Abstract
Global genomic approaches in cancer research have provided new and innovative strategies for the identification of signatures that differentiate various types of human cancers. Computational analysis of the promoter composition of the genes within these signatures may provide a powerful method for deducing the regulatory transcriptional networks that mediate their collective function. In this study we have systematically analyzed the promoter composition of gene classes derived from previously established genetic signatures that recently have been shown to reliably and reproducibly distinguish five molecular subtypes of breast cancer associated with distinct clinical outcomes. Inferences made from the trends of transcription factor binding site enrichment in the promoters of these gene groups led to the identification of regulatory pathways that implicate discrete transcriptional networks associated with specific molecular subtypes of breast cancer. One of these inferred pathways predicted a role for nuclear factor-κB in a novel feed-forward, self-amplifying, autoregulatory module regulated by the ERBB family of growth factor receptors. The existence of this pathway was verified in vivo by chromatin immunoprecipitation and shown to be deregulated in breast cancer cells overexpressing ERBB2. This analysis indicates that approaches of this type can provide unique insights into the differential regulatory molecular programs associated with breast cancer and will aid in identifying specific transcriptional networks and pathways as potential targets for tumor subtype-specific therapeutic intervention.
The application of high-throughput gene expression profiling to the study of cancer has broadened our thinking about the biology of neoplasia by providing deeper insights into the mechanisms underlying tumor promotion and progression.1,2,3,4 Numerous computational methods aimed at identifying trends and patterns of gene expression specific to different tumors and subtypes have lead to the discovery of several genetic patterns or molecular signatures that aid in distinguishing biologically relevant aspects of tumor behavior, function, and identity.3,5,6 These signatures provide a means of classifying tumors that would not otherwise be distinguished by conventional clinical parameters, such as size, location, and histological appearance, and are beginning to increase our basic understanding of tumor biology. Moreover, some of these molecular signatures have been found to have both therapeutic and prognostic significance, potentially enabling clinicians to identify those tumors that are likely to be most responsive to specific therapies.2,3,7
Recently, the application of hierarchical clustering to distinguish molecular phenotypes has identified five different molecular subtypes of breast cancer based on their differential expression of ∼534 genes.2,3 Three of these classes are characterized as having low to absent expression of estrogen receptor (ER) and other specific transcription factors when compared to the other subtypes.4 These three are referred to as 1) basal-like subtype, characterized by high expression of keratins 5 and 17, laminin, and fatty acid binding proteins, genes that are often more expressed in the basal cell of normal breast ascini; 2) the ERBB2+ subtype, characterized by higher expression of the epidermal growth factor (EGF) receptor family member and other genes associated with amplification of the ERBB2 locus at 17q22.24, which includes the growth factor receptor adaptor protein GRB7; and 3) the normal-like subtype, characterized by expression of a large number of genes normally expressed in adipose tissue and other tissues of nonepithelial origin and higher expression of genes associated more with basal epithelial cell expression than luminal epithelial cell expression. The last two remaining molecular phenotypes are breast tumor subtypes referred to as luminal A and luminal B. These two groups characteristically have the highest expression of ER. In addition, luminal A subtype is characterized by higher expression of the transcription factors GATA3, hepatocyte nuclear factor 3α, the estrogen-inducible secreted factor trefoil factor 3 (TFF3), and the estrogen-induced solute carrier SLC39A6/LIV-1. Luminal B is characterized by lower expression of luminal type genes. These markers have been shown repeatedly to reliably segregate breast carcinomas derived from independent data sets and samples into these five specific molecular subtypes.2,3
Recent attempts to identify the mechanism underlying the coordinated gene expression patterns observed in various molecular signatures of cancer are based on the rational assumption that co-expressed genes share similar properties of gene regulation. A large contribution to this coordinated expression occurs at the level of transcription; therefore, it is reasonable to presume that similarly regulated genes should have a higher probability of being regulated by similar transcription factors and transcriptional pathways.8,9
The major targets of these transcription factors and pathways are the noncoding sequences that reside primarily in the regulatory regions upstream of the start of transcription of target genes. An assembly of transcription factor binding sites (TFBSs) found to be nonrandomly shared by several members of a gene list associated with a molecular phenotype can therefore be readily considered to represent a regulatory signature of that phenotype. Identification of such regulatory signatures will help define the transcriptional pathways and molecular signaling events that are integrated to mediate the coordinated expression of multiple genes.8,9 The transcriptional networks elucidated by such an approach will provide important insight into the active molecular events responsible for the evolution of specific tumor subtypes of cancer and suggest new functional molecular targets for therapeutic intervention.
In this study we examined the promoter composition of the genetic signatures inferred from a previously published study that defined five different molecular subtypes of breast cancer that correlated with specific clinical outcomes.2,3 Using a position weight matrix scoring system, the relative enrichment of each of these tumor subtype-specific signatures for 409 different TFBS matrices was determined using a reference background model containing the proximal promoter region of 15,318 RefSeq genes. Comparison of the TFBS significance scoring by hierarchical clustering and principal component analysis (PCA) identified groups and clusters of enriched TFBS from which sets of transcriptional regulatory networks were inferred. One novel inference derived from this approach was the empirically validated identification of a positive autoregulatory loop through which ERBB2 utilizes nuclear factor (NF)-κB pathways to enforce its own expression. This approach represents a novel and powerful method through which the regulatory circuitry underlying breast cancer subtypes associated with specific patient outcomes can be distinguished and dissected in the context of functionally relevant molecular targets and pathways.
Materials and Methods
Promoter Analysis
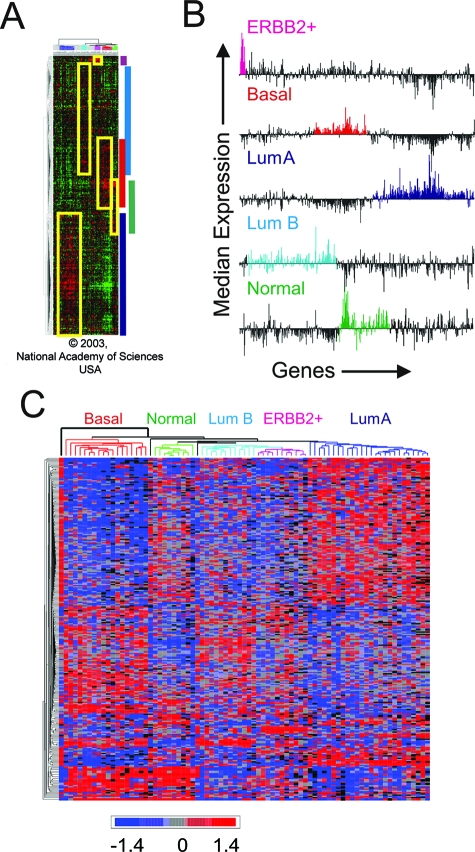
Hierarchical clustering data2 were used to construct an initial list of genes contained within the boundaries of the positive peaks of expression delimited by the gene order and clusters defined in Sorlie and colleagues2 (Figure 1, A and B). To do this the median expression of each of the 534 genes in each sample cluster was determined. This produced five median expression profiles for each of the 534 genes of each cluster (Figure 1B). The genes within the major contiguous positive peaks for each cluster were then broadly selected (color coded peaks in Figure 1B). This resulted in an initial overlapping list of genes containing: 136 genes from the basal subtype, 15 genes from the ERBB2+ subtype, 243 genes from the luminal A subtype, 217 genes from the luminal B subtype, and 108 genes from the normal-like subtype. These gene lists were then refined to remove overlapping and other noninformative genes by significance ranking for subtype discriminators using one-way analysis of variance and selecting genes with P values less than 0.01 (before correction for multiple comparisons) (see Supplementary Table S1 at http://ajp.amjpathol.org). The result was a final list of 221 genes: 95 unique genes for luminal A subtype, 21 unique genes for luminal B subtype, 66 unique genes for basal subtype, six unique genes for ERBB2+ tumor subtype, and 13 unique genes for the normal-like tumor subtype (see Supplementary Table S1 at http://ajp.amjpathol.org). Promoter sequences from three 600-bp promoter regions (proximal: −500 bp to +100 bp; upstream: −1100 bp to −500; and downstream: +100 bp to +700 bp, all relative to transcription start site) were retrieved for each gene from the five different tumor subtype-specific gene lists using the ProSpector web-based promoter annotation tool.10
Figure 1.
Inference of tumor subtype-associated genetic signatures from meta-analysis of gene expression profiles that reproducibly distinguish breast cancer subtypes. A: Gene lists for promoter analysis were derived from the data of Sorlie and colleagues,2 in which hierarchical clustering of 534 genes and 115 breast tumors showed five distinct gene expression signatures (boxed) subdividing tumors into five clinically relevant subgroups (Modified from Sorlie et al2 with permission, Copyright 2003, National Academy of Sciences, USA). Boxed regions overlap the starting list of genes used to construct tumor subtype-specific gene lists for proximal promoter analysis after significance ranking of subtype discriminators by analysis of variance (see Supplemental Table S1 at http://ajp.amjpathol.org). B: Median expression profiles of subtype-specific gene groups based on dendrogram boundaries. The genes are ordered from left to right according to the order generated by hierarchical clustering (top to bottom) in A. C: Rederivation of hierarchical clustering of tumor subtypes based on gene lists derived from significance ranking of subtype discriminators by analysis of variance. Subtype discriminators were then used as subtype-specific genetic signatures.
The promoter regions were analyzed for matches to 409 position weight matrices using the MatInpector module of the GEMS LauncheR 4.1 (Genomatix, Munich, Germany). The analysis was performed with an optimized matrix threshold and a threshold of 0.75 for the core similarity. A P value for each gene in each gene list (cluster) was obtained by comparing the number of matches per 1-kb promoter region in each of the tumor subtypes against the number of matches per 1-kb promoter region in a reference background model using a complemented Poisson distribution (pdtrc) in the Perl math (math-cephes) library. The reference background model was obtained by extracting all unique RefSeq IDs (24,704) from the UCSC genome browser (build hg17) and mapping them to 15,318 IDs using Prospector followed by promoter TFBS annotation using MatInspector as described above. A Perl script was used to calculate P values for each of the 409 position weight matrices in each of the gene lists. The Perl script processed an input file containing a list of the 409 matrices and the number of matches for each of the matrices in a particular gene list and a file containing a list of the 409 matrices and the number of matches for each of the matrices in the reference background model. The script produced a file containing the list of the 409 matrices and the P values associated with each of the matrices calculated using the complemented Poisson distribution from the Perl math (math-cephes) library.
Statistical Analysis
One-way analysis of variance, hierarchical clustering, PCA, and intensity plots were generated using Partek Pro 5.1. Principal component (PC) loading correlation values were calculated with Partek Pro 5.1. PCA biplot analysis was performed as previously described.11 For this analysis, TFBS matrices with a correlation value less than 0.75 in any of the first four PCs were removed resulting in 208 matrices. This list was filtered further for a P value ≤0.05 in one of any of the five subtypes leaving 44 matrices for Biplot analysis. Randomization was performed by taking 40,000 random gene list selections of 6, 13, 21, 66, and 95 genes from the reference background list of 15,318 RefSeq genes and analyzing for frequency of the 409 matrices in each of the 40,000 random iterations for each of the five gene list sizes using MatInspector. Perl scripts were used to create the 40,000 random gene lists, to calculate matches for each of the 409 matrices in each of the random gene lists, and to calculate P values for each of the 409 matrices in each of the random gene lists.
Pathway and Network Analysis
Gene lists were analyzed with the Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). Networks were constructed by overlaying the genes in the gene list, called Focus Genes, onto a global molecular network developed from information contained in the Ingenuity Pathways knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity. A network is a graphical representation of the molecular relationships between genes. Genes are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways knowledge base. P values for the enrichment of canonical pathways were generated based on the hypergeometric distribution and calculated with the right-tailed Fisher’s exact t-test for 2 × 2 contingency tables. The composite gene list constructed using TFBSs included all known genes cognate for the particular binding site.
Chromatin Immunoprecipitation
MCF-7 and MDA-MB-231 breast cancer cell lines (gifts from Dr. Alfred Johnson and Dr. Ira Pastan, respectively, from the National Cancer Institute, Bethesda MD) were grown in Dulbecco’s modified Eagle’s medium and 10% fetal calf serum at 37°C and 5% CO2. The cells were serum-starved overnight before a 1-hour stimulation with 12 ng/ml of EGF (Peprotech, Rocky Hill, NJ). Cells were then harvested, cross-linked with formalin, and chromatin immunoprecipitation was performed as previously described.12 The antibodies used were a 1:1 mixture containing 5 μg each of affinity-purified antibodies against p65 (sc-109; Santa Cruz Biotechnology, Santa Cruz, CA) and c-rel/Rel.13 Primers used for the ERBB2 promoter were 5′-TATTTTATCCTTGGTGTCGTGGCAGC-3′ and 5′-CATTGGCTGGCACTGGTCCC-3′.
Results
Derivation of Breast Cancer Subtype-Specific Gene Signatures
Breast cancer subtype-specific signatures were derived from a previously published hierarchical clustering of 534 genes (represented by 552 clones; 500 of these correspond to a single unique UniGene cluster) expressed in 115 breast cancer samples (Figure 1A).2 As noted by Sorlie and colleagues,2 specific core genes within these 534 genes showed particular discriminatory power. For our promoter analysis we sought to expand this core list of subtype-specific genes. Therefore lists of up-regulated genes, comprising the core genes grouped with the adjacent members within each cluster, as delimited by the dendrograms for each subtype (Figure 1A), were constructed based on the peak median gene expression for the 534 genes (Figure 1B). Although focusing exclusively on up-regulated expression creates a bias toward finding presumed positive correlations between TFBS enrichment and gene expression, this simplified approach results in minimizing noise that would result from attempts to distinguish and interpret both positive and negative discriminators during the promoter analysis. Based on this assumption, a preliminary list of subtype-specific genes was constructed from the peak of median expression for each subtype. To reduce stochastic noise in these lists we performed a one-way analysis of variance analysis to remove noninformative genes (P value >0.01, before correction for multiple comparisons) that did not contribute significantly to the classification of the five tumor subtypes (see Supplementary Table S1 at http://ajp.amjpathol.org). This reduced the original 534 genes to a total 201 RefSeq mapped genes comprising five nonoverlapping subtype-specific genetic signatures (95 for luminal A, 21 for luminal B, 66 for basal, 6 for ERBB2+, and 13 for normal-like subtype; see Supplementary Table S1 at http://ajp.amjpathol.org). As shown in Figure 1C, hierarchical clustering of this refined list resembles the basic partitioning of the tumor subtypes shown in the original hierarchical clustering analysis.2,3 A similar approach was recently described by Muggerud and colleagues.14
Promoter Analysis Reveals Tumor Subtype-Specific Enrichment of TFBSs
To search for potential common regulatory pathways associated with the five subtype-specific genetic signatures, the promoter regions of each gene were extracted from −500 bp upstream to +100 bp downstream of the transcription start site (see Materials and Methods). These promoter regions were then scored for matches to 409 different TFBS matrices using the MatInspector module of GEMS Launcher 4.1 (Genomatix). The observed frequency of the 409 matrices in the promoters of each subtype-specific signature was then compared to the expected frequency using a reference background model of 15,318 human RefSeq genes. The null hypothesis that the observed frequency of TFBS in the selected upstream sequences could be explained as a random fluctuation, as compared to the background frequency, was used to estimate a P value for significant enrichment. A complemented Poisson distribution was used to model the random processes governing the frequency of TFBS in the human genome. We verified this theoretical assumption by comparing the P values derived from the Poisson distribution to frequencies simulated in a random permutation test with 40,000 randomly selected lists of genes of the same size as the original lists. This analysis showed good agreement between the analytic and experimental distributions for each of the gene signatures (see Materials and Methods and Supplementary Figure S1 at http://ajp.amjpathol.org). The P value for each of the 409 matrices in each of the tumor subgroup gene lists was then analyzed by hierarchical clustering after −log2 transformation (Figure 2). Clustering of the matrices according to tumor subtype reveals multiple distinct groups of TFBSs that are enriched significantly and selectively within the five subtypes (Figure 2, Table 1). These patterns of TFBS enrichment are highly position-dependent because they are not recapitulated at contiguous 600-bp regions downstream or upstream of the proximal −500 to +100 regions characterized in this study (see Supplementary Figure S3 at http://ajp.amjpathol.org).
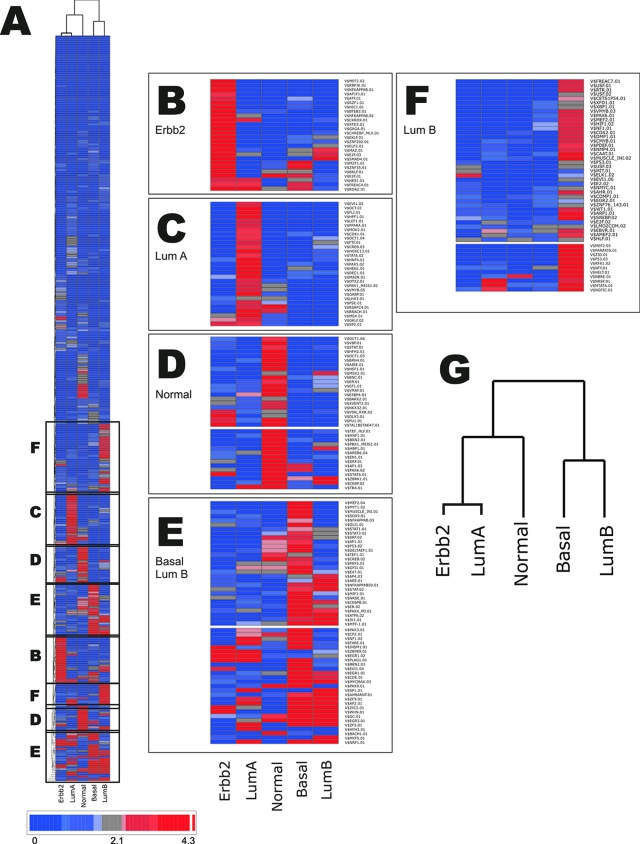
Figure 2.
Tumor subtypes show nonrandom enrichment for specific TFBS. A: Shown is reduced representation a two-way hierarchical clustering of 409 TFBS and the 5 tumor subtypes based on overrepresentation of TFBS matrices scored from P values derived using a background model of 15,318 RefSeq genes (see Materials and Methods). B–F: Magnifications of matrix cluster overrepresented in: ERBB2+ tumor subtype (B); luminal subtype A (C); normal-like breast tumor subtype (D); matrix clusters enriched in basal-like tumor subtype and luminal B subtype (E); and matrix clusters enriched preferentially in luminal B subtype (F). The scale bar represents the log2 transformation of the inverse of the P value (P value <0.05 corresponds to >4.3 on the scale). G: Dendrogram showing classification of tumor subtypes based on TFBS composition in the proximal promoter.
Table 1.
Top 15 Significantly Enriched TFBS Matrices in Each Subgroup
| Group | Matrix | E (freq) | O (freq) | P value | IUPAC |
|---|---|---|---|---|---|
| Erbb2 (n = 6) | V$INSM1.01 | 0.9588 | 2.7778 | 3.01E-03 | tgycwGGGGnnrn |
| V$PLAG1.01 | 1.8093 | 3.8889 | 7.22E-03 | GRGGsncnnnnnrggggknrn | |
| V$ZNF202.01 | 0.9778 | 2.5 | 1.02E-02 | ccnsccCCCAcccccaccccmmc | |
| V$EGR1.02 | 2.6865 | 5 | 1.05E-02 | ssskgnggGGGCgknnn | |
| V$GAGA.01 | 1.1729 | 2.7778 | 1.15E-02 | gnsngAGAGrgagrgrgagmgrnnn | |
| V$E2F.03 | 0.512 | 1.6667 | 1.15E-02 | nnngCGCGaaantkn | |
| V$SMAD4.01 | 0.2419 | 1.1111 | 1.21E-02 | GTCTmgncn | |
| V$XFD3.01 | 0.2431 | 1.1111 | 1.23E-02 | nnwnwgtmAACAwwmwn | |
| V$EKLF.01 | 0.5776 | 1.6667 | 1.96E-02 | ncnsnnaGGGTnn | |
| V$CHREBP_MLX.01 | 0.1804 | 0.8333 | 2.83E-02 | CAYGtggnasysncgtg | |
| V$ROAZ.01 | 0.6403 | 1.6667 | 3.03E-02 | nnGCACccawgggtgmn | |
| V$WHN.01 | 1.0039 | 2.2222 | 3.14E-02 | nngACGCtnnn | |
| V$NFKAPPAB.02 | 0.2116 | 0.8333 | 4.21E-02 | nnGGGActttccnnn | |
| V$E2F.01 | 0.5243 | 1.3889 | 4.31E-02 | twsgcgcGAAAaykr | |
| V$MAZ.01 | 1.8984 | 3.3333 | 4.62E-02 | nkgsGAGGggagn | |
| Basal (n = 66) | V$NRF1.01 | 3.5354 | 4.9242 | 6.49E-06 | nncGCGCangcgcnnnn |
| V$BRN2.03 | 0.2098 | 0.5303 | 1.56E-04 | nnnnnmttnATTWnmwtnn | |
| V$MYCMAX.03 | 0.5074 | 0.9091 | 8.68E-04 | nngcCAYGygsnnnn | |
| V$PAX9.01 | 0.4347 | 0.8081 | 9.10E-04 | nnnnnnsgCACCgatgsgtgannrcynnn | |
| V$CDE.01 | 1.0392 | 1.5404 | 2.25E-03 | kcnkCGCGgtwtn | |
| V$ZF5.01 | 2.2194 | 2.9293 | 2.36E-03 | gtgnGCGCnnn | |
| V$EVI1.04 | 0.3881 | 0.7071 | 2.40E-03 | nGATAnganwagatannnn | |
| V$EGR1.01 | 0.4133 | 0.7323 | 3.00E-03 | nnwtgcgtgGGCGknnn | |
| V$EGR3.01 | 0.5003 | 0.8081 | 7.13E-03 | nnntGCGTgggcgknnn | |
| V$CP2.01 | 0.4351 | 0.7071 | 1.04E-02 | nnCTKGktnkngcnnnnnn | |
| V$GC.01 | 1.2128 | 1.6414 | 1.13E-02 | nnrggGGCGgggcnk | |
| V$PLAG1.01 | 1.8093 | 2.3232 | 1.17E-02 | GRGGsncnnnnnrggggknrn | |
| V$PAX3.01 | 0.4666 | 0.7323 | 1.41E-02 | TCGTcacrcttnm | |
| V$AP2.01 | 1.7056 | 2.1717 | 1.71E-02 | mkCCCScnggcgn | |
| V$WHN.01 | 1.0039 | 1.3384 | 2.55E-02 | nngACGCtnnn | |
| Lum A (n = 95) | V$HFH3.01 | 0.0873 | 0.2632 | 2.14E-04 | nntaaayAAAYannnnn |
| V$ZF5.01 | 2.2194 | 2.7544 | 4.87E-03 | gtgnGCGCnnn | |
| V$ZBP89.01 | 1.4978 | 1.9298 | 5.90E-03 | nnnnnnnccnCCCCcnnnnnnnn | |
| V$PLAG1.01 | 1.8093 | 2.2632 | 7.74E-03 | GRGGsncnnnnnrggggknrn | |
| V$EGR1.02 | 2.6865 | 3.2105 | 1.03E-02 | ssskgnggGGGCgknnn | |
| V$SP1.01 | 2.4941 | 2.9825 | 1.26E-02 | nngggGGCGgggynn | |
| V$FXRE.01 | 0.2703 | 0.4386 | 1.50E-02 | gggtcamTGACcynnnn | |
| V$DEC1.01 | 0.1404 | 0.2632 | 1.73E-02 | nnystCACGtgannn | |
| V$NF1.02 | 0.2193 | 0.3684 | 1.73E-02 | nnnTGGCasnnngccaann | |
| V$TR4.01 | 0.1595 | 0.2807 | 2.39E-02 | nnnnaraGGTCarrknwsn | |
| V$STAF.01 | 0.3002 | 0.4561 | 2.70E-02 | nttwCCCAnmatgcayyrcgnyn | |
| V$ROAZ.01 | 0.6403 | 0.8596 | 2.76E-02 | nnGCACccawgggtgmn | |
| V$HEN1.01 | 0.2599 | 0.4035 | 2.92E-02 | nnggncnCAGCtgcgncccnn | |
| V$MTATA.01 | 0.4676 | 0.6491 | 3.32E-02 | nnntwTAAAncnnnnss | |
| V$NRSF.01 | 0.2659 | 0.4035 | 3.61E-02 | ttcAGCAccacggacagmgsc | |
| Lum B (n = 21) | V$MYF5.01 | 0.4415 | 1.5079 | 6.29E-06 | nmrgCARCwgswgnn |
| V$NRF1.01 | 3.5354 | 5.6349 | 1.57E-04 | nncGCGCangcgcnnnn | |
| V$EGR3.01 | 0.5003 | 1.2698 | 8.49E-04 | nnntGCGTgggcgknnn | |
| V$SP1.01 | 2.4941 | 3.8889 | 2.21E-03 | nngggGGCGgggynn | |
| V$AHRARNT.01 | 0.4511 | 1.1111 | 2.26E-03 | nnnnnnnntygCGTGcmsnnnnn | |
| V$HELT.01 | 0.7947 | 1.5079 | 7.28E-03 | nnsgCACGygacnnn | |
| V$AP2.01 | 1.7056 | 2.6984 | 7.64E-03 | mkCCCScnggcgn | |
| V$GC.01 | 1.2128 | 2.0635 | 7.73E-03 | nnrggGGCGgggcnk | |
| V$ZF5.01 | 2.2194 | 3.3333 | 7.85E-03 | gtgnGCGCnnn | |
| V$ZF9.01 | 2.1669 | 3.254 | 8.55E-03 | nnngrnsCCRCcccynnnnnnnn | |
| V$NBRE.01 | 0.3245 | 0.7937 | 9.36E-03 | nnnnrAAGGtcrnnnnnnn | |
| V$NFY.01 | 0.5114 | 1.0317 | 1.50E-02 | nnrrCCAAtsrgnnn | |
| V$ZIC2.01 | 0.624 | 1.1905 | 1.50E-02 | nnnaccaCCCCnnnn | |
| V$RFX1.02 | 0.3117 | 0.7143 | 1.93E-02 | nngtnrcnnnrGYAAcnnn | |
| V$WHN.01 | 1.0039 | 1.6667 | 1.94E-02 | nngACGCtnnn | |
| Normal (n = 13) | V$BACH1.01 | 0.1323 | 0.8974 | 1.01E-04 | nnnnnnnsaTGAGtcatgnynnnnn |
| V$EN1.01 | 0.4519 | 1.2821 | 3.48E-03 | raaTTTAattgaa | |
| V$AREB6.04 | 0.4606 | 1.2821 | 3.97E-03 | nnnnnGTTTsnnn | |
| V$STAT6.01 | 0.5733 | 1.4103 | 6.38E-03 | nnrnyTTCCyrrgaannnn | |
| V$SRF.01 | 0.4185 | 1.1538 | 6.46E-03 | atgcccaTATAtggwnnnn | |
| V$AP1.03 | 0.1037 | 0.5128 | 9.42E-03 | rsTGACtmann | |
| V$HNF1.01 | 0.3708 | 1.0256 | 9.74E-03 | gGTTAatnwttammnnn | |
| V$BRN2.01 | 0.3752 | 1.0256 | 1.04E-02 | nnnncatnnnWAATnmrnn | |
| V$PBX1_MEIS1.01 | 0.2414 | 0.7692 | 1.27E-02 | nnnwTGATtgacagstn | |
| V$TEF_HLF.01 | 0.5046 | 1.1538 | 1.95E-02 | nnnnaTTACgtaacnnn | |
| V$CEBP.02 | 0.2809 | 0.7692 | 2.45E-02 | ngwntkwnGYAAknm | |
| V$HBP1.01 | 0.6134 | 1.2821 | 2.47E-02 | nnnaatgAATGar | |
| V$PAX6.02 | 0.4608 | 1.0256 | 3.05E-02 | nnnnnagkkCCAGgnnmgn | |
| V$HSF1.01 | 0.4675 | 1.0256 | 3.28E-02 | RGAAnrttcnn | |
| V$E4BP4.01 | 0.6461 | 1.2821 | 3.33E-02 | nnnnnrttayGTAAynnnnnn |
E, expected frequency (per kb) from the reference background model; O, observed frequency in breast cancer subtype gene signatures (see Materials and Methods).
Many of the TFBSs enriched in the specific tumor subtypes (P value ≤0.05) bind factors that participate in well known transcriptional pathways. For example, binding sites for NF-κB (NF-κB) (P ≤ 0.042), E2F factors (P ≤ 0.01), EGR1 (early growth response protein 1) (P ≤ 0.01), and SMAD (P ≤ 0.01) are enriched in the ERBB2+ group indicating that signaling through NF-κB-, E2F-, EGR-, and transforming growth factor (TGF)-β-regulated pathways are likely to play functional roles in the biology of tumors classified by this molecular signature.15,16,17 The two groups of breast cancer that showed the greatest overlap in promoter composition were luminal B and basal-like subtype. A common feature of these groups is the relatively high GC content of many of the consensus sequences representing the matrices enriched in both groups despite the fact that the average GC content of the promoter regions of these groups are not substantially different from the other subtypes (Table 1 and Supplementary Table S1 at http://ajp.amjpathol.org). These include binding sites for early growth response factor-3, EGR3 (P = 0.0008/luminal B; P = 0.007/basal), and the nuclear respiratory factor NRF1 (P = 0.0001/luminal B; P value = 0.000006/basal).18 Distinguishing elements for luminal B include binding sites for the xenobiotic responsive aryl-hydrocarbon factor ARH/ARNT (P = 0.002) that plays a prominent role in estrogen metabolism and cross-talks significantly with ER in breast cancer cells,19 and a binding site for the Nurr1/Nur77 class orphan receptors NBRE (P = 0.009) which have differential roles in controlling cell death and proliferation.20 Elements that distinguish the basal-like group include multiple members of the PAX family of transcription factors known to play a role in cellular differentiation, stem cell biology, and cancer. These include PAX3 (P = 0.014), PAX9 (P = 0.0009), and PAX4/6 (PAX4_PD) (P = 0.047).21 Other highly significant and distinguishing pathways for the basal-like phenotype include the well known oncogene cMyc (P = 0.0008), a pathway commonly amplified in ER-negative breast cancer,22 and the BRN family of pou domain factors (P = 0.0001), which recently have been implicated to have a role in angiogenesis and cancer.23
Interestingly, the promoter signatures derived from this analysis show relationships among the different tumor subtypes that are different from those implied from the hierarchical clustering of the original gene expression data (Figure 1A). Dendrograms of the gene expression signature suggest a close relationship between the basal-like and ERBB2+ phenotypes whereas clustering of the regulatory signatures of these gene groups suggest a closer relationship between luminal B and the basal-like molecular phenotypes (Figure 2G). Although this may reflect differences in the clustering parameters (also note that the rederived gene list of 201 genes in Figure 1C has a slightly different relationship than that of Figure 1A), this finding suggests that such similarities in the molecular phenotypes may be derived from very different signaling pathways and may therefore implicate different regulatory and/or oncogenic origins despite the fact that they are both associated with a poor prognosis.
PCA of Tumor Subtype-Specific Regulatory Signatures Reveals Distinct Regulatory Trends and Minimizes Redundancies
In Figure 3A PCA was used to reduce the initial 409 variables or dimensions, represented by the 409 TFBS matrices, to three dimensions that capture most of the trends (variance) in the data set. As shown in Figure 3A, the tumor subgroups are separated in a three-dimensional space defined by the first three principal components (PCs) of the PCA representing 80.4% of the variance of the data set. Because greater than 80% of the trends in the data can be described in the three PCs, we focused on those patterns that were most apparent in three dimensions. Each PC is a composite derived from specific contribution from each of the original 409 matrices. Thus the relative position of the data points representing the five tumor subgroups are compared in three dimensional space based on their aggregate enrichment for the 409 TFBS. The distance between the data points for each tumor subtype in the three dimensions represents the relative similarity of their regulatory signatures. In other words, shorter distances between signatures indicate greater similarity in promoter composition. As assessed by this method, the tumor subtypes have regulatory signatures that are very distinct. Consistent with the hierarchical clustering in Figure 2, the greatest similarity exists between the basal-like and luminal B regulatory signatures.
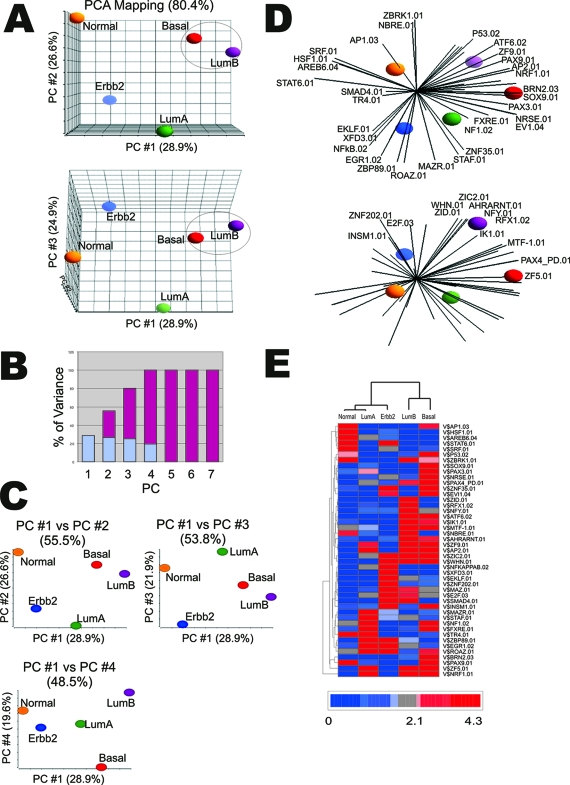
Figure 3.
Principal components analysis (PCA) of TFBS enrichment reduces redundancy and highlights the most effective tumor subtype discriminators. A: PCA profile of the promoter regions for the gene set in each of the tumor subtypes based on TFBS’s enrichment scores (P values). Each sphere represents the comparative statistical averaging of the TFBS enrichment profile for each tumor subtype based on the enrichment score of 409 matrices (ie, 409 vectors). The data are color-coded according to respective tumor subtype: red, basal-like tumor subtype; purple, luminal B subtype; green, luminal A subtype; blue, ERBB2+-overexpressed tumor subtype; orange, normal-like breast tumor subtype. B: Plot of the contribution of each principal component (blue) to the cumulative variance (red). C: Two-dimensional plotting of PCA principal components (PC): PC1 versus PC2, PC 1 versus PC3, and PC1 versus PC4. D: Forty-four matrices with PC loading correlations greater than 0.75 in at least one subtype and with a P value less than 0.05 in at least one subtype were chosen for PCA biplot analysis. E: Hierarchical clustering of the 44 matrices shown in D according to enrichment P values.
Figure 3B shows that the remaining trends in the dataset are captured by the fourth PC. Because 100% of the trends in the data can be explained by four composite dimensions, the relationship between the tumor subtypes were compared using multiple two dimensional plots to extract any additional information that may be inferred from the remaining PC (Figure 3C). Here the basic relationship between the subgroups shown in Figure 3A persists except for PC1 versus PC4, which demonstrates a greater separation between the basal-like and luminal B signature along PC4. This suggests that matrices contributing strongly to PC4 are significant discriminators for the basal-like versus luminal B regulatory signatures. To identify which matrices contributed to this discrimination, we analyzed the principal component (PC) loading.11,24 PC loading provides the correlation of each of the original 409 variables (TFBS dimension) with the principal component vectors (see Supplementary Table S2 at http://ajp.amjpathol.org). Those matrices that make the strongest contribution to the composition of the PC will show significantly higher loading correlations L (eg, L ≥ 0.75). Major contributors to PC4 are BRN2 (L = −0.83), EVI1 (EVI1-myleoid transforming protein) (L = −0.90), NBRE (L = 0.80), NRSE (L = −0.82), PAX3 (L = −0.86), PAX9 (L = −0.79), SOX9 (L = −0.82), and ZNF35 (L = −0.83). All but NBRE have negative correlations, suggesting they are reciprocally enriched in one of the two subtypes. In this case all but NBRE are significantly enriched in the basal-like tumor subtype compared to luminal B. Interestingly most of the factors that bind these sites play substantial roles in embryogenesis and differentiation (see Discussion). Also both three- and two-dimensional PCAs show persistent separation of the luminal B, basal-like, and luminal A subtypes as a common group, distinct from the normal-like and ERBB2+ subtypes, along PC1. An examination of the PC loading of PC1 shows a strong correlation with AP2 (L = 0.987) suggesting that enrichment of this GC-rich matrix is a major distinction separating luminal B, basal-like, and luminal A from the normal-like and ERBB2+ regulatory signatures.
The parameters of PC loading can also be used to filter the original variables to produce a set of reduced size that best characterizes the trends in the regulatory signatures, highlighting those matrices that are significant discriminators despite borderline or low-ranking P values. To do this the 409 matrices were screened by PC loading for those matrices showing a PC loading ( L ) ≥0.75 in any one of the four PCs and a P value ≤0.05 in any one of the tumor subtypes. This reduced the list of 409 matrices to a final list of 44 (see Supplementary Table S3 at http://ajp.amjpathol.org). In Figure 3D these 44 original variables are superimposed on the PCA separation of the subtypes in the form of a biplot image. This projection provides a graphic illustration of how the 44 matrices discriminate the different tumor subtype signatures. For instance, AP1 is a dominating discriminating factor and pathway for the normal- like tumor subtype. The ERBB2+ signature projects in the direction of the NF-κB and E2F matrices. The basal-like subtype projects along various PAX matrices, whereas the luminal B subtype projects along the AHR/ARANT pathways. Finally luminal A appears to project in a direction bounded by STAF and NF1 pathways. As shown in Figure 3E, these 44 matrices produced a separation of breast cancer subtypes that is similar, although not identical, to the hierarchical clustering in Figure 2G.
Tumor Subtype-Specific Genes and Transcriptional Regulators Are Associated with Specific Regulatory Networks
To search for regulatory networks or pathways that appear most associated with the regulatory signatures found in the tumor subtype promoters, the genes of each respective subtype were combined with the transcription factor genes cognate for the most significantly enriched TFBSs within each subtype. This created a composite list of the regulated and regulator for each genetic signature.10 These enhanced lists were then used to interrogate the Ingenuity Knowledge database25 to construct regulatory networks based on curated interactions between the genes in each respective list.11 Figure 4 shows the five networks that were most populated by the five composite gene lists described above. The basal-like group is dominated by a highly connected Myc node. Luminal A is characterized by multiple hormone and orphan receptor nodes and, interestingly, luminal B contains a central hub with p53. The normal-like signature is dominated by AP-1 pathways and components. Most notably the ERBB2+ signature contains multiple nodes linking NF-κB, E2F, EGR, Forkhead, and SMAD pathways, and show many linkages to canonical pathways that regulate cell-cycle progression.
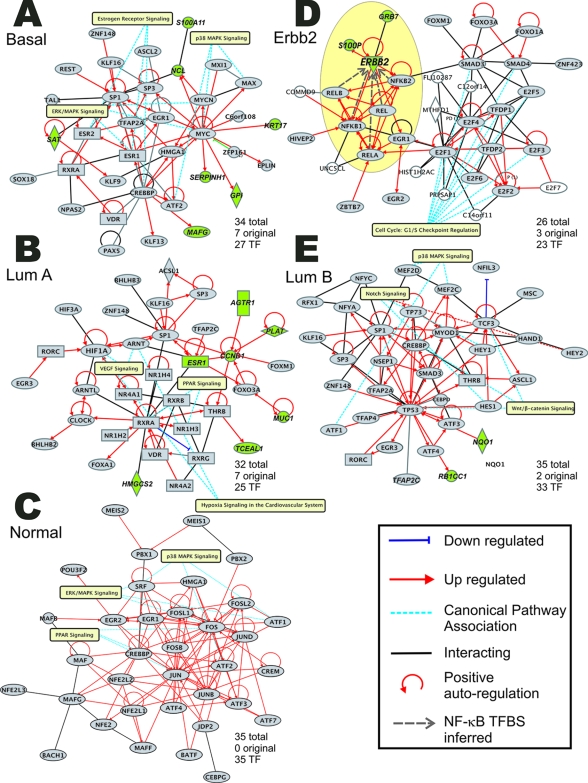
Figure 4.
Pathway analysis of composite list of expressed genes and enriched transcription factors implicates specific regulatory networks. The number of original genes is the number of genes in the network (highlighted in green) that were in the original gene list (excluding the genes encoding the transcription factors). The number of transcription factor genes is the number of genes in the network that encode significant transcription factors. The functions shown are the functions that are most significant for that network. A: Network derived from expressed genes and significant transcription factors in basal-like tumor subtype. B: Network derived from expressed genes and significant transcription factors in luminal subtype A. C: Network derived from expressed genes and significant transcription factors in normal-like breast tumor subtype. D: Network derived from expressed genes and significant transcription factors in ERBB2+ tumor subtype. Highlighted regions indicate feed forward autoregulatory loop inferred from the presence of NF-κB TFBS in the ERRB2 and GRB7 promoters (see Supplementary Figure S4 at http://ajp.amjpathol.org). E: Network derived from expressed genes and significant transcription factors in luminal subtype B.
The canonical regulatory pathways associated with the composite genetic and regulatory signatures of each subgroup are shown in Table 2. As demonstrated in Figure 4 the ERBB2+ signature is dominated by cell-cycle regulation pathways (Fisher exact t-test P = 9.6, E-13) and NF-κB-associated signaling (P = 4.27, E-03). The basal-like subtype composite signature is also enriched in cell-cycle regulatory pathways in addition to WNT/catenin signaling and endoplasmic reticulum/ER-pathways. Normal-like breast cancer subtype shows relative overpopulation with diverse cytokine- and growth factor-responsive pathways downstream of Jak/Stat and MAP kinase signaling circuitry. The luminal A tumor subtype signature shows a high association with pathways linked to metabolism of hydrophobic amino acids, VEGF and IGF-1 signaling, although IGF-1 signaling is overrepresented in three of the five signatures. Finally, the luminal B composite signature is characterized by sonic hedgehog signaling, notch signaling, sterol synthesis, and p38 MAPK and cell-cycle signaling. The latter two are also shared by the basal-like subtype signature.
Table 2.
Statistical Enrichment of Curated Canonical Pathways Based on the Ingenuity Database
| Canonical pathway | Significance |
|---|---|
| Erbb2 | |
| Cell cycle: G1/S checkpoint regulation | 9.68E-13 |
| Toll-like receptor signaling | 1.73E-04 |
| T-cell receptor signaling | 9.83E-04 |
| NF-κB signaling | 4.27E-03 |
| PPAR signaling | 7.87E-03 |
| P13K/AKT signaling | 1.82E-02 |
| B-cell receptor signaling | 3.55E-02 |
| Death receptor signaling | 4.23E-02 |
| TGF-β signaling | 5.51E-02 |
| IGF-1 signaling | 6.90E-02 |
| Basal-like | |
| Wnt/B-catenin signaling | 7.36E-04 |
| Endoplasmic reticulum stress pathway | 2.19E-03 |
| Cell cycle: G1/S checkpoint regulation | 4.39E-03 |
| p38 MAPK signaling | 1.51E-02 |
| Glycine, serine, and threonine metabolism | 3.96E-02 |
| Estrogen receptor signaling | 7.91E-02 |
| PPAR signaling | 8.31E-02 |
| Normal | |
| JAK/Stat signaling | 4.68E-09 |
| PDGF signaling | 3.61E-07 |
| EGF signaling | 8.45E-07 |
| PPAR signaling | 2.13E-05 |
| IL-2 signaling | 2.59E-05 |
| ERK/MAPK signaling | 8.51E-05 |
| p38 MAPK signaling | 2.23E-03 |
| IFG-1 signaling | 3.15E-03 |
| IL-6 signaling | 3.33E-03 |
| IL-4 signaling | 4.30E-03 |
| Luminal A | |
| Valine/leucine and isoleucine degradation | 1.28E-02 |
| Glutathione metabolism | 1.36E-02 |
| VEGF signaling | 1.68E-02 |
| IGF-1 signaling | 7.99E-02 |
| Luminal B | |
| p38 MAPK signaling | 2.95E-04 |
| Sonic Hedgehog signaling | 1.26E-03 |
| Notch signaling | 5.32E-03 |
| Cell cycle: G1/S checkpoint regulation | 8.76E-03 |
| Sterol biosynthesis | 1.85E-02 |
| TGF-β signaling | 2.07E-02 |
| Aminosugars metabolism | 3.61E-02 |
| Hypoxia signaling in the cardiovascular system | 7.21E-02 |
| FGF signaling | 9.31E-02 |
P values were generated by Fisher’s exact t-test (see Materials and Methods).
Inferred Identification of a Role for NF-κB and ERBB2 in a Self-Amplifying Regulatory Loop in Human Breast Cancer Cells
The selective enrichment of NF-κB TFBSs in the promoters of the ERBB2+ tumor subtype is consistent with observations that overexpression of ERBB2 is associated with increased activation of NF-κB.26,27,28,29,30 An intriguing aspect of this relationship is that in addition to ERRB2, the growth receptor adaptor protein, GRB7, a direct modulator of the ERBB2 receptor family31 (Figure 4D), is also a target of NF-κB pathways because both ERBB2 and GRB7 contain binding sites for NF-κB in their promoters (Table 1 and Supplementary Figure S4 at http://ajp.amjpathol.org). It is therefore very likely that these genes participate in a self-enhancing feed forward loop that amplifies NF-κB molecular signals driven by ERBB2 (see highlighted region in Figure 4D).
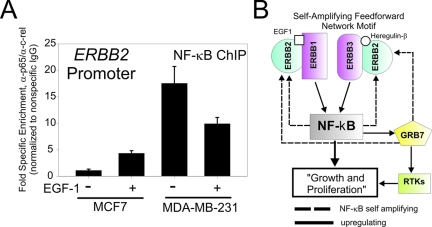
At high levels ERBB2 dimerizes with itself and becomes active in the absence of ligand.32 When present at physiological levels, ERBB2 readily heterodimerizes with all other members of the ERBB family, particularly EGFR, to produce ligand-specific complexes responsive to secreted EGF-1.33 To test whether or not an autoregulatory loop linking NF-κB to ERBB2 may exist in human breast cancer cells, we examined the in vivo association of NF-κB complexes with ERBB2 promoter before and after EGF-1 stimulation by chromatin immunoprecipitation (ChIP) in cells known to express normal (MCF-7) and amplified levels (MDA-MB-231) of ERBB2.33 An antibody cocktail containing affinity-purified antibodies specific for human p65/RelA and c-rel/Rel was used to perform ChIP in resting and EGF-1 stimulated MCF-7 and MDA-MB-231 breast cancer cells. As shown in Figure 5A, when normalized to nonspecific antibody and input DNA, there is a significant increase in NF-κB association with the ERBB2 promoter of MCF-7 cells after treatment with EGF-1. In contrast, NF-κB binding to ERRB2 in resting MDA-MB-231 is significantly higher than either resting or stimulated MCF-7. Moreover, the response to EGF-1 appears deregulated because the addition of EGF-1 fails to induce further NF-κB binding and instead shows some variable depression in MDA-MB-231. These novel data demonstrate that an autoregulatory loop, similar to what is schematically outlined in Figure 5B, exists in human breast cancer and is the first demonstration that NF-κB associates with the ERBB2 promoter in vivo in human breast-derived cells.
Figure 5.
NF-κB complexes associate with the ERBB2 promoter in vivo. A: Analysis of EGF-1-dependent association of NF-κB complexes with the ERBB2 promoter by chromatin immunoprecipitation in MCF-7 and the ERBB2 overexpressing cell line, MDA-MB-231, in the presence or absence of 12 ng/ml of EGF-1. Relative enrichment was determined by quantitative PCR and normalized to nonspecific IgG. Data represent the average of triplicate determinations and error bars show ±SEM. B: Schematic of NF-κB/ERBB feed-forward autoregulatory loop. ERBB1 = EGFR; ERBB2 = Neu/Her2; ERBB3 = Her3; RTK = receptor tyrosine kinases other than ERBB family. Dashed lines indicate interactions that have positive feedback or self-amplifying influence. Solid lines indicate regulatory influences that enhance expression or function.
Discussion
In this study we analyzed the promoter regions of genes representing specific genetic signatures of molecular subtypes of breast cancer that have been shown repeatedly in independent data sets to be predictive of clinical outcome. The analysis identified several transcriptional pathways and implicated multiple regulatory networks that characterize and classify the different molecular subtypes. Among the networks identified, two notable ones were found to be associated with molecular subtypes of breast cancer that predicted poor patient outcome. The first network, characterized by the ERBB2+ molecular subtype, was dominated by molecular signaling and possible cross-talking interactions between NF-κB, E2F, EGR1, and SMAD (TGF-β) transcriptional pathways. The second network, which was more highly associated with the basal-like molecular subtype of breast cancer, was dominated by PAX transcriptional circuitry. The implications of these inferred interactions forms the foundation for specific hypothesis generation with the goal of defining the underlying biology of these breast cancer subtypes and uncovering promising new therapeutic targets and prognostic molecular markers.
Defining functional promoter composition in complex organisms continues to be a daunting task.34,35,36 In the field of cancer research, a variety of approaches have been conceived and each has particular strengths and weaknesses.6,8,9 In this study we used position weight matrix scoring.37 A typical problem faced by this and many other similar promoter annotation approaches is the number of false positives generated in the analysis. To minimize this occurrence we chose to use a background model containing the promoter regions of 15,318 RefSeq genes to use as a reference for statistical enrichment assuming a Poisson distribution. This type of reference model has the advantage over using a reference of random DNA sequences in that it reflects and maintains the natural bias toward GC richness that exists in many promoter regions of the human genome.38,39,40,41 Thus GC-rich TFBS are not inappropriately overrepresented. The robustness of the enrichment analysis is illustrated by our postanalysis permutation testing which indicates that our significance scoring is conservative (Supplementary Figure S1 at http://ajp.amjpathol.org). A recent very interesting reexamination of human promoters suggests that mammalian promoters can be classified into four categories characterized by the GC content upstream and downstream of the transcription start site (combination of high or low GC content downstream or upstream of the transcription start site).41 By this classification all five genetic signatures analyzed in this study are GC-rich (>55%) upstream and downstream of the transcription start site (class A, according to Bajic and colleagues41) (see Supplementary Figure S2 at http://ajp.amjpathol.org). Thus the differences in promoter composition identified in this study are more likely to reflect true biology rather than asymmetric fluctuations in GC content.
Another significant feature of position-weighted TFBS matrices that hinders even the most focused promoter analysis is their inherent redundancy and degenerate nature. This is a property that is not readily handled within the limitations of the separation provided by hierarchical clustering. Although this feature insures the ability to detect subtle differences, it has the negative result of adding significant noise to any multivariate analysis. We used the method of PCA to address some of these flaws by reducing high dimensional variables into fewer dimensions that explain the most characteristic features or trends in the data sets.10,11,13,42 The noise reduction provided by this transformation had the net effect of limiting the negative contribution of the more degenerate and redundant matrices. In this way we feel we increased the likelihood that observed correlations will reflect true linkages with more informative biological significance.
When the results of hierarchical clustering and PCA are compared, the results were similar although not identical in several aspects. Each approach indicated that the basal-like and lumen B-like subtypes are more similar to each other than the other three suggesting common regulatory phenotype for these two subtypes. There was more variability for the relationship between the ERBB2+ subtype versus the normal-like and lumen A subtype. Interestingly, when top binding sites clustered to each subgroup by hierarchical clustering in Figure 2 was compared to the most discriminating TFBS motifs by PCA in Figure 3E, the agreement was 60% for the normal subtype, 56% for ERBB2+, 82% for luminal B, 86% for the basal-like subtype, and only 7% for the lumen A. The reason for major discrepancy between the two techniques for luminal A-type annotation is not clear. However, it may arise from the greater bias of the PCA analysis to strong enrichment signals because most of the TFBSs scored as PC loading discriminators are the most highly enriched TFBS in the group. It would be interesting in the future to compare different thresholds used in the analysis of the PCA and for position weight matrix scoring. In the current analysis we arbitrarily chose the default threshold cutoff of 0.75 for position weight matrix and we used 0.75 for the PC coefficients because these thresholds have performed well as discriminators in prior studies.10,11
One of the most compelling aspects of this study is the correct identification and subsequent empirical validation of a autoregulatory linkage of NF-κB pathways to ER-negative/ERBB2-positive breast cancer (Figures 2 to 5). Since its initial identification several years ago, the functional interaction of NF-κB pathways in ER-negative/ERBB2-positive breast cancer has been extensively examined.26,27,28,29,30 Now it is widely recognized that NF-κB plays a role in a variety of different human cancers.43 ERBB2 is a member of the ERBB superfamily of receptor tyrosine kinases (RTKs) that mediate growth signaling in many different cellular lineages. It is overexpressed in more than 20% of invasive breast cancers and a founding feature of the ERBB2+ tumor subtype signature that is associated with poor prognosis.44 There are four members of the ERBB family including epidermal growth factor (EGFR/ERBB1), ERBB2 (NEU/HER2), ERBB3 (HER3), and ERBB4.33 Feed forward loops are important network motifs that can act as biological switches to render cellular processes more sensitive to sustained, rather than transient stimuli.45,46 This is an essential property of the events that control epithelial growth and differentiation. This is particularly important because co-expression of EGFR (ERBB1) and ERBB2 is found in greater than 10% of patients with breast cancer and carries a poorer prognosis than elevated ERBB2 expression alone.47,48 Heterodimers between ERBB2 and ERBB3 are believed to be the most biologically active and tumor-promoting forms.32 Both the hetero- and homodimerization of ERBB2+ are thought to play a significant role in the activation of NF-κB protein complexes and the evolution of breast cancer.49,50 The identification of a feed forward network motif that drives signaling from the ERBB receptor network through NF-κB has important therapeutic and prognostic implications. As suggested in the schematic illustration in Figures 4D and 5B, NF-κB functions as a hub, connecting and self-amplifying ligand-dependent signals emanating from different combinations of the ligand-bound ERBB2 family receptors. The involvement of GRB7 indicates that this feed forward loop will influence the activity of many other growth factor receptors including c-Kit, PDGFR (platelet-derived growth factor receptor), and insulin receptor.31
The fact that multiple ERBB2 ligands and receptor dimers influence NF-κB signaling highlights the importance of ligand-receptor interactions in the pathophysiology of breast cancer and emphasizes an area important for therapeutic intervention. ERBB2 is targeted therapeutically in breast cancer patients by humanized anti-ERBB2 antibodies such as Herceptin.51 The interaction of the ERBB and NF-κB networks, inferred from our study, provides a rationale for the design of combinatorial therapeutic strategies that will simultaneously target the NF-κB, EGFR, and ERBB2 components in this regulatory network. A representative example would be the combination of agents such as Velcade (which targets NF-κB52), Herceptin (which targets ERBB2), and Iressa (which targets EGFR53) as a therapeutic regimen. It should be noted that while this work was in review, a report describing the combinatorial use of Velcade and Herceptin in a preclinical study was published.54 As we predicted, the combination of these compounds showed significant synergy against ERBB2+ tumors. It reasonable then to assume that one of the major underlying mechanisms for this synergy is the multicomponent disruption of the tumor’s addiction to reinforced signaling through NF-κB.
EGR1 was also inferred as an interacting member of the ERBB2+ regulatory signature (Figures 2, 4, and 5). EGR1 has been previously shown to play a major role in cell growth, differentiation, survival, and transformation of other epithelial tumors.55 Recent studies suggest that EGR1 regulates EGFR expression.56 Therefore it is possible that EGR1 may participate in a second self-amplifying feed forward response in the regulation of ERBB2 networks in breast cancer. The EGR family members of the ERBB2+ tumor subtype regulatory signature may therefore be therapeutic targets in this form of breast cancer.
A previously unrecognized regulatory limb in the ERBB2+ transcriptional network is the TGF-β-regulated pathway (Figure 2, 4, and 5). The SMAD transcription factors are major effectors of TGF-β signaling.57 Reports of a cross talk between NF-κB and TGF-β signaling had previously been described, but with conflicting outcomes. In some cases the cross-TGF-β signaling was inhibitory for NF-κB signaling58 whereas in others it was stimulatory.59 The precise role of the interaction between NF-κB and TGF-β in breast cancer will require further investigation. It is tempting to speculate that the tumor microenvironment may have a role in determining the influence TGF-β with inflammatory components within the tumor microenvironment possibly by producing interactions between NF-κB and TGF-β signaling that may act in synergy to promote more aggressive phenotypes.
Like ERBB2+, the basal-like molecular signature is also associated with a poor prognosis. The basal-like subtype is uniquely enriched with transcription factors linked to development and differentiation including Sox and several Pax transcription factors.21,60 Recent phenotypic characterization of basal-like tumors by immunohistochemistry indicates that they are typically negative for ERBB2 and ER with infrequent expression of myoepithelial markers.61 The lack of myoepithelial markers is contrary to their presumed basal-like cell origin.3 Other possibilities for the origin of these cells include epithelial-to-mesenchymal transition or derivation from breast epithelial stem cells.62,63 The high enrichment of the basal-like regulatory signature with binding sites for factors that control differentiation and development would argue in favor of either of these possibilities. A curious observation in the basal-like group is the enrichment for ER-binding sites and a marginal enrichment for p53-binding sites. The fact that both of these genes are mutated or absent in this tumor subtype3 suggests that several of the genes responsible for the basal-like molecular phenotype may be potential targets for repression by intact ER and p53 signaling. Given the regulatory similarity between the ER-positive, lumen B molecular signature, and the basal-like molecular signature, it would be reasonable to speculate that the basal-like phenotype could have originated from a more luminal B-like phenotype after loss of ER expression and mutation of p53. Similar to the insights gained from the analysis of the ERBB2+ tumor subtype, these inferences could be important for future pharmacological and gene therapeutic strategies that specifically target these types of breast cancers.
Finally, it must be stressed that this study does not use TFBSs as predictors of outcomes. It is more or less an annotation of the most enriched binding sites from gene sets that have been previously defined as predictive. These annotations are then used to define pathways that are associated with the signatures. Thus the flaws of this approach will be no fewer than that of the original study. One drawback that must be considered is that the original gene expression study is derived from samples of tumor that may be a mixture of many different tissue types including stromal elements and inflammatory components. Therefore the pathway inference interpretation should be approached with appropriate caution or expanded to consider that the pathway inferences could represent a composite signature of both the tumor and the tumor microenvironment. When comparable expression data from microdissected tissue samples become available it will be of interest to perform a similar analysis.
Acknowledgments
We thank the National Institutes of Health Fellow Editorial Board for editorial assistance in preparation of the manuscript.
Footnotes
Address reprint requests to Kevin Gardner, 41 Library Dr., Bldg 41/D305, Bethesda, MD 20892-5065. E-mail: gardnerk@mail.nih.gov.
Supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
G.I. and S.H.N. contributed equally to this study.
Supplemental material for this article can be found on http://ajp. amjpathol.org.
References
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein LP, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet. 2005;37:S38–S45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37:S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- McNutt MC, Tongbai R, Cui W, Collins I, Freebern WJ, Montano I, Haggerty CM, Chandramouli GV, Gardner K. Human promoter genomic composition demonstrates non-random groupings that reflect general cellular function. BMC Bioinformatics. 2005;6:259. doi: 10.1186/1471-2105-6-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freebern WJ, Haggerty CM, Montano I, McNutt MC, Collins I, Graham A, Chandramouli GV, Stewart DH, Biebuyck HA, Taub DD, Gardner K. Pharmacologic profiling of transcriptional targets deciphers promoter logic. Pharmacogenomics J. 2005;5:305–323. doi: 10.1038/sj.tpj.6500325. [DOI] [PubMed] [Google Scholar]
- Idelman G, Taylor JG, Tongbai R, Chen RA, Haggerty CM, Bilke S, Chanock SJ, Gardner K. Functional profiling of uncommon VCAM1 promoter polymorphisms prevalent in African American populations. Hum Mutat. 2007;28:824–829. doi: 10.1002/humu.20523. [DOI] [PubMed] [Google Scholar]
- Smith JL, Collins I, Chandramouli GV, Butscher WG, Zaitseva E, Freebern WJ, Haggerty CM, Doseeva V, Gardner K. Targeting combinatorial transcriptional complex assembly at specific modules within the IL-2 promoter by the immunosuppressant SB203580. J Biol Chem. 2003;278:41034–41046. doi: 10.1074/jbc.M305615200. [DOI] [PubMed] [Google Scholar]
- Muggerud AA, Johnsen H, Barnes DA, Steel A, Lonning PE, Naume B, Sorlie T, Borresen-Dale AL. Evaluation of MetriGenix custom 4D arrays applied for detection of breast cancer subtypes. BMC Cancer. 2006;6:59. doi: 10.1186/1471-2407-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death—a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer. 2005;41:2403–2414. doi: 10.1016/j.ejca.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Arteaga CL. Inhibition of TGFbeta signaling in cancer therapy. Curr Opin Genet Dev. 2006;16:30–37. doi: 10.1016/j.gde.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Wormke M, Samudio I. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J Mammary Gland Biol Neoplasia. 2000;5:295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- Hsu HC, Zhou T, Mountz JD. Nur77 family of nuclear hormone receptors. Curr Drug Targets Inflamm Allergy. 2004;3:413–423. doi: 10.2174/1568010042634523. [DOI] [PubMed] [Google Scholar]
- Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Persons DL, Borelli KA, Hsu PH. Quantitation of HER-2/neu and c-myc gene amplification in breast carcinoma using fluorescence in situ hybridization. Mod Pathol. 1997;10:720–727. [PubMed] [Google Scholar]
- Chiarugi V, Del Rosso M, Magnelli L. Brn-3a, a neuronal transcription factor of the POU gene family: indications for its involvement in cancer and angiogenesis. Mol Biotechnol. 2002;22:123–127. doi: 10.1385/MB:22:2:123. [DOI] [PubMed] [Google Scholar]
- Smith JL, Freebern WJ, Collins I, De Siervi A, Montano I, Haggerty CM, McNutt MC, Butscher WG, Dzekunova I, Petersen DW, Kawasaki E, Merchant JL, Gardner K. Kinetic profiles of p300 occupancy in vivo predict common features of promoter structure and coactivator recruitment. Proc Natl Acad Sci USA. 2004;101:11554–11559. doi: 10.1073/pnas.0402156101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil P, Iyer LK, Adelstein SJ, Kassis AI. A combined approach to data mining of textual and structured data to identify cancer-related targets. BMC Bioinformatics. 2006;7:354. doi: 10.1186/1471-2105-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianetti S, Arsura M, Romieu-Mourez R, Coffey RJ, Sonenshein GE. Her-2/neu overexpression induces NF-kappaB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IkappaB-alpha that can be inhibited by the tumor suppressor PTEN. Oncogene. 2001;20:1287–1299. doi: 10.1038/sj.onc.1204257. [DOI] [PubMed] [Google Scholar]
- Romieu-Mourez R, Landesman-Bollag E, Seldin DC, Traish AM, Mercurio F, Sonenshein GE. Roles of IKK kinases and protein kinase CK2 in activation of nuclear factor-kappaB in breast cancer. Cancer Res. 2001;61:3810–3818. [PubMed] [Google Scholar]
- Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor kappa B activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–8547. doi: 10.1073/pnas.97.15.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, Iglehart JD. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci USA. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas DK, Dai SC, Cruz A, Weiser B, Graner E, Pardee AB. The nuclear factor kappa B (NF-kappa B): a potential therapeutic target for estrogen receptor negative breast cancers. Proc Natl Acad Sci USA. 2001;98:10386–10391. doi: 10.1073/pnas.151257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DC, Shen TL, Guan JL. The Grb7 family proteins: structure, interactions with other signaling molecules and potential cellular functions. Oncogene. 2001;20:6315–6321. doi: 10.1038/sj.onc.1204775. [DOI] [PubMed] [Google Scholar]
- Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Michelson AM. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc Natl Acad Sci USA. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, Makeev VJ, Mironov AA, Noble WS, Pavesi G, Pesole G, Regnier M, Simonis N, Sinha S, Thijs G, van Helden J, Vandenbogaert M, Weng Z, Workman C, Ye C, Zhu Z. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Yamashita R, Suzuki Y, Sugano S, Nakai K. Genome-wide analysis reveals strong correlation between CpG islands with nearby transcription start sites of genes and their tissue specificity. Gene. 2005;350:129–136. doi: 10.1016/j.gene.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic VB, Tan SL, Christoffels A, Schonbach C, Lipovich L, Yang L, Hofmann O, Kruger A, Hide W, Kai C, Kawai J, Hume DA, Carninci P, Hayashizaki Y. Mice and men: their promoter properties. PLoS Genet. 2006;2:e54. doi: 10.1371/journal.pgen.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Karin M. NF-kappaB in cancer: a marked target. Semin Cancer Biol. 2003;13:107–114. doi: 10.1016/s1044-579x(02)00128-1. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Milo R, Itzkovitz S, Kashtan N, Levitt R, Shen-Orr S, Ayzenshtat I, Sheffer M, Alon U. Superfamilies of evolved and designed networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol. 2002;196:17–25. doi: 10.1002/path.1003. [DOI] [PubMed] [Google Scholar]
- DiGiovanna MP, Stern DF, Edgerton SM, Whalen SG, Moore D, Thor AD. Relationship of epidermal growth factor receptor expression to ErbB-2 signaling activity and prognosis in breast cancer patients. J Clin Oncol. 2005;23:1152–1160. doi: 10.1200/JCO.2005.09.055. [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Sweeney CJ, Nakshatri H. Identification of signal transduction pathways involved in constitutive NF-kappaB activation in breast cancer cells. Oncogene. 2002;21:2066–2078. doi: 10.1038/sj.onc.1205243. [DOI] [PubMed] [Google Scholar]
- Chen D, Xu LG, Chen L, Li L, Zhai Z, Shu HB. NIK is a component of the EGF/heregulin receptor signaling complexes. Oncogene. 2003;22:4348–4355. doi: 10.1038/sj.onc.1206532. [DOI] [PubMed] [Google Scholar]
- Nahta R, Esteva FJ. Herceptin: mechanisms of action and resistance. Cancer Lett. 2006;232:123–138. doi: 10.1016/j.canlet.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- Siegel-Lakhai WS, Beijnen JH, Schellens JH. Current knowledge and future directions of the selective epidermal growth factor receptor inhibitors erlotinib (Tarceva) and gefitinib (Iressa). Oncologist. 2005;10:579–589. doi: 10.1634/theoncologist.10-8-579. [DOI] [PubMed] [Google Scholar]
- Cardoso F, Durbecq V, Laes JF, Badran B, Lagneaux L, Bex F, Desmedt C, Willard-Gallo K, Ross JS, Burny A, Piccart M, Sotiriou C. Bortezomib (PS-341, Velcade) increases the efficacy of trastuzumab (Herceptin) in HER-2-positive breast cancer cells in a synergistic manner. Mol Cancer Ther. 2006;5:3042–3051. doi: 10.1158/1535-7163.MCT-06-0104. [DOI] [PubMed] [Google Scholar]
- Baron V, De Gregorio G, Krones-Herzig A, Virolle T, Calogero A, Urcis R, Mercola D. Inhibition of Egr-1 expression reverses transformation of prostate cancer cells in vitro and in vivo. Oncogene. 2003;22:4194–4204. doi: 10.1038/sj.onc.1206560. [DOI] [PubMed] [Google Scholar]
- Chen A, Xu J, Johnson AC. Curcumin inhibits human colon cancer cell growth by suppressing gene expression of epidermal growth factor receptor through reducing the activity of the transcription factor Egr-1. Oncogene. 2006;25:278–287. doi: 10.1038/sj.onc.1209019. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Bottinger EP. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Shigemori N, Hasegawa K, Sugita T. TGF-beta-activated kinase 1 stimulates NF-kappa B activation by an NF-kappa B-inducing kinase-independent mechanism. Biochem Biophys Res Commun. 1998;243:545–549. doi: 10.1006/bbrc.1998.8124. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Semin Cell Dev Biol. 2005;16:694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Ponti D, Zaffaroni N, Capelli C, Daidone MG. Breast cancer stem cells: an overview. Eur J Cancer. 2006;42:1219–1224. doi: 10.1016/j.ejca.2006.01.031. [DOI] [PubMed] [Google Scholar]