Abstract
The insulin signaling pathway is of pivotal importance in metabolic diseases, such as diabetes, and in cellular processes, such as aging. Insulin activates a tyrosine phosphorylation cascade that branches to create a complex network affecting multiple biological processes. To understand the full spectrum of the tyrosine phosphorylation cascade, we have defined the tyrosine-phosphoproteome of the insulin signaling pathway, using high resolution mass spectrometry in combination with phosphotyrosine immunoprecipitation and stable isotope labeling by amino acids in cell culture (SILAC) in differentiated brown adipocytes. Of 40 identified insulin-induced effectors, 7 have not previously been described in insulin signaling, including SDR, PKCδ binding protein, LRP-6, and PISP/PDZK11, a potential calcium ATPase binding protein. A proteomic interaction screen with PISP/PDZK11 identified the calcium transporting ATPase SERCA2, supporting a connection to calcium signaling. The combination of quantitative phosphoproteomics with cell culture models provides a powerful strategy to dissect the insulin signaling pathways in intact cells.
Keywords: diabetes, insulin action, tyrosine phosphorylation
Reversible phosphorylation is a major regulatory mechanism controlling the activity of proteins. Many signaling pathways, including the insulin/IGF-1 signaling pathway, transduce signals from the cell surface to downstream targets via tyrosine kinases and phosphatases (1). Insulin or IGF-1 binding initiates a complex cascade of events, starting with phosphorylation of specific tyrosine residues on the insulin and the IGF-1 receptors (2). Once activated, these receptors phosphorylate a number of docking proteins; the best characterized are the insulin receptor substrate (IRS) proteins 1–4 (3). IRS 1–4 interact with other intracellular signaling molecules primarily through SH2 domains leading to activation of several downstream pathways. These in turn coordinate and regulate vesicle trafficking, protein synthesis, and glucose uptake. The insulin receptor and its substrates, therefore, constitute the first critical node in the insulin signaling network (1) and thus define the full set of proteins that are tyrosine phosphorylated upon insulin stimulation and provide information that is central to determining the molecules involved in this signaling network.
Mass spectrometry (MS)-based proteomics has become increasingly powerful not only to identify complex protein mixtures but also regulated protein modifications (4). We have studied tyrosine phosphorylated effectors and interactors in the EGF pathway (5, 6) and others have used MS to study tyrosine phosphorylation of the insulin pathway in white adipocytes (7). To quantify the time course of stimulation, we employ stable isotope labeling with amino acids in cell culture (SILAC) (8). Labeling three cell states by SILAC allows measuring a time course of activation of tyrosine phosphorylation by quantifying the relative protein amounts after anti-phosphotyrosine immunoprecipitation. Here, we quantitatively assess temporal dynamics of tyrosine phosphorylation events upon insulin stimulation in differentiated brown adipocytes. We identify different protein “effector” classes that influence distinct branches of the insulin signaling pathway.
Results
SILAC Quantification Reveals 40 Insulin-Induced Effectors in Differentiated Brown Adipocytes.
To characterize specific signaling events downstream of the insulin/IGF-1 receptors, we stimulated differentiated brown adipocytes with insulin and quantitatively analyzed the tyrosine phosphoproteome by MS compared with unstimulated control samples. For accurate quantitation of tyrosine phosphorylated (pY) proteins, stable isotope labeling in cell culture (SILAC) was used (8). In each experiment, one cell population was grown in unlabeled medium as a control and a second population was grown in medium substituted with 13C615N4-Arg (Arg-10) and 13C615N2-Lys (Lys-8). Peptides derived from proteins of the two cell populations then occur in doublets, with either 10-Da or 8-Da mass differences. After complete incorporation of the heavy or light SILAC amino acids in brown preadipocytes, cells were allowed to differentiate into brown adipocytes as described in ref. 9 (Fig. 1) and stimulated with 100 nM insulin for 5 min. Tyrosine phosphorylated proteins were immunoprecipitated by anti-pY antibodies from the lysates of the combined cell populations, separated by 1D gel electrophoresis and measured by MS. Quantitative analysis of the peptides identifying each protein resulted in a fold change (“activation”) between control and insulin-stimulated cells. The relative standard deviation (%SD) of all proteins revealed a quantitation precision better than ±20% [supporting information (SI) Fig. 4]. As in the experiments reported in ref. 6, we chose a minimum of a 1.3-fold change as biologically significant. In four independent experiments, we identified 33 proteins precipitated by anti-pY antibodies that had a 1.3-fold or higher change in the peak intensities upon 5 min of insulin stimulation (Table 1 and SI Tables 2–5).
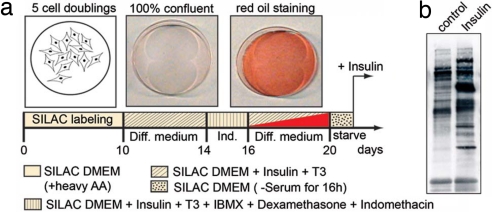
Fig. 1.
Schematic overview of the differentiation protocol for immortalized brown adipocytes. (a) After SILAC labeling, cells were grown until confluence and differentiation was induced. At the indicated days, cells were stained with red oil, which is an indicator for differentiation. After 20 days, the differentiated cells were stimulated with insulin (5 min) for Western blot analysis. (b) After immunoprecipitation with the antiphosphotyrosine antibody (4G10) and SDS/PAGE, blots were probed with the same antibody. Diff. medium, differentiation medium; Ind., induction medium.
Table 1.
Proteins activated after 5 min of insulin treatment
| Ac. Id. | Name | Ratio | SD | pTyr sites |
|---|---|---|---|---|
| Known effectors of the insulin pathway | ||||
| P15208 | Insulin receptor | 23.0 | 7.7 | pY1185 |
| Q60751 | IGF-1 receptor | 6.3 | 2.5 | — |
| P35569 | IRS-1 | 12.7 | 1.5 | pY891, pY983, pY1173 |
| P81122 | IRS-2 | 4.6 | 2.4 | pY970 |
| Q91VW7 | Gab-1 | 5.8 | 3.2 | pY259 |
| Q9JID9 | APS | 16.5 | 6.6 | pY618 |
| P98083 | Shc | 4.4 | 0.6 | pY423 |
| P22682 | cCbl | 3.9 | 0.5 | — |
| P49817 | Caveolin-1 | 4.7 | 1.4 | pY14 |
| Q9WVC3 | Caveolin-2 | 3.5 | 2.1 | — |
| O54724 | Caveolin-60 / PRTF | 2.2 | 0.4 | — |
| Q91ZX7 | LRP-1 | 3.1 | 0.8 | — |
| P26450 | PI3K p85 alpha | 1.8 | 0.6 | — |
| P42337 | PI3K p110 alpha | 1.7 | 0.5 | pY317 |
| O08908 | PI3k p85 beta | 2.8 | 1.0 | — |
| Q8BXD7 | PI3K p110 beta | 1.7 | 0.6 | — |
| Q80YB9 | SHIP-2 | 6.2 | 2.4 | — |
| Q7TMK9 | HnRNP Q | 2.6 | 1.0 | — |
| O54967 | Ack1 | 1.3 | 0.02 | pY842, pY533 |
| Q63844 | ERK-1 | 5.6 | 3.5 | pY185 |
| P27703 | ERK-2 | 6.7 | 2.6 | — |
| Q60631 | Grb2 | 1.3 | 0.02 | — |
| P35235 | SHP-2 | 2.6 | 1.2 | pY584 |
| P57780 | alpha-Actinin 4 | 1.7 | 0.3 | — |
| Q9ESJ4 | SPIN90 | 1.4 | 0.2 | — |
| P46935 | Nedd-4 | 1.3 | 0.1 | — |
| Proteins not previously reported in insulin signaling | ||||
| Q9CZG9 | PDZK11 | 10.5 | 1.1 | — |
| O88572 | LRP-6 | 3.2 | 0.7 | — |
| Q921C5 | Bicaudal D2 | 1.9 | 0.3 | — |
| Q63918 | SDR | 1.9 | 0.2 | — |
| Q9DBP6 | WW bdg. protein 2 | 1.8 | 0.4 | — |
| Q8C064 | PKC delta bdg. Prot. | 1.7 | 0.1 | — |
| P39447 | Tight junction ZO-1 | 1.6 | 0.2 | — |
| Downregulated proteins | ||||
| P20152 | Vimentin | 0.6 | 0.1 | — |
| Q9ES28 | Rho-Gef 7 | 0.6 | 0.1 | — |
| P07356 | Annexin A2 | 0.6 | 0.1 | — |
| Q8VI36 | Paxillin | 0.6 | 0.1 | pY117 |
| Q5F258 | GIT-1 | 0.7 | 0.1 | — |
| Q64727 | Vinculin | 0.7 | 0.0 | — |
| P34152 | FAK-1 | 0.7 | 0.1 | pY614 |
The detected tyrosine phosphorylation sites are listed in the right column. Ac. Id., Swiss-Prot/TrEMBL.
Using this immunoprecipitation protocol, both the tyrosine phosphorylated proteins and molecules that have bound tightly to them can be detected. Of the 33 proteins identified, in most cases, the protein was known to be tyrosine phosphorylated or to associate with such a protein. These included proteins in the insulin pathway with a wide variety of cellular functions, including the insulin receptor substrates IRS-1, IRS-2, APS, and Shc, which served as positive controls. The identification of other effectors from different branches of the pathway, such as MAP kinases, showed that experimental conditions were also suitable to detect downstream factors. For this and subsequent experiments, we term the up-regulated proteins as activated proteins, defined as proteins with a fold change >1.3 after insulin stimulation. Note that this activation could in principle down-regulate insulin signaling. Proteins with a fold change <0.7 were termed “deactivated.” The quantitative MS data were confirmed by standard Western blot analysis (Fig. 2).
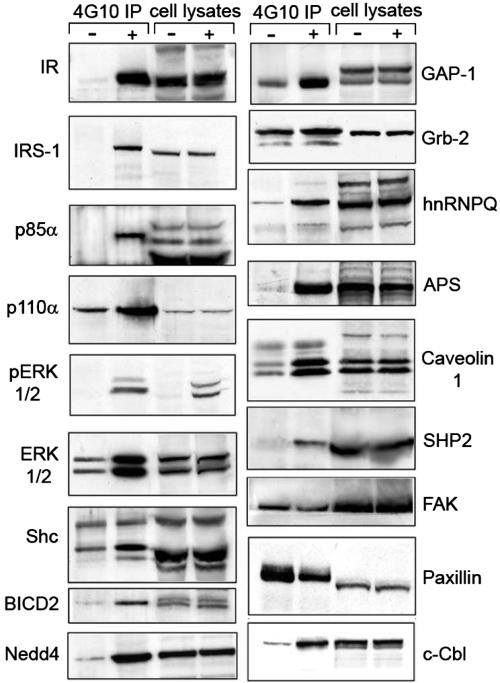
Fig. 2.
Western blot analysis of selected insulin-dependent effectors. Brown adipocytes were stimulated with insulin for 5 min. After immunoprecipitation with an anti-phosphotyrosine antibody and SDS/PAGE, blots were probed with antibodies against the indicated proteins (Right). (Left) Represents the analysis of whole cell lysates to demonstrate equal loading. −, control, unstimulated cells; +, insulin treatment; IP, immunoprecipitation.
Analysis of Temporal Activation Profiles Revealed Different Categories of Insulin-Induced Effectors.
Signaling events are not regulated by a simple “on” and “off” switch at a single time point, but require a highly coordinated kinetic cascade of effectors and events. To capture the temporal dynamics of tyrosine phosphorylated proteins, we used a triple labeling SILAC experiments in which three differentially labeled cell populations were stimulated for different times (see also SI Fig. 5). The temporal proteome is encoded by the SILAC state of each cell population and the relative intensities of peptides from each state are a direct measure of their presence in the pY immunoprecipitate (6). The combination of two three-state experiments allowed us to generate a time profile for 0, 1, 5, 10, and 20 min (Fig. 3). The kinetic analysis revealed distinctive temporal patterns of insulin signaling. Based on the similarity of patterns, we grouped known and unknown candidate effectors into different branches of the pathway. As expected, one of the first tyrosine phosphorylation events upon insulin stimulation occurred on the IR itself. The IR was activated ≈20-fold compared with unstimulated control within the first minute and remained fully activated for the next 5 min. After 20 min, IR phosphorylation/activation levels still remained at 85% of the maximum, reflecting the saturating insulin concentrations in this experiments (Fig. 3a and SI Tables 6 and 7). Insulin can also bind to the IGF-1 receptor and vice versa, albeit with a 10- to100-fold lower affinity (10). In addition, some IGF-1 and insulin receptors exist as heterodimer hybrids allowing cross-phosphorylation of their intracellular kinase domains (11). Reflecting these mechanisms, the IGF-1R showed an early activation profile, similar to the insulin receptor, but with a fivefold lower activation. Therefore, at high insulin concentrations, downstream effectors may potentially reflect activity of either or both receptors (see below for a dissection of these pathways).
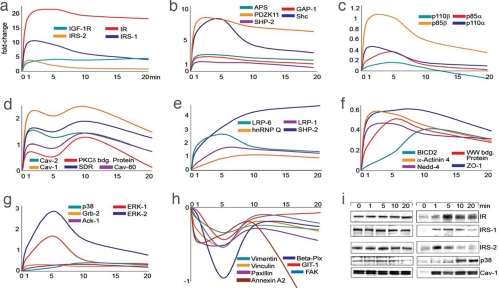
Fig. 3.
Activation profiles of different categories of insulin-induced effectors. (a–h) To directly compare the kinetic profile of activated and deactivated effectors, all fold changes were normalized to zero (x − 1). Normalized inverted ratios were calculated for ratios smaller than one [1 − (1/x)]. (i) Western blot analysis of selected effectors. Brown adipocytes were stimulated for the indicated time intervals. After immunoprecipitation with anti-phosphotyrosine antibodies and SDS/PAGE, blots were probed with antibodies against the indicated proteins.
To date, at least 11 substrates of the IR and IGF-1R have been identified in different cellular systems (1). Of these, we detected the activation of five substrates in brown adipocytes. These included IRS-1, IRS-2, Gab-1, Shc, APS, and c-Cbl. For most of these substrates, we also sequenced the tyrosine phosphorylated peptides (Table 1, SI Fig. 6, and SI Table 8). For an accurate assignment of the phosphorylation sites in phosphopeptides, we used the posttranslational modification (PTM) score. The PTM score is based on an algorithm that tests and scores all possible combination of serine, threonine, and tyrosine phosphorylations within a potential phosphopeptide (12). The substrate proteins IRS-1 and Shc showed a similar kinetics of activation as the insulin receptor (Fig. 3 a and b), suggesting physical proximity as has recently been found for Shc upon EGFR activation (13). In contrast, IRS-2 showed its highest activation after 1 min of stimulation, followed by rapid deactivation. This transient activation profile is consistent with the results of earlier experiments using L6 muscle cells (14). The rapid abrogation of the IRS-2 signal suggests fast negative feedback mechanisms, such as serine phosphorylation or activation of phosphotyrosine phosphatases. In support of this notion, we identified several IRS-2 peptides with phosphorylated S/T- sites (SI Fig. 6 and SI Table 8).
Tyrosine phosphorylation of substrate/adaptor proteins creates docking sites for several SH2-containing proteins that are central to insulin signaling (15), most importantly the regulatory subunit (p85) of the phosphatidylinositol 3-kinase (PI3K) (16). We detected the α and β isoforms of both the regulatory p85 and catalytic p110 subunit of PI3K (Fig. 3c) with the apparently most abundant species being p85β. PI3K activation leads to the generation of 3-phosphorylated lipids (PIP3) and their synthesis is counteracted by the SH2-domain-containing lipid phosphatase SHIP2 (17). The appearance of SHIP2 in the activation profile showed slower kinetics, being half-max at ≈3 min and reaching a fivefold maximum after ≈15 min (Fig. 3e).
The other major branch of the insulin signaling pathway is the activation of the Ras-MAP kinase cascade, which plays a role in control of cell growth and gene expression. Initial docking proteins in this cascade are the SH2-domain-containing protein Shc (18) and Gab-1 (19). After Shc activation (Fig. 3b) a strong activation of Erk-1 and Erk-2 was observed (Fig. 3g). Both MAP-kinases reached a maximum activation after 5 min and decreased at the 10- and 20-min time points. The stress induced MAP-kinase p38 showed only weak activation between 10 and 20 min (Fig. 3g).
An acute effect of insulin treatment is the rapid formation of caveolin-enriched lipid raft subdomains within the plasma membrane (20). Upon stimulation, the adapter protein containing a PH and SH2 domain (APS) (21) interacts with Cbl and CAP and relays the insulin signal to the accruing lipid raft structures. Caveolin-1 is known to become tyrosine phosphorylated and acts as a scaffolding protein to organize signaling molecules and receptor tyrosine kinases, such as the insulin receptor (20). Consistent with this finding, we detected Caveolin-1 (pY 14) and Caveolin-2 in the pY immunoprecipitated fraction (Fig. 3d). Both proteins showed interesting biphasic kinetics, with two maxima at 1 and 10 min and an intermittent decrease at the 5-min time point. This biphasic stimulation was supported by Western blot analysis (Fig. 3i). Polymerase I release transcription factor (PTRF) showed a similar activation profile to the caveolins. PTRF has been implicated in caveolar formation and is also known as Caveolin-60 (22). Two proteins not previously implicated in insulin signaling, serum deprivation response protein (Sdr) (23) and the PKCδ binding protein (also known as sdr-related gene product that binds to C-kinase) (24), had a biphasic activation profile, suggesting they might also associate with insulin dependent formation of lipid rafts.
The family of low density lipoprotein receptor-related proteins (LRP) is also known to be recruited to caveolae structures in response to insulin treatment (25). We identified LRP-1 as part of the activation response (Fig. 3d); however, in contrast to the caveolins, LRP-1 showed a slower increase and only one maximum at the 5-min time point, as did LRP-6, another lipoprotein that had not been implicated in insulin signaling (26). LRP-6 has also been reported to be a part of the Wnt/beta-catenin pathway (27) and could be an interesting candidate for cross-talk of the insulin and Wnt pathways.
Another effect of insulin treatment is the rapid reorganization of the actin cytoskeleton mediated mainly by small G proteins (28). Focal adhesion signaling complexes are also involved in this process. Fig. 3h (see also Fig. 2 for Western blot) shows a number of proteins involved in focal adhesion formation, such as the focal adhesion kinase FAK, paxillin, Beta-PIX, vinculin, and the G protein-coupled receptor kinase interactor 1 (GIT1). Somewhat surprisingly, these proteins showed deactivation in response to insulin. In most cases this was transient, reaching a nadir at 5 min. Annexin A2, a calcium-dependent phospholipid-binding protein, displayed a similarly rapid down regulation, followed by recovery and a second downward trend after 10 min. Other effectors, such as the Nck binding protein Spin 90, α-actinin-4, the cytoskeleton-like bicaudal D protein homolog 2, the tight junction ZO-1 protein, and the PDZ-containing protein PDZK11/PISP showed prominent activation and are likely further candidates to connect insulin signaling to cytoskeletal reorganization.
Signal attenuation is critical for the fine-tuning of signaling events. This attenuation can be achieved by serine/threonine phosphorylation or ubiquitination. A number of ubiquitin ligases have been identified as important for the fast ubiquitination and subsequent degradation of receptor tyrosine kinases (29). Both c-Cbl and Nedd4 (30), two ubiquitin ligases, were identified after 5 min, and the Nedd4 activation profile is shown in (Fig. 3f).
PISP/PDZK11-pY7 Peptide Pull-Down Revealed Interaction with Myosins and with the Calcium Transporting ATPase SERCA2A.
Among the proteins not previously described as insulin-induced candidate effectors, we were intrigued by the PDZ-containing protein PISP/PDZK11 because its dynamic profile and fold activation are very similar to IRS-1 (Fig. 3b). This ubiquitously expressed protein is named PISP for plasma membrane Ca2+ ATPase (PMCA)-interacting single-PDZ-protein (31). The early activation profile and the high fold change of PISP/PDZK11 suggest a direct function in the early steps of insulin signaling, which might link to the previously observed acute effects of insulin on calcium flux (32) To gain more insight into the function of the activation of the PISP/PDZK11 protein, we synthesized two tyrosine-containing desoxy-biotin linked peptides derived from PISP/PDZK11 (including peptides containing tyrosines pTyr7 and pTyr10 in a phosphorylated and nonphosphorylated version to find phosphorylation-dependent binding partners (33). These phosphorylated and nonphosphorylated peptides were used as bait and incubated with lysates of SILAC light and heavy labeled cells, respectively. Peptides and binding partners were purified by streptavidin-coated beads, and the eluates were combined for in solution digest and MS analysis. We found that the PISP/PDZK11-pY7-containing sequence specifically interacted with myosin regulatory light chain 2, and two calcium binding proteins—SERCA2 and S100 calcium-binding protein A4 (SI Fig. 6 and SI Tables 9 and 10). Because PISP/PDZK11 is a potential calcium ATPase binding partner and we find that it interacts with proteins involved in calcium homeostasis, it is a possible link between insulin and calcium signaling.
Discussion
Here, we used high resolution quantitative proteomics technology to discover candidate effectors in the insulin pathway to delineate their temporal activation profile. As our model systems, we used immortalized adipocytes derived from brown fat tissue of newborn mice. Using the SILAC technology and anti-pY immunoprecipitation to quantify differentially pY phosphorylated proteins, we identified at total of 40 effectors in differentiated brown fat cells. Seven of these had not been previous reported to be involved in insulin signaling, indicating the power of this approach to discover new components of this signaling system. For the majority of insulin-activated proteins, for which tyrosine phosphorylation had not been reported, we were also able to sequence and quantitate the pY-containing peptides. Thus, this approach is not only suitable for the detection, but also the quantitation of tyrosine phosphorylated proteins upon insulin treatment.
Signaling events via hormones and receptor tyrosine kinases are very fast processes. To resolve these phosphorylation events, we included in our analysis very early (60 s) and two later (10–20 min) time points. Such a five-time-point kinetic dependably resolved the diverse temporal dynamics of tyrosine phosphorylation in this very complex signaling cascade (Fig. 2). One example of accurate profiling was highlighted by the different kinetics of phosphorylation of IRS-1 (pY 983, pY1173, and pY891) and IRS-2 (pY970 and pY528). The steady elevation of IRS-1activation profile compared with the fast rise and fall of the IRS-2 activation reflects the different roles of these two key components of this insulin signaling nodes (1). Besides the identification of pY sites on IRS1/2, we detected several serine/threonine sites on IRS-1 and IRS-2. Studies reported that S/T phosphorylation on IRS proteins leads to inhibition of the insulin signal cascade (34). For example, phosphorylated serine 307 on IRS1 leads to attenuated insulin signaling in patients with type 2 diabetes (35). Therefore, the phosphorylation on serine/threonine residues seems to be a crucial step in inhibiting insulin signals. Although the physiological mechanisms remain poorly defined, the phosphorylation may block further tyrosine phosphorylation or induce intracellular degradation. Notably, we measured seven serine/threonine phosphorylation sites at these early time points. Although interpretation of their time course is complicated because the phosphotyrosine-containing protein was immunoprecipitated, they are potential new modulator sites for the fast inhibition of IRS-2 function. Future studies of the temporal activation profile of these phosphopeptides will help to make clear the dynamic of insulin-induced IRS2 activation and deactivation.
After this first layer of tyrosine phosphorylated proteins, further downstream targets, such as members of the MAP kinase pathway also become tyrosine phosphorylated. Activation for the MAP kinases Erk-1 and Erk-2 was very transient, rising rapidly up to 5 min then declining by >60–75%. In contrast, p38 was activated weakly and at later time points of 10–20 min.
Our unbiased phosphoproteomic approach allowed us to identify several components of the insulin signaling network. Among the novel candidate effectors, PISP/PDZK11, a protein with a single PDZ domain, showed the highest fold activation and similar kinetics as IRS-1. PDZK11 has been described as an interactor of the plasma membrane Ca2+-ATPase (PMCA) and as an interactor for the Menkes copper ATPase (AIPP1) (36). The dramatic increase in tyrosine phosphorylation of PDZK11 after insulin treatment strongly suggests its involvement in calcium signaling as part of the insulin cascade. There are several instances in which extracellular hormone stimulation leads to a rise of cytosolic Ca2+, which in turn is quickly removed by calcium-pumps that either eject the calcium ions into the lumen of the endoplasmic reticulum or into the extracellular space (37). Recently, another PDZ domain-containing protein, Enigma, was found to interact with APS upon insulin stimulation in 3T3-L1 adipocytes, and this complex was critical to insulin-induced action cytoskeleton remodeling and Glut4 translocation (38). Interestingly, APS and several other cytoskeletal components become highly tyrosine phosphorylated upon 5 min of insulin stimulation as revealed by the current study. Thus, this forms a phosphoproteome network of insulin signaling involving the immediate substrate of insulin receptor APS, the PDZ domain-containing protein PDZK11, calcium binging proteins, and the cytoskeletons. This network may mediate critical function of insulin, such as regulation of glucose uptake. Insulin increases the phosphorylation of the myosin regulatory light chain 2 in adipocytes via the Ca2+/Calmodulin-dependent myosin light chain kinase, which subsequently regulates GLUT4 translocation (39). With our pull down strategy, we identified the myosin regulatory light chain 2 as an interactor of the PISP/PDZK11 protein, and this may be a link to Ca2+ dependent phosphorylation and glucose uptake. Previously, the sarcoplasmatic Ca2+-ATPases SERCA1 and SERCA2, have been shown to interact with the IRS proteins (40). Using a recently developed peptide pull-down approach based on SILAC, we identified SERCA2 and the S-100 calcium binding protein as specific binding partners of the tyrosine phosphorylated PISP/PDZK11. It is known that the control of calcium levels is tightly connected to the insulin pathway, and our results implicate PISP/PDZK11 in this process. The peptide pull-down approach used here is generic for the determination of binding partners of candidate proteins from proteomic screens.
Another acute effect of insulin stimulation is the formation of caveolin-containing lipid rafts and targeting receptors to those structures (41). Caveolins 1 and 2 are known to be involved in insulin-induced lipid raft formation. In our experiments, other proteins, such as caveolin-60/PTRF, the serum deprivation response factor (SDR), and the PKCδ binding protein also showed a “caveolin-like” activation profile, based on temporal analysis. We hypothesize that the two proteins have a related function during insulin-induced organization of caveola structures as reflected by their similar biphasic activation profile. The SDR and PKCδ binding protein belong to a class of molecules called STICKs (substrates that interact with C-kinase) (42). SDR interacts with PKCα (43), whereas PKCδ binding protein with PKCδ. Both proteins have a primary function in targeting these two isoforms of PKC to caveolae. This targeting of PKCs to the cell membrane facilitates interactions with membrane lipids and with other signaling proteins, for example with IRS-1 (44). Oriente et al. (44) have shown that PKCα, and IRS-1 form a multiprotein complex with the adaptor protein 14-3-3ε. Although the presence of 14-3-3ε is not necessary for the formation of the complex, it modulates the PKCα activity, thereby regulating insulin signaling and degradation. In our study, we did not detect 14-3-3 adaptor proteins, probably because they are not tyrosine phosphorylated or bound to tyrosine phosphorylated sites or because the PKCδ-IRS-1 complex is regulated by a different mechanism.
The low density lipoprotein receptor-related proteins (LRPs) are another important protein family involved in formation of lipid rafts (45). Besides LRP-1, we also detected LRP-6, which was not known to be involved in insulin signaling. Recently, LRP-6 was linked to the Wnt canonical pathway (26) and reported to be an inhibitor of the glycogen synthase kinase 3 (GSK3) (46). Thus, the observed activation of LRP-6 upon insulin stimulation could be a potential link between the insulin and the Wnt canonical pathway.
Another important node within the insulin pathway is PI3K. Interestingly, the regulatory p85β isoform, which is less abundant in the cell, was detected with a higher fold activation compared with the p85α isoform. This observation raises two possibilities. One is that p85β has an increased binding via its SH2 domain to activated insulin receptor substrates due either to compartmentalization or higher affinity, and therefore a higher number of p85β subunits were detected compared with p85α in the activated state. The second possibility is that insulin stimulation results in an increased level of tyrosine phosphorylated p85β as a dimer and/or monomer. Because we did not detect any phosphorylation sites for the regulatory subunits, their exact phosphorylation status after insulin stimulation needs to be assessed by future experiments.
We detected several proteins that showed a significant decreased SILAC-ratio, and most of these factors are involved in the formation of focal adhesions (47). In this case, however, insulin treatment promoted a decrease in these proteins in the activation profile. We cannot determine whether the observed decreasing level of these proteins over time is due to a direct dephosphorylation of these focal adhesion proteins or whether dephosphorylation occurs secondary to insulin-induced cytoskeletal rearrangements (48). In any case, insulin stimulation clearly leads to rapid reorganization of the cytoskeleton, and our study revealed that the Beta-PIX, GIT1, FAK, and paxillin multiprotein complex plays a critical role in that process.
In summary, our data clearly demonstrate that the SILAC approach is well suited for the analysis and comparison of signaling events in cell lines. The quantitation of tyrosine phosphorylated proteins in response to insulin allowed us to discover several new candidate effectors and associate those proteins with different branches of the insulin pathway. The identification of effectors from diverse pathways, including the Wnt pathway, provides clues for studying cross-talk between signaling pathways. Future analysis of the global site-specific phosphoproteome (5) and studies in normal tissues of an intact mouse will allow us to define additional insulin-induced phosphorylation events and the cross-talk between different cascades. It would also be interesting to complement the study of genetic models by quantitative phosphoproteomics with similar analysis of the phosphoproteome in the presence of pharmacological agents. This could help to analyze physiological mechanism of the insulin pathway and to find new therapeutic targets for the treatment of diseases, such as type 2 diabetes mellitus.
Methods
Cell Culture and SILAC.
Brown preadipocyte cell lines were established from wild type and IRS-1 or IRS-2 deficient mice as described in ref. 9. After a 16-h starvation period, the cells were treated with insulin (1 μg/ml; Sigma). After hormone incubation for 1–20 min at 37°C, the media were removed, and the cells were immediately lysed with ice-cold modified RIPA buffer. For SILAC experiments, the DMEM was custom-made and was deficient for l-Arg, and l-Lys (Invitrogen). l-argine-6, l-arginine-10, l-lysine-4, and l-lysine-8 were from Sigma–Aldrich.
Antibodies and Immunoprecipitation.
Antibodies against the following proteins were used: immobilized anti-phosphotyrosine antibody (4G10; Upstate) and anti-pY antibody (P-Tyr-100; Cell Signaling); for Western blot analysis, insulin receptor β, IRS-1, p85α (Santa Cruz), IRS-2, p42/p44 MAP kinase, phospho-p42/p44 MAP kinase, phospho p38 (Cell Signaling), Shc, Grb-2, SHP2, Paxillin, Nedd4, c-CBL (BD Transduction ), FAK, Caveolin-1 (Upstate), hnRNP Q, GAP-1, APS (Abcam), and BICD2 (kindly provided by C. Hoogenraad, Erasmus University, Rotterdam, The Netherlands). Immunoprecipitation was performed as described in ref. 6.
Mass Spectrometric Analysis.
A detailed description of the mass spectrometric measurements and peptide pull downs can be found in SI Methods. Briefly, after SDS/PAGE and peptide extraction (49), samples were processed for liquid chromatography-mass spectrometry (GeLC-MS). Reverse phase nano-LC was performed on an Agilent 1100 nanoflow LC system (Agilent Technologies). The LC system was coupled to a 7-Tesla LTQ-FT instrument (Thermo Electron) equipped with a nanoeletrospray source (Proxeon). The following search parameters were used in all MASCOT searches: fully tryptic peptides, maximum of two missed trypsin cleavages, cysteine carbamidomethylation, methionine oxidation, pSTY, N-term protein acetyl, and SILAC labels Lys-D4, Lys-8, Arg-6, and Arg-10. The maximum initial mass tolerance for MS scans was 10 ppm and 0.5 Da for MS/MS and MS3 scans. Only proteins identified and quantified with at least two peptides with cores >20. MSQuant were considered for validation.
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of Center for Experimental BioInformatics (Odense, Denmark), the Department of Proteomics and Signal Transduction (Martinsried, Germany), and all Diabetes Genome Anatomy Project members for fruitful discussions. We thank S. Krueger for excellent technical assistance and M. S. Cambridge for constructive comments on the manuscript. The work in the Center for Experimental BioInformatics was supported by the Danish National Research foundation and National Institute of Health Grants DK60837 (to the Diabetes Genome Anatomy Project) and DK3320. This work was supported by a long-term fellowship of the European Molecular Biology Organization (to I.M.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711713105/DC1.
References
- 1.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signaling pathways: Insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 2.Nandi A, Kitamura Y, Kahn CR, Accili D. Mouse models of insulin resistance. Physiol Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 3.Saltiel AR, Kahn CR. Insulin signaling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 5.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 7.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes. 2006;55:2171–2179. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 8.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Fasshauer M, et al. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol Cell Biol. 2001;21:319–329. doi: 10.1128/MCB.21.1.319-329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Entingh-Pearsall A, Kahn CR. Differential roles of the insulin and insulin-like growth factor-I (IGF-I) receptors in response to insulin and IGF-I. J Biol Chem. 2004;279:38016–38024. doi: 10.1074/jbc.M313201200. [DOI] [PubMed] [Google Scholar]
- 11.Pessin JE, Frattali AL. Molecular dynamics of insulin/IGF-I receptor transmembrane signaling. Mol Reprod Dev. 1993;35:339–344. doi: 10.1002/mrd.1080350404. discussion 344–335. [DOI] [PubMed] [Google Scholar]
- 12.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci USA. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dengjel J, et al. Quantitative proteomic assessment of very early cellular signaling events. Nat Biotechnol. 2007;25:566–568. doi: 10.1038/nbt1301. [DOI] [PubMed] [Google Scholar]
- 14.Inoue G, Cheatham B, Emkey R, Kahn CR. Dynamics of insulin signaling in 3T3–L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J Biol Chem. 1998;273:11548–11555. doi: 10.1074/jbc.273.19.11548. [DOI] [PubMed] [Google Scholar]
- 15.White MF. The IRS-signaling system: A network of docking proteins that mediate insulin action. Mol Cell Biochem. 2007;182:3–11. [PubMed] [Google Scholar]
- 16.Ueki K, et al. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J Biol Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- 17.Clement S, et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 18.Sasaoka T, et al. Evidence for a functional role of Shc proteins in mitogenic signaling induced by insulin, insulin-like growth factor-1, and epidermal growth factor. J Biol Chem. 1994;269:13689–13694. [PubMed] [Google Scholar]
- 19.Holgado-Madruga M, Wong AJ. Role of the Grb2-associated binder 1/SHP-2 interaction in cell growth and transformation. Cancer Res. 2004;64:2007–2015. doi: 10.1158/0008-5472.can-03-2886. [DOI] [PubMed] [Google Scholar]
- 20.Kimura A, Mora S, Shigematsu S, Pessin JE, Saltiel AR. The insulin receptor catalyzes the tyrosine phosphorylation of caveolin-1. J Biol Chem. 2002;277:30153–30158. doi: 10.1074/jbc.M203375200. [DOI] [PubMed] [Google Scholar]
- 21.Moodie SA, Alleman-Sposeto J, Gustafson TA. Identification of the APS protein as a novel insulin receptor substrate. J Biol Chem. 1999;274:11186–11193. doi: 10.1074/jbc.274.16.11186. [DOI] [PubMed] [Google Scholar]
- 22.Vinten J, et al. A 60-kDa protein abundant in adipocyte caveolae. Cell Tissue Res. 2001;305:99–106. doi: 10.1007/s004410100389. [DOI] [PubMed] [Google Scholar]
- 23.Gustincich S, et al. The human serum deprivation response gene (SDPR) maps to 2q32–q33 and codes for a phosphatidylserine-binding protein. Genomics. 1999;57:120–129. doi: 10.1006/geno.1998.5733. [DOI] [PubMed] [Google Scholar]
- 24.Izumi Y, et al. Protein kinase Cdelta-binding protein SRBC whose expression is induced by serum starvation. J Biol Chem. 1997;272:7381–7389. doi: 10.1074/jbc.272.11.7381. [DOI] [PubMed] [Google Scholar]
- 25.Gotthardt M, et al. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- 26.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 27.Mi K, Johnson GV. Role of the intracellular domains of LRP5 and LRP6 in activating the Wnt canonical pathway. J Cell Biochem. 2005;95:328–338. doi: 10.1002/jcb.20400. [DOI] [PubMed] [Google Scholar]
- 28.Tsakiridis T, et al. Role of the actin cytoskeleton in insulin action. Microsc Res Tech. 1999;47:79–92. doi: 10.1002/(SICI)1097-0029(19991015)47:2<79::AID-JEMT1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 29.Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 30.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goellner GM, DeMarco SJ, Strehler EE. Characterization of PISP, a novel single-PDZ protein that binds to all plasma membrane Ca2+-ATPase b-splice variants. Ann NY Acad Sci. 2003;986:461–471. doi: 10.1111/j.1749-6632.2003.tb07230.x. [DOI] [PubMed] [Google Scholar]
- 32.Rosado JA, et al. Reduced plasma membrane Ca2+-ATPase function in platelets from patients with non-insulin-dependent diabetes mellitus. Haematologica. 2004;89:1142–1144. [PubMed] [Google Scholar]
- 33.Schulze WX, Mann M. A novel proteomic screen for peptide-protein interactions. J Biol Chem. 2004;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 34.De Fea K, Roth RA. Modulation of insulin receptor substrate-1 tyrosine phosphorylation and function by mitogen-activated protein kinase. J Biol Chem. 1997;272:31400–31406. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- 35.Danielsson A, Ost A, Nystrom FH, Stralfors P. Attenuation of insulin-stimulated insulin receptor substrate-1 serine 307 phosphorylation in insulin resistance of type 2 diabetes. J Biol Chem. 2005;280:34389–34392. doi: 10.1074/jbc.C500230200. [DOI] [PubMed] [Google Scholar]
- 36.Stephenson SE, Dubach D, Lim CM, Mercer JF, La Fontaine S. A single PDZ domain protein interacts with the Menkes copper ATPase, ATP7A A new protein implicated in copper homeostasis. J Biol Chem. 2005;280:33270–33279. doi: 10.1074/jbc.M505889200. [DOI] [PubMed] [Google Scholar]
- 37.Bruton JD, Katz A, Westerblad H. The role of Ca2+ and calmodulin in insulin signaling in mammalian skeletal muscle. Acta Physiol Scand. 2001;171:259–265. doi: 10.1046/j.1365-201x.2001.00828.x. [DOI] [PubMed] [Google Scholar]
- 38.Barres R, Gonzalez T, Le Marchand-Brustel Y, Tanti JF. The interaction between the adaptor protein APS, Enigma is involved in actin organization. Exp Cell Res. 2005;308:334–344. doi: 10.1016/j.yexcr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Choi YO, et al. Implication of phosphorylation of the myosin II regulatory light chain in insulin-stimulated GLUT4 translocation in 3T3–F442A adipocytes. Exp Mol Med. 2006;38:180–189. doi: 10.1038/emm.2006.22. [DOI] [PubMed] [Google Scholar]
- 40.Algenstaedt P, Antonetti DA, Yaffe MB, Kahn CR. Insulin receptor substrate proteins create a link between the tyrosine phosphorylation cascade and the Ca2+-ATPases in muscle and heart. J Biol Chem. 1997;272:23696–23702. doi: 10.1074/jbc.272.38.23696. [DOI] [PubMed] [Google Scholar]
- 41.Saltiel AR, Pessin JE. Insulin signaling in microdomains of the plasma membrane. Traffic. 2003;4:711–716. doi: 10.1034/j.1600-0854.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 42.Jaken S. Protein kinase C isozymes and substrates. Curr Opin Cell Biol. 1996;8:168–173. doi: 10.1016/s0955-0674(96)80062-7. [DOI] [PubMed] [Google Scholar]
- 43.Mineo C, Ying YS, Chapline C, Jaken S, Anderson RG. Targeting of protein kinase Calpha to caveolae. J Cell Biol. 1998;141:601–610. doi: 10.1083/jcb.141.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oriente F, et al. Protein kinase C-alpha regulates insulin action and degradation by interacting with insulin receptor substrate-1 and 14-3-3 epsilon. J Biol Chem. 2005;280:40642–40649. doi: 10.1074/jbc.M508570200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, et al. Localization of low density lipoprotein receptor-related protein 1 to caveolae in 3T3–L1 adipocytes in response to insulin treatment. J Biol Chem. 2004;279:2221–2230. doi: 10.1074/jbc.M310679200. [DOI] [PubMed] [Google Scholar]
- 46.Mi K, Dolan PJ, Johnson GV. The low density lipoprotein receptor-related protein 6 interacts with glycogen synthase kinase 3 and attenuates activity. J Biol Chem. 2006;281:4787–4794. doi: 10.1074/jbc.M508657200. [DOI] [PubMed] [Google Scholar]
- 47.Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 48.Wang Q, Bilan PJ, Klip A. Opposite effects of insulin on focal adhesion proteins in 3T3–L1 adipocytes and in cells overexpressing the insulin receptor. Mol Biol Cell. 1998;9:3057–3069. doi: 10.1091/mbc.9.11.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.