Abstract
Proteins able to participate in unrelated biological processes have been grouped under the generic name of moonlighting proteins. Work with different yeast species has uncovered a great number of moonlighting proteins and shown their importance for adequate functioning of the yeast cell. Moonlighting activities in yeasts include such diverse functions as control of gene expression, organelle assembly, and modification of the activity of metabolic pathways. In this review, we consider several well-studied moonlighting proteins in different yeast species, paying attention to the experimental approaches used to identify them and the evidence that supports their participation in the unexpected function. Usually, moonlighting activities have been uncovered unexpectedly, and up to now, no satisfactory way to predict moonlighting activities has been found. Among the well-characterized moonlighting proteins in yeasts, enzymes from the glycolytic pathway appear to be prominent. For some cases, it is shown that despite close phylogenetic relationships, moonlighting activities are not necessarily conserved among yeast species. Organisms may utilize moonlighting to add a new layer of regulation to conventional regulatory networks. The existence of this type of proteins in yeasts should be taken into account when designing mutant screens or in attempts to model or modify yeast metabolism.
INTRODUCTION
During a study of sugar fermentation by yeasts, Emil Fischer enunciated a principle that has had a lasting impact on biochemistry. His famous statement “… to use a metaphor, I would like to say that enzyme and glucoside have to fit together like lock and key in order to exert a chemical effect on each other” (49) deeply shaped the concept of proteins as monolithic entities able to perform a single strictly determined function. As knowledge about proteins progressed, new results appeared showing that enzymes were able to carry out more than one function. The discovery of allosterism revealed that certain proteins could also detect signals and modify their behavior accordingly, thereby becoming the conductors of the cellular orchestra (103). In addition, it was found that some proteins were endowed with more than one catalytic function in a single polypeptide chain (80). These multifunctional proteins are widespread among organisms ranging from bacteria to mammals (12, 22, 29, 68, 108), and usually their different activities remain limited to the realm of metabolism. In many cases, it has been demonstrated that the dual function resulted from the fusion of two genes that initially encoded proteins with single functions (12, 22, 29). Later on, new results uncovered the surprising fact that a significant number of proteins could perform apparently disparate functions; for example, phosphoglucose isomerase can act as an autocrine motility factor (58), aconitase can act as an iron-responsive element binding protein (79) or as a determinant of mitochondrial DNA stability (27), and glyceraldehyde-3-phosphate dehydrogenase participates in apoptosis and neurodegeneration (125). Although sensu stricto this behavior qualifies those proteins as multifunctional, the great difference in the type of functions performed sets them apart from conventional multifunctional proteins. To designate them as a group, Jeffery (71) coined the term “moonlighting proteins.” According to The Compact Oxford English Dictionary, one meaning of moonlighting is “to do [paid] work, [usually at night], in addition to one's regular employment,” and the word has been used in this sense since the late 1950s (129); therefore, “moonlighting proteins” is a quite appropriate designation for the collective. As indicated above, moonlighting activities of one protein are usually unrelated to its metabolic function and show great diversity; metabolic enzymes may double as transcription factors, participate in assembly or degradation of organelles, or contribute to maintenance of mitochondrial DNA, among others. As originally defined, the category of moonlighting proteins excludes proteins whose different functions are due to gene fusion or are consequences of splice variants as well as those proteins that are located in different subcellular compartments but perform the same function in each of them (70, 71).
Although moonlighting proteins are present in different organisms, more attention has been given to those found in higher eukaryotes, perhaps due to the implications that this multifunctionality may have on the interpretation of certain single-gene disorders whose symptoms or phenotype does not directly correlate with that expected from the relevant genotype (132). For lower eukaryotes, such as yeasts, several well-documented cases of moonlighting proteins exist; however, no systematic consideration has been given to this phenomenon. The genetic tractability of yeasts allows the creation and utilization of extensive sets of mutants, which facilitate an in-depth study of each moonlighting protein to relate structure and function for each displayed activity. This contrasts with the situation in higher eukaryotes, where this approach remains much more difficult.
In this review, we discuss in detail several instances of moonlighting proteins in different yeast species and examine the experimental evidence available to justify their inclusion in this category. Another goal of this review is to bring to the attention of the zymological community the importance of this phenomenon and its wide implications for both basic and applied research.
METABOLIC PROTEINS THAT PARTICIPATE IN GENE TRANSCRIPTION
Galactokinase and Transcription of the GAL Genes in Kluyveromyces lactis and Saccharomyces cerevisiae
Yeasts belonging to different genera use galactose as a carbon and energy source (10). Utilization of this sugar requires induction of the GAL genes, which encode a transporter and three intracellular enzymes that metabolize the sugar up to the glycolytic intermediate glucose-6-phosphate, which enters the common glycolytic trunk. One of these proteins, galactokinase, moonlights as a regulator of the transcription of the GAL genes. Although the moonlighting action of galactokinase occurs basically in Kluyveromyces lactis, we first consider the situation in Saccharomyces cerevisiae as a basis to understand the control mechanisms of the expression of those genes. The expression of the GAL genes is both induced by galactose and repressed by glucose (53, 73), but for the purposes of this review, only the induction process is relevant. Basically, the induction of the GAL genes is determined by the interplay of two proteins with opposite roles, the transcriptional activator Gal4 and its inhibitor, Gal80. In the presence of galactose, the inhibitory action of Gal80 is relieved and transcription of the GAL genes occurs, while in its absence, Gal80 prevents the interaction of the activation domain of Gal4 with the transcription machinery. Release of Gal4 inhibition by Gal80 is facilitated by the signal-transducing protein Gal3 (for reviews, see references 19 and 122). This 520-amino-acid protein exhibits great sequence similarity with galactokinase (9, 116), the first enzyme of the intracellular galactose utilization pathway, which phosphorylates galactose to galactose-1-P, encoded by GAL1. Gal3 has no detectable galactokinase activity (9), but insertion of two amino acids at position 164 in its sequence confers this activity on the protein (116). It has been found that Gal3 interacts with Gal80 (18), and this interaction is stabilized in the presence of galactose and ATP (150). Using an in vitro transcription system containing Gal80 and Gal4, Platt and Reece (115) found that the addition of Gal3 in the presence of galactose promoted transcription of the adenovirus E4 gene engineered with several tandem repeats of GAL4 binding sites upstream of its TATA box. In the same assay, it was found that galactokinase could substitute for Gal3, although in a much less effective manner, and that its mechanism of action appears to be the same as that of Gal3 (116). This finding is in line with in vivo results that showed that overexpression of GAL1 suppressed the phenotype of gal3 mutants (20). These mutants initiate growth in galactose several days after galactose addition, while a wild-type strain starts it within a few hours after addition of the sugar; this is the “long term adaptation” to galactose described by Winge and Roberts as early as 1948 (148). Therefore, under certain conditions, galactokinase may moonlight in S. cerevisiae as a protein that regulates the transcription of the GAL genes.
In K. lactis, the proteins KlGal4 and KlGal80 play roles in the induction of the KlGAL genes equivalent to those described for their homologs in S. cerevisiae (122, 154). However, no mutants with a phenotype like that of S. cerevisiae gal3 mutants have been found for this yeast (100). Interestingly, induction of transcription of the GAL genes requires expression of KlGAL1, but the sugar-phosphorylating activity of galactokinase was not needed to release the inhibitory action of KlGal80 on KlGal4 (100). In a Klgal80 mutant, the induction of the KlGAL genes occurred in the absence of KlGal1, and therefore a role for KlGal1 in the release of KlGal80 inhibition was suspected (153). Since KlGal1 complements an S. cerevisiae gal3 mutant (100), a plausible hypothesis is that a direct interaction between KlGal1 and KlGal80 could be the mechanism to release the inhibition of transcription caused by KlGal80. Zenke et al. (153) obtained experimental evidence for the formation of a complex between a tagged KlGal80 protein and KlGal1, using immunoblot analysis, and showed that the formation of the KlGal80-KlGal1 complex required galactose and ATP. Further support for the physiological importance of the KlGal1-KlGal80 interaction was provided by a study done with the following sets of mutant forms of KlGal1: mutants with kinase activity but unable to release KlGal80 inhibition, mutants without kinase activity but able to release the inhibition, or mutants lacking both functions. In vitro analysis of the interaction showed that KlGal1 from those mutants unable to release KlGal80 inhibition did not show interaction between KlGal1 and KlGal80, while the mutants without kinase activity but able to release inhibition did (143, 153). Using two-hybrid assays of the interaction, a similar result was obtained (143, 153). Thus, one of the roles of KlGal1 in K. lactis is functionally equivalent to the one performed by Gal3 in S. cerevisiae.
An interesting point in the regulation of GAL gene transcription is the difference in the subcellular compartment where the interaction between the regulatory proteins occurs: in S. cerevisiae, Gal3 and Gal80 interact in the cytosol (113, 114), whereas in K. lactis the interaction between KlGal80 and KlGal1 occurs in the nucleus (4).
Hexokinase and Glucose Catabolite Repression in S. cerevisiae
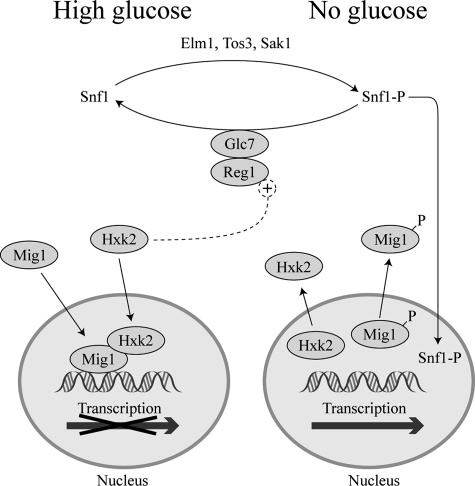
S. cerevisiae possesses three glucose-phosphorylating enzymes, namely, two hexokinases and a glucokinase. However, during growth in glucose, hexokinase 2 (Hxk2) is the predominantly expressed hexokinase and is therefore responsible for the initiation of the intracellular metabolism of glucose (61). Glucose causes repression of transcription of numerous genes in yeast, in a complex process called catabolite repression in which a plethora of factors and signals interact (16, 25, 53, 72). It has long been known that in hxk2 mutants the catabolite repression of some genes, such as SUC2, encoding invertase, or the GAL genes, encoding enzymes necessary for galactose metabolism, is abolished (46, 155). Several results indicate that the hexokinase protein itself participates in the regulatory circuitry of S. cerevisiae. Assays of enzymatic activity in isolated nuclei, immunoblotting, and the fluorescence distribution of a Hxk2-green fluorescent protein fusion have shown unequivocally that in yeast growing in 2 to 4% glucose, about 15% Hxk2 is localized in the nucleus (62, 119). In addition, the decapeptide Lys6-Met15 of Hxk2, dispensable for its enzymatic activity, is critically important for the enzyme to be localized in the nucleus and to mediate repression of SUC2, HXK1, or GLK1 (62, 121). Using two-hybrid assays, chromatin immunoprecipitation, and mobility shift analyses, it has also been demonstrated that Hxk2 interacts directly with the transcriptional repressor Mig1, a C2H2 zinc finger protein that binds to the promoters of most glucose-repressible genes, and that the Lys6-Met15 decapeptide of Hxk2 is needed for this interaction (1). The binding of Mig1 to Hxk2 is required for retention of Hxk2 within the nucleus; in fact, there is a correlation between the level of Mig1 and the amount of Hxk2 located in the nucleus (1). The nuclear localization of Hxk2 is glucose dependent (1), which may be explained by the requirement of glucose for nuclear localization of Mig1 (37). When glucose is low or absent, Mig1 is phosphorylated by the active form of the protein kinase Snf1, a protein required for derepression of glucose-repressed genes, and this phosphorylation abolishes the repressive capacity of Mig1 (131, 136). Phosphorylation occurs at Ser311, a residue that is critical for the interaction with Hxk2 (2). Once phosphorylated, Mig1 is exported to the cytosol, Hxk2 does not enter the nucleus, and repression is relieved (Fig. 1). In high glucose, Hxk2 and Mig1 interact in the nucleus, suggesting that an important role of Hxk2 in glucose repression may be to block phosphorylation of Mig1 by possible active molecules of the protein kinase Snf1, thus maintaining its repressing capacity (1, 2). The fact that Snf1 interacts with Hxk2 at both high and low glucose concentrations, while it interacts with Mig1 only at low concentrations, is compatible with the previous hypothesis.
FIG. 1.
Simplified scheme of the role of Hxk2 in catabolite repression. In the presence of high glucose, a fraction of Hxk2 enters the nucleus and binds to the repressor protein Mig1. Hxk2 hinders the phosphorylation of Mig1 by any active Snf1 that may be present and allows Mig1 to exert its repressive effect. Hxk2 also interacts with Reg1, and this interaction facilitates the action of the phosphatase Glc7 on Snf1 to maintain it in its inactive, dephosphorylated form; this, in turn, contributes to a low level of phosphorylation of Mig1. In the absence of glucose, Snf1 is phosphorylated by any of the kinases Elm1, Tos3, and Sak1; in this form, it phosphorylates Mig1, thus maintaining it and Hxk2 in the cytosol. For the sake of simplicity, other proteins that form complexes with Snf1, Mig1, or Hxk2 are not represented.
The interaction between Hxk2 and Mig1 could also be important to avoid cross talk between different signaling pathways, as it has been observed that in sodium-stressed cells growing in the presence of high glucose, Snf1 is activated but does not phosphorylate Mig1 (94).
Hxk2 also appears to be implicated in the regulation of the phosphorylation state of Snf1 through its interaction with Reg1, the regulatory subunit of the phosphatase Glc7. The precise mechanism of action of Hxk2 in this regulation is not completely elucidated. In the absence of Hxk2, Reg1 is dephosphorylated by Glc7, and in this form, Reg1 is unable to target Glc7 for dephosphorylation of Snf1 (124). Therefore, in an hxk2 mutant, Snf1 may remain active in its phosphorylated state even in the presence of glucose. de la Cera et al. (36) also showed an interaction between Hxk2 and the protein Med8, a protein of the Srb/mediator complex that binds to regulatory regions in the promoter of the SUC2 gene and in the coding region of HXK2 (26, 104, 111). This interaction may reinforce repression of transcription by hindering the activation of RNA polymerase.
It is therefore clear that the protein Hxk2 in S. cerevisiae is an element of the regulatory circuit of glucose catabolite repression for some genes. An additional effect of its metabolic activity may also be important in this process (89).
The Pleiotropic Transcriptional Regulator Arg82 Is an Inositol Phosphokinase in S. cerevisiae
The story of the moonlighting function of Arg82 differs from the usual path of discovery of this phenomenon: most often a well-characterized metabolic enzyme is at some time discovered to act in an unrelated way, but the contrary happened with Arg82. Using classical mutagenesis to generate mutations affecting the expression of enzymes related to arginine metabolism in S. cerevisiae, Bechet et al. (13) described one mutationargRIII, termed ArgRIII, that caused a loss of repression of biosynthetic enzymes and a lack of induction of the degradative ones by arginine. Further studies showed that a genomic disruption of the gene encoding ArgRIII (renamed Arg82) caused multiple problems for the cell, including defects in growth at 37°C, sterility in a MATα background, vacuolar fragmentation, decreases in transcription of both MATα- and MATa-specific genes, and defective sporulation when the disruption was homozygous in diploids (40). Work of the Messenguy and Dubois group showed that the coordinated regulation of the genes involved in arginine biosynthesis and degradation requires different proteins that form a complex and interact with specific sequences present in the arginine-regulated genes. Among these proteins were the MADS box proteins Arg80 and Mcm1 (96, 97). Arg82 is localized in the nucleus (17) and interacts with Mcm1 (44); since an arg82 mutant showed low levels of Mcm1 due to a decreased stability of the protein, it was concluded that the interaction of Arg82 with Mcm1 stabilizes this protein (44).
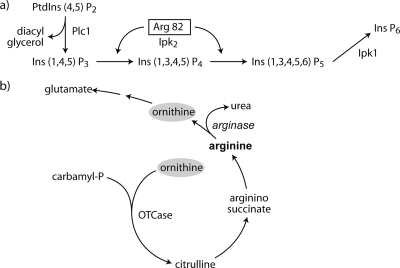
Two different groups reported a finding that placed Arg82 in a wider perspective. Saiardi et al. (123), searching for the gene encoding human inositol-P-6 kinase, defined a putative consensus sequence for inositol polyphosphate kinases and found it in the yeast proteins Arg82 and Kcs1. They purified a glutathione S-transferase (GST)-Arg82 fusion protein and showed that it possessed inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] and Ins(1,3,4,5)P4 kinase activity. Meanwhile, Odom et al. (106), studying the signaling roles of inositol polyphosphates and also using sequence alignments and enzymatic assays, found a similar result and renamed Arg82 Ipk2 (Fig. 2). The identification of Arg82 as a protein that produces inositol polyphosphates raised the question of their role in the regulation of transcription of the genes involved in arginine metabolism. The complex between Arg80, Arg82, Mcm1, and a DNA-specific sequence present in the promoter of ARG5,6 was not formed in vitro when extracts of an arg82 deletion mutant were used. Reintroduction of the ARG82 gene to the mutant restored the ability of the extracts to form the complex, confirming the necessity of the Arg82 protein for this function (106). However, its kinase activity was not necessary, as shown by the formation of the complex with a mutated form of Arg82 that lacked that activity (106). Since extracts from a plc1 mutant that lacks phospholipase C activity and therefore does not produce Ins(1,4,5)P3, the substrate for Ipk2/Arg82, could form the complex, it was concluded that inositol phosphates were not necessary for its formation (106). However, it could be possible that the complexes formed in the absence of inositol phosphates were inactive. If this were the case, mutants with these complexes would not be able to grow in a medium with arginine or ornithine as the sole nitrogen source because the arginine catabolic genes would not be induced. This was what Odom et al. (106) found when the growth of arg82 or plc1 mutants was tested on ornithine; they concluded that inositol phosphates were dispensable for the formation of the regulatory complexes but were required for them to be transcriptionally active. However, Dubois et al. (39) disputed this conclusion, arguing that it was based only on growth data for ornithine medium. They reported that a plc1 mutant grew as poorly with ammonium as a nitrogen source as with ornithine, indicating that Plc1 activity was required for normal growth, not specifically for the regulation of expression of arginine metabolic genes. This conclusion was reinforced by enzymatic measurements of ornithine transcarbamylase and arginase, which are anabolic and catabolic enzymes of arginine metabolism, respectively (Fig. 2). Deletion of ARG82 abolished repression of the transcarbamylase and induction of arginase, while the plc1 mutation did not affect the levels of these enzymes (39). El Alami et al. (43) further showed that stabilization of Mcm1 (and Arg80) by Arg82 did not involve its kinase activity, as a strain with a mutated version of Arg82 (D131A) that abolished the catalytic activity showed a normal expression pattern of a gene whose expression is Mcm1 dependent.
FIG. 2.
Moonlighting activities related to arginine metabolism. (a) Scheme of inositol polyphosphate biosynthesis. Arg82 was initially identified as a protein that participates in the regulation of the transcription of genes related to arginine metabolism (13). Further studies have shown that it also functions as an Ins(1,4,5)P3 and Ins(1,3,4,5)P4 kinase (106, 123). (b) Scheme of arginine biosynthesis and degradation. Ornithine is a common metabolite that participates in both processes. See the text for details. OTCase, ornithine transcarbamylase.
Since Arg82 is a pleiotropic regulator, an exploration of the requirement of its kinase activity for different functions could reveal unknown roles of inositol phosphates. Two pathways have been identified for which this activity is important, one that controls the response of yeast to nitrogen availability and another that regulates expression of phosphate-controlled genes. In an arg82 mutant, genes responsive to nitrogen availability were downregulated, while those normally repressed by phosphate were upregulated. This was due to a lack of kinase activity of the encoded protein, as ARG82 alleles mutated in each of several amino acid codons that abolished the inositol phosphate kinase activity but did not influence the stability of the protein showed the same phenotype. The results on the regulation of phosphate-controlled gene expression have been confirmed by Auesukaree et al. (6) and point to important roles of inositol phosphates in the regulation of several pathways.
Arg5,6 and Transcription of Mitochondrial and Nuclear Genes in S. cerevisiae
The question of direct participation of metabolic enzymes in the control of gene expression has been addressed by Hall et al. (59), who used proteome arrays and chromatin immunoprecipitation to identify proteins that specifically bind to DNA. Among the proteins identified which were not known to bind DNA was Arg5,6, a polypeptide that is cleaved in the mitochondrion to yield two proteins, Arg5 and Arg6, which participate in the synthesis of arginine (21). Arg5 corresponds to the C-terminal part of the original polypeptide and possesses acetyl glutamate kinase activity, while Arg6 corresponds to the N-terminal part of the polypeptide and has N-acetylglutamyl-P reductase activity. When myc-tagged Arg5,6 was used in chromatin immunoprecipitation assays and the immunoprecipitated DNA was used to probe an array of genomic intergenic regions and mitochondrial DNA, Arg5,6 was found bound to several DNA regions, mainly of mitochondrial but also of nuclear origin. Gel shift experiments using a GST-tagged version of Arg5,6 and two different regions from the mitochondrial COX1 gene showed that the protein associated with COX1 DNA in vitro. Comparing DNA sequences of the different Arg5,6 targets, a common motif for binding, with a 78% GC content, was identified. Crucial results supporting a role of Arg5,6 in the regulation of gene expression were obtained when the levels of mRNAs corresponding to different potential gene targets were compared in wild-type and arg5,6 mutant cells grown either in rich medium, in a medium with a limiting nitrogen source, or in one lacking amino acids. While there were no remarkable differences between the wild type and the mutant in rich medium, in medium lacking amino acids or limited for nitrogen, lower levels of mitochondrial COX1, nuclear YOR352W (3- to 6-fold), and nuclear PUF4 (2.2-fold) transcripts were observed.
METABOLIC PROTEINS THAT PLAY A ROLE IN PEROXISOMAL PHYSIOLOGY
Pyruvate Carboxylase Participates in the Assembly of Alcohol Oxidase in Hansenula polymorpha
A number of yeast species are able to use methanol as a carbon source. When growing in methanol, these methylotrophic yeasts induce the proliferation of peroxisomes, specialized organelles equipped with distinct enzymes that participate in several metabolic pathways. One such enzyme is alcohol oxidase (AOX), which oxidizes methanol to formaldehyde and hydrogen peroxide. AOX is a flavin adenine dinucleotide (FAD)-containing homo-octamer (48) located in the peroxisomal matrix; its assembly requires other proteins (109). Different types of mutants unable to grow in methanol have been isolated for Hansenula polymorpha (141). One mutant, a strain carrying the ass3 mutation, had a very low AOX activity but exhibited normal levels of AOX protein, which was basically located in the cytosol. Interestingly, these mutants also exhibited a severe growth defect in minimal glucose medium with ammonium as a nitrogen source, reminiscent of that of S. cerevisiae mutants lacking pyruvate carboxylase (Pyc) activity (134). The growth defect of the ass3 mutant could be corrected, as occurs with pyc mutants, by the addition of aspartate or glutamate; however, these amino acids did not restore AOX activity. Transformation of the ass3 mutant with a genomic library from H. polymorpha showed that all its defects were complemented by a DNA fragment encoding a putative Pyc. Disruption of the gene encoding Pyc in H. polymorpha produced the same phenotype as that of the ass3 mutant. Analysis of the progeny from a cross between an ass3 mutant and a pyc1 mutant revealed that both mutations were located in the same locus. It was thus concluded that a mutation in HpPYC1 produces a phenotype that affects AOX activity (109).
AOX from extracts of wild-type H. polymorpha induced in methanol sedimented as an octamer, while that from similar extracts of a pyc mutant sedimented as a monomer. Immunodetection with AOX-specific antibodies revealed that AOX was mostly localized in the cytoplasm of the pyc mutant, while other peroxisomal enzymes showed a normal localization. A point mutation that abolished Pyc activity by replacing an arginine with a glutamine in the active site of the enzyme caused a growth defect in glucose medium similar to that of the ass3 mutants. However, the mutant was able to grow in methanol and exhibited normal levels of AOX, indicating that the Pyc protein but not Pyc activity was needed for correct assembly of AOX (109). How could Pyc participate in AOX assembly? Since assembly of active AOX takes place in the peroxisome (133) and Pyc is cytosolic, the Pyc action should precede this step. It has been found that a riboflavin auxotrophic mutant of H. polymorpha accumulated AOX monomers lacking FAD in the cytosol, a feature also observed in the pyc mutants (48). Since Pyc interacts in vitro with both AOX and FAD, Ozimek et al. (109) proposed that Pyc mediates FAD binding to AOX monomers synthesized in the cytosol. Once charged with FAD, the monomers will be transported to the peroxisome, where AOX will spontaneously assemble into the active octameric form. The region of Pyc required for AOX assembly has been located in the transcarboxylation domain of Pyc, which participates in the transfer of the carboxyl group of carboxylated biotin to pyruvate (110).
Pyc may also play a similar role in other methylotrophic yeasts, since it has been observed that a Pichia pastoris mutant with a disrupted PYC gene (95) did not grow in methanol and failed to import AOX into the peroxisomes (109).
Phosphofructokinase and Microautophagy in Pichia pastoris
When the methylotrophic yeast P. pastoris is transferred from a medium with methanol as a carbon source to another one with glucose, vacuolar degradation of peroxisomes takes place (137, 138). This process is termed microautophagy. During a study of this process, Yuan et al. (151) isolated a mutant, the gsa1 mutant, which was unable to engulf peroxisomes in the vacuole. Transformation of the mutant with a genomic P. pastoris library revealed that the mutant phenotype was complemented by a DNA fragment encoding the putative α subunit of the glycolytic enzyme phosphofructokinase (Pfk), which catalyzes the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate. In most yeasts studied, Pfks are hetero-octamers composed of two types of subunits, α and β, with high sequence similarity. When the gene encoding the α subunit of P. pastoris Pfk was disrupted, the mutant obtained had a Pfk activity of 15% that of the wild type and showed a slow decay of AOX activity in the transition from methanol to glucose (151). Also, the morphology of the peroxisomes was similar for this mutant to that observed for the gsa1 mutant. Pfk activity in the gsa1 mutant was assayed and found to be reduced greatly; it was then concluded that the gsa1 mutant carries a mutation in the gene encoding Pfk. To determine whether the effect observed on microautophagy was due to a disturbance in sugar metabolism caused by low Pfk activity, a Pfkα mutant form with a D362S substitution in the binding site for the substrate fructose-6-P was constructed. While this mutant had a Pfk activity of only about 7% that of the wild type, microautophagy occurred normally in it, indicating that it was the Pfk protein itself, not its activity, that was needed for correct degradation of the peroxisomal enzymes (151). The precise mode of action of the Pfk subunit in the process has not yet been unraveled; also, no role in the process has been reported for the β subunit of Pfk.
It would be interesting to study the role, if any, of Pfk in peroxisomal degradation in a yeast such as Yarrowia lipolytica, which grows on substrates that induce peroxisome proliferation, such as alkanes or fats, and whose Pfk has a homo-octameric structure (50).
METABOLIC PROTEINS IMPLICATED IN MITOCHONDRIAL FUNCTIONS
Aconitase and Maintenance of Mitochondrial DNA in S. cerevisiae
Mitochondria have their own DNA (mtDNA) that is packaged together with multiple proteins in structures called nucleoids (55, 101). Some of the proteins found in mitochondrial nucleoids have functions related to the expression or maintenance of mtDNA. During a study of yeast mitochondrial nucleoids, Chen et al. (27) found that in addition to those proteins, several enzymes of the Krebs cycle were also associated with the nucleoid. To determine whether this association was due to some unknown function of these enzymes in nucleoids, the genes encoding those proteins were disrupted and the effects of the disruption on mtDNA maintenance were examined. S. cerevisiae is the organism of choice for this type of study due to its ability to live without functional mtDNA, as it is a facultative aerobe able to ferment and propagate by using the fermentative glycolytic pathway as an energy producer. Mutants affected in respiration or without functional mtDNA are viable but do not grow in respiratory substrates such as ethanol or glycerol; due to the small size of their colonies on glucose, they are named petites (47). Although disruption of different genes encoding proteins related to the Krebs cycle found in the mitochondrial nucleoid did not severely affect the stability of mtDNA, disruption of the ACO1 gene, encoding aconitase, caused a great instability. Aconitase catalyzes the conversion of citrate to isocitrate and is localized in the mitochondrial matrix; a yeast mutated in aconitase is unable to grow in nonfermentable carbon sources and has a requirement for glutamate (54, 107). However, the instability of mtDNA in the aco1 mutant was not due to the metabolic defect, since mutations in citrate synthase causing a similar growth phenotype did not influence mtDNA stability. To ascertain whether the enzymatic activity of aconitase was needed for mtDNA maintenance, Chen et al. (27) mutated some residues critical to its activity. Aconitase has an iron-sulfur center (4Fe-4S) (15), and in the yeast enzyme, residues Cys 382, 445, and 448 participate in iron coordination. Each of these residues was changed to Ser, and the ability of the corresponding mutants to maintain mtDNA was studied. All strains carrying these mutations expressed the mutant proteins to the same levels found for Aco1 in the wild type and, as expected, were glutamate auxotrophs. In contrast, mtDNA was maintained in those mutants, thus showing that aconitase activity is not necessary for this role (27).
Acetohydroxyacid Reductoisomerase and mtDNA Maintenance in S. cerevisiae
The syntheses of the branched-chain amino acids leucine and valine and that of isoleucine share an enzyme that catalyzes the same type of reaction using two different substrates. This enzyme, acetohydroxyacid reductoisomerase, is encoded in S. cerevisiae by the ILV5 gene. During a study of mtDNA stability, ILV5 was isolated as a multicopy suppressor of mtDNA instability in abf2 mutants, which lose their mtDNA during growth in glucose (152). A two- to threefold increase in ILV5 expression was sufficient to stabilize mtDNA in those mutants. On the other hand, the presence in the growth medium of leucine, valine, and isoleucine, amino acids that repress ILV5 expression, increased mtDNA instability in abf2 mutants relative to that in control cultures in which the amino acids were absent. These results suggested a possible role for Ilv5 in the maintenance of mtDNA.
Biosynthesis of amino acids in yeast is regulated by the general amino acid control circuit (63). A major player of this circuit is Gcn4, which activates the transcription of ILV5, as well as that of other genes encoding amino acid biosynthetic enzymes, when the yeast is starved for amino acids. Constitutive expression of GCN4 in abf2 mutants increased mtDNA stability in comparison with a control strain expressing the wild-type, regulated version of GCN4 (152). Further support for the participation of Ilv5 in the maintenance of mtDNA came from the observation that ilv5 mutants generated petites at a high frequency, thus showing significant mtDNA instability (152). All of these results are suggestive of and consistent with a role of the Ilv5 protein in the maintenance of mtDNA stability. However, they could also be explained as a result of a lack of synthesis of branched-chain amino acids. This possibility was eliminated by demonstrating that a mutation in ILV2, the gene encoding the enzyme catalyzing the step preceding that catalyzed by Ilv5, had no effect on mtDNA stability (152). Further evidence in favor of a role of Ilv5 independent of its catalytic activity was provided by Bateman et al. (11), who obtained mutations in ILV5 that could either abolish the metabolic activity of the protein and yet retain its role in mtDNA maintenance or vice versa. Both types of mutants were the result of single missense mutations in regions structurally conserved among acetohydroxyacid reductoisomerases from different organisms. It is interesting that the Escherichia coli ortholog of ILV5, when targeted to the mitochondria, complements the metabolic defect of an ilv5 mutant but is ineffective for mtDNA maintenance (11).
Mitochondrial nucleoids contain more than one equivalent of mtDNA per nucleoid (102, 147). MacAlpine et al. (90) found that the distribution of mtDNA in the nucleoids was under the control of the general amino acid control circuit. They also found that constitutive expression of GCN4 increased the number of nucleoids in wild-type cells but had no effect in ilv5 mutants. An ilv2 control showed the same behavior as the wild type in this aspect. These findings implicate Ilv5 not only in mtDNA maintenance but also in the distribution of mtDNA molecules into nucleoids (11).
METABOLIC PROTEINS THAT PARTICIPATE IN VACUOLAR FUNCTIONS
Enolase and Vacuolar Protein Traffic in S. cerevisiae
The yeast vacuole is an organelle that participates in diverse processes, such as the response to osmotic stress, turnover of proteins, or accumulation of amino acids, polyphosphates, or toxic compounds. In addition, the vacuole has an active relationship with the protein trafficking machinery (81). The number of vacuoles per yeast cell usually fluctuates between one and five (30). This number and the morphology of the organelle vary depending on environmental factors; while low levels of glucose or hypotonic conditions make vacuoles fuse, hypertonic media or formation of daughter vacuoles during cell division produces fragmentation of the organelle (30, 144).
The fusion of vacuoles is termed homotypic fusion to differentiate it from fusions between different types of vesicles during protein trafficking, called heterotypic fusion. Homotypic fusion is a complex process that may be studied in vitro using assays that monitor the appearance of some property, usually an enzymatic activity, when mixing of two different vacuole preparations results in vacuolar fusion (146). During an in vitro study of homotypic fusion, it was found that a high-molecular-weight, cytosolic component was necessary for the process to proceed (149). Further characterization by classical protein fractionation techniques led to its identification as enolase (35). Enolase is an abundant glycolytic enzyme that catalyzes the interconversion of 2-phosphoglycerate and phosphoenolpyruvate. In S. cerevisiae, there are two genes, ENO1 and ENO2, which encode two isoenzymes with >90% identity in amino acid sequence (93). The addition of exogenous, purified recombinant Eno1 or Eno2 to an in vitro vacuolar fusion reaction mixture stimulated fusion in a dose-dependent manner, thus showing that enolase was an actual stimulator, not a contaminant carried over with the unknown high-molecular-weight factor.
Remarkably, a recombinant enolase-GST protein with a point mutation that abolished enzymatic activity was also active in the fusion assay. Independent support for the physiological importance of enolase in homotypic vacuolar fusion derives from the observation that yeast mutants with a deleted ENO1 gene and a low level of ENO2 expression presented defects in vacuolar structure, exhibiting a picture of fragmented vacuoles. These vacuoles also had low levels of other proteins implicated in vacuolar fusion, suggesting a role for enolase in vacuolar protein trafficking (35).
Recombinant enolase stimulated in vitro vacuolar fusion when the components in the assay mixture came from a wild-type yeast but failed to do so when the components came from an enolase-deficient strain, thus indicating that other enolase-dependent factors are implicated in vacuole fusion. It was found that about 1% of total vacuolar protein is enolase that seems to bind to the vacuole through association with the external membrane. The association of enolase with the vacuole is dependent on the activity of the gene BOR1 (35), which encodes a protein with sequence similarity to human “band 3,” a protein implicated in the binding of glycolytic enzymes to erythrocyte membranes (24).
Aldolase and Assembly of Vacuolar H+-ATPase in S. cerevisiae
Vacuoles of yeast have a mild acidic pH of around 6 (23, 117) that is important for their function (81). The pH gradient with respect to the less acidic cytoplasm (23) is generated and maintained by the activity of a proton pump, the vacuolar H+-ATPase. This pump belongs to a family of ATPases, the V-ATPases, that hydrolyze ATP to generate an ion motive force. They are composed of two distinct macrodomains, V0 and V1, built in turn by the assembly of different subunits (51, 75). These domains exist in equilibrium with the assembled proton pump, which may partially dissociate under certain conditions. In the case of yeast V-ATPase, deprivation of glucose leads to a partial dissociation of the pump that is reversed by addition of the sugar (76, 112). This dissociation appears to be an important control mechanism of the activity of the yeast V-ATPase, although other mechanisms may also be operative (66, 76). In a search for proteins that could modulate the activity of V-ATPases, Lu et al. (87) used a yeast two-hybrid assay in which the bait was subunit E of the kidney V-ATPase, which does not present homology with subunits of F-ATPases, a family of structurally related ATPases that generally utilize proton motive force to generate ATP. Using a human kidney cDNA expression library, they identified an interaction between the bait subunit E and aldolase, the glycolytic enzyme that hydrolyzes fructose-1,6-bisphosphate to dihydroxyacetone-P and glyceraldehyde 3-P. Results of coimmunoprecipitation and immunocytochemistry experiments (87) supported the occurrence of this interaction in vivo (87). In view of this result, they studied the influence of aldolase in the dissociation of macrodomains V1 and V0 in response to glucose in an S. cerevisiae mutant lacking aldolase. For this purpose, they used either in situ staining with specific antibodies against components of the V1 and V0 macrodomains or immunoprecipitation assays. They found that in the aldolase-deficient mutant, the domains remained dissociated, while they were associated in the wild-type control under the same conditions. The dissociation resulted in a threefold reduction of V-ATPase activity; it was concluded that this effect was genuine and not due to the impairment of glycolysis caused by a lack of aldolase because the complex remained associated in a phosphoglucose isomerase that also exhibited impaired glycolysis (87). Lu et al. (88) showed that the phenotype of aldolase mutants, growth on rich medium containing 1% ethanol at pH 5.5 but not at pH 7.5, was similar to that observed for mutants with defects in some subunits of V-ATPase. Again, mutants defective in phosphoglucose isomerase grew in that medium at both pH values. To examine the interaction between aldolase and V-ATPase in vivo, assembled V-ATPase was immunoprecipitated from vacuolar membrane vesicles purified from cells growing in glucose or deprived of the sugar. Subsequently, the preparation was probed with antibodies against aldolase; these antibodies reacted strongly with the assembled V-ATPase when the vesicles were obtained from a glucose culture, but the reaction was decreased eight times when vesicles from glucose-deprived cells were used (88). Further support for the implication of aldolase in the assembly of the yeast V-ATPase was obtained using different aldolase mutants. A K323M mutant that conserves enzymatic activity but is unable to bind to the B subunit of V-ATPase produced disassembly and malfunction of V-ATPase and competed with wild-type aldolase when expressed in a wild-type yeast, resulting in decreased ATP hydrolysis and proton translocation activity (86). In contrast, an aldolase H108A mutant that has no aldolase activity but binds to V-ATPase did not affect those processes (86).
All of these results strongly argue in favor of aldolase participation in the regulation of the activity and assembly of the yeast V-ATPase in addition to its metabolic role in glycolysis.
A METABOLIC PROTEIN SWITCHES OFF A BIOSYNTHETIC PATHWAY
Control of Ornithine Transcarbamylase by Arginase in S. cerevisiae
Ornithine is one of the starting molecules for arginine biosynthesis and is also produced in the degradation of this amino acid (Fig. 2). When yeast uses arginine as a nitrogen source, arginase produces ornithine and urea; if unchecked, the simultaneous action of the arginine biosynthetic and degradative pathways would lead to a futile cycle. Therefore, a series of mechanisms exist to regulate the expression of the genes encoding enzymes that catalyze the catabolic and anabolic reactions and to control the activities of those enzymes (31); arginase itself participates in a peculiar regulatory phenomenon. Bechet and Wiame (14) observed that the addition of arginine to a yeast culture growing with ammonium as a nitrogen source caused a rapid disappearance of the activity of ornithine transcarbamylase (OTCase), the enzyme catalyzing the first step of arginine biosynthesis. This disappearance was observed when the enzyme was assayed in permeabilized cells; however, OTCase activity could be detected when the assay was performed with cell extracts. If an inhibitor of protein synthesis was present when arginine was added, the decrease in OTCase activity was not observed. They called this phenomenon “epiarginasic regulation” and proposed that the disappearance of OTCase activity was caused by a protein synthesized after arginine addition that acted directly on OTCase and whose operation was likely to be stoichiometric rather than catalytic (14). In vitro experiments showed that this was indeed the case and that the newly synthesized protein was arginase (99). Arginase, in the presence of arginine and ornithine, forms a complex with OTCase and completely inhibits its activity while remaining active itself (99); this complex is formed by the stoichiometric association of one molecule of the trimeric arginase with one of the trimeric OTCase (60, 98). The association is reversed by dilution or removal of the effectors arginine and ornithine, thus explaining the initial, puzzling finding of why OTCase activity could not be measured in permeabilized cells but could be detected in cell extracts. Mutations in OTCase that prevent its inhibition by arginase have been identified in two regions conserved in other OTCases and thought to be involved in ornithine binding (42). Binding of arginase appears to favor a transition from an open to a closed conformation of OTCase, thus eliminating its catalytic activity. Mutations in cysteines 321 and 326, located in the C-terminal portion of arginase, eliminated its regulatory function without significantly affecting its catalytic activity or its affinity for the substrate (42).
Epiarginasic regulation has been found in different yeast species; its presence or absence is correlated with the different subcellular locations of OTCase and arginase. Obviously, the interaction between the two proteins requires a common subcellular localization; this is the case for S. cerevisiae and related species, in which OTCase and arginase are cytosolic, but not in other yeast species, in which OTCase is mitochondrial and arginase is cytosolic (69, 140, 142).
OTHER CASES
Ure2, a Regulator of Nitrogen Catabolite Repression, Exhibits Glutathione Peroxidase Activity
The protein Ure2 is an important regulator of nitrogen catabolite repression, the process that controls the utilization of available nitrogen sources by S. cerevisiae (32). It has also received much attention due to its relationship with [URE3], a non-Mendelian, nonmitochondrial mutation originally isolated by its effects on nitrogen metabolism in yeast (3). The finding that [URE3] is the prion form of Ure2 (145) has prompted multiple studies to understand the processes that affect prion formation and maintenance (84). The ure2 mutant was generated by Lacroute (83) during a study of genes implicated in the regulation of pyrimidine biosynthesis. This mutant was able to transport ureidosuccinate in the presence of both poor and rich nitrogen sources, such as proline and ammonia, respectively, in contrast with the wild type, which could transport ureidosuccinate only under the first condition. Further studies showed that ure2 mutants exhibited a pleiotropic phenotype leading to loss of nitrogen catabolite repression of several genes. Work in Magasanik's laboratory demonstrated that Ure2 was a negative regulator of Gln3, a key player in nitrogen catabolite repression (34). It was found that Ure2 binds Gln3, a GATA factor that activates the transcription of genes subject to nitrogen catabolite repression, and maintains it in the cytoplasm (32). Coschigano and Magasanik (33) cloned and sequenced URE2 and found that its protein product showed important homology in the C-terminal region with GSTs of different organisms; later on, crystallographic studies showed that the structure of this fragment has the same fold as that found in the superfamily of GSTs (139). However, such an activity could not be detected in the protein (33). Umland et al. (139) showed that between residues 97 and 354 of Ure2, some conserved amino acids implicated in catalysis in other GSTs were absent, and they proposed that this could justify the observed lack of activity. GSTs are enzymes that conjugate glutathione to different substances and in this way decrease their toxicity. This property was used by Rai et al. (118) to test in vivo, indirectly, the putative GST activity of Ure2, bypassing possible problems encountered with the in vitro assay of that activity. They found that ure2 mutants were hypersensitive to cadmium and nickel ions as well as to hydrogen peroxide. Interestingly, ure2 mutants were not or were minimally more sensitive than wild-type cells to 1-chloro-2,4-dinitrobenzene, a widely used substrate of GSTs. Rai et al. showed that ure2 mutants have phenotypes similar to those of mutants affected in genes encoding GSTs in S. cerevisiae or Schizosaccharomyces pombe, and they concluded that their results were consistent with a role of Ure2 as a GST (118). Although 1-chloro-2,4-dinitrobenzene is considered an almost universal substrate for those enzymes, a number of them fail to show activity toward it (84), and this is the case for Ure2.
Some GSTs possess glutathione peroxidase activity (5). This type of activity was demonstrated for Ure2 by Bai et al. (8), who purified the protein and performed in vitro kinetic measurements to characterize the reaction of glutathione with several hydroperoxides. The results showed that Ure2 does not have a typical GST substrate specificity but belongs to a subset of these proteins that are active against different oxidants (84). The same authors showed that a truncated Ure2 protein lacking the N-terminal 90 amino acids, which are a determinant for conversion in the prion form (91), showed kinetic parameters similar to those of wild-type protein in the reactions tested and that fibrillar aggregates behaved similarly. Since the regulatory function of Ure2 in nitrogen catabolite repression is lost upon conversion to the prion form (URE3) (3), this result indicates that the enzymatic activity is not sufficient for the regulatory function. Recent results indicate that the prion domain is implicated in stabilization of the protein and its interaction with elements of the nitrogen catabolite repression system (128).
Enolase and Mitochondrial tRNA Import
Mitochondria are important organelles in eukaryotic cells. A majority of their proteins are encoded by nuclear genes, translated in the cytosol, and imported into the organelle. However, some mitochondrial proteins are synthesized in the organelle itself, and mitochondrial genomes encode the set of tRNAs used for this synthesis. In spite of this, mitochondria also import nucleus-encoded tRNAs (127). In S. cerevisiae, three nucleus-encoded tRNAs are imported, namely, tRNACUGGln, tRNAUUGGln (120), and tRNACUULys, termed tRK1 (92). The import of tRK1 into the mitochondrion was found to require interaction of the aminoacylated molecule with the cytosolic precursor of mitochondrial lysyl-tRNA synthetase, pre-Mks1 (135). In addition, other soluble components appeared to be necessary, since pre-Mks1 alone could not import tRK1 into the mitochondria in an in vitro assay. Fractionation of proteins from an extract identified enolase as a protein that specifically interacted with aminoacylated tRK1; no interaction was observed with either the deacylated form or with tRK2, a tRNAUUULys. The isoform Eno2 was the most effective, although Eno1 could act with about 10 to 20% import capacity. Using a three-hybrid assay, an interaction between Eno2 and tRK1, but not tRK2, was inferred from the growth behavior of the strains used (45). Moreover, in a gel retardation assay, only Eno2 was observed to form a complex with tRK1. Formation of a tRK1-Eno2-pre-Mks1 ternary complex was not seen in gel retardation assays. In the presence of pre-Mks1, the amount of Eno2-tRK1 complex decreased in favor of a tRK1-pre-Mks1 complex (45). In view of this result, it was hypothesized that Eno2 binds the aminoacylated tRK1 in the cytosol and directs it to the mitochondrial membrane, where it is transferred to pre-Mks1 and enters the mitochondrion in this way.
No parallelism between enzymatic activity and import capacity was found, since a mutated form of Eno2 with 1% activity still directed import with 30% of wild-type efficacy (45).
FINAL REMARKS
We have reviewed a number of well-authenticated cases of proteins that moonlight in different yeast species. The variety of proteins and the diversity of their moonlighting functions show the wide range of different processes in which yeast proteins originally identified as metabolic enzymes may participate. In this sense, the situation in yeasts does not differ from the picture found in higher eukaryotes (132). The fact that more cases have been reported for S. cerevisiae than for other yeast species does not necessarily mean that in other yeasts moonlighting is less frequent but likely reflects the fact that S. cerevisiae has been studied much more extensively than other yeast species. The attractiveness of the moonlighting designation may induce premature or unwarranted attributions of this characteristic to proteins that do not strictly conform to the original definition (71, 132). An important cautionary remark is that prior to the attribution of a moonlighting role to an enzyme, it should be ascertained that the effects associated with an enzymatic defect are not simply secondary effects derived from changes in the metabolite profile caused by the lack of enzymatic activity. In this review, only proteins whose multiplicity of functions has been demonstrated clearly by different experimental approaches have been considered; therefore, we have not included the increasing number of proteins that are being found associated with the yeast cell wall. These proteins, which are different from the intrinsic cell wall proteins (78, 82), form a heterogeneous group with a variety of original functions that range from glycolytic enzymes to translation elongation factors (105). Although in general their function in the cell wall has not been demonstrated clearly, in the case of pathogenic yeasts some of them have been reported to act in the invasion of host tissues (74, 105). The fact that the appearance of these proteins in the cell wall is nonrandom, at least in some cases (105), suggests that they might perform some unknown function in that location. Thus, although some of these proteins are potential candidates to be moonlighting proteins, there is not yet, in our opinion, sufficient information to qualify them as such.
One intriguing question is how moonlighting capabilities appeared. It seems plausible to assume that the enzymatic function of the protein is the primordial one. The sites implicated in the catalytic process are likely to be more fixed by the requirements of the chemical process but potentially occupy only a small area of the structure of the protein. Parts of the structure not strictly required for the metabolic function will be more tolerant of changes and offer possibilities to develop new activities. The new activities may or may not have a functional relationship with the original enzymatic function. In the case of KlGal1, certain characteristics of the protein, such as binding to galactose and ATP, are used in the moonlighting function, while in other cases the moonlighting role appears not to have such a relationship, like, for example, the participation of pyruvate carboxylase in the assembly of alcohol oxidase in H. polymorpha. The tinkerer's way of the evolutionary process (67) may account for these differences. Exon shuffling (56) followed by subsequent rearrangements may be another way to originate genes that encode proteins with different functions. With the data available, no sensible hypothesis about the origin of this type of proteins may be offered. However, it seems that the acquisition of moonlighting properties occurred at different moments along evolution, as shown by the differences and lack of conservation of these activities between similar proteins in various yeasts.
An interesting case of evolution is that of KlGal1 in K. lactis versus the pair of proteins Gal1-Gal3 in S.cerevisiae. KlGal1 has, in addition to its enzymatic function, a moonlighting regulatory activity, while in the second case one protein, ScGal3, has no enzymatic activity and is entirely devoted to the regulatory function and ScGal1 has mainly a metabolic role, with only marginal moonlighting activity. In the evolutionary history of yeasts, K. lactis did not undergo the whole genome duplication and subsequent gene loss that originated S. cerevisiae (41, 126). The duplication and subsequent evolution allowed a separation of functions; an interesting question is which alternative is more advantageous for the organism, i.e., partitioning two activities between two proteins or maintenance of both in one protein. The first solution may provide more flexibility, while the second one provides an economy of genetic material and concomitant energy expenditure. However, the second solution has the disadvantage that a mutation that affects the regulatory domain makes the organism unable to grow on galactose; there are no gal3-like mutants of K. lactis. What is quite curious is that the separation of functions has brought about a difference in the subcellular localization of the regulatory event. No explanation is available for this difference. Hittinger and Carroll (64) performed a detailed study of the evolution of the GAL1-GAL3 genes in S. cerevisiae. These genes evolved by changes in both their coding and promoter regions; this has allowed a very tight control on GAL1 expression that varies about 1,000 times depending on the presence or absence of galactose in the medium. It remains to be ascertained if a division of functions in the natural ecological environment of K. lactis would have increased its adaptive fitness.
A remarkable finding is the large number of proteins related with sugar metabolism that moonlight; for example, hexokinase, phosphofructokinase, aldolase, enolase, and pyruvate carboxylase perform some unrelated function in different yeasts. Among them, enolase appears to be a most versatile protein, as shown in this review. Also, one of its isoforms, Eno1, has been identified as heat shock protein HSP48 (65). This parallels findings with higher organisms, in which 7 of 10 enzymes from the glycolytic pathway appear to moonlight (132). It could be speculated that the antiquity of this pathway as well as the constitutive expression of its enzymes could have allowed extensive trial and selection of its proteins for multiple functions. Curiously, no moonlighting activity has been reported for glyceraldehyde-3-P dehydrogenase in yeast, even though it is implicated in several nonenzymatic functions in other organisms (130, 132). It shall be noted, however, that the moonlighting activities of an enzyme are usually organism specific; for example, aconitase in S. cerevisiae moonlights to maintain mtDNA, while in mammals its alternative function is to regulate the amount of mRNA encoding ferritin and the transferring receptor (11).
In almost all cases considered in this review, the catalytic metabolic activity of the corresponding protein was not required for its moonlighting function. Yeast hexokinase remains, up to now, an exception, since no bona fide mutants devoid of catalytic activity but retaining its regulatory function have been reported. This contrasts with the situation in plants, where hexokinase activity is not necessary for its glucose signaling role (28). It seems, therefore, that a protein's moonlighting activity may be dissociated from the metabolic pathway in which its enzymatic activity participates, suggesting that these proteins may receive signals from different pathways. Moonlighting activity may also be influenced by the cellular environment; changes in the concentrations of metabolites are important for this function, as seen by the requirement for galactose and ATP for the formation of the KlGal1-KlGal80 complex or by that for arginine and ornithine for the moonlighting function of arginase. There is not extensive information about the exclusivity of moonlighting functions; it is known that arginase activity is not abolished by its binding to OTCase (99), but similar information is not available for yeasts in other cases. In higher organisms, aconitase exerts its moonlighting role only when iron scarcity makes the protein enzymatically inactive (79).
An area that deserves consideration is that of the possible moonlighting function of some transporters as nutrient sensors. In S. cerevisiae or K. lactis, glucose transporters do not sense the sugar; instead, other proteins highly similar to them but without such activity act as sensors and control the expression of genes encoding the transporters (77). This raises the question of the possible evolutionary origin of the sensors from ancient moonlighting transporters. It would be interesting to study if in more ancient yeasts sugar transporters have the capacity to act as sugar sensors and in this role act on other pathways. In the case of certain transporters, such as Gap1 or Mep2, a role as sensors has been found. Upon interaction with amino acids, the amino acid permease Gap1 activates protein kinase A activity (38), and Mep2, an ammonium transporter, is required for pseudohyphal differentiation under nitrogen starvation conditions (85).
Up to now, the moonlighting functions of proteins have usually been found by chance. The identification of moonlighting activities is important for understanding cell function, for interpretation of unusual phenotypes, and for targeted modification of metabolic pathways. Therefore, it would be of great interest to develop tools to predict which proteins may moonlight and their physiological role. Unfortunately, this appears to be quite difficult, as shown by the following considerations. One guide for prediction could be to infer the moonlighting role from previously documented cases; for example, the influence of the pyc1 mutation on methanol metabolism was successfully predicted for P. pastoris from the findings with H. polymorpha (109). However, the same cannot be said of hexokinase and its participation in catabolite repression, since in most of the yeast species studied the protein does not play a role similar to that found in S. cerevisiae. Another unsuccessful case of prediction is that of the epiarginasic regulation of OTCase by arginase, as arginase moonlights in this role in only some yeast species (69, 140). A possible hint for a moonlighting activity could be an unusual pattern of expression of the coding gene, localization of the protein, or both. However, even if a suggestive case for moonlighting could be made, it will be quite difficult to predict what the unknown activity might be. Bioinformatics has not yet been of much help in predicting moonlighting functions. The difficulty in prediction is reflected by work done with known moonlighting proteins as a starting point to test different algorithms. Although domains for alternative functions were found by bioinformatic analysis, the predictive power appeared to be very low (57). The limitations currently associated with automated protein function prediction have been reviewed by Friedberg (52). It should also be considered that small changes in protein structure can trigger the appearance of unexpected activities, as shown for mammalian dihydrolipoamide dehydrogenase, in which certain mutations decrease the normal enzymatic activity and make the protein function as a protease (7). It therefore appears that discovery of moonlighting activities will remain a matter of serendipity for the immediate future.
The moonlighting phenomenon has revealed several previously unknown functional relationships between cellular processes and shows the importance of an integrated network of interactions for adequate functioning of an organism. Less than 10 years ago, moonlighting was an almost unknown word in biology; since the proposal of the concept (71), the number of recognized cases of moonlighting proteins has greatly increased, and it is likely that we know only a narrow segment of the whole. Given the importance of the roles identified for moonlighting proteins and the relevance of the phenomenon for both basic and applied problems, its existence ought to be widely recognized.
Acknowledgments
We thank Juana M. Gancedo (Instituto de Investigaciones Biomédicas Alberto Sols, CSIC-UAM, Madrid, Spain), J. M. Siverio (Department of Biochemistry and Molecular Biology, Universidad de La Laguna, Spain), and M. A. Blázquez (Institute for Molecular and Cell Biology of Plants, Valencia, Spain) for critical readings of the manuscript or comments and J. M. Siverio for calling to our attention the moonlighting activity of Ure2.
Work in our laboratory was partly supported by grant BFU2004-02855-CO2-01 from the Dirección General de Investigación Científica y Técnica of the Spanish Ministry of Educación y Ciencia.
REFERENCES
- 1.Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 27914440-14446. [DOI] [PubMed] [Google Scholar]
- 2.Ahuatzi, D., A. Riera, R. Pelaez, P. Herrero, and F. Moreno. 2007. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 2824485-4493. [DOI] [PubMed] [Google Scholar]
- 3.Aigle, M., and F. Lacroute. 1975. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136327-335. [DOI] [PubMed] [Google Scholar]
- 4.Anders, A., H. Lilie, K. Franke, L. Kapp, J. Stelling, E. D. Gilles, and K. D. Breunig. 2006. The galactose switch in Kluyveromyces lactis depends on nuclear competition between Gal4 and Gal1 for Gal80 binding. J. Biol. Chem. 28129337-29348. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong, R. N. 1997. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 102-18. [DOI] [PubMed] [Google Scholar]
- 6.Auesukaree, C., H. Tochio, M. Shirakawa, Y. Kaneko, and S. Harashima. 2005. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 28025127-25133. [DOI] [PubMed] [Google Scholar]
- 7.Babady, N. E., Y. P. Pang, O. Elpeleg, and G. Isaya. 2007. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc. Natl. Acad. Sci. USA 1046158-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai, M., J. M. Zhou, and S. Perrett. 2004. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J. Biol. Chem. 27950025-50030. [DOI] [PubMed] [Google Scholar]
- 9.Bajwa, W., T. E. Torchia, and J. E. Hopper. 1988. Yeast regulatory gene GAL3: carbon regulation; UASGal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol. Cell. Biol. 83439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett, J. A., R. W. Payne, and D. Yarrow. 1990. Yeasts. Characteristics and identification, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 11.Bateman, J. M., P. S. Perlman, and R. A. Butow. 2002. Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of ilv5p of budding yeast. Genetics 1611043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazan, J. F., R. J. Fletterick, and S. J. Pilkis. 1989. Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Proc. Natl. Acad. Sci. USA 869642-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bechet, J., M. Grenson, and J. M. Wiame. 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 1231-39. [DOI] [PubMed] [Google Scholar]
- 14.Bechet, J., and J. M. Wiame. 1965. Indication of a specific regulatory binding protein for ornithinetranscarbamylase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 21226-234. [DOI] [PubMed] [Google Scholar]
- 15.Beinert, H., and M. C. Kennedy. 1993. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 71442-1449. [DOI] [PubMed] [Google Scholar]
- 16.Belinchón, M. M., and J. M. Gancedo. 2007. Glucose controls multiple processes in Saccharomyces cerevisiae through diverse combinations of signaling pathways. FEMS Yeast Res. 8808-818. [DOI] [PubMed] [Google Scholar]
- 17.Bercy, J., E. Dubois, and F. Messenguy. 1987. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene 55277-285. [DOI] [PubMed] [Google Scholar]
- 18.Bhat, P. J., and J. E. Hopper. 1992. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: evidence for a new model of induction for the yeast GAL/MEL regulon. Mol. Cell. Biol. 122701-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat, P. J., and T. V. Murthy. 2001. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol. Microbiol. 401059-1066. [DOI] [PubMed] [Google Scholar]
- 20.Bhat, P. J., D. Oh, and J. E. Hopper. 1990. Analysis of the GAL3 signal transduction pathway activating GAL4 protein-dependent transcription in Saccharomyces cerevisiae. Genetics 125281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boonchird, C., F. Messenguy, and E. Dubois. 1991. Determination of amino acid sequences involved in the processing of the ARG5/ARG6 precursor in Saccharomyces cerevisiae. Eur. J. Biochem. 199325-335. [DOI] [PubMed] [Google Scholar]
- 22.Brilli, M., and R. Fani. 2004. The origin and evolution of eucaryal HIS7 genes: from metabolon to bifunctional proteins? Gene 339149-160. [DOI] [PubMed] [Google Scholar]
- 23.Calahorra, M., G. A. Martinez, A. Hernandez-Cruz, and A. Peña. 1998. Influence of monovalent cations on yeast cytoplasmic and vacuolar pH. Yeast 14501-515. [DOI] [PubMed] [Google Scholar]
- 24.Campanella, M. E., H. Chu, and P. S. Low. 2005. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl. Acad. Sci. USA 1022402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2202-207. [DOI] [PubMed] [Google Scholar]
- 26.Chaves, R. S., P. Herrero, and F. Moreno. 1999. Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun. 254345-350. [DOI] [PubMed] [Google Scholar]
- 27.Chen, X. J., X. Wang, B. A. Kaufman, and R. A. Butow. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307714-717. [DOI] [PubMed] [Google Scholar]
- 28.Cho, Y. H., S. D. Yoo, and J. Sheen. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127579-589. [DOI] [PubMed] [Google Scholar]
- 29.Clarke, J. L., D. A. Scopes, O. Sodeinde, and P. J. Mason. 2001. Glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase. A novel bifunctional enzyme in malaria parasites. Eur. J. Biochem. 2682013-2019. [DOI] [PubMed] [Google Scholar]
- 30.Conibear, E., and T. H. Stevens. 2002. Studying yeast vacuoles. Methods Enzymol. 351408-432. [DOI] [PubMed] [Google Scholar]
- 31.Cooper, T. G. 1982. Nitrogen metabolism in Saccharomyces cerevisiae, p. 39-100. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of Sacharomyces cerevisiae, 1st ed., vol. 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 32.Cooper, T. G. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coschigano, P. W., and B. Magasanik. 1991. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol. 11822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courchesne, W. E., and B. Magasanik. 1988. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol. 170708-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decker, B. L., and W. T. Wickner. 2006. Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J. Biol. Chem. 28114523-14528. [DOI] [PubMed] [Google Scholar]
- 36.de la Cera, T., P. Herrero, F. Moreno-Herrero, R. S. Chaves, and F. Moreno. 2002. Mediator factor Med8p interacts with the hexokinase 2: implication in the glucose signalling pathway of Saccharomyces cerevisiae. J. Mol. Biol. 319703-714. [DOI] [PubMed] [Google Scholar]
- 37.De Vit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 81603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donaton, M. C., I. Holsbeeks, O. Lagatie, G. Van Zeebroeck, M. Crauwels, J. Winderickx, and J. M. Thevelein. 2003. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50911-929. [DOI] [PubMed] [Google Scholar]
- 39.Dubois, E., V. Dewaste, C. Erneux, and F. Messenguy. 2000. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 486300-304. [DOI] [PubMed] [Google Scholar]
- 40.Dubois, E., and F. Messenguy. 1994. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol. Gen. Genet. 243315-324. [DOI] [PubMed] [Google Scholar]
- 41.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 43035-44. [DOI] [PubMed] [Google Scholar]
- 42.El Alami, M., E. Dubois, Y. Oudjama, C. Tricot, J. Wouters, V. Stalon, and F. Messenguy. 2003. Yeast epiarginase regulation, an enzyme-enzyme activity control: identification of residues of ornithine carbamoyltransferase and arginase responsible for enzyme catalytic and regulatory activities. J. Biol. Chem. 27821550-21558. [DOI] [PubMed] [Google Scholar]
- 43.El Alami, M., F. Messenguy, B. Scherens, and E. Dubois. 2003. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol. 49457-468. [DOI] [PubMed] [Google Scholar]
- 44.El Bakkoury, M., E. Dubois, and F. Messenguy. 2000. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Mol. Microbiol. 3515-31. [DOI] [PubMed] [Google Scholar]
- 45.Entelis, N., I. Brandina, P. Kamenski, I. A. Krasheninnikov, R. P. Martin, and I. Tarassov. 2006. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 201609-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Entian, K. D. 1980. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol. Gen. Genet. 178633-637. [DOI] [PubMed] [Google Scholar]
- 47.Ephrussi, B., and P. P. Slonimski. 1955. Yeast mitochondria. Subcellular units involved in the synthesis of respiratory enzymes in yeast. Nature 1761207-1208. [DOI] [PubMed] [Google Scholar]
- 48.Evers, M. E., V. Titorenko, W. Harder, I. ven der Klei, and M. Veenhuis. 1996. Flavin adenine dinucleotide binding is the crucial step in alcohol oxidase assembly in the yeast Hansenula polymorpha. Yeast 12917-923. [DOI] [PubMed] [Google Scholar]
- 49.Fischer, E. 1894. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dtsch. Chem. Ges. 272985-2993. [Google Scholar]
- 50.Flores, C. L., O. H. Martinez-Costa, V. Sanchez, C. Gancedo, and J. J. Aragon. 2005. The dimorphic yeast Yarrowia lipolytica possesses an atypical phosphofructokinase: characterization of the enzyme and its encoding gene. Microbiology 1511465-1474. [DOI] [PubMed] [Google Scholar]
- 51.Forgac, M. 1999. Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 27412951-12954. [DOI] [PubMed] [Google Scholar]
- 52.Friedberg, I. 2006. Automated protein function prediction—the genomic challenge. Brief. Bioinform. 7225-242. [DOI] [PubMed] [Google Scholar]
- 53.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gangloff, S. P., D. Marguet, and G. J. Lauquin. 1990. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol. 103551-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A. M. van der Bliek, and J. N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 141583-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert, W. 1985. Genes-in-pieces revisited. Science 228823-824. [DOI] [PubMed] [Google Scholar]
- 57.Gómez, A., N. Domedel, J. Cedano, J. Pinol, and E. Querol. 2003. Do current sequence analysis algorithms disclose multifunctional (moonlighting) proteins? Bioinformatics 19895-896. [DOI] [PubMed] [Google Scholar]
- 58.Gurney, M. E., B. R. Apatoff, G. T. Spear, M. J. Baumel, J. P. Antel, M. B. Bania, and A. T. Reder. 1986. Neuroleukin: a lymphokine product of lectin-stimulated T cells. Science 234574-581. [DOI] [PubMed] [Google Scholar]
- 59.Hall, D. A., H. Zhu, X. Zhu, T. Royce, M. Gerstein, and M. Snyder. 2004. Regulation of gene expression by a metabolic enzyme. Science 306482-484. [DOI] [PubMed] [Google Scholar]
- 60.Hensley, P. 1988. Ligand binding and multienzyme complex formation between ornithine carbamoyltransferase and arginase from Saccharomyces cerevisiae. Curr. Top. Cell Regul. 2935-75. [DOI] [PubMed] [Google Scholar]
- 61.Herrero, P., J. Galindez, N. Ruiz, C. Martinez-Campa, and F. Moreno. 1995. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast 11137-144. [DOI] [PubMed] [Google Scholar]
- 62.Herrero, P., C. Martinez-Campa, and F. Moreno. 1998. The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 43471-76. [DOI] [PubMed] [Google Scholar]
- 63.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59407-450. [DOI] [PubMed] [Google Scholar]
- 64.Hittinger, C. T., and S. B. Carroll. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449677-681. [DOI] [PubMed] [Google Scholar]
- 65.Iida, H., and I. Yahara. 1985. Yeast heat-shock protein of Mr 48,000 is an isoprotein of enolase. Nature 315688-690. [Google Scholar]
- 66.Inoue, T., Y. Wang, K. Jefferies, J. Qi, A. Hinton, and M. Forgac. 2005. Structure and regulation of the V-ATPases. J. Bioenerg. Biomembr. 37393-398. [DOI] [PubMed] [Google Scholar]
- 67.Jacob, F. 1977. Evolution and tinkering. Science 1961161-1166. [DOI] [PubMed] [Google Scholar]
- 68.James, C. L., and R. E. Viola. 2002. Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry 413726-3731. [DOI] [PubMed] [Google Scholar]
- 69.Jauniaux, J. C., L. A. Urrestarazu, and J. M. Wiame. 1978. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J. Bacteriol. 1331096-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeffery, C. J. 2004. Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14663-668. [DOI] [PubMed] [Google Scholar]
- 71.Jeffery, C. J. 1999. Moonlighting proteins. Trends Biochem. Sci. 248-11. [DOI] [PubMed] [Google Scholar]
- 72.Johnston, M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1529-33. [DOI] [PubMed] [Google Scholar]
- 73.Johnston, M., and M. Carlson. 1992. Regulation of carbon and phosphate metabolism utilization, p. 193-281. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 74.Jong, A. Y., S. H. Chen, M. F. Stins, K. S. Kim, T. L. Tuan, and S. H. Huang. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52615-622. [DOI] [PubMed] [Google Scholar]
- 75.Junge, W., and N. Nelson. 2005. Structural biology. Nature's rotary electromotors. Science 308642-644. [DOI] [PubMed] [Google Scholar]
- 76.Kane, P. M. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70177-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kapteyn, J. C., L. L. Hoyer, J. E. Hecht, W. H. Muller, A. Andel, A. J. Verkleij, M. Makarow, H. Van Den Ende, and F. M. Klis. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35601-611. [DOI] [PubMed] [Google Scholar]
- 79.Kennedy, M. C., L. Mende-Mueller, G. A. Blondin, and H. Beinert. 1992. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl. Acad. Sci. USA 8911730-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirschner, K., and H. Bisswanger. 1976. Multifunctional proteins. Annu. Rev. Biochem. 45143-166. [DOI] [PubMed] [Google Scholar]
- 81.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klis, F. M., A. Boorsma, and P. W. De Groot. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23185-202. [DOI] [PubMed] [Google Scholar]
- 83.Lacroute, F. 1968. Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. J. Bacteriol. 95824-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lian, H. Y., Y. Jiang, H. Zhang, G. W. Jones, and S. Perrett. 2006. The yeast prion protein Ure2: structure, function and folding. Biochim. Biophys. Acta 1764535-545. [DOI] [PubMed] [Google Scholar]
- 85.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 171236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu, M., D. Ammar, H. Ives, F. Albrecht, and S. L. Gluck. 2007. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 28224495-24503. [DOI] [PubMed] [Google Scholar]
- 87.Lu, M., L. S. Holliday, L. Zhang, W. A. Dunn, Jr., and S. L. Gluck. 2001. Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 27630407-30413. [DOI] [PubMed] [Google Scholar]
- 88.Lu, M., Y. Y. Sautin, L. S. Holliday, and S. L. Gluck. 2004. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 2798732-8739. [DOI] [PubMed] [Google Scholar]
- 89.Ma, H., L. M. Bloom, C. T. Walsh, and D. Botstein. 1989. The residual enzymatic phosphorylation activity of hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 95643-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maddelein, M. L., and R. B. Wickner. 1999. Two prion-inducing regions of Ure2p are nonoverlapping. Mol. Cell. Biol. 194516-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin, R. P., J. M. Schneller, A. J. Stahl, and G. Dirheimer. 1979. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 184600-4605. [DOI] [PubMed] [Google Scholar]
- 93.McAlister, L., and M. J. Holland. 1982. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isoenzymes. J. Biol. Chem. 2577181-7188. [PubMed] [Google Scholar]
- 94.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 27636460-36466. [DOI] [PubMed] [Google Scholar]
- 95.Menéndez, J., J. Delgado, and C. Gancedo. 1998. Isolation of the Pichia pastoris PYC1 gene encoding pyruvate carboxylase and identification of a suppressor of the pyc phenotype. Yeast 14647-654. [DOI] [PubMed] [Google Scholar]
- 96.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 132586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Messenguy, F., and E. Dubois. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 3161-21. [DOI] [PubMed] [Google Scholar]
- 98.Messenguy, F., M. Penninckx, and J. M. Wiame. 1971. Interaction between arginase and ornithine carbamoyltransferase in Saccharomyces cerevisiae. The regulatory site for ornithine. Eur. J. Biochem. 22277-286. [DOI] [PubMed] [Google Scholar]
- 99.Messenguy, F., and J. Wiame. 1969. The control of ornithinetranscarbamylase activity by arginase in Saccharomyces cerevisiae. FEBS Lett. 347-49. [DOI] [PubMed] [Google Scholar]
- 100.Meyer, J., A. Walker-Jonah, and C. P. Hollenberg. 1991. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol. Cell. Biol. 115454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miyakawa, I., S. Fumoto, T. Kuroiwa, and N. Sando. 1995. Characterization of DNA-binding proteins involved in the assembly of mitochondrial nucleoids in the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 361179-1188. [PubMed] [Google Scholar]
- 102.Miyakawa, I., M. Miyamoto, T. Kuroiwa, and R. Sando. 2004. DNA content of individual nucleoids varies depending on the culture conditions of the yeast Saccharomyces cerevisiae. Cytologia 69101-107. [Google Scholar]
- 103.Monod, J., and F. Jacob. 1961. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor Symp. Quant. Biol. 26389-401. [DOI] [PubMed] [Google Scholar]
- 104.Moreno-Herrero, F., P. Herrero, J. Colchero, A. M. Baro, and F. Moreno. 1999. Analysis by atomic force microscopy of Med8 binding to cis-acting regulatory elements of the SUC2 and HXK2 genes of Saccharomyces cerevisiae. FEBS Lett. 459427-432. [DOI] [PubMed] [Google Scholar]
- 105.Nombela, C., C. Gil, and W. L. Chaffin. 2006. Non-conventional protein secretion in yeast. Trends Microbiol. 1415-21. [DOI] [PubMed] [Google Scholar]
- 106.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 2872026-2029. [DOI] [PubMed] [Google Scholar]
- 107.Ogur, M., L. Coker, and S. Ogur. 1964. Glutamate auxotrophs in Saccharomyces 1. I. The biological lesion in the glt1 mutants-2. Biochem. Biophys. Res. Commun. 14193-197. [DOI] [PubMed] [Google Scholar]
- 108.Orita, I., H. Yurimoto, R. Hirai, Y. Kawarabayasi, Y. Sakai, and N. Kato. 2005. The archaeon Pyrococcus horikoshii possesses a bifunctional enzyme for formaldehyde fixation via the ribulose monophosphate pathway. J. Bacteriol. 1873636-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ozimek, P., R. van Dijk, K. Latchev, C. Gancedo, D. Y. Wang, I. J. van der Klei, and M. Veenhuis. 2003. Pyruvate carboxylase is an essential protein in the assembly of yeast peroxisomal oligomeric alcohol oxidase. Mol. Biol. Cell 14786-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ozimek, P. Z., S. H. Klompmaker, N. Visser, M. Veenhuis, and I. J. van der Klei. 2007. The transcarboxylase domain of pyruvate carboxylase is essential for assembly of the peroxisomal flavoenzyme alcohol oxidase. FEMS Yeast Res. 71082-1092. [DOI] [PubMed] [Google Scholar]
- 111.Palomino, A., P. Herrero, and F. Moreno. 2005. Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem. J. 388697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parra, K. J., and P. M. Kane. 1998. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 187064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng, G., and J. E. Hopper. 2000. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 205140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peng, G., and J. E. Hopper. 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. USA 998548-8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Platt, A., and R. J. Reece. 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 174086-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Platt, A., H. C. Ross, S. Hankin, and R. J. Reece. 2000. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc. Natl. Acad. Sci. USA 973154-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Preston, R. A., R. F. Murphy, and E. W. Jones. 1989. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc. Natl. Acad. Sci. USA 867027-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rai, R., J. J. Tate, and T. G. Cooper. 2003. Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity. J. Biol. Chem. 27812826-12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rández-Gil, F., P. Herrero, P. Sanz, J. A. Prieto, and F. Moreno. 1998. Hexokinase PII has a double cytosolic-nuclear localisation in Saccharomyces cerevisiae. FEBS Lett. 425475-478. [DOI] [PubMed] [Google Scholar]
- 120.Rinehart, J., B. Krett, M. A. Rubio, J. D. Alfonzo, and D. Soll. 2005. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 19583-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rodríguez, A., T. De La Cera, P. Herrero, and F. Moreno. 2001. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 355625-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rubio-Texeira, M. 2005. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis. FEMS Yeast Res. 51115-1128. [DOI] [PubMed] [Google Scholar]
- 123.Saiardi, A., H. Erdjument-Bromage, A. M. Snowman, P. Tempst, and S. H. Snyder. 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 91323-1326. [DOI] [PubMed] [Google Scholar]
- 124.Sanz, P., G. R. Alms, T. A. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 201321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sawa, A., A. A. Khan, L. D. Hester, and S. H. Snyder. 1997. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. USA 9411669-11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scannell, D. R., G. Butler, and K. H. Wolfe. 2007. Yeast genome evolution—the origin of the species. Yeast 24929-942. [DOI] [PubMed] [Google Scholar]
- 127.Schneider, A., and L. Marechal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 10509-513. [DOI] [PubMed] [Google Scholar]
- 128.Shewmaker, F., L. Mull, T. Nakayashiki, D. C. Masison, and R. B. Wickner. 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 1761557-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Simpson, J. A., and E. S. C. Weiner. 1992. The compact Oxford English dictionary, 2nd ed. Oxford University Press Inc., New York, NY.
- 130.Sirover, M. A. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432159-184. [DOI] [PubMed] [Google Scholar]
- 131.Smith, F. C., S. P. Davies, W. A. Wilson, D. Carling, and D. G. Hardie. 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453219-223. [DOI] [PubMed] [Google Scholar]
- 132.Sriram, G., J. A. Martinez, E. R. McCabe, J. C. Liao, and K. M. Dipple. 2005. Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 76911-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stewart, M. Q., R. D. Esposito, J. Gowani, and J. M. Goodman. 2001. Alcohol oxidase and dihydroxyacetone synthase, the abundant peroxisomal proteins of methylotrophic yeasts, assemble in different cellular compartments. J. Cell Sci. 1142863-2868. [DOI] [PubMed] [Google Scholar]
- 134.Stucka, R., S. Dequin, J. M. Salmon, and C. Gancedo. 1991. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 229307-315. [DOI] [PubMed] [Google Scholar]
- 135.Tarassov, I., N. Entelis, and R. P. Martin. 1995. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 143461-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 186273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tuttle, D. L., and W. A. Dunn, Jr. 1995. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 10825-35. [DOI] [PubMed] [Google Scholar]
- 138.Tuttle, D. L., A. S. Lewin, and W. A. Dunn, Jr. 1993. Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 60283-290. [PubMed] [Google Scholar]
- 139.Umland, T. C., K. L. Taylor, S. Rhee, R. B. Wickner, and D. R. Davies. 2001. The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p. Proc. Natl. Acad. Sci. USA 981459-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Urrestarazu, L. A., S. Vissers, and J. M. Wiame. 1977. Change in location of ornithine carbamoyltransferase and carbamoylphosphate synthetase among yeasts in relation to the arginase/ornithine carbamoyltransferase regulatory complex and the energy status of the cells. Eur. J. Biochem. 79473-481. [DOI] [PubMed] [Google Scholar]
- 141.van Dijk, R., K. L. Lahchev, A. M. Kram, I. J. van der Klei, and M. Veenhuis. 2002. Isolation of mutants of Hansenula polymorpha defective in the assembly of octameric alcohol oxidase. FEMS Yeast Res. 1257-263. [DOI] [PubMed] [Google Scholar]
- 142.Vissers, S., L. A. Urrestarazu, J. C. Jauniaux, and J. M. Wiame. 1982. Inhibition of ornithine carbamoyltransferase by arginase among yeasts: correlation with energy production, subcellular localization and enzyme synthesis. J. Gen. Microbiol. 1281235-1247. [Google Scholar]
- 143.Vollenbroich, V., J. Meyer, R. Engels, G. Cardinali, R. A. Menezes, and C. P. Hollenberg. 1999. Galactose induction in yeast involves association of Gal80p with Gal1p or Gal3p. Mol. Gen. Genet. 261495-507. [DOI] [PubMed] [Google Scholar]
- 144.Weisman, L. S. 2003. Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37435-460. [DOI] [PubMed] [Google Scholar]
- 145.Wickner, R. B. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264566-569. [DOI] [PubMed] [Google Scholar]
- 146.Wickner, W., and A. Haas. 2000. Yeast homotypic vacuole fusion: a window on organelle trafficking mechanisms. Annu. Rev. Biochem. 69247-275. [DOI] [PubMed] [Google Scholar]
- 147.Williamson, D. H., and D. J. Fennell. 1979. Visualization of yeast mitochondrial DNA with the fluorescent stain DAPI, p. 728-733. Methods Enzymol. 56728-733. [DOI] [PubMed] [Google Scholar]
- 148.Winge, O., and C. Roberts. 1948. Inheritance of enzymatic characters in yeast in the phenomenon of long term adaptation. C. R. Trav. Carlsberg Ser. Physiol. 24263-315. [Google Scholar]
- 149.Xu, Z., and W. Wickner. 1996. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J. Cell Biol. 132787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yano, K., and T. Fukasawa. 1997. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 941721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yuan, W., D. L. Tuttle, Y. J. Shi, G. S. Ralph, and W. A. Dunn, Jr. 1997. Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J. Cell Sci. 1101935-1945. [DOI] [PubMed] [Google Scholar]
- 152.Zelenaya-Troitskaya, O., P. S. Perlman, and R. A. Butow. 1995. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 143268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zenke, F. T., R. Engles, V. Vollenbroich, J. Meyer, C. P. Hollenberg, and K. D. Breunig. 1996. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 2721662-1665. [DOI] [PubMed] [Google Scholar]
- 154.Zenke, F. T., W. Zachariae, A. Lunkes, and K. D. Breunig. 1993. Gal80 proteins of Kluyveromyces lactis and Saccharomyces cerevisiae are highly conserved but contribute differently to glucose repression of the galactose regulon. Mol. Cell. Biol. 137566-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zimmermann, F. K., and I. Scheel. 1977. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol. Gen. Genet. 15475-82. [DOI] [PubMed] [Google Scholar]