Abstract
The spike (S) protein of SARS coronavirus (SARS-CoV) has been known to recognize and bind to host receptors, whose conformational changes then facilitate fusion between the viral envelope and host cell membrane, leading to viral entry into target cells. However, other functions of SARS-CoV S protein such as proteolytic cleavage and its implications to viral infection are incompletely understood. In this study, we demonstrated that the infection of SARS-CoV and a pseudovirus bearing the S protein of SARS-CoV was inhibited by a protease inhibitor Ben-HCl. Also, the protease Factor Xa, a target of Ben-HCl abundantly expressed in infected cells, was able to cleave the recombinant and pseudoviral S protein into S1 and S2 subunits, and the cleavage was inhibited by Ben-HCl. Furthermore, this cleavage correlated with the infectivity of the pseudovirus. Taken together, our study suggests a plausible mechanism by which SARS-CoV cleaves its S protein to facilitate viral infection.
Keywords: SARS-CoV, Spike protein, Protease Factor Xa, Protease inhibitor, Cleavage of S protein
Severe acute respiratory syndrome (SARS) is a novel life-threatening infectious disease caused by SARS coronavirus (SARS CoV) [1]. Viral spike (S) protein is a multifunctional protein that plays pivotal roles in the biology and pathogenesis of SARS-CoV. The N-terminal region of the S protein has been demonstrated to mediate viral infection, by binding through the receptor-binding domain (RBD) to a cellular receptor angiotensin-converting enzyme 2 (ACE2) [2], and subsequently inducing virus–cell membrane fusion, which involves two domains in the C-terminal region of the S protein named heptad repeat (HR) 1 and 2 [3]. However, other functions of the S protein that are important in viral infection have not yet been defined.
In most other members of the coronavirus family, such as human coronavirus HCoV-OC43 and bovine coronaviruses BCoV, the S protein may be post-translationally cleaved into two fragments, S1 (receptor-binding domain) and S2 (membrane fusion domain) [4], [5]. Studies on other coronaviruses have further elucidated that conformational changes of the S protein can be induced at 37 °C and pH 8.0, accompanied by the cleavage of S1 and S2, which triggers virus–cell membrane fusion [6], [7]. As a new member of the coronavirus family, SARS-CoV shares with other coronaviruses some similarity and common features in the amino acid sequences of the S protein [8]. It has been found that the S protein of SARS-CoV is post-translationally cleaved into S1 and S2 functional domains in the process of viral infection [9]. However, the impact of this cleavage on viral infectivity remains unclear [10].
Our previous study has shown that SARS-CoV infectivity was inhibited over 99.99% in cell cultures by pre-incubation with two peptides, which correspond to the sequences (amino acid residues 598–617 and 737–756) proximal to the potential S1 and S2 cleavage site, suggesting that these critical sites or regions might be influential in viral infection [11]. In this study, we aimed to further investigate which proteases may be important in the cleavage of the S protein and whether the cleavage of SARS-CoV S protein has an impact on viral infectivity.
Materials and methods
Inhibition assay of live SARS-CoV infection. Thirteen protease inhibitors, including Ben-HCl, 6-aminohexanoic acid, antipain, aprotinin, bestatin, chymostatin, E-64, EDTA disodium salt, N-ethylmaleimide, leupeptin, pepstatin, phosphoramidon, and trypsin inhibitor (Novagen, USA and Sigma, USA), were selected for testing the potential inhibition of SARS-CoV infection in cell cultures by plaque reduction assay [12] using SARS-CoV strain GZ50 (GenBank Accession No. AY304495).
Construction and preparation of pseudotyped SARS-CoV/HIV. Pseudotyped SARS-CoV/HIV (pseudovirus) bearing the codon-optimized full-length S protein of SARS-CoV and a defective HIV-1 genome expressing luciferase as reporter (pNL4-3.luc.RE) was prepared as previously described [13]. The produced pseudovirus was quantified by measuring p24 protein level using a Vironostika HIV-1 antigen MicroELISA kit (Biomerieux bv Boxtel, Netherlands).
Inhibition assay of pseudovirus infection. To assess inhibitory effect of Ben-HCl on the infection of pseudovirus, 293T cells expressing ACE2 (293T/ACE2) were pre-incubated with Ben-HCl at different concentrations for 30 min at 37 °C, followed by infection with varying concentrations of pseudovirus. The medium was replaced after overnight incubation or at indicated time points, and cells were continually incubated for an additional 48 h. The infectivity of the pseudovirus was determined by measuring relative luciferase activities (RLU) using a luciferase kit (Promega, USA) in a Wallac Microbeta 1420 counter (Perkin-Elmer, USA) according to the manufacture’s instruction.
Cleavage and inhibition assay of the S protein. The cleavage and inhibition assay of the full-length recombinant SARS-CoV S protein (Protein Sciences Corporation, USA) was performed using protease Factor Xa and its inhibitor Ben-HCl (Novagen, USA) according to the manufacturer’s instruction. In brief, the S protein was incubated with Factor Xa in the presence or absence of Ben-HCl for 6 h at room temperature. In the cleavage and inhibition assay of the pseudoviral S protein, pseudovirus was incubated with different concentrations of Factor Xa in the presence or absence of Ben-HCl at room temperature for 2 and 6 h, respectively. Pseudovirus was also added to 293T/ACE2 cells with or without pre-incubation with Ben-HCl at 37 °C for 30 min. The culture supernatant was collected at indicated time points.
SDS–PAGE and Western blot. The cleavage and inhibition of the full-length and pseudoviral S protein were detected by SDS–PAGE and Western blot according to the protocol described previously [14]. The cleaved S1 and S2 bands were confirmed using specific primary antibodies against SARS-CoV S1 (S1-121B8) and S2 (S2-102D7 or -119F6) subunits [15] and horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibody (DAKO, Denmark).
Detection of Factor Xa transcription. Transcription of Factor Xa in 293T/ACE2 cells was tested by reverse transcriptase polymerase chain reaction (RT-PCR). Total cellular RNA was extracted using RNeasy Mini kit (Qiagen, USA), and cDNA was subsequently synthesized using random primers and SuperScript II RT kit (Invitrogen, USA). Three pairs of primers for amplifying three fragments of the coding region of Factor Xa were: the forward primers 5′-GACACAGTACTCGGCCACACCATGG-3′, 5′-AATGAACGCAGGAAGAGGTCAGTG-3′, 5′-AGATTCAAGGTGAGGGTAGGGGACC-3′, respectively, and the reverse primers 5′-CGTCGTCTTGTCGCTGTCCTCAAAG-3′, 5′-TACCCTCACCTTGAATCTCTTGGCT-3′, 5′-GAGTGGGATCTCACTTTAATGGAGA-3′, respectively. As the internal control, β-actin was amplified with a forward primer 5′-GCACACTTAGCCGTGTTCTTTGCACTTTCT-3′ and a reverse primer 5′-AGGCGTACAGGGATAGCACAGCCTGGATAG-3′. PCR for amplifying specific gene fragments was performed with 40 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, followed by a final extension at 72 °C for 10 min in the GeneAmp PCR System 9700 (Applied Biosystems, USA).
Detection of Factor Xa protein expression. Expression of Factor Xa in 293T/ACE2 cells at the protein level was detected by immunoprecipitation (IP) and Western blot. β-Actin was included as a house-keeping protein control. IP was done using EZview Red Protein A Affinity Gel kit in accordance with the manufacturer’s instruction (Sigma, USA). Precipitated proteins (30 μg) were separated on SDS–PAGE and analyzed by Western blot as described above. The primary antibodies for IP and Western blot were anti-Factor Xa (R&D Systems, USA) and anti-β-actin (Abcam, USA).
Results
A protease inhibitor Ben-HCl inhibited SARS-CoV infection
To assess which protease inhibitor(s) would affect SARS-CoV infectivity in cell cultures, we screened 13 potential protease inhibitors, which are listed in Materials and methods, for their inhibitory effects on SARS-CoV infection. Among these inhibitors, only Ben-HCl was able to inhibit SARS-CoV infection in cell cultures. Fig. 1 A shows that Ben-HCl inhibited the SARS-CoV plaque formation in Vero E6 cells in a concentration-dependent manner. The inhibition rates of SARS-CoV were 1.5% at 1 mM, 40% at 2 mM, and 70% at 4 mM of Ben-HCl. The results indicate that at least one protease inhibitor can suppress SARS-CoV infection or replication.
Fig. 1.
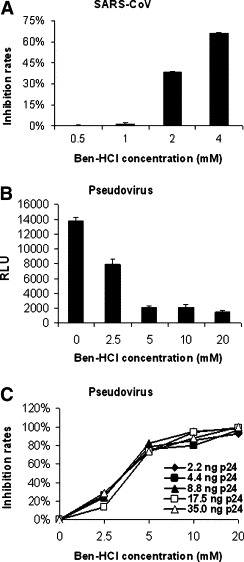
Ben-HCl inhibits the infection of SARS-CoV and pseudotyped SARS-CoV/HIV. (A) Inhibitory effect of Ben-HCl on Vero E6 cells infected with wild-type SARS-CoV. The mean values from three independent assays are shown with standard error (SE). (B) Inhibitory effect of Ben-HCl on viral entry. 293T/ACE2 cells were infected with pseudovirus in the presence or absence of Ben-HCl, and RLU was measured. The data are shown as mean + SE of three independent experiments. (C) Correlation of entry inhibitory activity of Ben-HCl on pseudovirus infection. The data are presented as mean values of three independent experiments.
To confirm that the suppression of SARS-CoV infection by Ben-HCl occurs at the level of viral entry but not at other steps of viral replication, the inhibitory effect of Ben-HCl on SARS-CoV S protein/ACE2-mediated cell entry was detected using a pseudovirus expressing the S protein of SARS-CoV. As shown in Fig. 1B, RLU decreased progressively with increased concentrations of Ben-HCl. Fig. 1C further shows that the inhibition rates of Ben-HCl were closely correlated to the concentration of Ben-HCl but were independent of the concentration of the pseudovirus. In addition, treatment with 20 mM of Ben-HCl resulted in nearly complete inhibition of the pseudovirus infection. The results further demonstrate that Ben-HCl indeed inhibited viral entry into the target cells.
SARS-CoV S protein was cleaved by protease Factor Xa and the cleavage was inhibited by its inhibitor Ben-HCl
Since Ben-HCl is an inhibitor of a panel of proteases including Factor Xa, thrombin, and trypsin, we tested these proteases for their activities to cleave the full-length recombinant S protein of SARS-CoV. We found that only Factor Xa was able to effectively cleave the S protein into S1 and S2 subunits, and the cleavage was inhibited by 20 mM of Ben-HCl (Fig. 2 A). We further demonstrate that Factor Xa also cleaved the S protein in pseudotyped SARS-CoV/HIV. When the pseudovirus was incubated with Factor Xa at room temperature for 2 h, the pseudoviral S protein was partially cleaved, and the cleavage increased with the concentration of Factor Xa (Fig. 2B). When the incubation time was extended to 6 h, the pseudoviral S protein could be almost completely cleaved by 0.5 U of Factor Xa (Fig. 2C). Furthermore, the cleavage of pseudoviral S protein by Factor Xa was inhibited by Ben-HCl at the concentration of 80 mM, showing the same pattern as untreated pseudovirus (Fig. 2). These results indicate that both recombinant and pseudoviral S protein can be cleaved by Factor Xa into S1 and S2 subunits, and that the cleavage can be prevented by the inhibitor Ben-HCl.
Fig. 2.
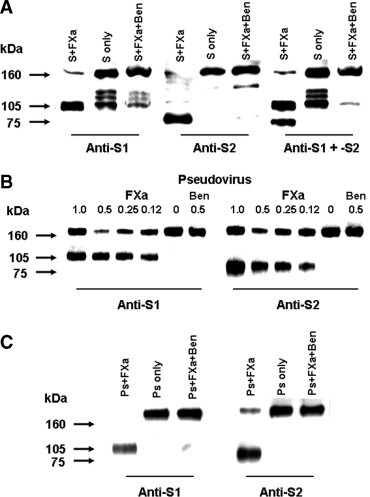
In vitro cleavage of SARS-CoV S protein by Factor Xa. (A) The full-length recombinant S protein (1.55 μg) was incubated with 0.5 U Factor Xa (S + FXa) for 6 h. Untreated S protein (S only), and S protein incubated with Factor Xa and 20 mM Ben-HCl (S + FXa + Ben) for 6 h, were detected by Western blot. The cleaved S1 (∼105 kDa) and S2 (∼75 kDa) fragments, which were respectively recognized by anti-S1 (121B8), anti-S2 (102D7), and both (anti-S1 + -S2) are indicated. (B) The pseudovirus (3 ng p24) was incubated with Factor Xa (FXa) at the indicated concentrations with or without 80 mM Ben-HCl (Ben) for 2 h, and detected by Western blot using anti-S1 (121B8) and anti-S2 (119F6), respectively. (C) The pseudovirus (Ps) was incubated with 0.5 U Factor Xa (Ps + FXa), with buffer only (Ps only), or with 0.5 U Factor Xa and 80 mM Ben-HCl (Ps + FXa + Ben) for 6 h. Proteins were detected by Western blot using the same antibodies as in (B). The molecular marker (kDa) is indicated on the left.
The S protein was cleaved when the pseudovirus was incubated with target cells
To determine if the infectivity of pseudovirus SARS-CoV/HIV is associated with the cleavage of the S protein by proteases on the target cell membrane, we tested the cleavage of the S protein in the culture supernatant by Western blot and the infectivity of the pseudovirus in cell lysate by luciferase assay, after the pseudovirus was incubated with 293T/ACE2 cells for 2 and 4 h in the presence or absence of Ben-HCl (20 mM). Fig. 3 A shows extra bands in the pseudovirus infected culture supernatant in the absence of the inhibitor Ben-HCl at 2 and 4 h, which reacted with anti-S1 (121B8) and anti-S2 (119F6) mAbs, respectively, indicating that the cleavage enhanced with the increase of incubation time. In contrast, no such bands were detected in the pseudovirus infected supernatant in the presence of Ben-HCl or the pseudovirus alone. Luciferase assay further demonstrates that the infectivity of the pseudovirus SARS-CoV/HIV increased with the extension of incubation and the cleavage of more S protein (Fig. 3B). The above results indicate that the infectivity or the infection process of the pseudovirus SARS-CoV/HIV was closely associated with the cleavage of the S protein into functional S1 and S2 subunits.
Fig. 3.
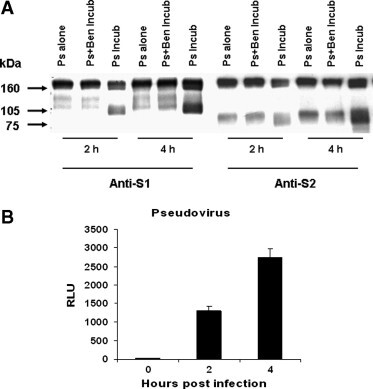
Association of pseudovirus infection with the cleavage of S protein. (A) Culture supernatant of 293T/ACE2 cells infected with pseudovirus (3 ng p24) in the presence (Ps + Ben Incub) and absence (Ps Incub) of Ben-HCl (20 mM) was collected at the indicated time points and detected for the cleavage of S protein by Western blot using anti-S1 (121B8) and anti-S2 (119F6), respectively. The pseudovirus alone (Ps alone) incubated at the same condition was included as a control. (B) After removing the pseudovirus at the indicated time points, the cells were continually cultured for 48 h and harvested for luciferase assay to determine pseudovirus infectivity, which is shown as mean + SE of RLU from three independent experiments.
The expression of protease Factor Xa in 293T/ACE2 cells
The expression of the protease Factor Xa in 293T/ACE2 cells was further confirmed by RT-PCR (Fig. 4 A) and Western blot (Fig. 4B). As shown in Fig. 4A, RT-PCR products of 255, 337, and 630 bp, which are respectively corresponding to nucleotide sequences 5–260, 552–889, and 872–1502 of Factor Xa transcript, were amplified from RNA extracts of 293T/ACE2 cells. β-Actin was also amplified in the same sample as the internal control. Fig. 4B shows that, biologically active 53 kDa Factor Xa protein was detected from 293T/ACE2 cell lysate by Western blot using mAb specific to Factor Xa, whose expression level was compatible with that of the internal control β-actin. The results at both RNA and protein levels clearly indicate the expression of the protease Factor Xa in 293T/ACE2 cells, implying that this protease could be at least one of the proteases responsible for the cleavage of the S protein during the course of pseudovirus infection.
Fig. 4.
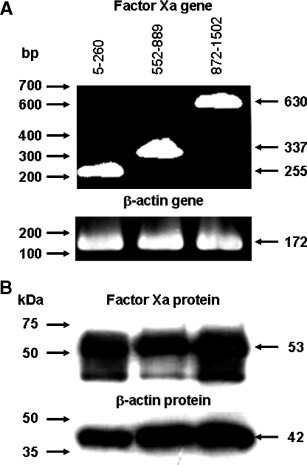
Expression of protease Factor Xa in 293T/ACE2 cells. (A) RT-PCR products of indicated sizes were amplified from total RNA samples extracted from 293T/ACE2 cells. The products correspond to nucleotide sequences 5–260 (255 bp), 552–889 (337 bp), and 872–1502 (630 bp) of Factor Xa transcript, respectively. The molecular marker (bp) is shown on the left. RT-PCR product of β-actin mRNA (172 bp) was amplified in the same RNA samples as an internal control. (B) Factor Xa protein expression was detected in 293T/ACE2 cell lysate by Western blot. Factor Xa of 53 kDa and β-actin of 42 kDa were recognized by their specific antibodies, respectively. The molecular marker (kDa) is indicated on the left.
Discussion
In this study, we initially screened 13 inhibitors of proteases which might potentially correspond to the cleavage of the S protein, for their effects on the suppression of SARS-CoV infection. This is based on a hypothesis that the inhibitors might inactivate proteases on the cell membrane and thus block the cleavage of the S protein, thereby suppressing viral infection. Our results show that one inhibitor, Ben-HCl, is able to effectively suppress SARS-CoV infection (Fig. 1A). Using the pseudovirus SARS-CoV/HIV, we further defined that Ben-HCl inhibits viral entry into the target cells but apparently does not suppress other steps of viral replication (Fig. 1B).
It has been known that Ben-HCl is an inhibitor of a series of proteases, including trypsin, trypsin-like enzymes, and serine proteases such as thrombin and Factor Xa [16], [17]. We therefore investigated if these proteases could cleave the S protein into functional S1 and S2 subunits. Our results indicate that one of these proteases, Factor Xa, is able to effectively cleave the full-length recombinant S protein into S1 (∼105 kDa) and S2 (∼75 KDa) subunits, and that the cleavage can be inhibited by its inhibitor Ben-HCl (Fig. 2A). However, the protease thrombin cannot cleave the full-length recombinant S protein, while trypsin cleaves the S protein into multiple small fragments (data not shown).
We further explored if the cleavage of the S protein indeed plays a critical role in SARS-CoV infection. Our results demonstrate that: (1) Factor Xa can effectively cleave the S protein in the pseudovirus SARS-CoV/HIV into S1 and S2 subunits and this cleavage is inhibited by Ben-HCl (Fig. 2B); (2) the S protein can be cleaved into S1 and S2 subunits when the pseudovrus is incubated with its target cells (293T/ACE2) and the level of the cleavage correlates with viral infectivity (Fig. 3); and (3) the target cells express Factor Xa (Fig. 4), a membrane-bound protease [18]. As a whole, our study suggests a plausible mechanism by which SARS-CoV cleaves the S protein through the protease Factor Xa of infected cells to facilitate viral infection. Thus, the infection of SARS-CoV not only involves the binding of RBD with its receptor ACE2 and the fusion between viral envelope and host cell membrane, but also is associated with the cleavage of the S protein by proteases on the cell membrane, such as Factor Xa. It has been reported that the S protein of SARS-CoV forms trimeric peplomer on the surface of the virion [11]. The cleavage of the S protein may trigger virus–cell fusion subsequent to the binding with ACE2 receptor and the dissociation of trimeric S protein to monomers.
Previous studies have explored several targets for vaccine and antiviral agent developments. For example, vaccine candidates targeting the RBD are able to induce strong neutralizing antibodies and thus provide protection against SARS-CoV infection [19], [20]. Antiviral peptides targeting the HR2 region of the S protein can block the entry of SARS-CoV into target cells [21]. Small molecules targeting SARS-CoV proteases, such as main proteases 3C-like protease (3CLpro) [22], papain-like protease 2 (PLP2) [23] and helicase [24], can inhibit viral replication. In addition to these targets, here we have provided the cleavage of the S protein as another crucial target for the development of vaccines and antiviral agents. Inhibition of the cleavage of the S protein into functional S1 and S2 subunits using agents such as Ben-HCl can effectively block viral entry.
Acknowledgments
This study was supported by the Research Fund for the Control of Infectious Diseases, the Health, Welfare and Food Bureau of the Hong Kong SAR Government, by the National 973 basic research program of China (2005CB523001), and by the National Institutes of Health (NIH) of the United States (RO1 AI68002).
References
- 1.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mounir S., Talbot P.J. Molecular characterization of the S protein gene of human coronavirus OC43. J. Gen. Virol. 1993;74(Pt. 9):1981–1987. doi: 10.1099/0022-1317-74-9-1981. [DOI] [PubMed] [Google Scholar]
- 5.Abraham S., Kienzle T.E., Lapps W., Brian D.A. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176:296–301. doi: 10.1016/0042-6822(90)90257-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelus B.D., Schickli J.H., Blau D.M., Weiss S.R., Holmes K.V. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 degrees C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 2003;77:830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturman L.S., Ricard C.S., Holmes K.V. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37 degrees C correlates with virus aggregation and virus-induced cell fusion. J. Virol. 1990;64:3042–3050. doi: 10.1128/jvi.64.6.3042-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 9.Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng B.J., Guan Y., Hez M.L., Sun H., Du L., Zheng Y., Wong K.L., Chen H., Chen Y., Lu L., Tanner J.A., Watt R.M., Niccolai N., Bernini A., Spiga O., Woo P.C., Kung H.F., Yuen K.Y., Huang J.D. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir. Ther. 2005;10:393–403. [PubMed] [Google Scholar]
- 12.Kao R.Y., Tsui W.H., Lee T.S., Tanner J.A., Watt R.M., Huang J.D., Hu L., Chen G., Chen Z., Zhang L., He T., Chan K.H., Tse H., To A.P., Ng L.W., Wong B.C., Tsoi H.W., Yang D., Ho D.D., Yuen K.Y. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006;176:6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- 14.Du L., He Y., Wang Y., Zhang H., Ma S., Wong C.K., Wu S.H.W., Ng F., Huang J.D., Yuen K.Y., Jiang S., Zhou Y., Zheng B.J. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: implication for developing SARS vaccines. Virology. 2006;353:6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J. Virol. 2006;80:5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Crich D., Wink D., Lam M., Rheingold A.L., Case D.A., Fu W., Zhou Y., Rao M., Olson A.J., Johnson M.E. Design and synthesis of highly constrained factor Xa inhibitors: amidine-substituted bis(benzoyl)-diazepan-2-ones and bis(benzylidene)-bis(gem-dimethyl)cycloketones. Bioorg. Med. Chem. 2003;11:3379–3392. doi: 10.1016/s0968-0896(03)00332-8. [DOI] [PubMed] [Google Scholar]
- 17.Quan M.L., Wexler R.R. The design and synthesis of noncovalent factor Xa inhibitors. Curr. Top. Med. Chem. 2001;1:137–149. doi: 10.2174/1568026013395407. [DOI] [PubMed] [Google Scholar]
- 18.Hung H.L., Pollak E.S., Kudaravalli R.D., Arruda V., Chu K., High K.A. Regulation of human coagulation factor X gene expression by GATA-4 and the Sp family of transcription factors. Blood. 2001;97:946–951. doi: 10.1182/blood.v97.4.946. [DOI] [PubMed] [Google Scholar]
- 19.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 20.Du L., Zhao G., He Y., Guo Y., Zheng B.J., Jiang S., Zhou Y. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25:2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sha Y., Wu Y., Cao Z., Xu X., Wu W., Jiang D., Mao X., Liu H., Zhu Y., Gong R., Li W. A convenient cell fusion assay for the study of SARS-CoV entry and inhibition. IUBMB Life. 2006;58:480–486. doi: 10.1080/15216540600820974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Gui C., Luo X., Yang Q., Gunther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Y.S., Chang G.G., Juo C.G., Lee H.J., Yeh S.H., Hsu J.T., Chen X. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry. 2005;44:10349–10359. doi: 10.1021/bi0504761. [DOI] [PubMed] [Google Scholar]
- 24.Frick D.N., Lam A.M. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]


