Abstract
DUSP6 (dual-specificity phosphatase 6), also known as MKP-3 [MAPK (mitogen-activated protein kinase) phosphatase-3] specifically inactivates ERK1/2 (extracellular-signal-regulated kinase 1/2) in vitro and in vivo. DUSP6/MKP-3 is inducible by FGF (fibroblast growth factor) signalling and acts as a negative regulator of ERK activity in key and discrete signalling centres that direct outgrowth and patterning in early vertebrate embryos. However, the molecular mechanism by which FGFs induce DUSP6/MKP-3 expression and hence help to set ERK1/2 signalling levels is unknown. In the present study, we demonstrate, using pharmacological inhibitors and analysis of the murine DUSP6/MKP-3 gene promoter, that the ERK pathway is critical for FGF-induced DUSP6/MKP-3 transcription. Furthermore, we show that this response is mediated by a conserved binding site for the Ets (E twenty-six) family of transcriptional regulators and that the Ets2 protein, a known target of ERK signalling, binds to the endogenous DUSP6/MKP-3 promoter. Finally, the murine DUSP6/MKP-3 promoter coupled to EGFP (enhanced green fluorescent protein) recapitulates the specific pattern of endogenous DUSP6/MKP-3 mRNA expression in the chicken neural plate, where its activity depends on FGFR (FGF receptor) and MAPK signalling and an intact Ets-binding site. These findings identify a conserved Ets-factor-dependent mechanism by which ERK signalling activates DUSP6/MKP-3 transcription to deliver ERK1/2-specific negative-feedback control of FGF signalling.
Keywords: dual-specificity phosphatase 6 (DUSP6), fibroblast growth factor (FGF), mitogen-activated protein kinase (MAPK), mitogen-activated protein kinase phosphatase-3 (MKP-3), phosphatase, transcription
Abbreviations: AER, apical ectodermal ridge; ChIP, chromatin immunoprecipitation; ConA, concanavalin A; DMEM, Dulbecco's modified Eagle's medium; DUSP, dual-specificity phosphatase; EGFP, enhanced green fluorescent protein; Elk1, Ets-like kinase 1; EMSA, electrophoretic mobility-shift assay; ER, oestrogen receptor; ERK, extracellular-signal-regulated kinase; Ets, E twenty-six; FBS, fetal bovine serum; FGF, fibroblast growth factor; FGFR, FGF receptor; FKHD, Forkhead mutant; βGal, β-galactosidase; HA, haemagglutinin; HH10, Hamburger and Hamilton stage 10; 4-HT, 4-hydroxytamoxifen; IκBα, inhibitor of nuclear factor κB α; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; MKK, MAPK kinase; MKKK, MAPK kinase kinase; MKP, MAPK phosphatase; mRFP, monomeric red fluorescent protein; NF-κB, nuclear factor κB; PBX1, pre-B-cell leukaemia transcription factor 1; PI3K, phosphoinositide 3-kinase; RSV, rous sarcoma virus; SRF, serum-response factor; SRY, sex-determining region Y; TNFα, tumour necrosis factor α
INTRODUCTION
DUSP (dual-specificity phosphatase) 6 or MKP [MAPK (mitogen-activated protein kinase) phosphatase]-3 is the prototypical member of a subfamily of three cytoplasmic MKPs, which also includes DUSP7/MKP-X and DUSP9/MKP-4 [1,2]. These enzymes all display a high degree of substrate selectivity for the ERKs (extracellular-signal-regulated kinases) 1 and 2 in vitro and in vivo [3,4]. Specific recognition and binding to ERK2 is mediated by a conserved KIM (kinase interaction motif) within the N-terminal non-catalytic domain of DUSP6/MKP-3 and this region of the protein also contains a conserved NES (nuclear export signal), which is responsible for the cytoplasmic localization of this phosphatase [5,6]. The specificity of DUSP6/MKP-3 for dephosphorylation and inactivation of the ERK1 and ERK2 MAPKs is enhanced further by ERK-induced conformational change within the catalytic domain of MKP-3, which leads to greatly enhanced phosphatase activity in vitro [7,8].
The first clues as to the physiological role of MKP-3 came from the observation that DUSP6/MKP-3 mRNA is expressed at many sites of FGF (fibroblast growth factor) signalling in developing mouse and chicken embryos. These include the limb bud and branchial arch mesenchyme, midbrain/hindbrain isthmus, hair and mammary placodes [9], and early neural plate [10]. Further experiments involving tissue ablation and transplantation in chicken embryos identified the AER (apical ectodermal ridge) and Hensen's node as tissue sources of FGF which are essential for the expression of DUSP6/MKP-3 in the developing limb bud and neural plate respectively [10,11]. In addition, FGF signalling is also responsible for the expression of DUSP6/MKP-3 in the murine isthmic organizer during neural tube development and in developing chick somites [12,13]. These studies suggest that DUSP6/MKP-3 is a negative regulator of FGF signalling during vertebrate development, which may work to set the levels of ERK signalling downstream of this signalling pathway. This conclusion is supported by the results of a recent mouse knockout experiment. DUSP6/MKP-3-null embryos display elevated ERK phosphorylation in limb bud mesenchyme and present with variably penetrant skeletal dwarfism, premature fusion of cranial sutures (craniosynostoses) and deafness, all of which are consistent with increased levels of FGF signalling [14].
Although the link between FGF signalling and DUSP6/MKP-3 expression is now well established, the precise molecular mechanism by which this occurs is unknown. In particular, it is unclear which of the intracellular signalling pathways that lie downstream of the FGFR (FGF receptor) is responsible for mediating DUSP6/MKP-3 transcription, with essential roles proposed for both the ERK and PI3K (phosphoinositide 3-kinase) pathways [10–13,15]. The majority of these data were obtained in a variety of embryonic tissues often using different pharmacological inhibitors of these pathways and this may account for some of the contradictory data obtained [16].
In the present study, we have used a cell culture model to overcome the limitations of drug delivery using bead implantation in chicken embryos to address the nature of the intracellular signalling pathways involved in FGF-mediated DUSP6/MKP-3 expression. This has been combined with a bioinformatic and functional dissection of the DUSP6/MKP-3 gene promoter and has enabled us to define a mechanism by which signalling though the ERK MAPK pathway interacts with a conserved regulatory region within the proximal promoter of the gene to effect negative-feedback control of FGF signalling in vitro and in the developing chick embryo.
EXPERIMENTAL
Reagents
Recombinant human FGF2 (basic FGF), human FGF4 and mouse FGF8b were purchased from R&D Systems. SU5402 and LY294002 were from Calbiochem. PD184352 was kindly provided by Professor Sir Philip Cohen (MRC Protein Phosphorylation Unit, University of Dundee). Antibodies against ERK, phospho-ERK, p38, phospho-p38, JNK (c-Jun N-terminal kinase), phospho-JNK and phospho-Akt were purchased from Cell Signaling Technology. The antisera raised against Ets (E twenty-six) family proteins were from Santa Cruz Biotechnology. The polyclonal antibody against DUSP6/MKP-3 was raised in sheep using purified recombinant DUSP6/MKP-3 protein as an antigen. The specificity and sensitivity of this antiserum was verified by immunoblotting of recombinant DUSP6/MKP-3, its ability to recognize DUSP6/MKP-3, but not the related phosphatases encoded by DUSP7 and DUSP9, when expressed in Cos-1 cells and Western blotting of protein lysates from wild-type and DUSP6-knockout mouse embryos (results not shown). Reagents were purchased from Sigma, unless indicated otherwise. All tissue culture reagents were obtained from Invitrogen.
Cell lines and tissue culture
NIH 3T3 cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% newborn calf serum, penicillin (100 units/ml) and streptomycin (100 units/ml). Δ-Raf-1:ER* NIH 3T3 cells [NIH 3T3 fibroblasts that stably express a fusion between a constitutively active form of the Raf-1 MKKK (MAPK kinase kinase) and a mutant form of the ER (oestrogen receptor)] were maintained in Phenol Red-free DMEM containing glucose (4500 mg/l), penicillin (100 units/ml), streptomycin (100 units/ml), 2 mM glutamine and 10% FBS (fetal bovine serum) supplemented with 2 μg/ml puromycin. Δ-Raf-1:ER* NIH 3T3 cells were induced with either 10 nM or 100 nM 4-HT (4-hydroxytamoxifen) as indicated. For inhibitor studies, cells were cultured in Petri dishes for 48 h and were serum-starved overnight. Inhibitors SU5402 (50 μM), PD184352 (2 μM) and LY294002 (10 μM) were added 30 min before the addition of FGFs (30 ng/ml) and cells were then incubated for an additional 5 h. Cells were lysed, and proteins were analysed using the NuPAGE electrophoresis system (4–12%) (Invitrogen) and Western blotting. For analysis of DUSP6/MKP-3 mRNA levels, RNA was isolated from cells using an RNeasy kit (Qiagen) according to the manufacturer's instructions, and 200 ng of RNA was reverse-transcribed in a final volume of 50 μl using Taqman reverse transcription reagents (Applied Biosystems). A 4 ng sample of cDNA was analysed by quantitative PCR using pre-developed assay primers and probes for DUSP6/MKP-3 (Mm00650255_g1; Applied Biosystems). Real-time PCR was performed in the presence of 0.6× Taqman Universal PCR mix (Applied Biosystems) under the following conditions: 50 °C for 2 min, 95 °C for 10 min, 92 °C for 15 s and 60 °C for 1 min using an ABI Prism 7700 sequence detector. Fluorescence output was directly proportional to the concentration of input cDNA and was normalized against β-actin (Mm00607939_s1).
Analysis of the DUSP6/MKP-3 promoter in silico
The upstream genomic sequence of DUSP6/MKP-3 was inspected within the February 2006 (NCBI build 36) assembly of the murine genome using the UCSC genome browser [17], and a candidate region was selected such that the maximum number of vertebrate species genomes aligned to the mouse. The corresponding multiple species alignment was extracted using the Vertebrate Multiz Alignment & Conservation track [18] within the UCSC genome browser. The alignments were then screened for conserved transcription factor-binding sites using MatInspector [19] and a vertebrate factor subset of Genomatix's proprietary database.
Luciferase reporter assays
Cells were transfected with luciferase reporters using Lipofectamine™ Plus (Invitrogen). At 3 h post-transfection, cells were starved overnight in 0.5% serum. The following day, cells were stimulated with FGFs (all at 30 ng/ml) either alone or together with the indicated inhibitors and then incubated for an additional 24 h before the DLR (Dual-Luciferase® Reporter) Assay (Promega) was performed according to the manufacturer's instructions. pGL3Basic was used as a negative (no promoter) control and pRL-TK Renilla was used to normalize for transfection efficiency (Promega). A 6400 bp BamHI/BamHI genomic fragment lying upstream of the DUSP6/MKP-3 start codon and including the putative transcriptional start site was cloned into pGL3Basic. This full-length promoter was used as a template in PCRs to generate deletion mutants, which were subsequently cloned into pGL3Basic as KpnI/BamHI fragments. Point mutations within the DUSP6/MKP-3 promoter were introduced into the 508 bp promoter–reporter construct. Ets-binding mutants were made using complementary primers 5′-CTTATCCGGAGCGGAAATTCCTTTC-3′ in which the central core Ets-binding site GGA (underlined) was mutated to TTC (triple mutant) as described by Withers and Hakomori [20], or as single mutants TGA (first base), GTA (second base) or GGC (third base) respectively. The palindromic Ets-binding site TCC (underlined) was altered to GAA using complementary primers 5′-GGAGCGGAAATGAATTTCCGTTTTTG-3′. The Forkhead mutant (FKHD) was made using complementary primers 5′-GTTGCAGCTTGTTTGCACTGGG-3′ in which the central FKHD-binding site (underlined) was altered to CCCC. Wild-type and mutant (Ets2 Δ410–425 and Ets2 T72A) forms of Ets2 were expressed as HA (haemagglutinin) epitope-tagged proteins in the mammalian expression vector pcDNA3. The NF-κB (nuclear factor κB) reporter construct 3× κB–ConA (concanavalin A)–luciferase (3× κB), its control (lacking the NF-κB-binding sites) ConA–luciferase (ConA) and the RSV (rous sarcoma virus) expression vectors encoding the dominant-negative IκBα (inhibitor of NF-κB α) mutant (RSV-Mad3) and β-galactosidase (RSV-βGal) were kindly provided by Dr Neil Perkins (University of Dundee).
EMSA (electrophoretic mobility-shift assay) and ChIP (chromatin immunoprecipitation) assays
32P-labelled probes were generated by annealing complementary oligonucleotide pairs which generate 5′→3′ overhangs and subsequent radiolabelling by incubation with Klenow polymerase and [α-32P]dCTP. Probes were purified on Probequant G-50 columns (GE Healthcare) according to the manufacturer's instructions. Oligonucleotides used were MKP-3 (wild-type), 5′-GGGCTTATCCGGAGCGGAAATTCCTTT-3′ and 5′-CGGAAAGGAATTTCCGCTCCGGATAAG-3′; MKP-3m (Ets-site mutant), 5′-GGGCTTATCCGGAGCTTCAATTCCT-3′ and 5′-CGGAAAGGAATTGAAGCTCCGGATAG-3′; E74 (canonical wild-type Ets site), 5′-GGAGCTGAATAACCGGAAGTAACTCAT-3′ and 5′-GGGATGAGTTACTTCCGGTTATTCAGC-3′; E74m (mutant Ets site), 5′-GGAGCTGAATAACCGTAAGTAACTCAT-3′ and 5′-GGGATGAGTTACTTACGGTTATTCAGC-3′. Nuclear extracts were prepared and EMSAs were performed exactly as described by Feng et al. [21].
ChIP assays were performed exactly as described by Boyd et al. [22]. Briefly, approx. 107 NIH 3T3 cells were used per immunoprecipitation. Following sonication, 100 μl of chromatin samples were diluted in 1 ml of buffer containing 0.1% SDS, 167 mM NaCl, 1.1% (v/v) Triton X-100, 1 mM EDTA, 16.7 mM Tris/HCl (pH 8.0) with protease inhibitors and pre-cleared twice for 1 h at 4 °C with 25 μl of Protein G saturated with BSA and salmon sperm DNA (Upstate). Pre-cleared samples were then immunoprecipitated overnight with 2 μg of the anti-Ets1 (sc350x), anti-Ets2 (sc351x), anti-Erm81 (sc22807x) and the unrelated anti-HA (sc805x) antibodies (all Santa Cruz Biotechnology) in the presence of 20 μl of pre-blocked Protein G. Immune complexes were washed five times in modified RIPA buffer [20 mM Tris/HCl (pH 8.0), 1 mM EDTA, 1% (v/v) Nonidet P40, 0.4 M LiCl, 0.7% sodium deoxycholate and protease inhibitors]. The DNA was then eluted in TE (10 mM Tris/HCl and 1 mM EDTA, pH 7.5) containing 1% (w/v) SDS, incubated overnight at 55 °C in the presence of proteinase K (100 μg/ml) and purified using a PCR purification column (Qiagen). PCRs (40 μl volumes) consisting of 30 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 30 s were carried out using 3 μl of eluted DNA as the template and either ChIPFO2, 5′-TGTGAACTCTAAACAGAAGGAAACAC-3′, and ChIPRE2, 5′-ACAGGTTGTGTTGATGAATTGTTAAT-3′, which amplify nucleotides −555 to −764 containing the putative Ets site, or, as a control, ChIPFO3, 5′-CAGCGACTGGAATGAGAACA-3′, and ChIPRE3, 5′-GGTGCCTGATTAACCCTTGA-3′, which amplify nucleotides +330 to +550 lying downstream of the Ets-binding site. PCR products were analysed using a 2% agarose gel run in 0.5× TBE (Tris/borate/EDTA) and stained with ethidium bromide.
Embryo culture and electroporation
The wild-type 508 bp DUSP6/MKP-3 promoter fragment, and the corresponding Ets site (GGA to TTC) and FKHD (TGTT to CCCC) were subcloned into an EGFP (enhanced green fluorescent protein)-expressing plasmid ptkd2EGFP [23] as KpnI/BglII fragments. Fertile hen's eggs (High Sex×Rhode Island Red) were incubated at 38 °C to yield embryos of Hamburger and Hamilton stage 10 (HH10, ten somite) [23a]. Electroporation with appropriate constructs was carried out in ovo using standard techniques [24] and fluorescence was monitored after 4 h. For bead experiments, embryos were put in culture [25], electroporated as described in [10] and incubated for 1 h before beads (pre-soaked for 1 h) presenting inhibitors (5 mM SU5402, 20 mM PD184352, 20 mM LY294002 or DMSO vehicle) were implanted next to the caudal neural plate. Embryos were left to develop for 4–5 h in a humidified 38 °C incubator before being checked for fluorescence and photographed before fixation in 4% (w/v) paraformaldehyde in PBS overnight at 4 °C.
In situ hybridization and immunocytochemistry of chick embryos
A standard in situ hybridization protocol was used to detect DUSP6/MKP-3 mRNA in the chick embryo [10]. Whole-embryo immunocytochemistry to detect dual-phosphorylated and activated ERK1/2 was carried out as described previously using an anti-dpERK1/2 (dual-phosphorylated ERK1/2) antibody from Cell Signaling Technology [26].
RESULTS
DUSP6/MKP-3 is an FGF-inducible protein in NIH 3T3 cells
Studies in vertebrate embryos have provided compelling evidence for the involvement of FGFR-mediated signalling in the induction of DUSP6/MKP-3 mRNA expression during early development. However, it is uncertain exactly how this occurs with studies invoking essential roles for both ERK and PI3K in mediating this response.
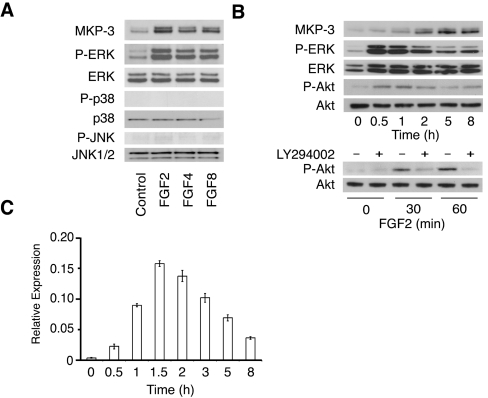
To dissect the molecular mechanisms underlying FGF-inducible DUSP6/MKP-3 expression in a highly tractable system, we screened a panel of mammalian cell lines for expression of the protein. Detectable levels of DUSP6/MKP-3 were seen in HepG2 (human hepatoma) cells and mouse NIH 3T3 fibroblasts. In contrast, DUSP6/MKP-3 was absent in Cos-1, HeLa and HEK-293T (human embryonic kidney) cells (results not shown). Because NIH 3T3 cells have been widely used in studies of inducible gene expression, we determined whether levels of DUSP6/MKP-3 are increased following FGF treatment (Figure 1A). Elevated levels of DUSP6/MKP-3 protein were detected following exposure to FGF2, FGF4 and FGF8, and this correlated with increased phosphorylation of ERK2. In contrast, none of the FGF treatments led to phosphorylation of either p38 or JNK. We note that DUSP6/MKP-3 is resolved as a doublet in these immunoblots. This reflects the use of alternative translational start sites (at codons 1 and 14) within the DUSP6/MKP-3 mRNA [27]. Exposure of mouse fibroblasts to FGF was shown previously to result in activation of both ERK1/2 and PI3K [28]. Consistent with this, we observed increased phosphorylation of both ERK1/2 and the Akt protein kinase, a downstream target of the PI3K pathway, in response to FGF2. Phosphorylation of both ERK and Akt was apparent 30 min after addition of FGF2, reached peak levels after 1 h and declined thereafter (Figure 1B, upper panels). As expected, Akt phosphorylation was abolished by LY294002 (10 μM), a specific inhibitor of PI3K signalling (Figure 1B, lower panel). Elevated levels of DUSP6/MKP-3 protein were detected approx. 1 h after exposure to FGF2 and persisted for at least 8 h (Figure 1B, upper panel) while DUSP6/MKP-3 mRNA levels were increased within 30 min of FGF exposure, reached maximum levels after approx. 90 min and declined thereafter (Figure 1C).
Figure 1. The expression of DUSP6/MKP-3 mRNA and protein is inducible by FGF in NIH 3T3 cells.
(A) NIH 3T3 cells were serum-starved overnight before exposure to FGF2, FGF4 or FGF8 (all at 30 ng/ml) for 5 h. Cells were then lysed, and proteins were analysed by SDS/PAGE and Western blotting using antisera against MKP-3, phospho-ERK (P-ERK), ERK, phospho-p38 (P-p38), p38, phospho-JNK (P-JNK) and JNK. (B) NIH 3T3 cells were serum-starved overnight and then exposed to FGF2 (30 ng/ml) for the indicated time before lysis and analysis of proteins by SDS/PAGE and Western blotting using antisera against MKP-3, phospho-ERK (P-ERK), ERK, phospho-Akt (P-Akt) or Akt (upper panels). NIH 3T3 cells were serum-starved overnight and then exposed to FGF2 (30 ng/ml) for the times indicated in either the absence or the presence of the PI3K inhibitor LY294002 (10 μM) before lysis and analysis of proteins by SDS/PAGE and Western blotting using antisera against phospho-Akt (P-Akt) or Akt (lower panel). (C) NIH 3T3 cells were serum-starved overnight before exposure to FGF2 (30 ng/ml) for the times indicated. Cells were then lysed, and cellular RNA was prepared. DUSP6/MKP-3 mRNA levels were then analysed using real-time PCR. Assays were performed in triplicate and relative DUSP6/MKP-3 mRNA levels are presented as means±S.E.M.
FGF-dependent activation of ERK2, but not PI3K, is essential for induction of DUSP6/MKP-3 mRNA and protein
To dissect the signalling events responsible for FGF-inducible DUSP6/MKP-3 expression, we have employed chemical inhibitors at concentrations where specificity is optimal. First, cells were treated with SU5402 (50 μM), a specific FGFR inhibitor [29], and then exposed to FGFs for 5 h (Figure 2A). As expected, this drug blocked both the FGF-dependent phosphorylation of ERK and phosphorylation of the Akt protein kinase, a target of the PI3K pathway. SU5402 also blocked the induction of DUSP6/MKP-3 protein by FGF2, FGF4 and FGF8, clearly demonstrating a requirement for FGFR activation in this response. We next employed the specific MEK (MAPK/ERK kinase) inhibitor PD184352 (2 μM), which is a more specific and potent inhibitor of the MAPK pathway than PD98059 [30,31]. In contrast with SU5402, this drug did not affect phosphorylation of Akt, but completely blocked the FGF-mediated activation of ERK1/2 and the induction of DUSP6/MKP-3 protein (Figure 2B).
Figure 2. The induction of DUSP6/MKP-3 protein and mRNA by FGFs is blocked by chemical inhibitors of either the FGFR tyrosine kinase or MEK, but not by a specific inhibitor of PI3K activity in NIH 3T3 cells.
NIH 3T3 cells were serum-starved overnight before exposure to FGF2, FGF4 or FGF8 (all at 30 ng/ml) for 5 h either in the absence (DMSO vehicle only) or presence of (A) the FGFR inhibitor SU5402 (50 μM), (B) the MEK inhibitor PD184352 (2 μM) or (C) the PI3K inhibitor LY294002 (10 μM). Cell lysates were analysed by SDS/PAGE and Western blotting using antisera against MKP-3, phospho-ERK (P-ERK), ERK, phospho-Akt (P-Akt) and Akt. (D) NIH 3T3 cells were serum-starved overnight before exposure to FGF2, FGF4 or FGF8 (all at 30 ng/ml) for 90 min in either the absence (DMSO vehicle only) or the presence of the indicated inhibitor. Cells were then lysed, and cellular RNA was prepared. DUSP6/MKP-3 mRNA levels were then analysed using real-time PCR. Assays were performed in triplicate and relative DUSP6/MKP-3 mRNA levels are presented as means±S.E.M.
LY294002 (10 μM) had no effect on either FGF-induced ERK activation or DUSP6/MKP-3 expression, but, as expected, it caused a dramatic reduction in the levels of phospho-Akt (Figure 2C). In addition to these inhibitors, we also determined that FGF-mediated induction of DUSP6/MKP-3 was unaffected by inhibition of either the TOR (target of rapamycin) pathway using rapamycin (100 nM) or phospholipase C activity using U-73122 (4 μM) (results not shown). Finally, we used real-time PCR to measure relative levels of DUSP6/MKP-3 mRNA in FGF-treated NIH 3T3 cells in the absence and presence of either PD184352 or LY294002. Clearly, inhibition of the ERK MAPK pathway, but not PI3K activity, blocks FGF-inducible DUSP6/MKP-3 mRNA expression (Figure 2D).
ERK1/2 activation is both necessary and sufficient for induction of DUSP6/MKP-3
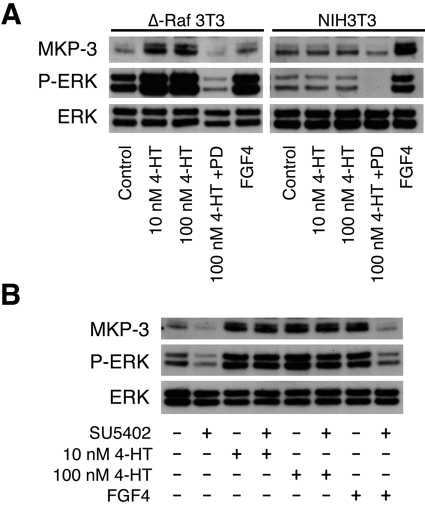
Although the experiments described above indicate that ERK signalling is essential for FGF-mediated induction of DUSP6/MKP-3, we wished to know whether activation of ERK alone is sufficient to trigger expression of this gene. To address this question, we obtained mouse NIH 3T3 fibroblasts that stably express a fusion between a constitutively active form of the Raf-1 MKKK and a mutant form of the ER (Δ-Raf-1:ER* NIH 3T3 cells). These Δ-Raf-1:ER* NIH 3T3 cells respond to 4-HT by activation of Raf, which selectively activates MKK (MAPK kinase), thus causing activation of ERK [32]. Treatment of Δ-Raf-1:ER* NIH 3T3 cells, but not parental wild-type NIH 3T3 cells with 4-HT (10 or 100 nM) leads to ERK activation and induction of DUSP6/MKP-3, whereas both cell lines respond normally to FGF4 (Figure 3A). As expected, both the induction of DUSP6/MKP-3 and activation of ERK in response to 4-HT are blocked by PD184352, but are insensitive to inhibition of FGFR activity by SU5402 (Figure 3). We conclude that activation of ERK is both necessary and sufficient to trigger expression of DUSP6/MKP-3.
Figure 3. ERK activation is both necessary and sufficient for the induction of DUSP6/MKP-3 protein in NIH 3T3 cells.
(A) Either Δ-Raf-1:ER* NIH 3T3 cells (Δ-Raf 3T3) or NIH 3T3 cells were serum-starved overnight and then exposed to the indicated concentration of 4-HT in either the absence or the presence of PD184352 (PD; 2 μM). As a control, cells were exposed to FGF4 (30 ng/ml). After 5 h of incubation, cells were lysed, and proteins were analysed by SDS/PAGE and Western blotting using antisera against MKP-3, phospho-ERK (P-ERK) and ERK. (B) Δ-Raf-1:ER* NIH 3T3 cells were serum-starved overnight and then exposed to either the indicated concentration of 4-HT or FGF4 (30 ng/ml) in the absence or presence of the FGFR inhibitor SU5402 (50 μM). After 5 h of incubation, cells were lysed, and proteins were analysed by SDS/PAGE and Western blotting using antisera against MKP-3, phospho-ERK (P-ERK) and ERK.
The murine DUSP6/MKP-3 promoter contains a conserved region that is required for FGF-responsive transcription
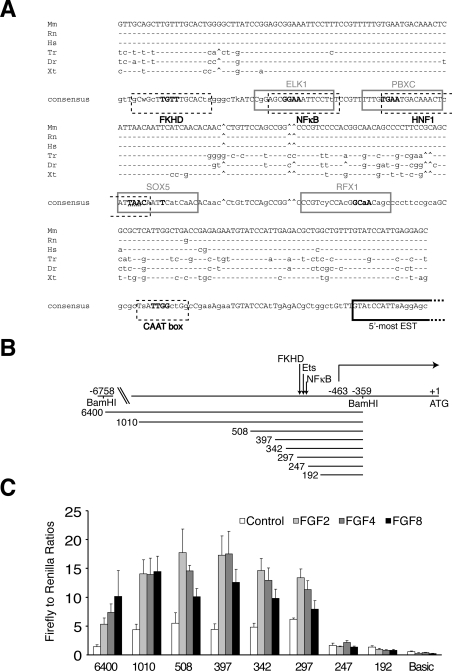
The use of comparative genome analysis to detect non-coding DNA sequence conservation within the 5′-flanking regions of orthologous genes across species is a useful tool in the identification of key regulatory elements [33]. An analysis of this region of the DUSP6/MKP-3 genes from mouse, rat, human, Xenopus, zebrafish and Fugu identified a highly conserved region extending approx. 250 bp upstream of the putative transcriptional start site for MKP-3 (Figure 4A). This putative promoter does not contain a TATA box, but, in common with many TATA-less promoters, it does contain a CAAT box in the reverse (ATTG) orientation. Interestingly, this region also contains consensus-binding sites for a number of different transcription factor families that occur in all five species in the same order and orientation. These include sites for Forkhead, the Ets family of transcription factors, NF-κB, PBX1 (pre-B-cell leukaemia transcription factor 1)-related homeobox factors and the SRY (sex-determining region Y)-box containing factor SOX5 (Figure 4A). Of these, the Ets-binding site is of particular interest as a number of Ets family members, including Elk1 (Ets-like kinase 1), Ets1 and Ets2, are known targets of ERK1/2 signalling [34,35]. In addition, Forkhead is a downstream target of PI3K signalling, and Meis1, which can form complexes with PBX1, is a retinoic acid-inducible protein in the proximal domain of the developing limb [36,37].
Figure 4. Bioinformatic and functional analysis of the murine DUSP6/MKP-3 gene promoter.
(A) DNA sequence alignment of a conserved region identified within the proximal promoter of the DUSP6/MKP-3 gene in vertebrates: Mm (Mus musculus), Rn (Rattus norvegicus), Hs (Homo sapiens), Tr (Takifugu rubripes), Dr (Danio rerio) and Xt (Xenopus tropicalis). Conserved transcription factor-binding sites are boxed, either grey-lined (forward strand) or black broken-lined (reverse strand) and identified as follows: FKHD (Forkhead family), ELK1 (Ets-binding site), NF-κB, PBXC (PBX1–Meis1 complexes), HNF1 (hepatic nuclear factor 1), SOX5 [Sox/SRY-sex/testis determining and related HMG (high-mobility group) box factors], RFX1 (regulatory factor X1), CAAT box (CAAT box: promoter element in some genes located approx. 75–80 bp upstream of the start site for transcription). Core bases are shown in bold, and, where these overlap, the nucleotides belonging to each site are identified to indicate orientation (either bold grey or hatched underlined). The putative transcriptional start site is also indicated (boxed in black). The latter is based on mapping the 5′-most extent of annotated ESTs (expressed sequence tags) for DUSP6/MKP-3. (B) Schematic diagram showing the 5′ boundaries of the DUSP6/MKP-3 promoter–reporter constructs employed in the deletion analysis. Nucleotides are numbered with the A of the ATG start codon designated as +1. The putative transcriptional start site (−463) is indicated as are the BamHI restriction enzyme sites used to subclone the longest (6400 bp) genomic fragment into the firefly luciferase reporter plasmid. The position of the conserved region containing the putative transcription factor-binding sites is also indicated. (C) The constructs indicated in (B) were co-transfected into NIH 3T3 cells along with pRL-TK Renilla to normalize for transfection efficiency. Cells were then starved overnight in 0.5% serum. The following day, cells were either left untreated or stimulated with the indicated FGF (all at 30 ng/ml) for an additional 24 h, before cells were lysed, and luciferase assays were performed. pGL3Basic acted as a negative (promoterless) control. Luciferase assays were performed in quadruplicate, and results are mean±S.E.M. firefly/Renilla ratios.
To perform a functional analysis of the DUSP6/MKP-3 promoter, we first isolated a 6.4 kb BamHI fragment containing nucleotides −6758 to −359 (where the A of the ATG start codon is +1) of the 5′ flanking sequence from the murine gene. This included the putative transcriptional start site and approx. 100 bp of the 5′-UTR (untranslated region). This was cloned into a luciferase-reporter construct, and, following transfection into NIH 3T3 cells, transcriptional activity was assayed in either the absence or presence of FGF2, FGF4 or FGF8 (Figures 4B and 4C). A robust increase in promoter activity was seen in response to all three FGFs. We then performed a deletion analysis of this FGF-responsive promoter. Reporter constructs containing sequences from −1368 to −359 (1010 bp), −866 to −359 (508 bp), −755 to −359 (397 bp), −700 to −359 (342 bp) and −655 to −359 (297 bp) demonstrated an increased transcriptional response following exposure of cells to FGFs. In contrast, reporters containing sequences from −605 to −359 (247 bp) and −550 to −359 (192 bp) were completely unresponsive to FGF treatment (Figures 4B and 4C). Interestingly, the 50 bp interval between constructs 297 and 247 falls within the conserved region identified in our in silico promoter analysis, indicating that it contains regulatory sequences essential for FGF-inducible DUSP6/MKP-3 transcription.
FGF-dependent DUSP6/MKP-3 reporter activity is mediated by activation of ERK and not by activation of the PI3K pathway
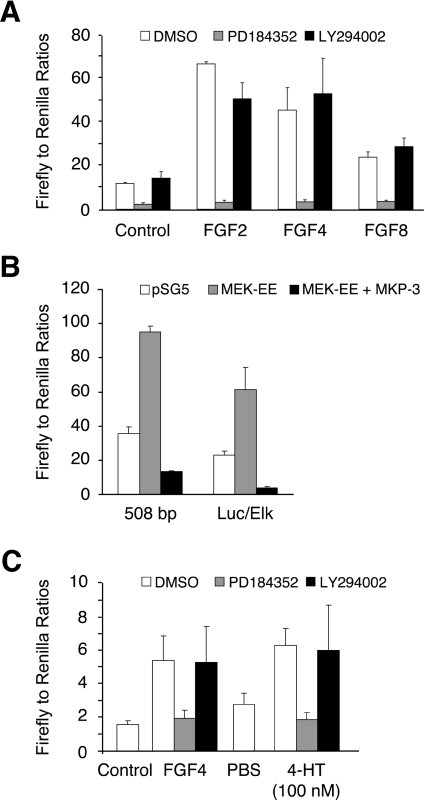
We next assessed whether FGF-responsive DUSP6/MKP-3 transcription was dependent on signalling through either the ERK or the PI3K pathway. The 508 bp DUSP6/MKP-3 reporter (nucleotides −866 to −359) was selected for these experiments as this is both FGF-responsive and contains the whole of the conserved region identified in our in silico analysis which includes the putative Forkhead-, Ets- and NF-κB-binding sites. This plasmid was transfected into NIH 3T3 cells, which were then stimulated with FGF2, FGF4 or FGF8 either in the absence or presence of PD184352 or LY294002. FGF-inducible promoter activity was strongly inhibited by the MEK inhibitor PD184352, while being refractory to inhibition of PI3K signalling (Figure 5A). Similar results were obtained with the longer (6400 bp) reporter construct containing nucleotides −6758 to −359 (results not shown). In an alternative approach, we co-transfected the 508 bp reporter with either MEK-EE, a constitutively active mutant of MKK, alone or together with human DUSP6/MKP-3 itself. The activated form of MKK caused a significant increase in reporter activity and this was abrogated by co-expression of DUSP6/MKP-3. In this regard, our MKP-3 promoter behaved similarly to a model ERK-responsive transcriptional readout in the form of a GAL4-dependent luciferase reporter co-transfected with a GAL4–Elk1 fusion protein (Figure 5B). Finally, we transfected Δ-Raf-1:ER* NIH 3T3 cells with the 508 bp reporter and stimulated these cells with either FGF4 or 4-HT. Both agonists caused a significant increase in DUSP6/MKP-3 reporter activity, and this was blocked by the MEK inhibitor PD184352, but not by the PI3K inhibitor LY294002 (Figure 5C). We conclude that the FGF-dependent activation of the DUSP6/MKP-3 gene promoter is dependent on the activation of ERK1/2, but not PI3K, in NIH 3T3 cells and that this activity accurately reflects the ERK-dependence of FGF-mediated induction of both endogenous DUSP6/MKP-3 protein and mRNA.
Figure 5. FGF-dependent DUSP6/MKP-3 promoter activity is mediated by activation of ERK and not by activation of the PI3K pathway.
(A) The 508 bp DUSP6/MKP-3 reporter construct was co-transfected into NIH 3T3 cells along with pRL-TK Renilla to normalize for transfection efficiency. Cells were then starved overnight in 0.5% serum. The following day, cells were either left untreated or stimulated with the indicated FGF (all at 30 ng/ml) in either the absence or the presence of vehicle (DMSO), PD184352 (2 μM) or LY294002 (10 μM) for an additional 24 h. Cells were then lysed, and luciferase assays were performed in quadruplicate. (B) Either the 508 bp DUSP6/MKP-3 reporter construct or a plasmid encoding a GAL4–Elk1 fusion protein together with a GAL4-dependent luciferase reporter (Luc/Elk) was co-transfected into NIH 3T3 cells along with pRL-TK Renilla to normalize for transfection efficiency and either empty pSG5 expression vector or pSG5 encoding either a constitutively active mutant of MEK (MEK-EE) or DUSP6/MKP-3 itself. After 24 h, cells were lysed, and luciferase assays were performed in quadruplicate. (C) Δ-Raf-1:ER* NIH 3T3 cells were co-transfected with the 508 bp DUSP6/MKP-3 reporter construct and pRL-TK Renilla to normalize for transfection efficiency. Cells were then serum-starved overnight and either left untreated or treated with either FGF4 (30 ng/ml) or 4-HT (100 nM) in the presence or absence of vehicle (DMSO), PD184352 (2 μM) or LY294002 (10 μM). After 24 h, cells were lysed, and luciferase assays were performed in quadruplicate. Results are mean±S.E.M. firefly/Renilla ratios.
ERK-dependent DUSP6/MKP-3 transcription requires a conserved Ets-binding site
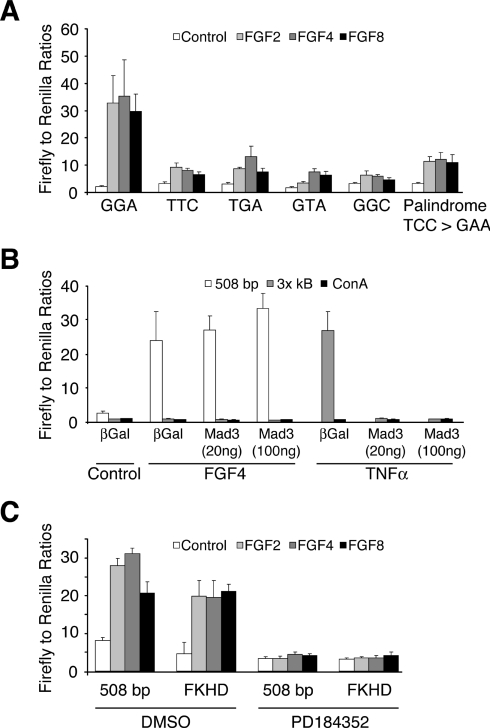
As noted previously, the region of the DUSP6/MKP-3 promoter identified in our deletion analysis contains putative binding sites for a number of transcription factors. Of these, the most obvious target for signalling through the ERK pathway is an Ets site, which overlaps with a potential binding site for NF-κB (Figure 4A). To investigate the importance of this sequence, we mutated the three core bases of the Ets site either singly or in combination within the context of the 508 bp reporter. All of these changes led to a significant reduction in promoter activity in response to FGF2, FGF4 and FGF8 (Figure 6A). In addition, we noticed that the Ets site is actually palindromic (CGGAAATTCCT) and mutation of this second motif (TCC to GAA) also causes a significant reduction in DUSP6/MKP-3 promoter activity (Figure 6A).
Figure 6. A conserved Ets-binding site is required for FGF-inducible DUSP6/MKP-3 promoter activity.
(A) NIH 3T3 cells were co-transfected with either the wild-type 508 bp DUSP6/MKP-3 reporter construct or the indicated mutants together with pRL-TK Renilla to normalize for transfection efficiency. Cells were then starved overnight in 0.5% serum. The following day, cells were either left untreated or stimulated with the indicated FGF (all at 30 ng/ml). After 24 h, cells were lysed, and luciferase assays were performed in quadruplicate. (B) NIH 3T3 cells were co-transfected with the indicated reporter constructs along with either RSV-Mad3 (20 or 100 ng) or as a control RSV-βGal and pRL-TK Renilla to normalize for transfection efficiency. Cells were then serum-starved overnight (0.5% FBS) and either left untreated or exposed to either FGF4 (30 ng/ml) for 24 h or TNFα (10 ng/ml) for 6 h. Cells were then lysed, and luciferase assays were performed in quadruplicate. (C) NIH 3T3 cells were co-transfected with either the wild-type 508 bp DUSP6/MKP-3 reporter construct or a 508 bp reporter in which the Forkhead-binding site was mutated (FKHD) together with pRL-TK Renilla to normalize for transfection efficiency. Cells were then starved overnight in 0.5% FBS. The following day, cells were either left untreated or stimulated with the indicated FGF (all at 30 ng/ml) either in the absence (DMSO) or presence of PD184352 (2 μM). After 24 h, cells were lysed, and luciferase assays were performed in quadruplicate. Results are mean±S.E.M. firefly/Renilla ratios.
Because the conserved Ets-binding site overlaps with a putative site for the NF-κB transcription factor (Figure 4A), it was important to investigate a possible role for this signalling pathway in mediating the response to FGF. We first determined whether expression of a dominant-negative mutant of IκBα (Mad3 super-repressor) affected the FGF-mediated activation of the DUSP6/MKP-3 promoter. Co-expression of this protein had no effect on FGF-inducible DUSP6/MKP-3 transcription (Figure 6B). In addition, an NF-κB-responsive reporter containing three NF-κB-binding sites (3× κB) was completely unresponsive to FGF. In contrast, TNFα (tumour necrosis factor α) stimulated this reporter, and its activity was completely inhibited by co-expression of the Mad3 super-repressor (Figure 6B). Finally, a putative site for the Forkhead (FOXO) family of transcription factors lies within the 5′ boundary of our FGF-responsive 297 bp DUSP6/MKP-3 reporter. However, mutation of this site within our 508 bp reporter had no effect on FGF-mediated activation, and transcriptional activity remained sensitive to inhibition of the ERK1/2 MAPK pathway by PD184352 (Figure 6C). We conclude that a functional Ets site is required for FGF-inducible DUSP6/MKP-3 transcription and that neither NF-κB nor Forkhead plays a role in mediating this response.
ERK-responsive transcription factors Ets1 and Ets2 bind to the endogenous DUSP6/MKP-3 gene promoter
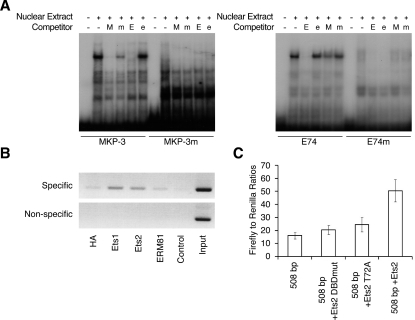
Our results thus far implicate a member or members of the Ets group of transcription factors in mediating ERK-dependent DUSP6/MKP-3 transcription in response to FGFs. To explore this further, we performed EMSAs using a labelled double-stranded oligonucleotide probe spanning the conserved Ets site (MKP-3). These experiments revealed a specific protein complex, which was not seen when labelled mutant oligonucleotide (MKP-3m) is used as a probe (Figure 7A). This complex is effectively competed away by unlabelled wild-type MKP-3 oligonucleotide (M), but not by a mutant oligonucleotide with a single substitution in the Ets site (m). This complex is also effectively competed by a wild-type (E) but not a mutant (e) oligonucleotide containing a canonical Ets-binding site. A complex that co-migrated with that seen using the wild-type MKP-3 oligonucleotide, was also observed using the wild-type canonical Ets site (E74), but not a mutant oligonucleotide (E74m) as a probe (Figure 7A). This complex was competed away by both wild-type Ets (E) oligonucleotide and, albeit less effectively, by the wild-type MKP-3 oligonucleotide (M). Further experiments showed that the complex detected using the MKP-3 oligonucleotide was present when nuclear extracts from either control or FGF-treated cells was used, indicating that protein(s) may be constitutively bound to the DUSP6/MKP-3 promoter (results not shown). With this information in hand, we decided to take a candidate approach to examine the binding of individual Ets factors to the endogenous DUSP6/MKP-3 promoter using ChIP assays.
Figure 7. Ets-family transcription factors bind to the endogenous DUSP6/MKP-3 gene promoter.
(A) EMSAs were performed using the following labelled oligonucleotide probes: wild-type MKP-3 or an Ets site mutant MKP-3 (MKP-3m), and wild-type canonical Ets-binding site (E74) or its corresponding Ets-binding site mutant (E74m). Labelled probes were then incubated with nuclear extract from NIH 3T3 cells in either the absence or the presence of the following unlabelled competitor DNAs: wild-type MKP-3 (M), mutant MKP-3 (m), wild-type E74 (E) or the corresponding Ets site mutant (e). Following incubation, mobility-shifts were visualized by electrophoresis and autoradiography. (B) ChIP assays were performed using an unrelated control antibody (HA) and antibodies specific for Ets1, Ets2 or ERM81. Specific PCR products corresponding to the region of the DUSP6/MKP-3 promoter containing the putative Ets-binding site and the non-specific control reactions are shown, as are the controls lacking template and the results of PCRs performed using the input chromatin as template. (C) NIH 3T3 cells were either transfected with the 508 bp DUSP6/MKP-3 reporter alone or together with either a deletion mutant of Ets2 lacking a functional DNA-binding domain (Ets2 DBDmut), a mutant Ets2 protein lacking a conserved MAPK phosphorylation site (Ets2 T72A) or wild-type Ets2. Following transfection, cells were cultured overnight in medium containing 10% FBS. Cells were then lysed, and luciferase assays were performed in triplicate. Results are mean±S.E.M. firefly/Renilla ratios.
For these experiments, NIH 3T3 cells were cultured in 10% serum before treatment with 1% formaldehyde for 10 min to cross-link DNA and proteins in vivo, followed by cell lysis and the preparation of chromatin. Chromatin from approx. 2×107 cells was sonicated to an average length of 600 bp before immunoprecipitation with either a non-specific (HA) antibody or specific antisera against Ets factors. Precipitated DNA was amplified by PCR, separated by agarose gel electrophoresis, and visualized by ethidium bromide staining (Figure 7B). Two sets of oligonucleotide PCR primers were used, one of which was specific for the Ets-binding site and, as a control, primers that annealed to sequences upstream of the Ets-binding site. Non-precipitated (input) chromatin was a positive control for these PCRs. We could detect significant association of both Ets1 and Ets2 with the Ets-binding site, whereas no detectable signal was seen using either the control antibody or an antibody against ERM81, a member of the closely related Pea3 subfamily of Ets factors. Additional ChIP experiments using antibodies specific for Elk1 also resulted in a failure to detect binding to the DUSP6/MKP-3 promoter (results not shown). Finally, we assessed the impact of expressing wild-type and mutant forms of Ets2 on the activity of the 508 bp DUSP6/MKP-3 luciferase reporter. Co-transfection of wild-type Ets2 caused a significant increase in reporter activity. In contrast, co-expression of either a deletion mutant of Ets2 (Δ410–425) lacking a functional DNA-binding domain (Ets2 DBDmut) or a mutant form of Ets2 (Ets2 T72A) lacking a critical MAPK phosphorylation site [38] did not increase DUSP6/MKP-3 reporter activity (Figure 7C).
The murine DUSP6/MKP-3 promoter is regulated appropriately by endogenous signals which depend on the FGF/MAPK pathway
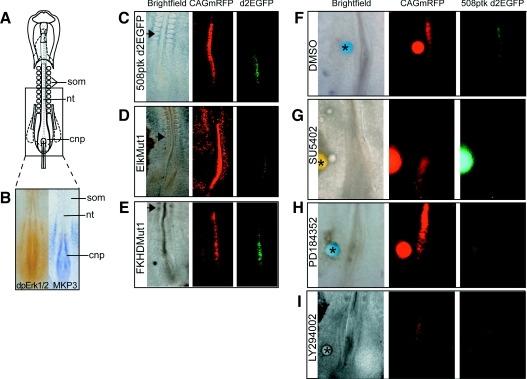
To test whether the 508 bp fragment of the DUSP6/MKP-3 promoter is appropriately activated in the context of a developing embryo, we placed this sequence into a reporter construct driving expression of destabilized EGFP (508ptkd2EGFP) [23]. HH10 (ten somite) embryos (Figure 8A) were then co-electroporated with this plasmid and a plasmid encoding CMV (cytomegalovirus) promoter-driven mRFP (monomeric red fluorescent protein) (CAGmRFP), which identifies all electroporated cells. At this stage DUSP6/MKP-3 is expressed in the caudal neural plate, a region of high ERK1/2 activity, but not more rostrally in the neural tube flanked by somites (Figures 8A and 8B) [26]. Introduction of these constructs into the neuroepithelium revealed that this DUSP6/MKP-3 promoter region drives EGFP expression within the endogenous DUSP6/MKP-3 domain (where cells co-expressed EGFP and mRFP) and not in the rostral neural tube, where only mRFP was detected (after 4–6 h of incubation) (Figure 8C, 14 of 14 cases). This 508 bp sequence is thus sufficient to recapitulate the expression pattern of the endogenous DUSP6/MKP-3 gene. When the Ets-binding site is mutated, expression of EGFP is either much reduced (Figure 8D, six of 11 cases) or absent (five of 11 cases). In contrast, mutation of the putative Forkhead-binding site does not reduce EGFP expression (Figure 8E, six of six cases).
Figure 8. The 508 bp DUSP6/MKP-3 promoter fragment directs transgene expression in an endogenous DUSP6/MKP-3 domain.
(A) Schematic diagram of the HH10 chick embryo. The boxed area indicates approximate field in (B–I). (B) Comparison of ERK1/2 phosphorylation and DUSP6/MKP-3 mRNA expression in the caudal region of the HH10 chick embryo. dpErk1/2, dual-phosphorylated ERK1/2. (C) HH10 chick embryo co-electroporated with a plasmid constitutively expressing mRFP (CAGmRFP) and the 508 bp fragment of the DUSP6/MKP-3 promoter driving expression of EGFP (508ptkd2EGFP) only within the endogenous DUSP6/MKP-3 domain. (D) Embryo co-electroporated with plasmid 508ptkd2EGFP in which the Ets-binding site is mutated (EtsMut1) and CAGmRFP shows reduced EGFP expression. (E) Co-electroporation of plasmid 508ptkd2EGFP in which the Forkhead-binding site is mutated (FKHDMut1) and CAGmRFP. (F–H). Embryos were co-electroporated with the wild-type DUSP6/MKP-3 promoter fragment (508ptkd2EGFP) and CAGmRFP before beads were soaked in DMSO (F), 5 mM SU5402 (G), 20 mM PD184352 (H) or 20 mM LY294002 (I) were placed next to the electroporation site. Both SU5402 and PD184352 cause a local down-regulation of DUSP6/MKP-3 promoter activity. In contrast, neither LY294002 nor DMSO alone have any significant effect on DUSP6/MKP-3 promoter activity. Asterisks (*) indicates grafted bead, and the arrowhead indicates the position of the last formed somite. nt, neural tube; cnp, caudal neural plate; som, somite.
We next tested whether the 508 bp DUSP6/MKP-3 promoter fragment is regulated by FGF-mediated ERK1/2 activation in caudal neural tissue. Embryos were co-electroporated with the 508ptkd2EGFP and CAGmRFP constructs, then cultured in the presence of locally applied beads soaked in either DMSO vehicle control (Figure 8F), the FGFR inhibitor SU5402 (Figure 8G) or the MEK inhibitor PD184352 (Figure 8H) (see the Experimental section). Blocking activation of either FGFRs or just ERK1/2 inhibited MKP-3 promoter activity (six of six and eight of eight cases respectively), whereas control beads did not attenuate EGFP expression. Finally, to test whether the 508 bp promoter is regulated by PI3K signalling, we electroporated embryos with the 508ptkd2EGFP and CAGmRFP constructs, then cultured them in the presence of beads soaked in the PI3K inhibitor LY294002 (Figure 8I). In all cases, there was no loss of EGFP, suggesting that DUSP6/MKP-3 expression in the caudal neural plate is not downstream of this signalling pathway (five of five cases).
DISCUSSION
In vertebrates, the induction of DUSP6/MKP-3 in response to FGF during early development represents the best-characterized link between a specific growth factor signalling axis and expression of an MKP [9,10]. However, whether DUSP6/MKP-3 induction represents a negative-feedback control or an example of regulated cross-talk between two intracellular signalling pathways is controversial, as essential roles have been proposed for both ERK MAPK and PI3K signalling in regulating DUSP6/MKP-3 expression [10–13,15]. The majority of these data were obtained using two different pharmacological inhibitors of MEK (PD98059 or PD184352) and LY294002, a specific inhibitor of PI3K. Furthermore, chemicals were delivered by implantation of beads pre-soaked in 10–20 mM solutions of drug into chicken embryos [10,11]. This makes the effective concentrations of inhibitor delivered to tissues difficult to assess and raises questions about both the potency and specificity of pathway inhibition in these studies [16]. In the present study, we have utilized cultured NIH 3T3 cells to study FGF-inducible expression of DUSP6/MKP-3, allowing a more precise delivery of drug and direct biochemical readout of its effects.
Our results clearly show that FGF treatment of NIH 3T3 fibroblasts induces DUSP6/MKP-3 expression at the level of both mRNA and protein. Furthermore, although inhibition of ERK1/2 activity blocks this expression completely, we can find no evidence for the involvement of PI3K in this process. These results are reinforced by the observation that conditional activation of the ERK1/2 MAPK pathway in Δ-Raf-1:ER* NIH 3T3 cells also increases levels of DUSP6/MKP-3 protein, indicating that ERK activation is both necessary and sufficient to trigger expression of this phosphatase. To probe the molecular mechanism by which ERK activation might influence DUSP6/MKP-3 expression levels, we performed both a bioinformatic and a functional analysis of the murine DUSP6/MKP-3 gene promoter.
Comparative genome analysis revealed a conserved region within the proximal promoter of MKP-3/DUSP6 containing a number of transcription factor-binding sites. This analysis included sequences from mammal, amphibian and fish genomes. Most of the work performed previously to analyse the signalling pathways involved in the regulation of DUSP6/MKP-3 by FGF has been performed in chicken embryos [10,11]. However, we were unable to include the chicken DUSP6/MKP-3 homologue in our analysis, as the 5′-flanking region of the gene is lacking from the current genome sequence release (http://www.ensembl.org/Gallus_gallus/index.html).
Of the putative regulatory elements within this conserved region, the most obvious candidate with respect to an interface with the ERK MAPK pathway is a conserved Ets-binding site. Many growth factor-activated genes such as c-Fos are regulated by SRF (serum-response factor), a MADS-box transcription factor. SRF forms a ternary complex with Ets family members such as Elk1, also known as TCFs (ternary complex factors), whose activity is controlled by MAPK signalling [34,39]. However, close examination of the DUSP6/MKP-3 promoter reveals no cognate binding sites for SRF, indicating that the Ets site is functioning autonomously.
We can detect the constitutive binding of protein(s) to this Ets consensus sequence in EMSAs and, by using ChIP analysis, the Ets family proteins Ets1 and Ets2 were found to be associated with this binding site in vivo. The latter observation provides a mechanistic link between DUSP6/MKP-3 transcription and ERK1/2 activity. The observation that two distinct Ets family members can associate with the DUSP6/MKP-3 promoter in NIH 3T3 cells also raises questions of binding specificity. A recent genomewide analysis of Ets protein binding revealed unexpected levels of redundant occupancy [40]. Furthermore, this degree of redundancy was increased if the Ets-binding site was proximal to the transcriptional start site, as is the case in DUSP6/MKP-3. The Ets site within DUSP6/MKP-3 is also palindromic, and core bases within both half-sites are required for FGF-responsive DUSP6/MKP-3 transcription. Interestingly, the stromelysin-1 gene promoter also contains a head-to-head Ets-binding site palindrome [41]. This site has recently been demonstrated to mediate stromelysin-1 expression in response to the tumour promoter PMA, and the Ets1 transcription factor is found to be constitutively associated with this site in vivo [42]. It will be interesting to explore further mechanistic similarities in the regulation of these two growth factor-regulated genes, both of which involve the Ets family of transcription factors acting independently of SRF as a conduit for ERK1/2 signalling.
The expression of the DUSP6/MKP-3 gene in response to tissue sources of FGF is observed in mouse, chicken and zebrafish embryos, and this is probably a reflection of the conservation of the regulatory sequences revealed by our promoter analysis. Scanning genomic fragments for enhancer activity utilizing embryo electroporation in the chicken can identify such regions. This method makes use of reporter constructs in which expression of EGFP is driven by DNA sequences from the locus of interest [23]. Using this approach, we have clearly shown that the region of the murine DUSP6/MKP-3 promoter encompassing the conserved Ets-binding site recapitulates the endogenous domain of DUSP6/MKP-3 expression in chicken embryo caudal neural tissue. Furthermore, this expression is dependent on the integrity of the Ets-binding site and is sensitive to inhibition of the ERK MAPK pathway. We have identified Ets1 and Ets2 as binding to the DUSP6/MKP-3 promoter in NIH 3T3 cells, and the question arises as to their expression in the tissues we have analysed in the present study. We have not determined the pattern of Ets1 and Ets2 mRNA expression in the chicken embryo. However, between day 8.5 and day 10.5 of mouse embryonic development, Ets2 mRNA is expressed in tail bud, pre-somitic mesenchyme and limb bud mesenchyme in a pattern which is almost indistinguishable from that which we observe for DUSP6/MKP-3 mRNA, which in turn correlates with sites of active FGF signalling [9,43].
In conclusion, we have provided compelling evidence that activation of the ERK MAPK pathway and not the PI3K pathway drives expression of DUSP6/MKP-3 mRNA and protein in response to FGFs in mammalian cells. This, together with our previous studies in developing mouse and chicken embryos [10,16], strongly supports the idea that DUSP6/MKP-3 performs a key role as a negative-feedback regulator of FGF-activated ERK1/2 signalling. In addition, our functional studies of the mouse gene promoter provide the first evidence of a molecular mechanism by which ERK1/2 signalling impinges on transcription factors that are responsible for mediating DUSP6/MKP-3 expression. DUSP6/MKP-3, along with genes such as Sprouty and Sef, belong to the FGF synexpression group of FGF antagonists [44]. Although the FGF-inducible expression of Sprouty and Sef has also been linked with activation of ERK1/2 [45,46], both the mechanism by which this occurs and their mode of action as inhibitors of FGF signalling remain unclear [47–50]. Our present results thus make DUSP6/MKP-3 the best understood of the FGF synexpression group in terms of a molecular mechanism for negative-feedback control of FGF signalling. Finally, DUSP6/MKP-3 gene expression is also reported to be responsive to other agonists during early development, including maternal β-catenin signalling and retinoic acid [51,52]. The tools we have developed in the present study should facilitate a more extensive analysis of the signalling pathways that interact with the transcription machinery to regulate DUSP6/MKP-3 expression.
Acknowledgments
We thank Professor Andy Sharrocks (School of Biological Sciences, University of Manchester, Manchester, U.K.) for reagents and advice on performing ChIP analysis, Dr Neil Perkins (School of Life Sciences, University of Dundee) for reagents and advice on the NF-κB signalling pathway, Dr Simon J. Cook (Babraham Institute, Cambridge, U.K.) for providing the NIH 3T3 cell line expressing Δ-Raf-1:ER* and Professor H. Kondoh (Graduate School of Frontier Biosciences, University of Osaka, Osaka, Japan) for the ptkd2EGFP plasmid. We are also grateful to Professor Cheryll Tickle (School of Life Sciences, University of Dundee) for many helpful discussions throughout the course of this work. K. G. S. and research in the Storey Laboratory is supported by grants from the MRC (Medical Research Council) (G0200220 and G0600234). M. P. S. holds a Collaborative Career Development Fellowship in Stem Cell Research (G113/18) in the Storey Laboratory. S. M. K. and work in the Cancer Research UK Stress Response Laboratory are supported by Cancer Research UK and by an MRC Ph.D. studentship (to C. S.).
References
- 1.Camps M., Nichols A., Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 2.Dickinson R. J., Keyse S. M. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J. Cell Sci. 2006;119:4607–4615. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- 3.Groom L. A., Sneddon A. A., Alessi D. R., Dowd S., Keyse S. M. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 4.Muda M., Theodosiou A., Rodrigues N., Boschert U., Camps M., Gillieron C., Davies K., Ashworth A., Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J. Biol. Chem. 1996;271:27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 5.Nichols A., Camps M., Gillieron C., Chabert C., Brunet A., Wilsbacher J., Cobb M., Pouyssegur J., Shaw J. P., Arkinstall S. Substrate recognition domains within extracellular signal-regulated kinase mediate binding and catalytic activation of mitogen-activated protein kinase phosphatase-3. J. Biol. Chem. 2000;275:24613–24621. doi: 10.1074/jbc.M001515200. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson M., Mathers J., Dickinson R. J., Mandl M., Keyse S. M. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal. J. Biol. Chem. 2004;279:41882–41891. doi: 10.1074/jbc.M406720200. [DOI] [PubMed] [Google Scholar]
- 7.Camps M., Nichols A., Gillieron C., Antonsson B., Muda M., Chabert C., Boschert U., Arkinstall S. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science. 1998;280:1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 8.Stewart A. E., Dowd S., Keyse S. M., McDonald N. Q. Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat. Struct. Biol. 1999;6:174–181. doi: 10.1038/5861. [DOI] [PubMed] [Google Scholar]
- 9.Dickinson R. J., Eblaghie M. C., Keyse S. M., Morriss-Kay G. M. Expression of the ERK-specific MAP kinase phosphatase PYST1/MKP3 in mouse embryos during morphogenesis and early organogenesis. Mech. Dev. 2002;113:193–196. doi: 10.1016/s0925-4773(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 10.Eblaghie M. C., Lunn J. S., Dickinson R. J., Munsterberg A. E., Sanz-Ezquerro J. J., Farrell E. R., Mathers J., Keyse S. M., Storey K., Tickle C. Negative feedback regulation of FGF signalling levels by Pyst1/MKP3 in chick embryos. Curr. Biol. 2003;13:1009–1018. doi: 10.1016/s0960-9822(03)00381-6. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami Y., Rodriguez-Leon J., Koth C. M., Buscher D., Itoh T., Raya A., Ng J. K., Esteban C. R., Takahashi S., Henrique D., et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- 12.Smith T. G., Sweetman D., Patterson M., Keyse S. M., Munsterberg A. Feedback interactions between MKP3 and ERK MAP kinase control scleraxis expression and the specification of rib progenitors in the developing chick somite. Development. 2005;132:1305–1314. doi: 10.1242/dev.01699. [DOI] [PubMed] [Google Scholar]
- 13.Echevarria D., Martinez S., Marques S., Lucas-Teixeira V., Belo J. A. Mkp3 is a negative feedback modulator of Fgf8 signalling in the mammalian isthmic organizer. Dev. Biol. 2005;277:114–128. doi: 10.1016/j.ydbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Li C., Scott D. A., Hatch E., Tian X., Mansour S. L. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signalling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez A. R., Lopez-Varea A., Molnar C., de la Calle-Mustienes E., Ruiz-Gomez M., Gomez-Skarmeta J. L., de Celis J. F. Conserved cross-interactions in Drosophila and Xenopus between Ras/MAPK signalling and the dual-specificity phosphatase MKP3. Dev. Dyn. 2005;232:695–708. doi: 10.1002/dvdy.20227. [DOI] [PubMed] [Google Scholar]
- 16.Smith T. G., Karlsson M., Lunn J. S., Eblaghie M. C., Keenan I. D., Farrell E. R., Tickle C., Storey K. G., Keyse S. M. Negative feedback predominates over cross-regulation to control ERK MAPK activity in response to FGF signalling in embryos. FEBS Lett. 2006;580:4242–4245. doi: 10.1016/j.febslet.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 17.Karolchik D., Baertsch R., Diekhans M., Furey T. S., Hinrichs A., Lu Y. T., Roskin K. M., Schwartz M., Sugnet C. W., Thomas D. J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., Baertsch R., Rosenbloom K., Clawson H., Green E. D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 20.Withers D. A., Hakomori S. I. Human α(1,3)-fucosyltransferase IV (FUTIV) gene expression is regulated by elk-1 in the U937 cell line. J. Biol. Chem. 2000;275:40588–40593. doi: 10.1074/jbc.M007262200. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y., Goulet A. C., Nelson M. A. Identification and characterization of the human Cdc2l2 gene promoter. Gene. 2004;330:75–84. doi: 10.1016/j.gene.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Boyd K. E., Wells J., Gutman J., Bartley S. M., Farnham P. J. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 23a.Hamburger V., Hamilton H. A series of normal stages in the development of the chick embryo. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 24.Itasaki N., Bel-Vialar S., Krumlauf R. ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- 25.Chapman S. C., Collignon J., Schoenwolf G. C., Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Lunn J. S., Fishwick K. J., Halley P. A., Storey K. G. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev. Biol. 2007;302:536–552. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Dowd S., Sneddon A. A., Keyse S. M. Isolation of the human genes encoding the Pyst1 and Pyst2 phosphatases: characterisation of Pyst2 as a cytosolic dual-specificity MAP kinase phosphatase and its catalytic activation by both MAP and SAP kinases. J. Cell Sci. 1998;111:3389–3399. doi: 10.1242/jcs.111.22.3389. [DOI] [PubMed] [Google Scholar]
- 28.Hadari Y. R., Gotoh N., Kouhara H., Lax I., Schlessinger J. Critical role for the docking-protein FRS2α in FGF receptor-mediated signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 30.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 31.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuels M. L., Weber M. J., Bishop J. M., McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human raf-1 protein kinase. Mol. Cell. Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aparicio S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S. H., Sharrocks A. D., Whitmarsh A. J. Transcriptional regulation by the MAP kinase signalling cascades. Gene. 2003;320:3–21. doi: 10.1016/s0378-1119(03)00816-3. [DOI] [PubMed] [Google Scholar]
- 35.Sharrocks A. D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 36.Arden K. C., Biggs W. H., 3rd Regulation of the FoxO family of transcription factors by phosphatidylinositol-3 kinase-activated signalling. Arch. Biochem. Biophys. 2002;403:292–298. doi: 10.1016/s0003-9861(02)00207-2. [DOI] [PubMed] [Google Scholar]
- 37.Mercader N., Leonardo E., Piedra M. E., Martinez A. C., Ros M. A., Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy S. A., Chen D., Yang B. S., Garcia Ramirez J. J., Cherwinski H., Chen X. R., Klagsbrun M., Hauser C. A., Ostrowski M. C., McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol. Cell. Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollenhorst P. C., Shah A. A., Hopkins C., Graves B. J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baillat D., Begue A., Stehelin D., Aumercier M. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J. Biol. Chem. 2002;277:29386–29398. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- 42.Baillat D., Leprivier G., Régnier D., Vintonenko N., Bègue A., Stéhelin D., Aumercier M. Stromelysin-1 expression is activated in vivo by Ets-1 through palindromic head-to-head Ets binding sites present in the promoter. Oncogene. 2006;25:5764–5776. doi: 10.1038/sj.onc.1209583. [DOI] [PubMed] [Google Scholar]
- 43.Ristevski S., Tam P. P., Hertzog P. J., Kola I. Ets2 is expressed during morphogenesis of the somite and limb in the mouse embryo. Mech. Dev. 2002;116:165–168. doi: 10.1016/s0925-4773(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 44.Niehrs C., Meinhardt H. Modular feedback. Nature. 2002;417:35–36. doi: 10.1038/417035a. [DOI] [PubMed] [Google Scholar]
- 45.Ozaki K., Kadomoto R., Asato K., Tanimura S., Itoh N., Kohno M. ERK pathway positively regulates the expression of Sprouty genes. Biochem. Biophys. Res. Commun. 2001;285:1084–1088. doi: 10.1006/bbrc.2001.5295. [DOI] [PubMed] [Google Scholar]
- 46.Furthauer M., Lin W., Ang S. L., Thisse B., Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat. Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 47.Kovalenko D., Yang X., Chen P. Y., Nadeau R. J., Zubanova O., Pigeon K., Friesel R. A role for extracellular and transmembrane domains of Sef in Sef-mediated inhibition of FGF signalling. Cell. Signalling. 2006;18:1958–1966. doi: 10.1016/j.cellsig.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Ren Y., Cheng L., Rong Z., Li Z., Li Y., Li H., Wang Z., Chang Z. hSef co-localizes and interacts with Ras in the inhibition of Ras/MAPK signalling pathway. Biochem. Biophys. Res. Commun. 2006;347:988–993. doi: 10.1016/j.bbrc.2006.06.193. [DOI] [PubMed] [Google Scholar]
- 49.Ren Y., Li Z., Rong Z., Cheng L., Li Y., Wang Z., Chang Z. Tyrosine 330 in hSef is critical for the localization and the inhibitory effect on FGF signalling. Biochem. Biophys. Res. Commun. 2007;354:741–746. doi: 10.1016/j.bbrc.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 50.Martinez N., Garcia-Dominguez C. A., Domingo B., Oliva J. L., Zarich N., Sanchez A., Gutierrez-Eisman S., Llopis J., Rojas J. M. Sprouty2 binds Grb2 at two different proline-rich regions, and the mechanism of ERK inhibition is independent of this interaction. Cell. Signalling. 2007;19:2277–2285. doi: 10.1016/j.cellsig.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Moreno T. A., Kintner C. Regulation of segmental patterning by retinoic acid signalling during Xenopus somitogenesis. Dev. Cell. 2004;6:205–218. doi: 10.1016/s1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 52.Tsang M., Maegawa S., Kiang A., Habas R., Weinberg E., Dawid I. B. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]