Abstract
Antimicrobial levels of reactive oxygen species (ROS) are produced by the mammalian host defense to kill invading bacteria and limit bacterial colonization. One main in vivo target of ROS is the thiol group of proteins. We have developed a quantitative thiol trapping technique termed OxICAT to identify physiologically important target proteins of hydrogen peroxide (H2O2) and hypochlorite (NaOCl) stress in vivo. OxICAT allows the precise quantification of oxidative thiol modifications in hundreds of different proteins in a single experiment. It also identifies the affected proteins and defines their redox-sensitive cysteine(s). Using this technique, we identified a group of Escherichia coli proteins with significantly (30–90%) oxidatively modified thiol groups, which appear to be specifically sensitive to either H2O2 or NaOCl stress. These results indicate that individual oxidants target distinct proteins in vivo. Conditionally essential E. coli genes encode one-third of redox-sensitive proteins, a finding that might explain the bacteriostatic effect of oxidative stress treatment. We identified a select group of redox-regulated proteins, which protect E. coli against oxidative stress conditions. These experiments illustrate that OxICAT, which can be used in a variety of different cell types and organisms, is a powerful tool to identify, quantify, and monitor oxidative thiol modifications in vivo.
Keywords: chaperone, proteomics, thiol modification
Many physiological and pathological conditions are associated with the accumulation of ROS, a condition termed oxidative stress (1). Prokaryotes experience severe oxidative stress conditions during the host defense in macrophages and neutrophils, where high levels of H2O2 and hypochlorite (NaOCl) are generated to kill the invading microorganisms (2, 3).
ROS readily react with and damage vital cellular structures, among them lipids, DNA, and proteins (4). Over the past years, an increasing number of proteins have been identified that are not damaged by oxidative stress conditions but use ROS-mediated thiol modifications to specifically regulate their protein function (5). Reversible oxidation of structurally or functionally important cysteine residues enables these redox-regulated proteins to rapidly regulate such diverse processes as gene expression (e.g., OxyR), protein quality control (e.g., Hsp33) and metabolic fluxes (e.g., GapDH) in response to accumulating oxidants (6–8). Many of these redox-mediated functional changes have been found to significantly contribute to the oxidative stress survival of pro- and eukaryotic cells.
Identification of proteins with redox-active cysteines has traditionally been a time-consuming endeavor, typically done by biochemical characterization of proteins with highly conserved cysteines that were selected based on genetic experiments (9). The recent interest in redox-regulated proteins fueled by the increasing recognition of their important physiological roles however, led to the development of several global thiol trapping techniques (10–13). These methods, albeit capable of detecting proteins with redox-sensitive cysteines in lysates and intact cells, often lack the ability to directly identify the proteins or cysteines involved, and more importantly, to quantify the extent of oxidative thiol modifications. The ability to distinguish oxidation of nonconserved cysteines in a small subpopulation of a protein from the modification of one or more highly conserved cysteines in the majority of the protein population is, however, absolutely critical for evaluating the potential physiological significance of the identified oxidative thiol modifications. This is crucial for identifying redox-regulated proteins that might play important roles in the oxidative stress protection of pro- and eukaryotic cells, and to determine the target proteins of ROS, whose altered functional state might explain the observed oxidative stress-induced changes in the physiology of an organism.
Here we provide a quantitative snapshot of in vivo thiol modifications in cells treated with sublethal concentrations of either H2O2 or hypochlorite, using a powerful mass spectrometric approach that we have developed and termed OxICAT. We identified a specific set of redox-sensitive target proteins, defined the affected cysteine(s) and determined the precise extent of their oxidative thiol modifications. Analysis of the oxidative stress resistance of mutant E. coli strains lacking these redox-sensitive proteins allowed us to identify a number of novel redox-regulated proteins, which have not previously been shown to be involved in the oxidative stress protection of E. coli.
Results and Discussion
The Concept of OxICAT.
To quantitatively describe the extent of oxidative thiol modifications in proteins, we combined a mass spectrometric global approach using isotope coded affinity tag (ICAT) technology (14–16) with our previously established differential thiol trapping technique (10). Both methods rely on the rapid and irreversible modification of accessible cysteines in proteins with the thiol-trapping reagent iodoacetamide (IAM). The ICAT reagent consists of the IAM-moiety, a cleavable biotin affinity tag, and a 9-carbon linker, which exists in an isotopically light 12C-form (herein referred to as light ICAT) and a 9-Da-heavier isotopically heavy 13C-form (herein referred to as heavy ICAT) (14). The differential thiol trapping technique is based on the sequential reaction of a nonradioactive and radioactive variant of IAM with accessible cysteines in proteins and subsequent visualization using 2D gels (10).
We realized that combining ICAT chemistry with our differential thiol trapping technique should allow us to precisely determine the oxidation state of proteins both in vitro and in vivo using mass spectrometry (MS), and therefore termed this method OxICAT (Fig. 1). In the first step of OxICAT, we denature the proteins to gain access to all reduced cysteines and label them irreversibly with light ICAT. We then reduce all reversible oxidative thiol modifications within the same sample using the strong thiol reductant Tris(2-carboxyethyl)phosphine (TCEP) and modify all newly accessible cysteines with heavy ICAT. Importantly, this procedure generates chemically identical proteins, which only differ in the specific mass of their ICAT-label (9 Da additional mass per heavy ICAT) depending on their previous redox state. After trypsin digest and affinity-purification of the ICAT-labeled peptides, MS for quantification and tandem MS/MS for peptide identification is conducted. Based on this scheme, the OxICAT method detects all reversibly oxidized cysteines, whereas higher oxidation states, such as sulfinic and sulfonic acids, will not be detected. However, substitution of the nonspecific thiol reductant TCEP with more specific reductants such as glutaredoxin or ascorbate will allow the use of OxICAT to specifically detect glutathionylations or nitrosylations, respectively (17, 18).
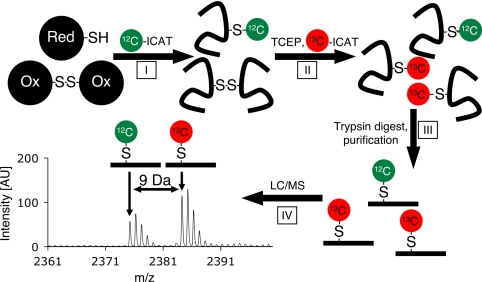
Fig. 1.
Determining the oxidation state of protein thiol using ICAT technology (OxICAT). A hypothetical cellular protein, which exists in either the reduced (Upper) or disulfide-linked form (Lower), is incubated under denaturing conditions to expose all of its cysteine side chains. In step I, isotopically light 12C- ICAT reagent (green ovals) is added, which irreversibly modifies all reduced cysteines in the protein. In step II, all oxidized cysteines are reduced with Tris(2-carboxyethyl)phosphine (TCEP) and subsequently modified with isotopically heavy 13C- ICAT reagent (red ovals). In step III, the protein mixture is digested and ICAT-labeled peptides are purified by using the biotin-affinity tag. In step IV, quantitative MS of the protein mixture reveals the extent of thiol modification in any given peptide. Peptide sequence and identity of the modified cysteine are determined by MS/MS.
Peptides that contain originally only reduced cysteines are predicted to yield single mass peaks corresponding to the light ICAT-labeled form. Peptides containing originally oxidized cysteines will exert masses that are exactly 9 Da higher (or multiples thereof) than the corresponding light ICAT peptides, depending on the number of oxidized cysteines present. Many cellular proteins are expected to be in equilibrium with their cellular redox environment and will be present in both oxidized and reduced form. The corresponding peptides should thus be present both in the light and heavy ICAT-modified form (i.e., ICAT pairs). Because our ICAT-labeled peptides are chemically identical, they will ionize to the same extent. We can thus precisely determine their relative ion intensities, which correlate directly to their relative abundance. Therefore, the ratio of oxidized and reduced peptide can be calculated, which reflects the extent of oxidative thiol modifications present in the protein population. MS/MS analysis of the peptide is used to identify the respective protein and, at the same time, the oxidation sensitive cysteine(s). Although Sethuraman and coworkers have previously used ICAT technology to determine relative changes in the availability of free protein thiols in two separate samples (13, 15, 16), the OxICAT method provides the absolute ratio of reduced to oxidized protein within a single sample. This makes OxICAT also ideally suited for analyzing changes in the redox status of proteins in cells exposed to stress conditions, because neither changes in protein expression nor protein stability influence the OxICAT result.
Using OxICAT to Monitor Oxidative Thiol Modifications in Vitro.
To investigate whether OxICAT can be used to quantitatively describe oxidative thiol modifications, we decided to test it on the redox-regulated chaperone Hsp33 (6). Oxidative stress-induced formation of two intramolecular disulfide bonds converts reduced, inactive Hsp33 into a potent chaperone that protects bacteria against oxidative stress (19). Recent studies suggested that although one disulfide bond in Hsp33 forms very quickly, formation of the other disulfide bond (Cys-232–Cys-234) is slow and might represent the rate-determining step in Hsp33's activation (20). However, appropriate quantitative methods to test this model were not available.
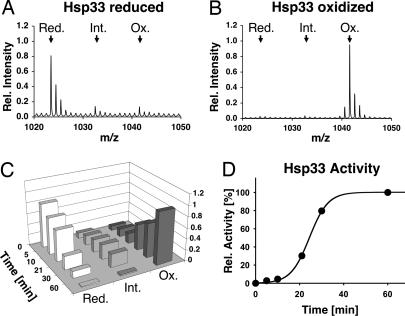
To monitor the kinetics of thiol modifications during Hsp33's activation process, we incubated reduced Hsp33 with H2O2 at 43°C (20). At defined time points, we removed aliquots and either tested them for chaperone activity or used them for our OxICAT labeling. To monitor the oxidation state of the two critical cysteines Cys-232 and Cys-234, we focused our OxICAT analysis on the Hsp33–232-236 peptide. We expected a mass peak at m/z = 1023.4710 if both cysteines were originally reduced or an 18 Da heavier mass peak (m/z = 1041.5314) if both cysteines were originally oxidized. In reduced Hsp33, the majority of the Hsp33–232-236 peptide revealed an m/z value of 1023.46 (Fig. 2A), demonstrating that both cysteines were predominantly reduced before Hsp33's activation. Upon exposure of Hsp33 to oxidizing conditions, we observed an initial lag phase, in which the thiol status of the two critical cysteines remained largely unchanged. This lag phase correlated very well with the observed lag phase in Hsp33's activation process (Fig. 2D). Upon further incubation of Hsp33 under oxidizing conditions, the reduced mass peak gradually decreased while two new mass peaks appeared (Fig. 2C). One of these mass peaks corresponded to the fully oxidized peptide (observed m/z = 1041.55) (Fig. 2B), whereas the other mass peak corresponded to an oxidation intermediate of Hsp33, in which only one of the two cysteines is oxidized whereas the other one is still reduced (observed m/z = 1032.51). Although this intermediate has been observed before, its physiological relevance was unclear, in part due to an inability to quantify its abundance. Using the OxICAT method, we now demonstrated that this oxidation intermediate is indeed an on-pathway intermediate. It slowly converts into the fully oxidized species with a rate that correlates well with Hsp33's rate of activation (Fig. 2 C and D). We concluded from these results that the formation of the Cys-232–Cys-234 disulfide bond is most likely the rate-determining step in Hsp33's activation. These results illustrate the unique ability of OxICAT to precisely quantify oxidative thiol modifications at distinct cysteines within a protein.
Fig. 2.
Using OxICAT to visualize disulfide bond formation in vitro. Reduced, zinc-reconstituted Hsp33 (50 μM) is incubated in the presence of 2 mM H2O2 at 43°C. Samples are taken at specific time points during the oxidation process, and either TCA precipitated to stop all thiol-disulfide exchange reactions and subjected to the OxICAT method (A–C), or analyzed for chaperone activity (D). (A) Detail of the mass spectrum of OxICAT-treated reduced Hsp33. The two cysteines in peptide 232–236 are almost exclusively labeled with light ICAT (calculated m/z = 1023.4710). (B) Detail of the mass spectrum of OxICAT labeled Hsp33 after 60 min of activation. The two cysteines in peptide 232–236 are mostly labeled with heavy ICAT, leading to a mass shift of 18 Da (calculated m/z = 1041.5314). (C) The monoisotope peak corresponding to the Hsp33 peptide 232–236 in the reduced (red) (m/z = 1023.46), partially oxidized (int) (m/z = 1032.51), or fully oxidized (ox) (m/z = 1041.55) form was plotted against incubation time. (D) Influence of Hsp33 (0.3 μM) on the aggregation of chemically denatured citrate synthase (75 nM) at 30°C. The light scattering signal 4 min after addition of citrate synthase was plotted against incubation time. The light scattering signal of citrate synthase in the absence of Hsp33 was used as 0% chaperone activity, whereas the signal in the presence of fully active Hsp33 was set to 100%.
Visualizing Oxidative Thiol Modifications in Vivo.
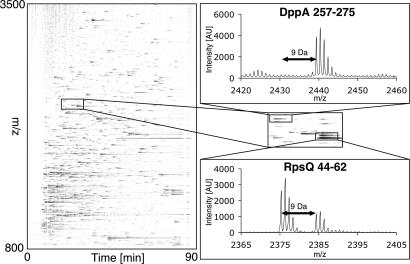
Our Hsp33 results indicated that the OxICAT method should be well suited to identify other proteins that use redox-sensitive cysteines to respond to and possibly protect against oxidative stress. As a first step toward this goal, we used OxICAT to quantitatively assess the global redox status of proteins in exponentially growing E. coli cells. Because of the high sample complexity, we separated the ICAT-labeled peptides by liquid chromatography (LC) and analyzed the resulting 192 fractions by MS. The 8 highest intensity peaks in the parental mass spectrum of each fraction were sequenced by MS/MS. To analyze the LC-MS data, we used the open-source software msInspect, which describes the series of parental mass spectra as image representation (21) (Fig. 3). Whereas ICAT-labeled peptides are represented as black signals, ICAT-pairs are represented by two signals that show the same elution profile but are 9 Da (or multiples thereof) apart from each other on the y axis (Fig. 3). In an average experiment, we identified ≈120 unique ICAT-modified peptides with an ion-score of 95% or higher (see data deposition footnote). About 10% of the identified peptides incorporated exclusively heavy ICAT-label, indicating that they are fully oxidized in vivo [supporting information (SI) Table 2 in SI Appendix], whereas a subpopulation of peptides eluted as ICAT pairs, indicating that the proteins are only partially oxidized in vivo. The remaining peptides were almost exclusively labeled with light ICAT reagent. This result was in excellent agreement with earlier studies that showed that the majority of cellular proteins harbors reduced cysteines in vivo (10).
Fig. 3.
Visualization of OxICAT results. Graphical representation of the LC-MS analysis of aerobically growing E. coli cells. Intensity of each mass signal is given as fraction of blackness. Each mass signal corresponds to one OxICAT labeled peptide. Peptide 44–62 of ribosomal protein RpsQ elutes as ICAT pair with a mass difference of 9.0 Da. Mass signal with lower m/z has incorporated light ICAT, representing the reduced form of the peptide whereas mass signal with higher m/z has incorporated heavy ICAT and represents the oxidized form of the peptide. DppA 257–275 contains a cysteine that is part of a structural disulfide bond and elutes as single mass peak labeled with heavy ICAT.
OxICAT Provides a Quantitative Assessment of the Cellular Redox Status.
Analysis of the ICAT pairs revealed a number of cytoplasmic proteins that are partially (>20%) oxidized under our aerobic growth conditions. We ascertained that we are not introducing any thiol modifications during the sample preparation (SI Fig. 4) and concluded from these results that E. coli cells are exposed to a considerable amount of ROS during aerobic growth. This observation agrees with earlier reports in both E. coli (10) and yeast cells (22) and provides an excellent explanation for the essential roles that the thioredoxin and glutaredoxin systems play in maintaining the reducing character of pro- and eukaryotic cytoplasm (23).
Proteins that are already significantly oxidatively modified in exponentially growing E. coli cells include the antioxidant proteins alkyl hydroperoxidase AhpC (≈38% oxidatively modified) and thiol peroxidase Tpx (≈35% oxidatively modified). Both proteins use disulfide bond formation as catalytic mechanism to detoxify ROS in vivo (24, 25). Another significantly oxidized cytoplasmic protein is the redox-sensitive glyceraldehyde-3-P-dehydrogenase (GapA). Although GapA's redox regulation has been suggested to involve S-glutathionylation of its active site cysteine Cys-149 (26), we found <10% of molecules to be modified at Cys-149 alone (SI Fig. 4 in SI Appendix). The majority of oxidized GapA molecules appeared to be modified at both Cys-149 and the equally conserved Cys-153, suggesting that disulfide bond formation might be the predominant redox-regulatory mechanism of GapA. The high percentage of oxidation (≈55%) and thereby partial inactivation (26) of this crucial glycolytic enzyme under aerobic growth conditions makes it tempting to speculate that ROS-mediated attenuation of GapA activity is a strategy to regulate glycolysis under conditions that produce considerable amounts of ROS. This mechanism might contribute to the Pasteur effect in bacteria, which describes the influence of oxygen on the glucose metabolism (27). Similar observations have recently also been made in aerobically growing yeast cells, where a large proportion of glycolytic enzymes including GapA were shown to be oxidatively modified in vivo (22).
Using OxICAT to Identify Redox-Sensitive E. coli Proteins.
Although the cellular response to H2O2 has been extensively studied, very little is known about the cellular effects of hypochlorite. This is very surprising, given that hypochlorite, an active ingredient of household bleach, is the most widely used disinfectant known. Moreover, it has been recently suggested that NaOCl is also involved in limiting bacterial colonization at mucosal barrier epithelia (3). This is particularly important for noninvasive enterobacteriae, which are not likely to encounter the oxidative burst of phagocytic cells but will be frequently exposed to oxidants in the intestine and other mucosal surfaces. Our ability to precisely quantify oxidative thiol modifications in proteins enables us now to globally determine physiologically relevant target proteins of defined ROS in vivo.
To identify both H2O2 andNaOCl redox-sensitive proteins, we treated exponentially growing E. coli cells with oxidant concentrations that transiently inhibit their growth without reducing the viable cell count (SI Fig. 5 in SI Appendix). Similar concentrations of H2O2, for instance, have been used in earlier proteomic studies designed to investigate the E. coli oxidative stress response (28). To detect proteins that are rapidly oxidatively modified in vivo, we then harvested cells 10 min after addition of the oxidants and conducted our OxICAT thiol trapping experiments. To directly compare individual LC/MS runs, we designed and implemented new software (see SI Appendix), which is available as extension for msInspect (see data deposition footnote). This program automatically identifies and matches peptides across multiple LC runs and calculates precise changes in the oxidation status of hundreds of different proteins. To be considered redox-sensitive, only peptides that showed a more than 1.5-fold increase in oxidative thiol modification upon exposure to ROS were selected.
We identified almost twice as many redox-sensitive peptides under NaOCl-stress conditions (Table 1) than under peroxide-stress (SI Table 3 in SI Appendix), confirming earlier observations that hypochlorite is a significantly more potent oxidant (29). Interestingly, only eight of the identified proteins (Table 1) and two nonidentified peptides (data not shown) harbored cysteines that reacted with both oxidants. These proteins included the previously identified Tpx, GapA, and proteins that were found to contain ROS-sensitive cysteines in other organisms (e.g., Mdh, GlyA) (30). Importantly, although these proteins have been previously shown to harbor redox-cysteines, it was unclear what proportion of these proteins became oxidatively modified under stress conditions. With OxICAT, we could now demonstrate that at least 30% and up to 90% of each of our identified protein populations is oxidatively modified at one or more cysteines. This apparently can have significant physiological consequences as has been shown for GapA, whose oxidative stress-induced inactivation was found to cause a massive decrease in intracellular ATP levels (19, 26). Additionally, OxICAT analysis provides directly the identity of the cysteine(s) affected by the oxidative stress treatment (Table 1, column 3). This, in turn, allows predictions about potentially significant functional or structural changes that are introduced by the observed thiol modifications.
Table 1.
Identified E. coli proteins harboring hypochlorite-mediated thiol modifications
| Peptide | Protein | ICAT-modified cysteine | % oxidized before stress | % oxidized after NaOCl | Fold change | NaOCl sensitivity of mutant* | Remarks |
|---|---|---|---|---|---|---|---|
| AcnB (397–407) | Aconitase B | C404 | 37.1 | 68.0 | 1.8 | ++++ | Oxidation sensitive (29) |
| AhpC (154–169) | Thiol peroxidase | C166 | 38.0 | 63.2 | 1.7 | (+) | Cys166 forms disulfide bond in catalytic cycle (21) |
| AlaS (398–414) | Alanyl-tRNA synthetase | C413 | 16.9 | 38.6 | 2.3 | - | |
| GapA (133–160)† | Glyceraldehyde-3-P- dehydrogenase | C150/ C154 | 55.6 | 92.7 | 1.7 | NA | C150 redox-sensitive, active site residue (23) |
| GlcB (488–499) | Malate synthase | C496 | 26.9 | 54.2 | 2.0 | - | |
| GlyA (64–81)† | Serine | C66 | 22.7 | 44.7 | 2.0 | - | Diamide sensitive protein in B. subtilis (11) |
| GlyA (64–83)† | Hydroxymethyltransferase | 22.7 | 49.5 | 2.2 | |||
| GrxC (51–75) | Glutaredoxin C | C66 | 29.4 | 63.7 | 2.2 | - | C12 in active site (33) |
| GuaA (254–266) | GMP synthetase | C257 | 30.1 | 57.2 | 1.9 | ++ | C86 in active site (34) |
| HflB (243–258) | Heat shock protease | C246 | 22.9 | 48.3 | 2.1 | NA | |
| InfC (41–66) | Initiation factor 3 | C65 | 30.3 | 59.7 | 2.0 | NA | C65 single cysteine |
| Mdh (241–262)† | Malate dehydrogenase | C251 | 21.5 | 47.6 | 2.2 | ++ | |
| Ndk (128–141) | Diphosphate kinase | C139 | 31.9 | 54.7 | 1.7 | ++ | C139 single cysteine |
| PepA (15–26) | Amino peptidase | C17 | 32.5 | 61.2 | 1.9 | - | |
| Pgk (245–258) | Phosphoglycerate kinase | C249 | 25.8 | 49.6 | 1.9 | NA | Redox sensitive (31) |
| ProQ (81–100)† | Regulator of ProP | C88 | 27.8 | 57.1 | 2.1 | + | |
| RplN (79–98)† | Ribosomal protein L14 | C84 | 32.0 | 66.8 | 2.1 | NA | |
| RpsQ (44–62) | Ribosomal protein S17 | C53 | 32.4 | 62.6 | 1.9 | NA | |
| ThrB (286–299) | Homoserine kinase | C298C270 | 36.5 | 59.7 | 1.6 | (+) | |
| ThrB (247–278) | 35.4 | 63.3 | 1.8 | ||||
| Tpx (94–110)† | Thiol Peroxidase | C95 | 34.1 | 56.7 | 1.7 | - | C61 in active site (22);H2O2 sensitive in B. subtilis (11) |
| Tpx (49–66)† | C61 | 35.2 | 64.0 | 1.8 | |||
| Tpx (48–66)† | C61 | 34.3 | 63.7 | 1.9 | |||
| TufA (254–263) | Elongation Factor TU | C256 | 35.3 | 61.1 | 1.7 | +++ | NO sensitive in E. coli (32) |
| YafJ (1–25)† | Putative amidotransferase | C2/C16 | 24.9 | 55.7 | 2.2 | - | C2 invariant |
| Yggx (4–12)† | Antioxidant Protein | C7 | 23.6 | 52.5 | 2.2 | +++ | C7 single and invariant (28) |
| YhiF (80–91)‡ | Transcriptional regulator | C80 | 33.2 | 61.5 | 1.8 | ++++ |
*NA, not available (27), essential E. coli gene under our growth conditions. For sensitivity, refer SI Fig. 7 in SI Appendix.
†Peptide also identified under H2O2 stress.
‡At this mass, we additionally identified the membrane protein ugpE.
Apart from the known redox-sensitive proteins that we identified as target proteins of both H2O2 and NaOCl stress, we also identified a number of proteins that have not been previously shown to have redox-sensitive cysteines. These proteins included ProQ, a regulator protein involved in osmoregulation, the 50S ribosomal protein L14, and a putative amidotransferase YafJ.
The majority of our proteins with NaOCl-sensitive cysteines (24 identified proteins, 10 nonidentified peptides) differed significantly from our proteins with H2O2-sensitive cysteines, indicating that the two physiological oxidants have a largely distinct set of in vivo target proteins (Table 1, SI Table 3 in SI Appendix, and data not shown). This specificity might reflect the specific mode of action of H2O2 and NaOCl and/or a distinct reactivity of thiol groups in individual proteins. Although H2O2 is considered to be a rather mild oxidant, which is thought to cause cysteine oxidation via sulfenic acid formation, NaOCl rapidly forms sulfenyl chlorides, which then produce either disulfide bonds or oxy acids (29). So far, it is unclear what determines the reactivity of individual cysteines toward specific reactive oxygen or nitrogen species (RNS). Our OxICAT method provides us now with a sensitive technique to identify the proteins that are redox-sensitive in response to treatment with any given oxidant. Our future challenge will be to use these data to obtain a comprehensive view about what it is that makes individual proteins redox-sensitive.
It is interesting to note that ROS and RNS appear to preferentially target thiol groups of proteins, whose genes are conditionally essential for growth. Although we found that 25 to 30% of our H2O2 and NaOCl-sensitive proteins are encoded by conditionally essential genes, more than 60% of RNS-target proteins in Mycobacterium tuberculosis and E. coli where encoded by genes that are essential for growth (31, 32). Altering the functional state of one or more of these proteins is the likely cause for the observed growth inhibition of E. coli upon treatment with distinct ROS or RNS. Identification of the proteins that are responsible for the observed oxidative-stress induced growth inhibition might reveal attractive targets for new antimicrobial therapies.
The hallmark of cysteine-mediated redox-regulation is the in vivo reversibility of thiol modifications. Treatment of E. coli cells with NaOCl in the presence of chloramphenicol and analysis of the thiol oxidation status after 60 min of incubation revealed a significant reduction in the oxidative thiol status of our proteins (SI Fig. 6 in SI Appendix). This result suggested that the thioredoxin and/or glutaredoxin systems are able to reduce NaOCl-induced thiol modifications and revealed that we have indeed identified a number of novel redox-regulated proteins in E. coli.
Identification of Redox-Regulated Proteins Involved in Oxidative Stress Defense.
Over the past years, a number of redox-sensitive proteins have been discovered that use oxidative thiol modifications to protect organisms against oxidative stress-induced cell death (9). To investigate whether some of our newly identified proteins are involved in oxidative stress protection of E. coli, we tested the H2O2 and NaOCl sensitivity of E. coli mutant strains, which individually lack the genes encoding our nonessential H2O2 and NaOCl-sensitive proteins. Of the 10 mutant strains that we tested for H2O2 sensitivity and the 17 that we analyzed for NaOCl sensitivity, 3 and 7 strains, respectively revealed a significant increase in oxidative stress sensitivity (Table 1 and SI Table 3 and SI Fig. 7 in SI Appendix). Deletion of either the mdh gene or the yggX gene was found to cause an increase in both H2O2 and NaOCl stress sensitivity, indicating that both proteins generally protect E. coli against oxidative stress. Interestingly, a NAD+-dependent MDH homologue from higher plants has recently been identified as a thioredoxin substrate (33). It is therefore tempting to speculate that the abundant NAD+-dependent E. coli MDH might exert oxidative stress protection by increasing the in vivo turnover of oxidants using thioredoxin-dependent redox-cycling.
Deletion of yggX decreased viability of E. coli cells by more than three orders of magnitudes over a wide range of different oxidant concentrations (SI Fig. 7 in SI Appendix). This observation, together with the fact that the single Cys-7 in YggX is absolutely conserved, suggests that redox-regulation of YggX plays a crucial role in the oxidative stress protection. This conclusion agrees with previous studies in Salmonella enterica, which showed that YggX protects bacteria against superoxide stress and requires its single invariant cysteine (Cys-7) to reduce Fe2+-induced oxidative damage of DNA (34). It remains to be determined to what extent oxidative modification of Cys-7 contributes to functionally important changes in the oligomerization state of YggX, its metal-binding properties or its potential detoxifying activities.
E. coli mutant strains that we found to be particularly sensitive toward NaOCl stress, had deletions in either tufA, acnB or the yhiF gene. Although the increased sensitivity of tufA deletion strains is probably due to oxidative stress-induced changes in Ef-TU activity, which have a larger impact in cells that only express Ef-TU from the tufB gene, the measures for the increased sensitivity of acnB mutants is less clear. Aconitase is a well-known, superoxide-sensitive enzyme, whose oxidative inactivation has been extensively used as landmark of aging cells and organisms (35). The third gene yhiF, whose deletion renders E. coli highly susceptible to NaOCl stress, encodes a putative transcriptional regulator. This result makes it tempting to speculate that YhiF might be a transcriptional regulator that is responsible for the NaOCl response in E. coli.
Conclusions
Accumulation of ROS and RNS causes severe oxidative damage. As such, oxidative stress is not only a powerful antimicrobal strategy used by the mammalian host defense but also the proposed underlying culprit of aging, diabetes, and various neurodegenerative diseases (36). One of the most sensitive targets in proteins are the thiol groups, which react rapidly with both ROS and RNS. It is via reversible thiol modifications that transcriptional (e.g., OxyR, Yap1) and posttranslational (e.g., Hsp33, Prx-2) responses are triggered and that physiological processes (e.g., GapA) start to change during oxidative stress conditions (9). To identify target proteins that use redox-sensitive thiol groups to modulate their protein activity, the monitoring of relative changes in the overall thiol oxidation state (10, 11, 13, 15, 22) or the selective identification of proteins with distinct thiol modifications (12, 37) is an invaluable approach. Previously established methods, however, were unable to directly quantify the extent of thiol-modified protein and identify the cysteines affected. These are, however, crucial pieces of information needed to determine the physiological and evolutionary significance of thiol modifications. For these precise reasons, we have now developed the differential thiol trapping technique termed OxICAT. Our method provides one with a quantitative assessment of the global in vivo thiol status of proteins during nonstress and distinct oxidative stress conditions. OxICAT, which is applicable to a variety of different cell types and organisms, is a highly powerful tool to precisely monitor physiologically relevant redox-mediated changes as organisms experience oxidative stress. In combination with large-scale genetic screens, we identified redox-regulated proteins, which play important roles in the oxidative stress protection of the organism. These proteins in microbes might provide attractive targets for antimicrobial therapies, whereas equivalent studies in eukaryotic organisms might allow the development of more effective antioxidants.
Materials and Methods
Bacterial Strains, Cell Growth, and Harvesting.
E. coli DHB4 (F′ lac-pro lacIQ/Δ(ara-leu)7697 araD139 ΔlacX74 galE galK rpsL phoR Δ(phoA)PvuII ΔmalF3 thi) was grown aerobically in glucose Mops minimal medium (38) containing 40 μg/ml l-leucine and 10 μM thiamine at 37°C until OD600 of 0.4 was reached. Then, the cultures were split and incubated with either 4 mM hydrogen peroxide (H2O2) or 500 μM sodium hypochlorite (NaOCl) for 10 min. For OxICAT analysis, 1.7 ml of cell culture were directly harvested either before (control) or after addition of the oxidants onto 200 μl ice cold 100% (wt/vol) trichloro acetic acid (TCA) to stop all thiol-disulfide exchange reactions (10). To test the reversibility of thiol modifications introduced by NaOCl, cells were supplemented with 320 mg/ml chloramphenicol just before the addition of NaOCl. Aliquots were removed after 10 and 60 min. Strain list and viability assays are described in SI Appendix.
Preparation of Reduced and Oxidized Hsp33.
Reduced and oxidized Hsp33 was prepared as described (20). To activate Hsp33, reduced Hsp33 was incubated with 2 mM H2O2 for 1 h at 43°C. Aliquots were removed and the influence of Hsp33 on the aggregation of chemically unfolded citrate synthase at 30°C was assayed as described in ref. 20. For OxICAT analysis, proteins were precipitated with 10% (wt/vol) TCA.
Differential Thiol Trapping Using ICAT.
The TCA precipitates were centrifuged (13,000 × g, 4°C, 30 min) and the pellet was washed with 500 μl ice cold 10% (wt/vol) TCA and 200 μl ice cold 5% (wt/vol) TCA. The pellet was then dissolved in 80 μl of denaturing alkylation buffer (DAB), consisting of 6 M urea, 0.5% (wt/vol) SDS, 10 mM EDTA, 200 mM Tris·HCl, pH 8.5, and the contents of one vial of cleavable light ICAT reagent (Applied Biosystems, Foster City, CA) dissolved in 20 μl of acetonitrile (ACN). The sample was incubated at 900 rpm for 1 h at 37°C in the dark. To remove light ICAT, the proteins were precipitated with 400 μl of prechilled (−20°C) acetone for 4 h at −20°C. After centrifugation (13,000 × g, 4°C, 30 min), the pellet was washed twice with 500 μl of prechilled acetone. Then, the protein pellet was dissolved in a mixture of 80 μl of DAB, 2 μl of 50 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and the contents of one vial of cleavable heavy ICAT dissolved in 20 μl of ACN. The sample was incubated at 900 rpm for 1 h at 37°C in the dark. The protein was acetone-precipitated and washed as described. Methods to control for labeling efficiency are described in supplemental information. Tryptic digest of the ICAT-labeled peptides, their enrichment on streptavidin columns, cleavage of the biotin tag and the LC/MS analysis are described in SI Appendix.
Data Analysis.
Our OxICAT data were analyzed by using an extension of the existing open-source program msInspect (see SI Appendix). Mass signals with identical elution profiles and a mass difference of 9 Da or multiples thereof were combined to generate “ICAT pairs.” MS/MS identification was assumed significant if an ion score of 95% or higher was reached. Identifications of just one isotopically labeled form were assigned to the complete ICAT pair. If an ICAT pair was found in more than one fraction, the ICAT pair in the fraction with the highest intensity was used for the calculation of the percentage of oxidized peptide. For the calculation of the percentage of oxidation of any given peptide, the average percentage of oxidized peptide of three independent experiments was used. Peptides that revealed an increase of at least 1.5-fold in the fraction of oxidized peptide upon oxidative stress treatment when compared with control conditions were considered as potentially redox-sensitive. In the case that no oxidized peptide was detected under control conditions, the fraction of oxidized peptide under oxidative stress conditions had to be at least 10% of the total protein to be considered physiologically relevant.
Acknowledgments.
LC-MS/MS and MS analysis was performed by the Michigan Proteome Consortium (www.proteomeconsortium.org), which is supported in part by funds from the Michigan Life Sciences Corridor. We thank Maureen Kachman for help with the MS analysis of Hsp33 and Dr. James Bardwell for critically reading this manuscript. This work was supported by National Institutes of Health Grants GM065318 and AG027349 and an Office of the Vice President for Research Faculty grant (to U.J.) and National Resource for Proteomics and Pathways Grant P41-RR018627 funded by the National Center of Competence in Research (to P.C.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data from this paper were deposited in the Tranche Project database, http://www.proteomecommons.org/dev/dfs (hash string d7Veeim+Eqtj0Ejdi/whi7oaQSDLv0 / pjRzVU1oEyG8lK5D+PYrrl+vmQDqdD7dYrtpRaYV3yq0vEeBhrDEaG3gOpD0AAAAAABPlfA==)
This article contains supporting information online at www.pnas.org/cgi/content/full/0707723105/DCSupplemental.
References
- 1.Sies H. Klin Wochenschr. 1991;69:965–968. doi: 10.1007/BF01645140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller RA, Britigan BE. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha EM, Oh CT, Bae YS, Lee WJ. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 5.Kiley PJ, Storz G. PLoS Biol. 2004;2:e400. doi: 10.1371/journal.pbio.0020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakob U, Muse W, Eser M, Bardwell JC. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 7.Zheng M, Aslund F, Storz G. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 8.Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linke K, Jakob U. Antioxid Redox Signal. 2003;5:425–434. doi: 10.1089/152308603768295168. [DOI] [PubMed] [Google Scholar]
- 10.Leichert LI, Jakob U. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochgrafe F, Mostertz J, Albrecht D, Hecker M. Mol Microbiol. 2005;58:409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 12.Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 13.Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Mol Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 15.Kozarova A, Sliskovic I, Mutus B, Simon ES, Andrews PC, Vacratsis PO. J Am Soc Mass Spectrom. 2007;18:260–269. doi: 10.1016/j.jasms.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Free Radic Biol Med. 2007;42:823–829. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, Cotgreave IA. Arch Biochem Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 18.Jaffrey SR, Snyder SH. Sci STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 19.Winter J, Linke K, Jatzek A, Jakob U. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin C, et al. Bioinformatics. 2006;22:1902–1909. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- 22.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 23.Toledano MB, Kumar C, Le Moan N, Spector D, Tacnet F. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Ellis HR, Poole LB. Biochemistry. 1997;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- 25.Baker LM, Poole LB. J Biol Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotgreave IA, Gerdes R, Schuppe-Koistinen I, Lind C. Methods Enzymol. 2002;348:175–182. doi: 10.1016/s0076-6879(02)48636-3. [DOI] [PubMed] [Google Scholar]
- 27.Krebs HA. Essays Biochem. 1972;8:1–34. [PubMed] [Google Scholar]
- 28.VanBogelen RA, Kelley PM, Neidhardt FC. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattison DI, Davies MJ. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 30.Fliss H. Mol Cell Biochem. 1988;84:177–188. doi: 10.1007/BF00421053. [DOI] [PubMed] [Google Scholar]
- 31.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Mol Syst Biol. 2006;2:20060008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandes N, Rinck A, Leichert LI, Jakob U. Mol Microbiol. 2007;66:901–914. doi: 10.1111/j.1365-2958.2007.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara S, Motohashi K, Arisaka F, Romano PG, Hosoya-Matsuda N, Kikuchi N, Fusada N, Hisabori T. J Biol Chem. 2006;281:32065–32071. doi: 10.1074/jbc.M605784200. [DOI] [PubMed] [Google Scholar]
- 34.Gralnick J, Downs D. Proc Natl Acad Sci USA. 2001;98:8030–8035. doi: 10.1073/pnas.151243198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan LJ, Levine RL, Sohal RS. Proc Natl Acad Sci USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berlett BS, Stadtman ER. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 37.Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, Bonetto V, Mengozzi M, Duffieux F, Miclet E, et al. Proc Natl Acad Sci USA. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neidhardt FC, Bloch PL, Smith DF. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]