Abstract
Calbindin D28 encodes a calcium binding protein that is expressed in the cerebellum exclusively in Purkinje cells. We have used biolistic transfection of organotypic slices of P12 cerebellum to identify a 40-bp element from the calbindin promoter that is necessary and sufficient for Purkinje cell specific expression in this transient in situ assay. This element (PCE1) is also present in the calmodulin II promoter, which regulates expression of a second Purkinje cell Ca2+ binding protein. Expression of high levels of exogenous calbindin or calretinin decreased transcription mediated by PCE1 in Purkinje cells 2.5- to 3-fold, whereas the presence of 1 μM ionomycin in the extracellular medium increased expression. These results demonstrate that PCE1 is a component of a cell-specific and Ca2+-sensitive transcriptional regulatory mechanism that may play a key role in setting the Ca2+ buffering capacity of Purkinje cells.
Keywords: transcription, biolistic transfection, Ca2+ responses, calbindin, calmodulin II
The identification of transcriptional control elements and the subsequent cloning of their cognate transcription factors has led to the elucidation of many important intracellular signaling pathways (1, 2). This strategy has proved difficult to apply to cells in the central nervous system (CNS), because the distinct behavior of specific cell types cannot be reproduced in cell lines.
Cerebellar Purkinje cells arise between days 11 and 13 of mouse embryogenesis (3), and shortly thereafter begin expressing high levels of the Ca2+ binding protein calbindin D28k (4). Calbindin expression in the cerebellum is restricted to Purkinje cells although it is present in several neuronal cell types in other regions of the CNS (5). It is one of three Ca2+ binding proteins, along with calretinin and parvalbumin, that are expressed throughout the brain and have not been found to modulate the activity of enzymes or ion channels. Consequently, they are thought to act mainly as buffers of intracellular free Ca2+ (6).
The very early and high-level expression of calbindin in Purkinje cells suggests that establishing the Ca2+ buffering capacity of these neurons may be crucial for their function. That calbindin is an effective Ca2+ buffer in vivo has been established in several systems. Calbindin has been shown to protect against excitotoxic cell death in hippocampal neurons in culture (7); its expression in dorsal root ganglion neurons reduces free Ca2+ concentrations after depolarization (8); and when transfected into hippocampal pyramidal cells calbindin blocks posttetanic potentiation (9). Furthermore, focal stimulation of the perforant pathway in rats results in a 3-fold rise in calbindin mRNA within 6 hr (10). These results suggest both that the Ca2+ buffering capacity of neurons is important for their normal physiologic function, and that this buffering capacity can be dynamically regulated at the transcriptional level in response to electrical stimulation.
To discover possible molecular mechanisms that regulate the Ca2+ buffering capacity of cerebellar Purkinje cells, we have examined transcriptional regulation of the mouse calbindin gene. Previous studies of calbindin transcriptional control in nonneuronal cells resulted in identification of a vitamin D response element that mediates transcription in kidney fibroblasts (11). This element is not likely to influence calbindin transcription in the CNS, because the level of calbindin in the brain does not change in response to vitamin D (12). More recent studies of calbindin transcriptional control in transgenic mice showed that expression in Purkinje cells could be obtained using 1 kb of 5′ flanking DNA (13). However, only 20% of the transgenic lines expressed the marker gene in Purkinje cells, and there was considerable variation in the patterns of expression between these lines. This variability was presumably due to the fact that the transgene integrated at different sites in each transgenic line and effectively precluded fine dissection of this regulatory region.
We previously have introduced the use of biolistic transfection of acute cerebellar slices as a means of analyzing transcriptional control in the CNS (14). We demonstrated that cell-specific transcription of both neuronal and glial specific promoters can be achieved. Moreover, nearly 100% cotransfection of multiple DNA constructs into single cells is possible, allowing the design of internal controls for variations in transfection efficiency between slices. In this study, we have used this methodology to map transcriptional regulatory elements in the calbindin promoter.
MATERIALS AND METHODS
Parasagittal slices 250 μm in thickness were prepared from P11 to P12 mouse cerebellum using a tissue chopper (McIlwain, Mickle Laboratory Engineering, Surrey, United Kingdom). The slices then were cultured in the same manner as previously described (14) except that a serum-free medium consisting of Earle’s salts (GIBCO), basal medium eagle (BME) amino acids (Sigma), BME vitamins (GIBCO), 20 mM NaHCO3, 10 mg/ml BSA (Sigma), 32 mM glucose (Sigma), 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, 20 units/ml penicillin/streptomycin, and 2 mM l-glutamine was used. After 4 hr in culture, the slices were transfected using the biolistic particle delivery system (Bio-Rad) (14). Plasmids were precipitated onto 1.0 μm in diameter gold beads by placing them in a solution of 1.25 M CaCl2 and 20 mM spermidine free base. The particle delivery system created a helium shock wave with a pressure gradient of 1,300 psi to accelerate the coated beads into a cerebellar slice that had been placed in a vacuum of 25 in of Hg (84 kPa). When cotransfected, all plasmids were mixed before precipitation onto the gold beads. Subsequent to transfection, slices were cultured for 48 hr, after which they were fixed and stained. Quantitations of expression levels were done using either immunocytochemistry or chromogenic color reactions. The latter were done as in ref. 14.
Immunocytochemistry was performed on the slices following procedures very similar to those in ref. 15. Anti-β-galactosidase Gal-13 clone (Sigma) at 1:100 and rabbit anti-alkaline phosphatase (Zymed) at 1:50 as well as fluorescein isothiocyanate-conjugated goat anti-rabbit at 1:300 and CY3-conjugated goat anti-mouse at 1:300 (Jackson ImmunoResearch) were used.
The calbindin, calmodulin, and major late promoter (pML) constructs were all made with the pnLacf vector (16), which contains LacZ fused to a nuclear localization sequence from the simian virus 40 large T antigen. The GW1-calbindin construct was made by ligating a rat cDNA clone of the calbindin D28k gene, which was a gift from M. Thomassett (Hôpital Robert Debre, Paris), into the GW1 expression vector, which was a gift from the British Biotechnology Corporation. Calretinin was amplified from a rat cDNA library and ligated into the GW1 expression vector in a similar manner to calbindin. All constructs were verified by cleavage with restriction enzymes and by sequencing. Cytomegalovirus (CMV) immediate early promoter driving alkaline phosphatase (CMVAP) was constructed by inserting the human placental alkaline phosphatase gene into the pCMVβ expression vector (Stratagene) in the NotI sites.
RESULTS
Cotransfection of CMVAP and the same promoter driving nuclear targeted β-galactosidase (CMVnLacZ) resulted in coexpression of these markers in virtually all transfected cells (Fig. 1A). In contrast, when CMVAP and ≈1 kb of 5′ flanking DNA from the calbindin gene driving nLacZ were cotransfected into the slices, only Purkinje cells expressed nLacZ (ref. 14, Fig. 1B).
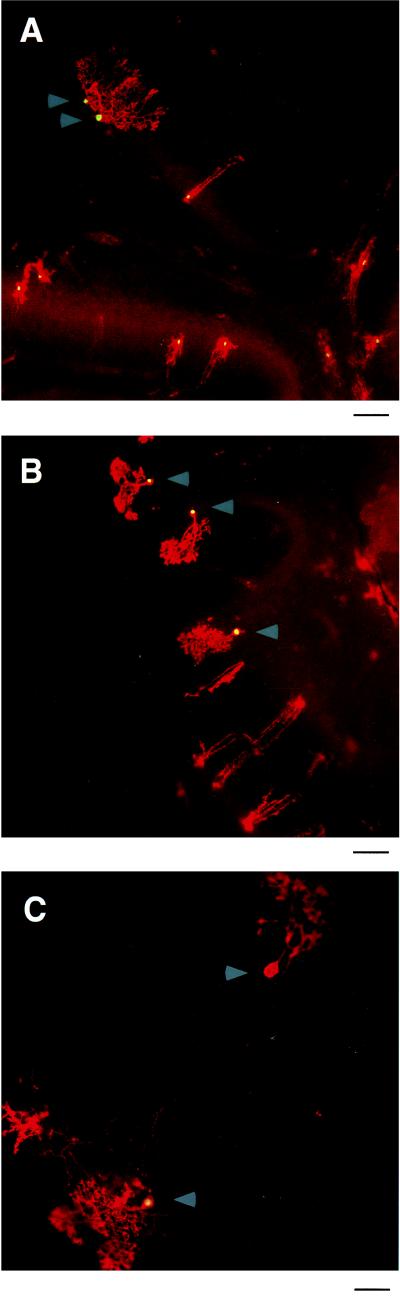
Figure 1.
P12 cerebellar slices cotransfected with calbindin and CMV promoter constructs. (A) P12 parasagittal slices of murine cerebellum were cotransfected with CMVAP and CMV-nLacZ, incubated for 48 hr, and then fixed and stained with fluorescence-conjugated antibodies. Alkaline phosphatase (red) is located throughout the entire cell, whereas β-galactosidase (yellow) is located only in the nucleus. Purkinje cells are marked with an arrow. (Bar, 100 μm.) (B) Cerebellar slices were cotransfected with CMVAP and a calbindin promoter construct extending from −1,010 to +105 (with respect to the transcription start site) driving nLacZ. Note that while many different cell types express alkaline phosphatase, only Purkinje cells (denoted by arrows) also express β-galactosidase. (Bar, 100 μm.) (C) Two Purkinje cells that illustrate the discrete nature of nLacZ expression in cotransfected cells. One Purkinje cell expresses β-galactosidase in addition to alkaline phosphatase (as indicated by the yellow nucleus) whereas the other expresses only alkaline phosphatase. (Bar, 50 μm.)
After cotransfection with CMVAP and the 1-kb calbindin promoter driving nLacZ, Purkinje cells were found to express nLacZ at one of two levels: either at a relatively high level, or at such a low level as to be undetectable (Fig. 1C). This “all or nothing” pattern of expression of nLacZ in individual transfected cells was seen after each transfection in this study. It allowed simple quantitation of promoter activity as the percentage of transfected Purkinje cells expressing the nLacZ marker, and promoter specificity as the ratio of the promoter activity in transfected Purkinje cells versus that in transfected non-Purkinje cells.
The 1-kb calbindin promoter fragment drove expression of nLacZ in greater than 90% of the transfected Purkinje cells but in less than 2% of the transfected non-Purkinje cells (Fig. 2A). Thus, the activity of the 1-kb calbindin promoter in Purkinje cells is ≈90% and its ratio of specificity is at least 50-fold. The −155-bp calbindin promoter construct drove expression in 80% of the transfected Purkinje cells and in less than 3% of the transfected non-Purkinje cells (30-fold specificity). Thus, the vitamin D response element, which drives expression of the calbindin gene in non-CNS cells, is not required for Purkinje cell specific expression. The −85-bp calbindin promoter construct showed both high transcription activity (80%) and cell specificity (>50-fold). However, deletion of this promoter to −45 bp completely destroyed its activity (Fig. 2A). These results allow two important conclusions. First, it is evident that the sequence between −85 bp and −45 bp, termed the Purkinje cell element (PCE1), is required for Purkinje cell specific expression in situ because transcription activity was lost by deleting these sequences. Second, it is likely that the calbindin promoter does not contain elements that repress transcription in non-Purkinje cells in situ, because the cell specificity of the calbindin promoter was retained in all active constructs.
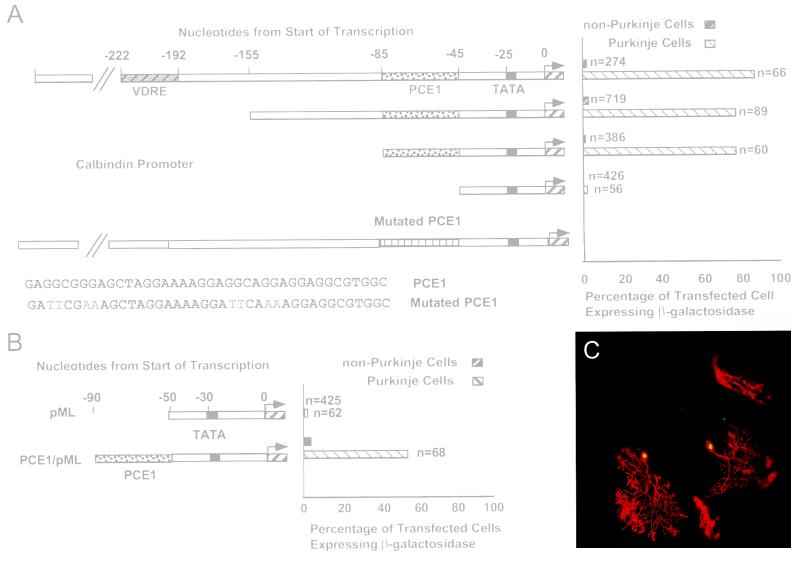
Figure 2.
Identification of PCE1 by transfection of truncated calbindin promoter constructs into P12 cerebellar slices. (A) Calbindin promoter constructs extending from −1,010, −155, −85, and −45 to +105 and driving expression of nLacZ each were cotransfected along with CMVAP into P12 cerebellar slices. After incubation for 48 hr they were fixed and stained for presence of β-galactosidase and alkaline phosphatase using either immunocytochemistry or chromogenic enzymatic reactions. The percentage of transfected cells expressing β-galactosidase refers to the ratio of the number of cells expressing β-galactosidase to the number expressing alkaline phosphatase both for Purkinje and non-Purkinje cells. n represents the number of transfected cells in each case. Mutations in the −1,010 promoter were introduced into PCE1 in two of 10-bp repeats to prepare the construct named mutated PCE1. The sequence of the mutated region and the results of these assays also are included. (B) One construct consisting of the minimal adeno major late promoter (pML), which extends from −40 to +66 of the adenovirus major late promoter, driving nLacZ, and another consisting of pML with PCE1 (see A for sequence) added at the 5′ end (PCE1/pML) and also driving nLacZ, each were cotransfected along with CMVAP into cerebellar slices. (C) Slices were cotransfected with PCE1/pML and CMVAP and then immunostained as in Fig. 1. Purkinje cells (marked by arrows) express both β-galactosidase and alkaline phosphatase, whereas non-Purkinje cells express only alkaline phosphatase.
PCE1 was found to contain three repeats of a GA-rich 10-bp motif. Because transcriptional regulation often involves relatively simple DNA elements that are repeated in the 5′ flanking regions of specific genes, we tested whether these repeats are essential for Purkinje cell specific expression of the calbindin promoter. When two of the repeats were mutated, the 1-kb promoter could no longer mediate transcription (Fig. 2A). These results clearly demonstrate that PCE1 is required for calbindin transcription in situ.
To determine whether PCE1 is sufficient to drive Purkinje cell specific expression, a single copy was inserted 20 bp upstream from the TATA box of the minimal adenovirus major late promoter (pML) and assayed in slice preparations. The pML has been extensively used as a minimal promoter containing all of the basal elements required for transcription initiation in vitro but none that can program efficient or cell-specific transcription in vivo (17). This was confirmed in cerebellar slice preparations, because pML was not expressed in any cell type after biolistic transformation (Fig. 2B). In contrast, pML plus PCE1 (PCE1/pML) caused expression in 54% of the transfected Purkinje cells and <4% of the non-Purkinje cells (Fig. 2 B and C). Thus, PCE1 conferred both high activity (54%) and cell specificity (≈20-fold) to pML. These results are comparable to the activity of other well characterized transcriptional regulatory elements, allowing us to conclude that PCE1 is both necessary and sufficient for Purkinje cell specific expression in organotypic slices of P12 cerebellum.
PCE1 appears to be novel, because it does not match any of the known transcription factor binding sites (18). However, a sequence containing three 10 nucleotide repeats very similar to those of PCE1 (referred to as PCE1*) was found in the 5′ flanking region of calmodulin II. Like calbindin, calmodulin II is a Ca2+ binding protein that is expressed abundantly in Purkinje cells (19), although unlike calbindin, it can activate enzymes and modulate activity of ion channels when it is in a calcium-bound state (6). Furthermore, it has been shown that 360 bp of DNA upstream from the calmodulin II gene, which includes PCE1*, can mediate expression in cerebellar Purkinje cells in transgenic mice (20).
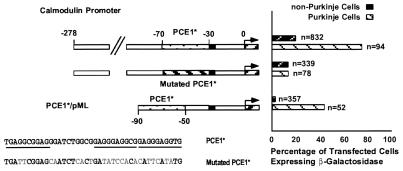
Three-hundred-thirty bp of 5′ flanking DNA of calmodulin II was found to cause expression in 82% of the transfected Purkinje cells and in 22% of the transfected non-Purkinje cells (Fig. 3). Thus, this promoter is approximately equally active for Purkinje cell transcription, but much less specific than the calbindin promoter (≈4-fold vs. >50-fold). This is in agreement with the in vivo expression pattern of calmodulin II, which is expressed strongly in cerebellar Purkinje cells and at much lower levels in other cerebellar cell types (20). When PCE1* was mutated in the context of the 330-bp promoter fragment (Fig. 3) it caused expression in 14% of both Purkinje and non-Purkinje cells. Thus, PCE1* is required for the Purkinje cell specificity of the calmodulin II promoter in situ, and it has very little effect on transcription of this gene in other cell types. Apparently, a second element that is not cell specific is responsible for the residual activity of this promoter. While we have not mapped this element, it has been reported that sequences between −280 bp and −260 bp are required for the activity of the calmodulin II promoter in 3T3 cells (20), suggesting that the same region is responsible for the remaining transcriptional activity of this promoter after mutation of PCE1. Finally, when the PCE1* was inserted upstream of pML, this construct was expressed in ≈40% of the transfected Purkinje cells and was not expressed in other cerebellar cell types (Fig. 3). We conclude, therefore, that PCE1* also acts as a Purkinje cell specific transcription element in the calmodulin II promoter.
Figure 3.
Two constructs, the first consisting of the calmodulin II promoter construct extending from −278 to +54, and the second consisting of the same construct, except that 20 of the 40 nucleotides comprising its PCE1 sequence (PCE1*) were altered (as shown in the sequence labeled mutated PCE1*), were cotransfected along with CMVAP into cerebellar slices. In addition, a construct consisting of pML with PCE1* added to its 5′ end (PCE1*/pML) was also cotransfected with CMVAP into cerebellar slices. The three 10 nucleotide repeats are underlined in the sequence labeled PCE1*.
PCE1 is conserved in the calbindin and calmodulin II promoters from different species, which is consistent with PCE1 being important for transcriptional control (Fig. 4). It was found that the 10-nucleotide repeats from the calbindin and calmodulin promoters of different species have the consensus sequence 5′-GAGGG(G/A)GGAG-3′, with the two nucleotides in the fifth and sixth positions more variable than the others. In addition, the first four nucleotides of the most 5′ repeat and the last four nucleotides of the most 3′ repeat tended to show more variation (Fig. 4). In both the mouse calbindin and calmodulin II promoters the three 10-bp repeats are arranged so that they are on the same face of the double helix as the TATA box, although there is 20 bp of sequence between the TATA box and the calbindin repeats, whereas the calmodulin II elements are immediately adjacent to this sequence (Fig. 4). This spacing also is conserved between different species, indicating that it may be important for the function of PCE1. It is for this reason that in the constructs PCE1/pML and PCE1*/pML, the elements were placed exactly 20 nucleotides upstream of the TATA box of pML. It is interesting to note that the 10 bp of sequence in each promoter that separates the first two repeats is different between calbindin and calmodulin II, however it is conserved between different species, indicating that it may have functional significance. Also note that the second repeat is not present in the dog and human calmodulin II promoters despite the fact that the first, the third, and the intervening sequence are preserved (Fig. 4).
Figure 4.
Comparison of PCE1 in the calbindin and calmodulin II promoters of different species. Comparison of the promoter sequence immediately 5′ of the TATA box for mouse calbindin D28k (11), human calbindin D27k (21), mouse calmodulin II (20), dog calmodulin II (20), and human calmodulin II (J. D. Karkera and F. Friedberg, unpublished results). Repeated motifs are surrounded by boxes. Comparison of individual motifs comprising PCE1 from the different promoters shown in part was used to derive the consensus sequence.
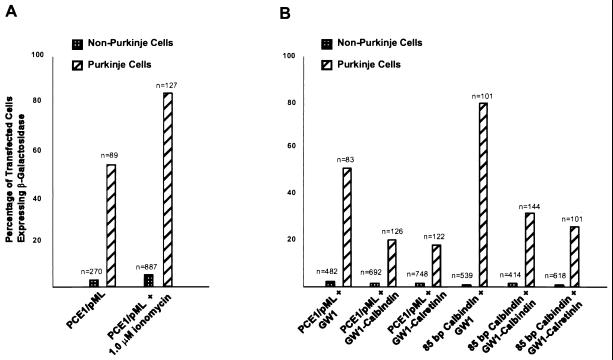
To see whether Ca2+ could modulate PCE1-mediated transcription, the ability of two calbindin promoter constructs to mediate transcription in the context of different levels of intracellular Ca2+ was assessed. When the intracellular free Ca2+ concentration was increased by supplementing the medium with 1 μM calcium ionophore ionomycin, a reproducible increase in PCE1 mediated Purkinje cell-specific expression was observed without an associated increase in the intensity of pCMV-mediated transcription. It was found in three experiments that PCE1/pML expressed in ≈50% more transfected Purkinje cells when ionomycin was added to the medium (Fig. 5A), a difference that is statistically significant (P < 0.01, t test). Consequently, the presence of ionomycin can increase the activity of PCE1/pML to roughly the level of the native 1-kb calbindin promoter, consistent with the hypothesis that increasing free Ca2+ can increase the efficiency with which PCE1 mediates transcription. The addition of ionomycin did not appreciably change the expression level in non-Purkinje cells (5% vs. 3%) (Fig. 5A), indicating that while Purkinje cell specific expression was significantly increased, nonspecific expression levels remained very low.
Figure 5.
The effect of modulating the level of intracellular free Ca2+ on expression mediated by PCE1. (A) Increasing the concentration of intracellular free Ca2+ causes an increase in the activity of PCE1. Cerebellar slices were cotransfected with CMVAP and PCE1/pML driving nLacZ with and without 1.0 μM ionomycin in the medium. The resulting percentages of transfected cells expressing β-galactosidase were calculated and graphed as in Fig. 2A. (B) Decreasing the concentration of intracellular free Ca2+ by cotransfection with calcium buffers causes a decrease in the activity of PCE1, both in the context of the native promoter and when combined with pML. Cerebellar slices were cotransfected with CMVAP, either PCE1/pML driving nLacZ or the 85-bp calbindin promoter driving nLacZ, and one of either the GW1 promoter alone, GW1 driving calbindin, or GW1 driving calretinin. The resulting percentages of transfected cells expressing β-galactosidase were calculated and graphed as in Fig. 2.
If PCE1-mediated transcription is Ca2+ sensitive, we reasoned that lowering the intracellular level of free Ca2+ should decrease PCE1 activity. To investigate this, we took advantage of the demonstrated Ca2+ buffering capacity of the calbindin and calretinin proteins and the ability to achieve nearly 100% efficiency of cotransfection of multiple DNA constructs using the biolistic methodology. In these experiments the biolistic carriers were coated with three different DNAs: the CMVAP marker gene to identify transfected cells; the test promoter construct to measure Purkinje cell specific transcription mediated by PCE1; and either an empty CMV vector control, or the CMV vector driving expression of the calbindin or calretinin proteins to lower free intracellular Ca2+ in the transfected cells. This experimental approach allowed us to test the effects of buffering free Ca2+ in individual cells rather than trying to alter intracellular Ca2+ levels using pharmacologic agents to treat the entire organotypic slice preparation. As shown in Fig. 5B, cotransfection of the empty vector (GW1) with either the 85-bp calbindin promoter construct (80%) or the PCE1/pML construct (50%) did not change their activity from that observed in the absence of the CMV empty vector (see also Fig. 2 A and B). In contrast, when the 85-bp calbindin promoter was cotransfected with the CMV promoter driving expression of calbindin (GW1-calbindin) (30%) or calretinin (GW1-Calretinin) (25%) there was a dramatic decrease in its activity (Fig. 5B). A similar ≈2.5-fold decrease in activity was observed when these Ca2+ buffering proteins were cotransfected with the PCE1/pML test promoter (Fig. 5B). These results demonstrate that transcription via PCE1 is sensitive to free intracellular Ca2+ concentrations, suggesting that the regulation of the Ca2+ buffering capacity of cerebellar Purkinje cells is at least partially regulated at the transcriptional level.
DISCUSSION
The identification of PCE1 provides an example of a transcriptional regulatory element that is both necessary and sufficient for expression in a specific CNS neuron in situ. Previously, the only precisely defined DNA sequence elements mediating neural specific gene expression were the inhibitory elements, RE-1 or NRSE 1, which were originally identified in the promoters of the type II sodium channel (22) and SCG10 (23), respectively. In combination with activator elements, repressor elements similar to RE-1 and NRSE 1 have been shown to cause neuronal-specific expression of numerous genes (24–26). The fact that PCE1 can mediate Purkinje cell-specific expression in the absence of any silencer elements indicates that it is part of a mechanism for achieving neuron-specific gene expression that is different from the mechanism involving RE-1 or NRSE 1.
Although PCE1 was found to mediate Purkinje cell-specific expression in both the calbindin and calmodulin II promoters, it is not involved in all cases of Purkinje cell specific expression. For example, PCE1 is not found in the promoters of either calmodulin I or calmodulin III. While this may seem unexpected because all three isoforms of calmodulin have similar expression patterns and, specifically, all three express at high levels in Purkinje cells, previous experiments have suggested that expression of these three isoforms is mediated through different mechanisms. It was shown that in transgenic mice, expression in Purkinje cells could be driven both by 360 bp of 5′ flanking DNA of the calmodulin II promoter (20) and 980 bp of 5′ flanking DNA of the calmodulin III promoter (27); however, there is no sequence identity between these two promoters or with that of calmodulin I. PCE1 is also absent from the 5′ flanking DNA of L7 (28) and of aldolase C (Zebrin II) (29), which is not surprising because the expression patterns of the two genes differ from that of calbindin both spatially and temporally. Specifically, calbindin is expressed in all Purkinje cells both during development and through adulthood, whereas L7 is expressed only in subsets of Purkinje cells during development (30) and Zebrin II is expressed in subsets of Purkinje cells through adulthood (31).
Because the calbindin and calmodulin II genes are expressed early and abundantly in Purkinje cells, it is probable that PCE1 and its cognate transcription factor(s) are critical to the function of these neurons. Identification of PCE1 provides an avenue for identification of this transcription factor and investigation of its biological role in Purkinje cell development. Furthermore, it is likely that the PCE1 pathway plays an important role in several other neuronal cell types. For example, both calbindin and calmodulin II are expressed at very high levels in dentate gyrus granule cells (20), and we expect that PCE1 also will be involved in directing expression of these genes in the hippocampus. On the other hand, both of these genes are expressed elsewhere in the CNS, and calbindin is expressed in various places outside the brain. Our results indicate that sequences controlling expression at these sites are distinct from PCE1.
The facts that PCE1 is required for the expression of calbindin, which participates in homeostatic control of intracellular Ca2+, and that the activity of this element is sensitive to intracellular Ca2+ levels, strongly suggest that PCE1 is part of a pathway that determines the Ca2+ buffering capacity of cerebellar Purkinje cells. Two properties of Ca2+-sensitive transcription via PCE1 suggest that regulation of the Ca2+ buffering capacity of Purkinje cells is distinct from these previously characterized Ca2+-sensitive transcriptional control mechanisms. First, PCE1 bears no significant sequence similarity to either the serum response element or cyclic AMP response element/calcium response element (Fig. 4; ref. 32). Given the strong conservation of both of these previously characterized calcium response elements in target genes, and the high homology of PCE1 across species in both the calbindin and calmodulin II promoters (Fig. 4), it is very unlikely that either serum response factor (SRF) or cyclic AMP response element binding factor (CREB) plays a role in transcriptional activation through PCE1. Second, unlike the SRF and CREB pathways, which are active in many different cell types in many different physiologic situations, the activity of PCE1 is cell specific. It seems probable, therefore, that PCE1 is a component of a novel Ca2+-sensitive signaling mechanism. Identification of transcription factors that operate through PCE1 and direct measurement of their sensitivity to free intracellular Ca2+ levels will be a critical next step in the analysis of this pathway.
Acknowledgments
We thank Claude Desplan, Morgan Sheng, Emily Liman, and Lisa Dailey for critical comments on the manuscript. Research in the laboratory of N.H. is supported by the Howard Hughes Medical Institute and a grant from the National Institutes of Health, National Institutes of Neurological Disorders and Stroke (2 P01 NS30532–06). D.B.A. is also supported by this grant.
ABBREVIATIONS
- PCE
Purkinje cell element
- CNS
central nervous system
- CMV
cytomegalovirus
- CMVAP
CMV immediate early promoter driving alkaline phosphatase
References
- 1.Darnell J E, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Treisman R. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miale I, Sidman R. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- 4.Enderlin S, Norman A W, Celio M R. Anat Embryol. 1987;177:15–28. doi: 10.1007/BF00325286. [DOI] [PubMed] [Google Scholar]
- 5.Garcia Segura L M, Baetens D, Roth J, Norman A W, Orci L. Brain Res. 1984;296:75–86. doi: 10.1016/0006-8993(84)90512-2. [DOI] [PubMed] [Google Scholar]
- 6.Baimbridge K G, Celio M R, Rogers J H. Trends Neurosci. 1992;15:303–308. doi: 10.1016/0166-2236(92)90081-i. [DOI] [PubMed] [Google Scholar]
- 7.Mattson M P, Rychlik B, Chu C, Christakos S. Neuron. 1991;6:41–51. doi: 10.1016/0896-6273(91)90120-o. [DOI] [PubMed] [Google Scholar]
- 8.Chard P S, Bleakman D, Christakos S, Fullmer C S, Miller R J. J Physiol. 1993;472:341–357. doi: 10.1113/jphysiol.1993.sp019950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chard P S, Jordan J, Marcuccilli C J, Miller R J, Leiden J M, Roos R P, Ghadge G D. Proc Natl Acad Sci USA. 1995;92:5144–5148. doi: 10.1073/pnas.92.11.5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowenstein D H, Miles M F, Hatam F, McCabe T. Neuron. 1991;6:627–633. doi: 10.1016/0896-6273(91)90065-8. [DOI] [PubMed] [Google Scholar]
- 11.Gill R K, Christakos S. Proc Natl Acad Sci USA. 1993;90:2984–2988. doi: 10.1073/pnas.90.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varghese S, Lee S, Huang Y C, Christakos S. J Biol Chem. 1988;263:9976–9984. [PubMed] [Google Scholar]
- 13.Pavlou O, Ehlenfeldt R, Horn S, Orr H T. Mol Brain Res. 1996;36:268–279. doi: 10.1016/0169-328x(95)00259-u. [DOI] [PubMed] [Google Scholar]
- 14.Arnold D, Feng L, Kim J, Heintz N. Proc Natl Acad Sci USA. 1994;91:9970–9974. doi: 10.1073/pnas.91.21.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo D C, McAllister A K, Katz L C. Neuron. 1994;13:1263–1268. doi: 10.1016/0896-6273(94)90412-x. [DOI] [PubMed] [Google Scholar]
- 16.Mercer E H, Hoyle G W, Kapur R P, Brinster R L, Palmiter R D. Neuron. 1991;7:703–716. doi: 10.1016/0896-6273(91)90274-4. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto N G, Moncollin V, Wintzerith M, Egly J M, Chambon P. Nucleic Acids Res. 1984;12:8779–8799. doi: 10.1093/nar/12.23.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulikas T. Crit Rev Eukaryotic Gene Expression. 1994;4:117–321. doi: 10.1615/critreveukargeneexpr.v4.i2-3.10. [DOI] [PubMed] [Google Scholar]
- 19.Ikeshima H, Yuasa S, Matsuo K, Kawamura K, Hata J, Takano T. J Neurosci Res. 1993;36:111–119. doi: 10.1002/jnr.490360112. [DOI] [PubMed] [Google Scholar]
- 20.Matsuo K, Ikeshima H, Shimoda K, Umezawa A, Hata J, Maejima K, Nojima H, Takano T. Mol Brain Res. 1993;20:9–20. doi: 10.1016/0169-328x(93)90106-y. [DOI] [PubMed] [Google Scholar]
- 21.Parmentier M, De Vijlder J J, Muir E, Szpirer C, Islam M Q, Guerts van Kessel A, Lawson D E, Vassart G. Genomics. 1989;4:309–319. doi: 10.1016/0888-7543(89)90335-2. [DOI] [PubMed] [Google Scholar]
- 22.Kraner S D, Chong J A, Tsay H-J, Mandel G. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 23.Mori N, Schoenherr C, Vandenburgh D J, Anderson D J. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 24.Schoch S, Cibelli G, Thiel G. J Biol Chem. 1996;271:3317–3323. doi: 10.1074/jbc.271.6.3317. [DOI] [PubMed] [Google Scholar]
- 25.Miede M, Haga T, Saffen D W. J Biol Chem. 1996;271:5177–5182. doi: 10.1074/jbc.271.9.5177. [DOI] [PubMed] [Google Scholar]
- 26.Bessis A, Salmon A M, Zoli M, Le Novere N, Picciotto M, Changeux J P. Neuroscience. 1995;69:807–819. doi: 10.1016/0306-4522(95)00303-z. [DOI] [PubMed] [Google Scholar]
- 27.Shimoda K, Ikeshima H, Matsuo K, Hata J, Maejima K, Takano T. Mol Brain Res. 1995;31:61–70. doi: 10.1016/0169-328x(95)00032-n. [DOI] [PubMed] [Google Scholar]
- 28.Oberdick J, Smeyne R J, Mann J R, Zackson S, Morgan J I. Science. 1990;248:223–226. doi: 10.1126/science.2109351. [DOI] [PubMed] [Google Scholar]
- 29.Vibert M, Henry J, Kahn A, Skala A. Eur J Biochem. 1989;181:33–39. doi: 10.1111/j.1432-1033.1989.tb14690.x. [DOI] [PubMed] [Google Scholar]
- 30.Smeyne R J, Oberdick J, Schilling K, Berrebi A S, Mugnaini E, Morgan J I. Science. 1991;254:719–721. doi: 10.1126/science.1948052. [DOI] [PubMed] [Google Scholar]
- 31.Hawkes R, Leclerc N. Brain Res. 1989;476:279–290. doi: 10.1016/0006-8993(89)91248-1. [DOI] [PubMed] [Google Scholar]
- 32.Gallin W J, Greenberg M E. Curr Opin Neurobiol. 1995;5:367–374. doi: 10.1016/0959-4388(95)80050-6. [DOI] [PubMed] [Google Scholar]