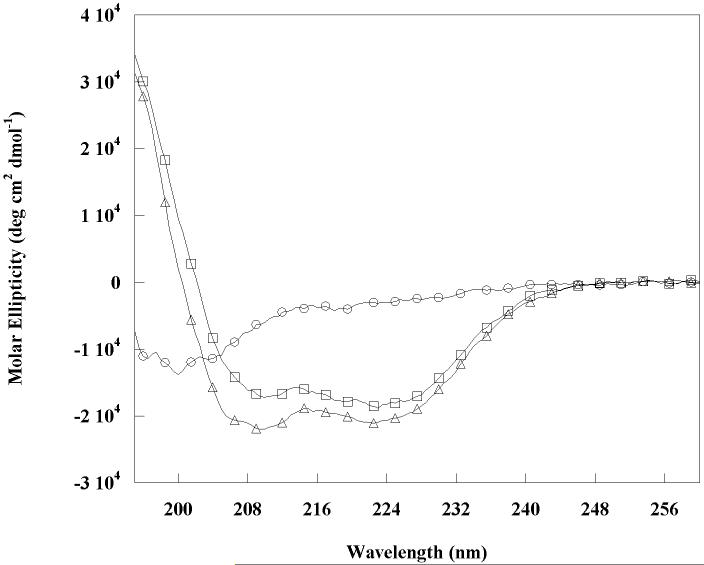
The far-ultraviolet CD spectra of RH50 and C-terminal truncation mutant RHCC showed two ellipticity minima around 208 nm and 222 nm consistent with an α-helical structure (Figure 7). Assuming that the ellipticity at 222 nm is -33400 deg cm2 dmol-1 for 100% α-helix (29), the calculated helical content was 63% for RH50 and 55% for RHCC. However, the difference is not likely to be significant given the probable errors in measuring protein concentrations and small differences in dynamics. The degree of coiled-coil formation was estimated from the Θ222/Θ208 ratio. The ratio is 0.83±0.03 for non-interacting α-helices and 1.03±0.03 for two-stranded coiled-coils (30). The Θ222/Θ208 ratio changes because upon formation of the two-stranded coiled-coil, the parallel-polarized amide π-π* electronic transition at 208 nm is reduced, while the n-π* transition at 222 nm remains the same (31). The Θ222/Θ208 ratio was 1.14 for RHCC and 0.99 for RH50. A reason that the ratio for RHCC was higher than 1.03 may be due to a sizeable contribution by an aromatic side-chain such as one of the tyrosines to the far-UV CD spectrum, an unexpected change in the π-π* or n-π* transitions of the peptide bond, or differences in dynamics (32, 33).
Figure 7.
Comparison of different constructs by circular dichroism. Data shows circular dichroism spectra of RH50 (triangles), RHCC (squares), and RHNQ (circles). Spectra were recorded at 25°C with a peptide monomer concentration of 11 μM in 10 mM Tris buffer, pH 7.3.
To define the boundaries of the coiled-coil more tightly, two additional constructs were studied. One was truncated by 8 residues at the N-terminus of RH50 (RHNQ) and the other 5 residues at the C-terminus of RHCC (RHCN). In the far-UV CD spectrum of RHNQ (Figure 7), the intensity of the molecular ellipticity at both 222 nm and 208 nm was dramatically reduced, indicating a poorly structured peptide. The non-dimeric nature of RHNQ is supported by BS3 cross-linking experiments in which it did not cross-link (Figure 1S). This suggests the first eight residues in RH50 were crucial for α-helix and coiled-coil dimer formation. RHCN was also not structured in CD measurements. The structural conclusions from the CD data are supported by NMR data in which the one-dimensional NMR spectrum of RH50 showed good resonance dispersion, which indicates a structured protein, while that of RHNQ showed poor resonance dispersion, corresponding to random coil (Figure 2S). Therefore, RHCC is the minimal structural core of the coiled-coil domain thus far defined.
Isothermal Titration Calorimetry Studies
Having defined a structural core of the FIP2 coiled-coil domain, we then investigated the relationship between structure and the ability to bind Rab11. To test interactions with Rab11, various constructs were subjected to ITC experiments (Figure 6). The binding constant (Kd), stoichiometry (n), enthalpy (ΔH), and entropy (ΔS) were obtained by curve-fitting the binding isotherms using a one-site model and calculating FIP2 concentrations for the dimeric form. The data showed that RH50 and RHNQ have similar binding affinity with Kd values of about 840 nM. These values are 20-fold lower than observed with larger constructs including ∼100 residues N-terminal to RH50, suggesting interactions between Rab11 and FIP2 outside the RBD (17). The binding stoichiometry values were 0.37±0.02 for RH50 and 0.36±0.02 for RHNQ. Assuming that the active Rab11 concentration may be over-estimated given that the purity of GMP-PNP used was 92%, these values are consistent with previous studies indicating that the molecular complex of FIP2 and Rab11 is a heterotetramer comprising a dimer of FIP2 and a dimer of Rab11 (17). Although appearing as a dimer in complex with FIP2, the oligomeric state of Rab11 alone is less well defined. While it was monomeric in solution by gel filtration experiments (17), it has been observed to be dimeric crystallographically (34) and diffusion measurements on Rab11 at 13 mg/ml using NMR spectroscopy show that it is a dimer (G.D. Henry & J.D. Baleja, unpublished results). The discrepancy may reflect an equilibrium between dimer and monomer that favors dimer formation at the high concentration found in crystals and NMR but a favored monomer at lower concentrations.
Figure 6.
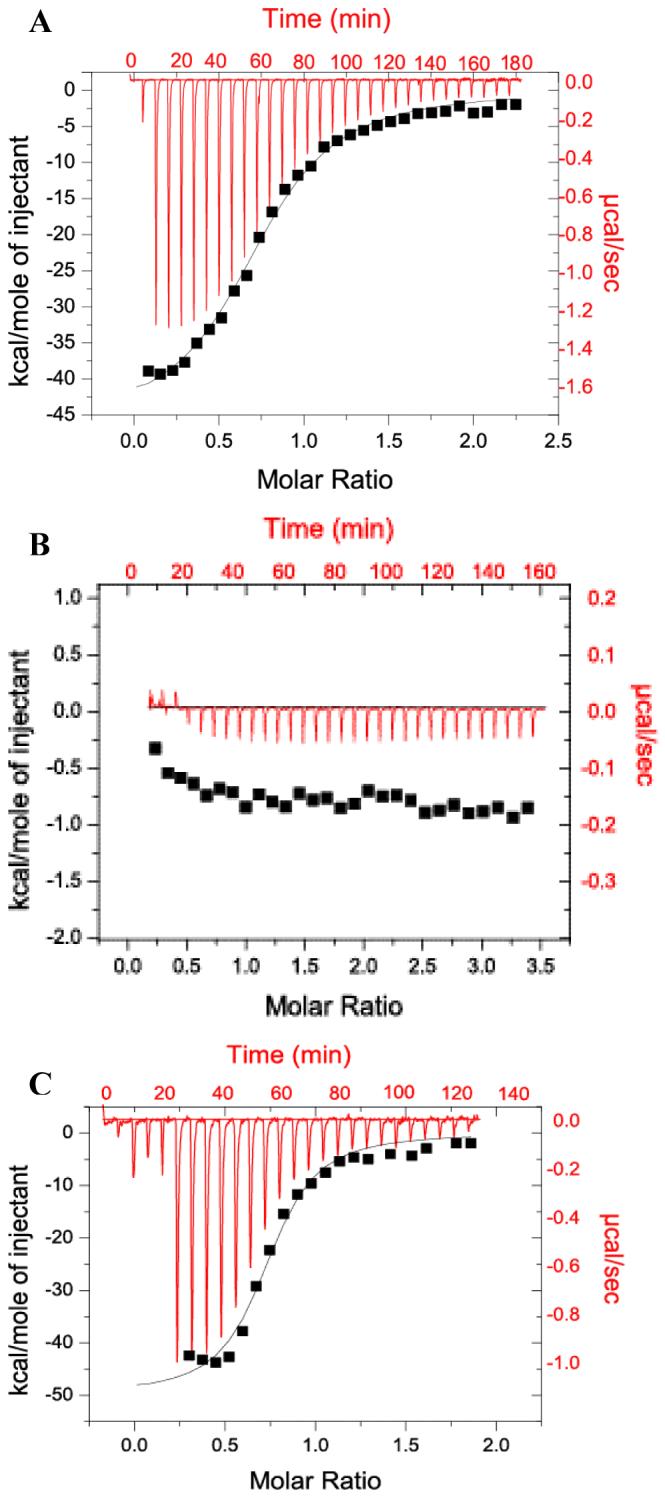
Isothermal titration calorimetry of various forms of the FIP2 coiled-coil domain with Rab11. Each experiment was conducted with 100 μM FIP2 domains injected into 10 μM Rab11. The raw data is shown in red and the integrated area of each peak and the non-linear, least square fit curve representing the heat released is shown in black. (A) RH50 injected into Rab11-GMP-PNP. Ka = (1.18 ± 0.25) × 106 M-1, ΔH = -39.5 ± 1.0 kcal/mol, TΔS = -31.1 kcal/mol, n=0.37. (B) RHCC injected into Rab11-GMppNp. (C) RHNQ injected into Rab11-GMP-PNP. Ka = (1.21± 0.25) × 106 M-1 ΔH = -25.8 ± 1.1 kcal/mol, TΔS = -18.4 kcal/mol, n=0.36.
When the RHCC construct lacking C-terminal amino acids relative to RH50 was used as a titrant, there was no detectable exothermic heat response indicating no detectable interaction. Although the CD spectra indicated that unlike RH50, RHNQ was not well structured, it had the same binding affinity as RH50. On the other hand, RHCC was as well structured as RH50, but did not bind Rab11 with appreciable affinity. Therefore, we conclude that a preformed coiled-coil is not required for Rab11 binding.
Association of Rab11 with the FIP2 Coiled-coil Domain Induces a Conformational Change
CD spectra of RH50 alone, RHNQ alone, Rab11 alone, and the mixtures each peptide with Rab11 and the calculated sum of the relevant intensities are shown in Figure 8. The intensity of the mixture of Rab11 and RH50 was 11% higher than the calculated sum of the separated components. For the mixture of Rab11 and RHNQ, the intensity was 20% higher. The higher intensities at 208 and 222 nm indicate additional α-helical structure in the Rab11-RHNQ complex is especially apparent and is likely to mostly derive from coiled-coil formation within FIP2 rather than a conformational change in Rab11. This suggests that the coiled-coil conformation may stabilize the complex, even though it is not required for direct interaction.
Figure 8.
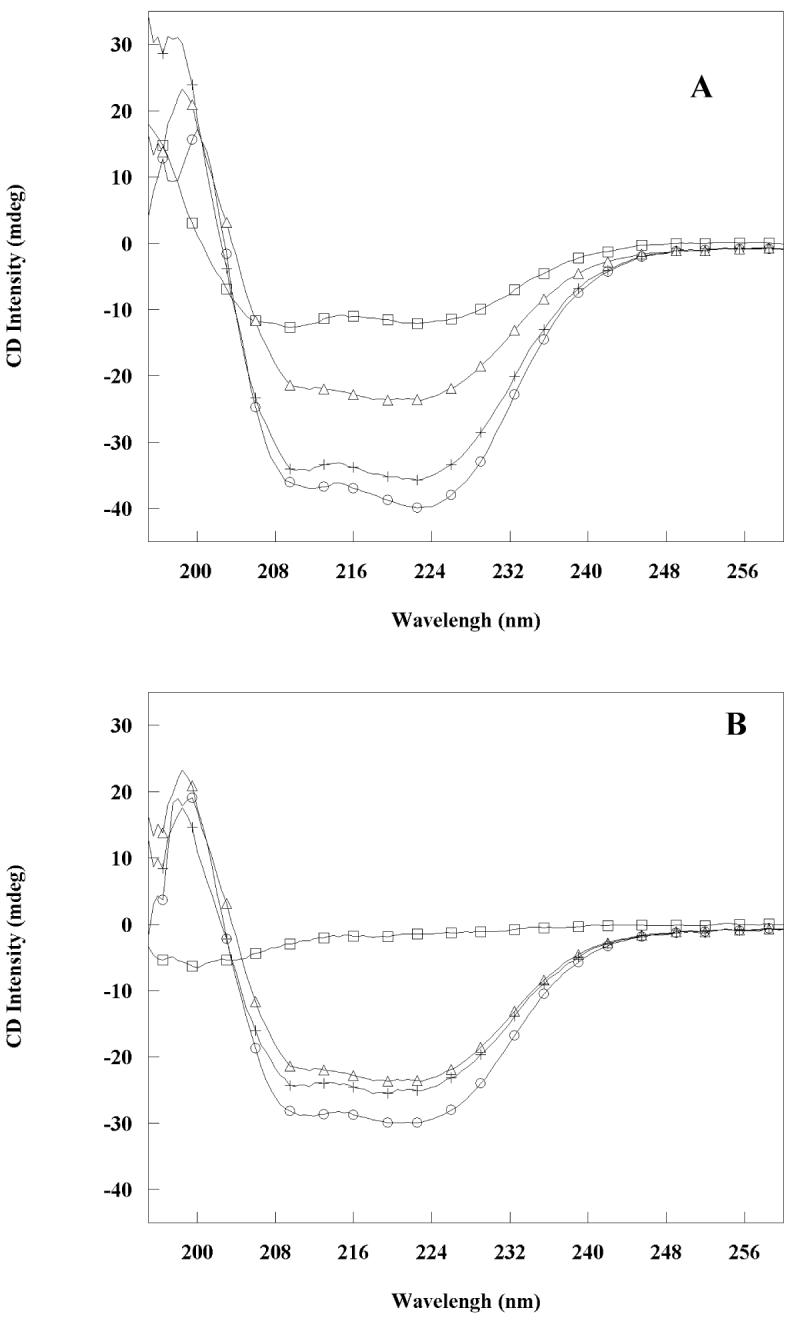
Circular dichroism spectra show induced helical content in the complexes formed by FIP2 coiled-coil domains and Rab11. (A: RH50, B: RHNQ). Squares represent the peptides, circles represent the 1:1 molar mixture of Rab11 and peptide, triangles represent Rab11, and plus signs represent the sum of the CD signals from peptide and Rab11. All spectra were recorded at 25°C in 10 mM Tris buffer, pH 7.3.
DISCUSSION
During the past few years, six members of a novel family of Rab11-interacting proteins have been identified: FIP1 (5), FIP2 (5), and FIP3 (5), Rip11 (35), Rab Coupling Protein (RCP) (6), and FIP4 (11). One of the common features shared by the family members is a highly conserved C-terminal region that mediates self-association (36). The protein-protein interactions associated with oligomerization can mediate formation of a complex network that segregates proteins into defined membrane domains (1). Oligomerization is a common phenomenon in the early endocytic pathway. For example, the Rab5 effectors, Rabaptin-5 and EEA-1, have been shown to form dimers (37, 38). The Rab5-binding protein RIN2 comprises two parallel dimers arranged in an anti-parallel manner (39). Eps15, a substrate of the epidermal growth factor receptor kinase, forms a similar tetramer (40). Because the C-terminal coiled-coil domain is shared among Rab11-FIP family members, oligomerization may be important for clustering Rab11 in the proper membrane location to insure the accurate fusion of the vesicles that Rab11 mediates.
The Rab11 family of interacting proteins share a highly homologous Rab11-binding domain (RBD) at their C-termini (5), which overlaps a conserved region that has been predicted to mediate coiled-coil formation (5). For the first time, these studies provide experimental evidence for coiled-coil formation and define the structured core of the coiled-coil domain in FIP2 by limited proteolysis. We demonstrated that the FIP2 coiled-coil domain forms a dimer from sedimentation and chemical cross-linking experiments. The RBD is the region reported to code specificity for the binding of Rab11 (7). The RBD overlaps with the last two heptads of the defined coiled-coil structural domain. ITC and CD measurements showed that the structured RH50 (containing the full RBD) bound to Rab11 with the same affinity as poorly structured RHNQ, while the well structured RHCC missing the last 8 residues of RBD did not. This suggests that the RBD is necessary for Rab11 binding, but it may not need to be in a preformed helical conformation to interact efficiently. Although the last 8 residues of the RBD domain are not important for coiled-coil formation considering that RHCC still forms a stable dimer, they are crucial for Rab11 interaction. These data allow a model in which the binding of FIP-family proteins to Rab11 is coupled to conformational changes (Figure 9). These in vitro results are consistent with mutagenesis data on FIP2 that suggest Rab11 binding and oligomerization are separable in a cellular context and represent distinct mechanisms of regulation (14).
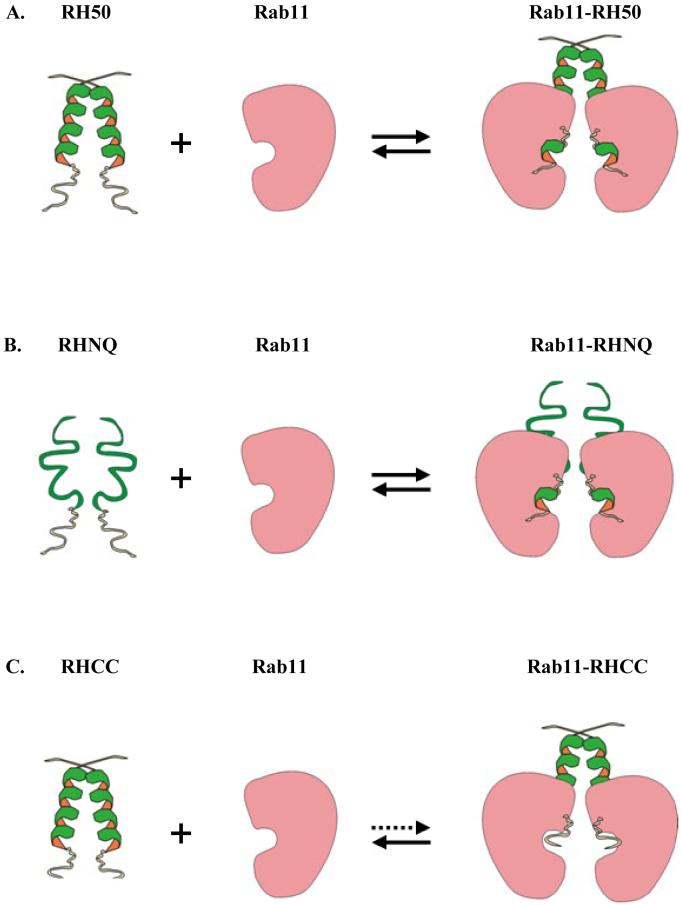
Figure 9.
A model of FIP2 binding to Rab11. The models show the interactions between various FIP2 coiled-coil domains and Rab11. A: RH50. The pre-formed dimer of RH50 binds Rab11; B: RHNQ. RHNQ does not have a pre-formed coiled-coil, but binds Rab11 as well as RH50 with significant conformational changes; C: RHCC. RHCC is a well-structured coiled-coil that does not have appreciable affinity for Rab11.
The formation of the Rab11-RH50 and Rab11-RHNQ complexes has favorable enthalpies and unfavorable entropies (Figure 6), suggesting an enthalpy-driven interaction. This is consistent with the studies by the Prekeris group (17). RH50 binding to Rab11 released more heat (ΔH=-39.5 kcal/mol) than RHNQ binding to Rab11 (ΔH=-25.8 kcal/mol), suggesting more polar interactions such as formation of hydrogen bonds or salt bridges. Horton and Lewis have shown that there is linear correlation between free enthalpies of dissociation and the interface area based on studies on protein-protein complexes (41). Secondary structure information obtained through CD experiments showed that RH50 has much higher degree of α-helicity than RHNQ. Higher degree of α-helicity, together with the fact RH50 has eight more residues than RHNQ, may provide more contact surface, therefore more opportunities for forming hydrogen bonds and salt bridges. However RH50 binding to Rab11 was more entropically unfavorable (TΔS=-31.1 kcal/mol) than RHNQ binding to Rab11 (TΔS=-18.4 kcal/mol), so the overall Gibbs free energy changes in both cases were very close, giving very similar Kd values.
The unfavorable entropy changes in both cases are particularly interesting. They indicate that the proteins undergo a disorder-order transition upon binding, which has been observed in the interaction of the serum protease thrombin and Hirudin, where the disordered C-terminal tail of free Hirudin becomes ordered upon binding to thrombin (42, 43). The differences in entropies in both cases may be caused by the different conformations of RH50 and RHNQ, complicated by the possibility that they may form slightly different complexes with Rab11. NMR studies on RH50 suggest that the C-terminal RBD region is more flexible than the N-terminal half (data not shown). Less structure suggests less rigidity that may facilitate binding.
The circular dichroism results indicate an increase in α-helical content upon complex formation between FIP2 and Rab11. This conformational change is likely to originate from FIP2 and may present a new binding site for other membrane trafficking regulators.
The data presented in this paper provide significant insight into the interaction between Rab11 and the FIP2 family of interacting proteins complementing the work of others (17). While they provide much information on the molecular details of the interaction, the details remain to be answered at an atomic level. Although the high-resolution three-dimensional structure of Rab11 has been determined (34), the structures of the RBD and its complex remain to be solved. Not only would such structures precisely define the conformational change suggested by this study, they would also begin to explain why the FIP2 family members preferentially form homodimers rather than heterodimers. Biophysical characterization of the FIP2 coiled-coil domain provides the necessary background to start such studies.
Supplementary Material
ACKNOWLEDGMENT
We thank Gillian Henry for the critical reading of the manuscript.
Footnotes
This work was supported in part by Grant GM067985 from the National Institutes of Health
- FIP
- family interacting protein
- RBD
- Rab-11 binding domain
- MDCK
- Madin-Darby canine kidney
- AEBSF
- 4-(2-Aminoethyl)-benzenesulfonylfluoride
- MWS
- Molecular Weight Standards
- DTT
- dithiothreitol
- GST
- glutathione-S-transferase
- GMP-PNP
- nonhydrolyzableGTP analog guanylyl-(β,γ-imido) diphosphonate
- TCEP
- Tris(2-Carboxyethyl) Phosphine
- ITC
- isothermal titration calorimeter
- CD
- circular dichroism spectroscopy
- HEPES
- N-(2-hydroxyethyl) piperazine-N-2-ethanesulfonic acid
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
REFERENCES
- 1.Zerial M, McBride H. Rab proteins as membrane organizers. Nature Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR. Regulation of vesicle trafficking in Madin-Darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 2000;275:29138–29146. doi: 10.1074/jbc.M004410200. [DOI] [PubMed] [Google Scholar]
- 3.Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J. Cell. Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell. 2000;6:1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 5.Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab Coupling Protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- 7.Prekeris R, Davies JM, Scheller RH. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- 8.Mammoto A, Ohtsuka T, Hotta I, Sasaki T, Takai Y. Rab11BP/Rabphilin-11, a downstream target of Rab11 small G Protein implicated in vesicle recycling. J. Biol. Chem. 1999;274:25517–25524. doi: 10.1074/jbc.274.36.25517. [DOI] [PubMed] [Google Scholar]
- 9.Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DW, Jr., Mercer JA, Bahler M, Goldenring JR. Myosin Vb is associated with plasma membrane recycling systems. Mol. Biol. Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamelin E, Theriault C, Laroche G, Parent J-L. The Intracellular Trafficking of the G Protein-coupled receptor TP{beta} depends on a direct interaction with Rab11. J. Biol. Chem. 2005;280:36195–36205. doi: 10.1074/jbc.M503438200. [DOI] [PubMed] [Google Scholar]
- 11.Wallace DM. The Novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem. Biophys. Res. Commun. 2002:909–915. doi: 10.1006/bbrc.2002.6736. [DOI] [PubMed] [Google Scholar]
- 12.Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J. Biol. Chem. 2002;277:49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- 13.Prekeris R. Rabs, Rips, FIPs, and endocytic membrane traffic. ScientificWorldJournal. 2003;3:870–880. doi: 10.1100/tsw.2003.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of Epidermal Growth Factor receptors. J. Biol. Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 15.Hales CM, Vaerman J-P, Goldenring JR. Rab11 family interacting Protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- 16.Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J. Biol. Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 17.Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- 18.Cullis DN. Ph. D. Thesis. Tufts University; Boston, MA: 2002. pp. 73–104. [Google Scholar]
- 19.Christoforidis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- 20.Yang C-S, Skiba NP, Mazzoni MR, Hamm HE. Conformational changes at the Carboxyl terminus of Galpha occur during G protein activation. J. Biol. Chem. 1999;274:2379–2385. doi: 10.1074/jbc.274.4.2379. [DOI] [PubMed] [Google Scholar]
- 21.Lavigne P, Kondejewski LH, Houston ME, Jr, Sonnichsen FD, Lix B, Sykes BD, Hodges RS, Kay CM. Preferential heterodimeric parallel coiled-coil formation by synthetic Max and c-Myc leucine zippers: A description of putative electrostatic interactions responsible for the specificity of heterodimerization. J. Mol. Biol. 1995;254:505–520. doi: 10.1006/jmbi.1995.0634. [DOI] [PubMed] [Google Scholar]
- 22.Lu SM, Hodges RS. Defining the minimum size of a hydrophobic cluster in two-stranded {alpha}-helical coiled-coils: Effects on protein stability. Protein Sci. 2004;13:714–726. doi: 10.1110/ps.03443204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steif C, Weber P, Hinz HJ, Flossdorf J, Cesareni G, Kokkinidis M. Subunit interactions provide a significant contribution to the stability of the dimeric four-alpha-helical-bundle protein ROP. Biochemistry. 1993;32:3867–3876. doi: 10.1021/bi00066a005. [DOI] [PubMed] [Google Scholar]
- 24.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 25.Yphantis DA, Johnson ML, Lary JW. WINNONLIN v. 1.06. University of Connecticut; 1997. [Google Scholar]
- 26.Kim H, Deonier RC, Williams JW. The investigation of self-association reactions. Chem. Rev. 1977;77:659–690. [Google Scholar]
- 27.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 28.O’Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 29.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 30.Zhou N, Kay C, Hodges R. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 31.Cooper TM, Woody RW. The effect of conformation on the CD of interacting helices: a theoretical study of tropomyosin. Biopolymers. 1990;30:657–676. doi: 10.1002/bip.360300703. [DOI] [PubMed] [Google Scholar]
- 32.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 33.Reisbig RR, Woody RW. Characterization of a long-wavelength feature in the absorption and circular dichroism spectra of beta-nicotinamide adenine dinucleotide. Evidence for a charge transfer transition. Biochemistry. 1978;17:1974–1984. doi: 10.1021/bi00603a028. [DOI] [PubMed] [Google Scholar]
- 34.Pasqualato S, Senic-Matuglia F, Renault L, Goud B, Salamero J, Cherfils J. The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J. Biol. Chem. 2004;279:11480–11488. doi: 10.1074/jbc.M310558200. [DOI] [PubMed] [Google Scholar]
- 35.Hickson GR, Matheson J, Riggs B, Maier VH, Fielding AB, Prekeris R, Sullivan W, Barr FA, Gould GW. Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol. Biol. Cell. 2003;14:2908–2920. doi: 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agou F, Ye F, Goffinont S, Courtois G, Yamaoka S, Israel A, Veron M. NEMO trimerizes through its coiled-coil C-terminal domain. J. Biol. Chem. 2002;277:17464–17475. doi: 10.1074/jbc.M201964200. [DOI] [PubMed] [Google Scholar]
- 37.Vitale G, Alexandrov K, Ullrich O, Horiuchi H, Giner A, Dobson C, Baykova O, Gournier H, Stenmark H, Zerial M. The GDP/GTP cycle of Rab5 in the regulation of endocytotic membrane traffic. Cold Spring Harb. Symp. Quant. Biol. 1995;60:211–220. doi: 10.1101/sqb.1995.060.01.024. [DOI] [PubMed] [Google Scholar]
- 38.Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H. The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem. J. 1999;338:539–543. [PMC free article] [PubMed] [Google Scholar]
- 39.Saito K, Murai J, Kajiho H, Kontani K, Kurosu H, Katada T. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J. Biol. Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 40.Cupers P, ter Haar E, Boll W, Kirchhausen T. Parallel dimers and Anti-parallel tetramers formed by Epidermal Growth Factor receptor pathway substrate clone 15 (EPS15) J. Biol. Chem. 1997;272:33430–33434. doi: 10.1074/jbc.272.52.33430. [DOI] [PubMed] [Google Scholar]
- 41.Horton N, Lewis M. Calculation of the free energy of association for protein complexes. Protein Sci. 1992;1:169–181. doi: 10.1002/pro.5560010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rydel TJ, Tulinsky A, Bode W, Huber R. Refined structure of the Hirudin-thrombin complex. J. Mol. Biol. 1991;221:583–601. doi: 10.1016/0022-2836(91)80074-5. [DOI] [PubMed] [Google Scholar]
- 43.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.