Abstract
We have developed an improved and rapid genomic engineering procedure for the construction of custom-designed microorganisms. This method, which can be performed in 2 days, permits restructuring of the Escherichia coli genome via markerless deletion of selected genomic regions. The deletion process was mediated by a special plasmid, pREDI, which carries two independent inducible promoters: (i) an arabinose-inducible promoter that drives expression of λ-Red recombination proteins, which carry out the replacement of a target genomic region with a marker-containing linear DNA cassette, and (ii) a rhamnose-inducible promoter that drives expression of I-SceI endonuclease, which stimulates deletion of the introduced marker by double-strand breakage-mediated intramolecular recombination. This genomic deletion was performed successively with only one plasmid, pREDI, simply by changing the carbon source in the bacterial growth medium from arabinose to rhamnose. The efficiencies of targeted region replacement and deletion of the inserted linear DNA cassette were nearly 70 and 100%, respectively. This rapid and efficient procedure can be adapted for use in generating a variety of genome modifications.
INTRODUCTION
The complete genome sequences of a rapidly growing number of bacterial strains have provided a wealth of information on the molecular structure and organization of myriad genes and open reading frames. This vast amount of information has been used in the construction of microorganisms with restructured, custom-designed genomes. These designer microbes have therapeutic and industrial applications, as well as a role in basic research into the origin and evolution of life and mechanisms of cellular metabolism (1–9). One of the most common approaches for the restructuring of a microbial genome to create custom-designed microorganisms is sequence-specific deletion or insertion of target genes or DNA sequences. For the precise modification of a genome, various methods have been developed based on RecA-dependent homologous recombination (10–12).
In addition to the RecA-dependent homologous recombination system in microbes, the λ-Red or RecET recombination system has also been exploited for the modification of large DNA constructs, including bacterial chromosomes and BAC clones (13,14). In these recombination events, selection markers are necessary to confirm the insertion or deletion of targeted regions. But the inserted selection markers prevent further modifications of the genome. To avoid having residual selection markers or foreign DNA sequences within the engineered chromosomes after genome modification, the Flp recombinase target (FRT) and the loxP-mediated site-specific recombination systems have been used for the precise excision of selection markers with the corresponding recombinase (Flp and Cre, respectively) (14–18). However, even with these site-specific recombination systems, at least one copy of the FRT site or the loxP site remains after excision of the selective markers, which limits the repeated use of these procedures (19).
Therefore, a more efficient method to delete target genes or genomic regions without leaving selection markers or foreign DNA sequences behind has been developed. This procedure involves the use of the intron-encoded homing endonuclease enzyme I-SceI as a counter-selection tool, which introduces a double-stranded break (DSB) in the genome (20–22). This DSB is a potent substrate for a microbial host recombination system that can repair the break by homologous recombination within the regions of sequence homology that flank the ends of the break. With the help of the host DSB-mediated repair system, several markerless modifications have been introduced into BAC clones as well as into the genomes of Gram-negative bacteria, such as Escherichia coli and Salmonella typhimurium (1,12,23–27).
Although the above methods have been used successfully to produce markerless modifications in genomes, several drawbacks remain. For example, these methods are time-consuming and labor-intensive, taking more than a week to delete a single targeted region, because of the repeated plasmid transformation and curing required for each deletion step (1,6). Here, we describe a highly efficient and rapid genomic engineering procedure that allows researchers to perform markerless deletion of a selected genomic region in 2 days.
MATERIALS AND METHODS
Bacterial strains, plasmids, enzymes and chemicals
Escherichia coli K-12 strain MG1655 was used in all markerless deletion experiments. E. coli strain DH5α was used as a cloning host. Plasmids pKD3, pKD4 and pKD46, were obtained from B. L. Wanner (15), pST76-K and pST76-ASceP were from G. Posfai (12), and pKO3 was from G. M. Church (11). Plasmids pSCI and pSKI were constructed as follows and used as a template to provide selection markers (CmR, KmR, sacB gene and I-SceI site) for markerless deletion cassettes. To construct pSCI, we cloned the KpnI fragment containing the chloramphenicol resistance gene (CmR) from plasmid pKD3 and the BamHI fragment containing the sacB gene from plasmid pKO3 into the KpnI and BamHI sites of pST76-K, respectively. Similarly, to construct pSKI, we cloned the KpnI fragment with the kanamycin resistance gene (KmR) from plasmid pKD4 and the BamHI fragment with the sacB gene from pKO3 into the KpnI and BamHI sites of pST76-K, respectively. All enzymes were purchased from New England BioLabs (Beverly, MA, USA), except for Taq polymerase, which was from Takara Bio Inc. (Shiga, Japan), and primers were synthesized by Bioneer (Daejeon, Korea). All antibiotics and chemicals were from Sigma-Aldrich (St Louis, MO, USA). Antibiotics were added to bacterial cultures at the following concentrations: ampicillin (Ap) 50 μg/ml, chloramphenicol (Cm) 17 μg/ml, kanamycin (Km) 25 μg/ml. l-arabinose, l-rhamnose and sucrose were added to culture media at concentrations of 10 mM, 10 mM and 5% (w/v), respectively.
Construction of pREDI
Plasmid pREDI, which expresses both the λ-Red proteins and the I-SceI endonuclease upon the appropriate induction, was constructed as follows. In order to introduce, into pKD46, the I-SceI endonuclease gene under the control of the rhamnose-inducible promoter PrhaB, a 2.0-kb DNA fragment that contained the rhaRS regulator genes and PrhaB was amplified by polymerase chain reaction (PCR) from the MG1655 genome with the following forward (NcoI-rha) and reverse (Prha) primers: NcoI-rha, 5′-CAT GCC ATG GGG CAT GGC GAA TTA ATC TTT CTG CG-3′ and Prha, 5′-CAT GCC ATG GGG CAT GGC GAA TTA ATC TTT CTG CG-3′. Next, a 0.7-kb DNA fragment that contained the I-SceI endonuclease gene was amplified by PCR from the plasmid pST76-ASceP with the following forward (I-SceI-F) and reverse (NcoI-I-SceI) primers: I-SceI-F, 5′-TTA GAC TGG TCG TAA TGA AAT TCA GCA GGA TCA CAT AAT GCA TCA AAA AAA CCA GGT AAT GAA CCT GGG TC-3′ and NcoI-I-SceI, 5′-CAT GCC ATG GGT CGA CTT ATT ATT TCA GGA AAG TTT CGG AGG AGA TAG TG-3′. The amplified 0.7-kb fragment contained a 50-bp flanking sequence on its 5′-end, that overlapped with the 3′-end of the 2.0-kb fragment described above. The 2.0- and 0.7-kb fragments were combined by recombinant PCR using the following forward (NcoI-rha) and reverse (NcoI-I-SceI) primers described above, to produce a 2.7-kb DNA fragment (rhaRS-PrhaB-I-SceI). The amplified 2.7-kb fragment was digested with NcoI and cloned into the NcoI site of pKD46, generating pREDI (Figure 1A).
Figure 1.
Description of rapid markerless deletion with pREDI. (A) Plasmid pREDI provides (i) arabinose-inducible (promoter = ParaB) λ-Red recombinase functions [gam (γ), bet (β), and exo] necessary for the replacement of a target genomic region with a linear DNA cassette, and (ii) rhamnose-inducible (promoter = PrhaB) I-SceI expression required for DSB-mediated markerless deletion. (B) Schematic representation of markerless deletion system with pREDI. To delete the E. coli chromosomal targeted region between homology boxes A and C, a linear DNA cassette containing a positive selective marker (CmR), a negative selective marker (sacB), a I-SceI endonuclease recognition site (S) and three homology boxes (A–C) was generated by recombinant PCR using pSCI and the E. coli genome as templates (refer to Materials and methods section). The linear DNA cassette was electroporated into pREDI-containing E. coli cells, where the cassette could replace a target genomic segment with the help of the λ-Red proteins (Red proteins) encoded by pREDI. Next, to remove the introduced selection markers, expression of the pREDI-encoded I-SceI endonuclease was induced by changing the carbon source in the media from 10 mM arabinose to 10 mM rhamnose. As a result, the chromosome was cleaved at the I-SceI endonuclease recognition site (S) present on the integrated DNA cassette, inducing the DSB repair function. Then, the DSB-mediated intramolecular recombination between the two homology arms (box C) resulted in the removal of the inserted deletion cassette, producing a clean, markerless deletion.
Markerless deletion of a genomic region
To delete the selected target region of the E. coli genome, which is housed between homology boxes A and C (Figure 1B), we constructed a 3.5-kb deletion cassette fragment (A-C-CmR-sacB-I-SceI-B, Figure 1B) that contained three homology regions (A–C, Figure 1B), a positive selection marker (CmR), a negative selection marker (sacB) and an I-SceI endonuclease recognition site by recombinant PCR as follows. First, a 3.0-kb DNA fragment that contained a CmR, a sacB gene, an I-SceI endonuclease recognition site and the 50-bp homology region B was amplified by PCR from plasmid pSCI [see Figure 1B for the forward (sc) and reverse (b) PCR primers], and a 0.5-kb homology fragment that contained homology regions A and C was amplified from the genomic DNA of MG1655 with the forward (a) and reverse (c) primers shown in Figure 1B. The amplified 0.5-kb fragment contained a short, 20-bp flanking sequence on its 3′-end that overlapped with the 5′-end of the 3.0-kb fragment described above. The 3.0- and 0.5-kb PCR products were combined by recombinant PCR with the forward (a) and reverse (b) primers. The resulting 3.5-kb linear DNA fragment (A-C-CmR-sacB-I-SceI-B) was purified with the Qiagen PCR purification kit (Hilden, Germany). Similarly, a markerless deletion cassette (A-C-KmR-sacB-I-SceI-B) was constructed as described above, except that plasmid pSKI was used as a template for the selection marker KmR instead of plasmid pSCI.
For the preparation of electro-competent cells, E. coli MG1655 cells harboring pREDI were grown at 30°C in 100 ml of LB medium supplemented with Ap and 10 mM l-arabinose. The cells were harvested in early log phase (OD600 = 0.4) by centrifugation at 2500g for 10 min and washed three times with ice-cold 10% glycerol.
For replacement of the targeted region of the E. coli genome, the appropriate markerless deletion cassette (400 ng) was electrotransformed into 50 μl of the electro-competent E. coli MG1655 cells harboring pREDI. The electrotransformed E. coli cells were incubated in 1 ml of SOC medium at 30°C for 1 h, spread onto LB plates containing Ap and either Cm or Km, and incubated at 30°C for an additional 16 h. Correct replacement of the target genomic region by the markerless deletion cassette was verified by PCR using a pair of primers that flanked the endpoints of the targeted region (primers If and MD in Figure 1B) and a pair of primers that were specific to genes that reside within the targeted region (primers sc and b in Figure 1B). The selection markers (CmR/KmR-sacB-I-SceI) that were introduced into the genome as described above were then excised from the recombinant E. coli strains by DSB repair mediated by the I-SceI endonuclease expressed from pREDI. Briefly, the recombinant strains were grown to OD600 = 0.4 at 30°C in 3 ml of LB liquid medium containing Ap and 10 mM rhamnose, then diluted 10-fold into 3 ml of fresh LB liquid medium containing Ap, 10mM rhamnose and 5% sucrose and grown to OD600 = 0.4 at 30°C. After three rounds of serial culture with 10-fold dilution, the cells were plated on LB plates containing Ap, 10 mM rhamnose and 5% sucrose. Then markerless deletion mutants (colonies that were sucrose-resistant and either Cm- or Km-sensitive) were selected by replica plating the recombinants on LB plates containing either Cm or Km versus LB plates containing 5% sucrose. The excision of the selection markers was verified by PCR using a pair of specific primers that flanked the endpoints of the genomic target region (primers If and MD in Figure 1B).
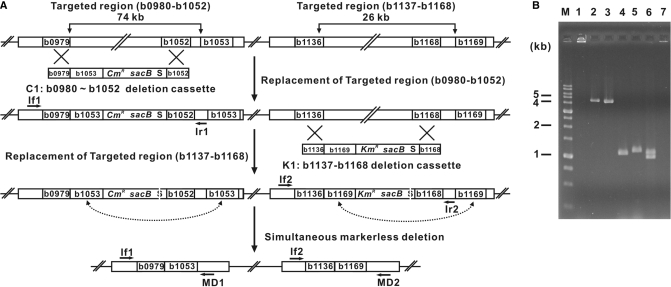
Simultaneous deletion of two separate regions of the E. coli genome (b0980–b1052 and b1137–b1168)
To delete simultaneously two targeted regions that are not adjacent to each other (b0980–b1052 and b1137–b1168) from the microbial genome, we constructed two markerless deletion cassettes, b0979-b1053-CmR-sacB-I-SceI-b1052 (C1) for deletion of the first target genomic region (b0980–b1052), and b1136-b1169-KmR-sacB-I-SceI-b1168 (K1) for deletion of the second target genomic region (b1137–b1168) (Figure 2A). The two targeted regions (b0980–b1052 and b1137–b1168) were then sequentially replaced with markerless deletion cassettes C1 and K1, respectively, as described above. Correct replacement of both targeted regions with the corresponding markerless deletion cassettes (C1 and K1) was verified by PCR as outlined above. The inserted selection markers were excised from the recombinant strains by I-SceI-mediated DSB repair as described above. The markerless deletion mutants (colonies that were sucrose-resistant and both Cm- and Km-sensitive) were selected by replica plating the recombinants on LB plates containing both Cm and Km versus LB plates containing 5% sucrose. Excision of the inserted markerless deletion cassettes was verified by PCR using a pair of specific primers that flanked the endpoints of each targeted region (If1 and MD1, and If2 and MD2; Figure 2A).
Figure 2.
Simultaneous deletion of two nonadjacent genomic targeted regions. To simultaneously delete two separate genomic regions (b0980–b1052 and b1137–b1168), we constructed two linear DNA cassettes: (i) b0979-b1053-CmR-sacB-I-SceI-b1052 (C1), for deletion of the first target genomic region (b0980–b1052), and (ii) b1136-b1169-KmR-sacB-I-SceI-b1168 (K1), for deletion of the second target genomic region (b1137–b1168) (refer to Materials and methods section). (A) The b0980–b1052 genomic region was replaced with deletion cassette C1, generating E. coli deletion strain Δb0980–b1052::C1. Then, the b1137–b1168 genomic region was replaced with the deletion cassette K1, producing E. coli deletion strain Δb0980–b1052::C1 Δb1137–b1168::K1. The subsequent expression of the I-SceI endonuclease in the double-replaced strain resulted in the simultaneous removal of the integrated DNA cassettes, generating the E. coli Δb0980–b1052 Δb1137–b1168 markerless double-deletion strain. (B) Markerless deletion of the two targeted regions was confirmed by PCR using three pairs of primers (If1/Ir1, If2/Ir2 and MD1/2) specific to both ends of the targeted regions and two pairs of primers (b1014F/R and b1150F/R) specific to the internal genes of each targeted region. PCR primers are indicated with arrows in (A). For the sequences of the PCR primers, see Material and methods section. Lane 1 shows the multiplex-PCR results obtained with the E. coli MG1655 wild-type genome and specific primers If1/Ir1 and If2/Ir2. Lanes 2 and 3 display PCR products obtained with the E. coli Δb0980–b1052::C1 genome and the E. coli Δb0980–b1052::C1 Δb1137–b1168::K1 genome and specific primers If1/Ir1 and If2/Ir2, respectively. Lanes 4 and 5 show PCR results obtained with the E. coli Δb0980–b1052 Δb1137–b1168 genome using primers If1/MD1 and If2/MD2 after simultaneous markerless deletions were carried out. Lanes 6 and 7 indicate multiplex PCR results obtained with the E. coli MG1655 wild-type genome and the E. coli Δb0980–b1052 Δb1137–b1168 genome and two pairs of primers specific to the internal sites (genes) of two deletion regions (internal genes selected for primers: b1014 and b1150, respectively). M indicates the lane containing DNA size markers.
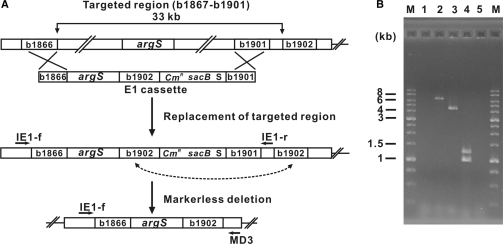
Markerless deletion of a genomic region that contained an essential gene(s)
To delete the targeted region of the E. coli genome (b1867–b1901) that contained the essential argS gene, which encodes arginyl-tRNA synthetase, we constructed a markerless deletion cassette [b1866-argS-b1902-CmR-sacB-I-SceI-b1901 (E1)] (Figure 3A) and electrotransformed into 50 μl of the electro-competent E. coli MG1655 cells harboring pREDI, as described before. The electrotransformed E. coli cells were incubated in 1 ml of SOC medium at 30°C for 1 h, spread onto LB plates containing Cm and Ap and incubated at 30°C for an additional 16 h. Correct replacement of the targeted region by E1 was verified by PCR as described before. The inserted selection markers were excised from the recombinant strains by I-SceI-mediated DSB repair as described above. The markerless deletion mutants (colonies that were Cm-sensitive and sucrose-resistant) were selected by replica plating the recombinants on LB plates containing Cm versus LB plates containing 5% sucrose. Excision of the inserted markerless deletion cassette was verified by PCR using a pair of specific primers that flanked the endpoints of the targeted region (IE1-f and MD3 in Figure 3A).
Figure 3.
Deletion of an E. coli genomic region that contains an essential gene. Deletion of the E. coli target genomic region that contained the essential gene argS was performed with a pREDI-containing strain of E. coli. (A) To delete the b1867–b1901 targeted region, that contained the essential gene argS (b1876) (which encodes arginyl-tRNA synthetase), we generated the linear DNA cassette b1866-argS-b1902-CmR-sacB-I-SceI-b1901 (E1), which carried the argS gene, and used this reagent to replace the selected genomic targeted region with the deletion cassette E1. Markerless deletion of the introduced selection markers was carried out as described in Figure 2. (B) Correct replacement of the genomic targeted region and complete removal of the inserted deletion cassette (E1) were confirmed by PCR using a pair of primers (IE1-f and IE1-r) specific to the ends of E1, and two pairs of primers (b1871F/R and b1897F/R) specific to the internal genes flanking argS. All PCR primers are indicated with arrows in (A). For the sequences of the PCR primers, see Material and methods section. Lanes 1 and 2 show PCR products obtained with the E. coli MG1655 wild-type genome and the E. coli Δb1867–b1901::E1 genome, respectively, using specific primers IE1-f and IE1-r. Lane 3 displays PCR products obtained with the E. coli Δb1867–b1901::argS genome and pair of primers IE1-f and MD3. Lanes 4 and 5 show the multiplex-PCR products obtained with the E. coli MG1655 wild-type genome and the E. coli Δb1867–b1901::argS genome, respectively, using two pairs of primers (b1871F/R and b1897F/R) specific to the internal sites (genes b1871 and b1897, respectively) of the deleted genomic region. M indicates the lane containing DNA size markers.
RESULTS
Construction of pREDI
As shown in Figure 1A, pREDI contains λ-Red genes under the control of the arabinose-inducible promoter ParaB and an I-SceI endonuclease gene under the control of the rhamnose-inducible promoter PrhaB. The replication origin of pREDI was derived from the plasmid pSC101, which is permissive at 30°C, but inactive at 42°C. Therefore, pREDI is easily curable after deletion of a targeted region of the E. coli genome.
The E. coli AraC protein acts as a transcriptional repressor in the absence of l-arabinose and a transcriptional activator of the ParaB promoter in the presence of this sugar (28,29). Therefore, addition of 10 mM arabinose to the culture medium inactivates the AraC repressor and subsequently induces the expression of λ-Red proteins, which mediate homologous recombination between the targeted region and the markerless deletion cassette. Without the addition of arabinose to the medium, replacement of the targeted region of the genome by the appropriate linear DNA cassette was not observed (data not shown).
In the presence of l-rhamnose, the E. coli RhaR activates the transcription of the rhaRS genes. Subsequent to rhaRS expression, RhaS protein activates transcription from the Prha promoter (30,31), and thus promotes expression of the I-SceI endonuclease, which forms the DSB necessary for activation of the host recombination system. Markerless deletion of the targeted region of the E. coli genome is then accomplished by homologous recombination in the regions of sequence homology that flank the ends of the DSB.
To examine the cleavage efficiency of I-SceI expressed from pREDI in E. coli, we transformed pREDI-containing E. coli with pSCI, a plasmid that contains an I-SceI endonuclease recognition site and a CmR gene. The transformants were grown at 30°C for 12 h in LB liquid medium supplemented with 10 mM rhamnose and Ap, and the resulting cells were spread on LB plates containing Ap. The cleavage efficiency of I-SceI was estimated by replica plating 200 colonies on LB plates with Ap versus LB plates with Ap and Cm. The fraction of surviving colonies on LB with Ap and Cm was <5%, suggesting that > 95% of the pSCI plasmids were cleaved by I-SceI expressed from pREDI in the presence of 10 mM rhamnose (data not shown).
Rapid markerless deletion of selected regions of the E. coli genome using pREDI
Sixteen large deletion mutants were constructed using pREDI (Table 1). The length of the markerless deletion cassettes was ∼3.5 kb. The sizes of homology regions A, B and C were 50-, 50- and 400- to 500-bp, respectively. Typically, 400 to 600 ng of the markerless deletion cassettes was electroporated into the E. coli cells harboring pREDI, from which 4–52 recombinant (CmR) colonies were obtained. Correct replacement of the targeted region with the markerless deletion cassette occurred in 70–100% of the recombinants (Table 1). The sizes of the deleted targeted regions ranged from 23- to 117-kb. We observed no significant correlation between the efficiency of replacement and the size of the targeted genomic region. After induction of I-SceI expression and sacB/sucrose counter-selection, almost all of the surviving cells exhibited the desired deletions. Indeed, as summarized in Table 1, the overall efficiency of the markerless deletion process ranged from 70% to 100%.
Table 1.
Efficiency of pREDI-mediated markerless deletions
| Target genomic region | Size (kb) | Positive colonies/Tested colonies (%) |
Overall efficiency (%) | ||
|---|---|---|---|---|---|
| First replacementa | Positive selection marker excisionb | Second DSB repairc | |||
| b0272–b0294 | 23 | 46/52 (88) | 100/100 (100) | 20/20 (100) | 88 |
| b0323–b0356 | 38 | 21/25 (84) | 100/100 (100) | 20/20 (100) | 84 |
| b0484–b0519 | 35 | 24/29 (83) | 100/100 (100) | 20/20 (100) | 83 |
| b0537–b0582 | 45 | 9/9 (100) | 100/100 (100) | 20/20 (100) | 100 |
| b0835–b0879 | 45 | 6/7 (86) | 99/100 (99) | 20/20 (100) | 85 |
| b1389–b1489 | 117 | 4/5 (80) | 100/100 (100) | 20/20 (100) | 80 |
| b1616–b1653 | 42 | 14/20 (70) | 100/100 (100) | 20/20 (100) | 70 |
| b1920–b1950 | 46 | 19/20 (95) | 100/100 (100) | 20/20 (100) | 95 |
| b2011–b2073 | 70 | 7/7 (100) | 100/100 (100) | 20/20 (100) | 100 |
| b2119–b2147 | 32 | 3/4 (75) | 100/100 (100) | 20/20 (100) | 75 |
| b2237–b2275 | 41 | 17/18 (94) | 100/100 (100) | 20/20 (100) | 94 |
| b2632–b2690 | 50 | 5/5 (100) | 100/100 (100) | 20/20 (100) | 100 |
| b2829–b2890 | 67 | 10/10 (100) | 100/100 (100) | 20/20 (100) | 100 |
| b3480–b3552 | 100 | 12/15 (80) | 100/100 (100) | 20/20 (100) | 80 |
| b4070–b4125 | 63 | 18/23 (78) | 100/100 (100) | 20/20 (100) | 78 |
| b4271–b4326 | 59 | 9/11 (82) | 100/100 (100) | 20/20 (100) | 82 |
aPCR screening of Cm-resistant colonies.
bCm-sensitive colonies/sucrose-resistant colonies.
cAs determined by PCR and sequencing.
Simultaneous deletion of multiple targeted regions
For the simultaneous deletion of two separate regions of the E. coli genome (b0980–b1052 and b1137–b1168, see Figure 2), we prepared two kinds of markerless deletion cassettes, C1 and K1. C1 carried a CmR gene as a positive selection marker and was used for the deletion of b0980–b1052. K1 contained a KmR gene as a positive selection marker and was used for the deletion of b1137–b1168. First, the b0980–b1052 region was replaced with the C1 cassette by λ-Red recombination (Figure 2A); next, the b1137–b1168 region was replaced with the K1 cassette by the same procedure. Subsequent expression of the I-SceI endonuclease resulted in the simultaneous removal of the introduced selection markers, generating an E. coli Δb0980–b1052 Δb1137–b1168 strain. Markerless deletion of the two targeted regions was confirmed by PCR using pairs of primers specific to the endpoints of the targeted regions (Figure 2B). The overall efficiency of simultaneous deletion of the two targeted genomic regions was ∼95%, and the complete process took only 72 h.
Deletion of genomic regions containing an essential gene(s)
Next, we sought to delete the targeted region b1867–b1901, which contained the essential gene argS (b1876); argS encodes arginyl-tRNA synthetase. To this end, we prepared a markerless deletion cassette that housed argS (E1) and used it to replace the targeted E. coli genomic region (Figure 3A). Correct replacement of the targeted region was confirmed by PCR using three pairs of primers (IE1-f and IE1-r, b1871F/R and b1897F/R) specific to internal genes that flanked the essential argS gene (Figure 3B). Of the 80 colonies grown on LB plates containing Cm, 10 colonies displayed correct replacement of the targeted region by the E1 cassette. To achieve markerless deletion, we removed the selection markers from the 10 recombinant strains as described previously.
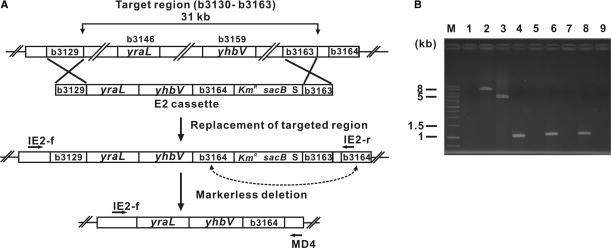
Our next step was to use the same procedure to delete a region of the E. coli genome (b3130–b3163) that contained two separated essential genes, yraL (b3146) and yhbV (b3159) (Figure 4). Using the λ-Red recombination procedure, we replaced the target genomic region with a linear markerless deletion cassette (E2) that contained yraL and yhbV. Accurate replacement of the targeted region with the E2 cassette was confirmed by PCR using a pair of primers (IE2-f and IE2-r) specific to both ends of the targeted region and three pairs of primers (b3138F/R, b3152F/R and b3162F/R) specific to the internal sequences of the targeted region that contained yraL and yhbV (Figure 4B). Of the 134 colonies grown on LB plates containing Cm, correct replacement of the targeted region by the E2 cassette occurred in 12 colonies. We then removed the remaining selection marker from the recombinant E. coli strains that housed the E2 cassette by the procedure described in Materials and methods section. The overall efficiency of markerless deletion of the genomic regions containing the essential gene(s) was 9–12.5%.
Figure 4.
Deletion of an E. coli genomic region that contains two essential genes. We performed deletion of an E. coli targeted region (b3130–b3163) that contained two nonadjacent essential genes, yraL (b3146) and yhbV (b3159). (A) The targeted genomic region b3130–b3163 was replaced with a linear DNA cassette, b3129-yraL-yhbV-b3164-KmR-sacB-I-SceI-b3163 (E2). Markerless deletion of the introduced selection markers was carried out as described in Figure 2. (B) Correct replacement of the target genomic region and complete removal of E2 were confirmed by PCR using a pair of primers (IE2-f and IE2-r) specific to the ends of E2 and three pairs of primers (b3138F/R, b3152F/R and b3162F/R) specific to the internal genes located in the targeted region. All PCR primers are indicated with arrows in (A). For the sequences of the PCR primers, see Material and methods section. Lanes 1 and 2 indicate PCR products obtained with the E. coli MG1655 genome and the E. coli Δb3130–b3163::E2 genome, respectively, and primers IE2-f and IE2-r. Lane 3 shows a PCR product obtained with the E. coli Δb3130–b3163::yraL yhbV genome and a primers IE2-f and MD4. Lanes 4, 6 and 8 indicate PCR products obtained with the E. coli MG1655 wild-type genome and three pairs of primers (b3138F/R, b3152F/R and b3162F/R) that were specific to internal genes in the targeted region (b3138, b3152 and b3162, respectively). Lanes 5, 7 and 9 also indicate PCR products obtained with the E. coli Δb3130–b3163::yraL yhbV genome and the same primers as in lanes 4, 6 and 8. M indicates the lane that contains the DNA size markers.
DISCUSSION
For markerless deletion of a specific region of the E. coli genome, a two-step procedure using two different plasmids typically has been employed by researchers. Step 1 includes the transformation of microbes with the first plasmid for the targeting of a selected gene/genomic region and then curing of the first plasmid from the cells. Step 2 involves retransformation of the microbes with a second plasmid for the markerless deletion of the selection markers introduced in step 1, followed by curing of the second plasmid (1,6,23,26). This procedure is time-consuming and labor-intensive. The new markerless deletion system described herein is rapid and efficient and thus represents an improvement over the currently used technique. Our procedure uses a single plasmid, pREDI, to induce the λ-Red recombination proteins and the DSB-repair system mediated by I-SceI endonuclease. With our method, targeting of a selected genomic region and markerless deletion of the introduced cassette can be accomplished without repeating the transformation and plasmid curing steps. These omissions allow markerless deletion to be completed in 2 days, rather than in the 7 days required by the currently used procedures (1,6,23,26).
In our deletion procedure, expression of the λ-Red recombinase genes (gam, bet and exo) and the I-SceI endonuclease gene are under the control of the tightly regulated promoters ParaB and PrhaB, respectively, and the arabinose and rhamnose added to cultures for the induction of ParaB and PrhaB are used and depleted by the bacteria (30). Therefore, for bacteria that contain pREDI, replacement of a target genomic region with a markerless deletion cassette and subsequent deletion of the introduced selection markers could be accomplished simply by changing the carbon source in the media from arabinose to rhamnose.
In addition, the basal expression level of the PrhaB promoter is ∼10-fold lower than that of ParaB in the absence of the corresponding inducers (32). This allows one to tightly regulate expression of the I-SceI endonuclease by PrhaB during replacement of the targeted genomic region with the markerless deletion cassette, thus minimizing unwanted cleavage of the cassette and resulting in increased targeting efficiency.
Upon the induction, the I-SceI endonuclease cleaves the unique I-SceI recognition site introduced by the markerless deletion cassette on the genome, causing the cell death. Cells can survive only by repairing the DSB or by deleting the I-SceI site by homologous recombination, becoming the correct markerless deletion mutants. Furthermore, the sacB/sucrose counter-selection procedure eliminates cells with the genomes not digested by the I-SceI endonuclease, increasing the selection efficiency of the markerless deletion mutants. To further improve the selection efficiency of the markerless deletion mutants, serial culture with the appropriate dilution is needed (12). One round of serial culture with 10-fold dilution in the selective medium showed <50% selection efficiency of the correct deletion mutants. However, the selection efficiency of the correct deletion mutants was close to 100% with three rounds of serial culture with 10-fold dilution in the selective medium (Table 1). Therefore, our overall efficiency of markerless deletion of a targeted region was much higher than those of the current procedures (1,23,26).
With our method, the simultaneous deletion of multiple nonadjacent genome segments and elimination of a genomic region that contains an essential gene(s) also could be performed rapidly. However, markerless deletion of target genomic regions that contained an essential gene(s) was not as efficient as that of nonessential targeted regions. In addition, replacement of a targeted region with two essential genes (b3130–b3163, yraL and yhbV) was less efficient than replacement of a targeted region that harbored only one essential gene (b1867–b1901, argS). This is because the essential gene(s) in the markerless deletion cassette serve as the substrates for homologous recombination rather than the short 50-bp homology arms, which results in incorrect replacement of the targeted region, decreasing the deletion efficiency. It also has been reported that the homologous recombination efficiency of DNA fragments <100 bp is 10-fold lower than that observed with DNA fragments of >1.0 kb (33). Despite this lower efficiency of deletion observed for essential genomic regions, recombinants were easily isolated using positive and negative selection markers and our single-deletion procedure, which provides a great advantage over having to perform two or more individual deletions for the same targeted region.
Finally, with appropriate modification of the markerless deletion cassettes, our system can be adapted for a variety of genome modifications. These include the introduction of point mutations and the insertion of genes or sequences into the genomes of E. coli and other Gram-negative bacterial species.
ACKNOWLEDGEMENTS
This work was supported by a grant from the 21C Frontier R&D Program (MG05-02-4-1-0), the Molecular & Cellular BioDiscovery Research Program (M10748000314-07N4800-31410) from the Ministry of Education, Science and Technology, the Korea Science & Engineering Foundation Grant (R01-2005-000-11010-0) and the Korea Research Foundation Grant (KRF-2004-042-D00072). The Open Access publication charges for this article were waived.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kolisnychenko V, Plunkett G., III, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G. Engineering a reduced Escherichia coli genome. Genome Res. 2002;12:640–647. doi: 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002;20:1018–1023. doi: 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- 3.Goryshin IY, Naumann TA, Apodaca J, Reznikoff WS. Chromosomal deletion formation system based on Tn5 double transposition: use for making minimal genomes and essential gene analysis. Genome Res. 2003;13:644–653. doi: 10.1101/gr.611403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westers H, Dorenbos R, van Diji JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, et al. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 2003;20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 5.Fukiya S, Mizoguchi H, Mori H. An improved method for deleting large regions of Escherichia coli K-12 chromosome using a combination of Cre/loxP and λ Red. FEMS Microbiol. Lett. 2004;234:325–331. doi: 10.1016/j.femsle.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki N, Okayama S, Nonaka H, Tsuge Y, Inui M, Yakawa H. Large-scale engineering of the Corynebacterium glutamicum genome. Appl. Environ. Microbiol. 2005;71:3369–3372. doi: 10.1128/AEM.71.6.3369-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 9.Posfai G, Plunkett G., III, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. Emergent properties of reduced genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 17.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 18.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 20.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt-Puchta W, Orel N, Kyryk A, Puchta H. Intrachromosomal homologous recombination in Arabidopsis thaliana. Methods Mol. Biol. 2004;262:25–34. doi: 10.1385/1-59259-761-0:025. [DOI] [PubMed] [Google Scholar]
- 23.Jamsai D, Orford M, Nefedov M, Fucharoen S, Williamson R, Ioannou PA. Targeted modification of a human beta-globin locus BAC clone using GET recombination and an I-SceI counter selection cassette. Genomics. 2003;82:68–77. doi: 10.1016/s0888-7543(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung BH, Lee CH, Yu BJ, Lee JH, Lee JY, Kim MS, Blattner FR, Kim SC. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 2006;72:3336–3342. doi: 10.1128/AEM.72.5.3336-3342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 27.Cox MM, Layton SL, Jiang T, Cole K, Hargis BM, Berghman LR, Bottje WG, Kwon YM. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 2007;7:59. doi: 10.1186/1472-6750-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 29.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan SM, Schleif RF. A regulatory cascade in the induction of rhaBAD. J. Mol. Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 31.Egan SM, Schleif RF. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J. Mol. Biol. 1994;243:821–829. doi: 10.1006/jmbi.1994.1684. [DOI] [PubMed] [Google Scholar]
- 32.Haldimann A, Daniels LL, Wanner BL. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]