Abstract
Phospho.ELM is a manually curated database of eukaryotic phosphorylation sites. The resource includes data collected from published literature as well as high-throughput data sets.
The current release of Phospho.ELM (version 7.0, July 2007) contains 4078 phospho-protein sequences covering 12 025 phospho-serine, 2362 phospho-threonine and 2083 phospho-tyrosine sites. The entries provide information about the phosphorylated proteins and the exact position of known phosphorylated instances, the kinases responsible for the modification (where known) and links to bibliographic references. The database entries have hyperlinks to easily access further information from UniProt, PubMed, SMART, ELM, MSD as well as links to the protein interaction databases MINT and STRING.
A new BLAST search tool, complementary to retrieval by keyword and UniProt accession number, allows users to submit a protein query (by sequence or UniProt accession) to search against the curated data set of phosphorylated peptides.
Phospho.ELM is available on line at: http://phospho.elm.eu.org
INTRODUCTION
Protein phosphorylation is one of the most-studied post-translational modifications: it has been estimated that up to one-third of the proteins may be modified by protein kinases (1). This ubiquitous regulatory mechanism controls many biological processes, including cellular growth, differentiation and DNA repair (2).
Knowing the phosphorylated residues in proteins is central to understanding the various signaling events in which they partake; therefore much effort has been invested in trying to identify and characterize phosphorylation sites. Traditional methods for measuring protein phosphorylation, such as mutational analysis and Edman degradation chemistry on phosphopeptides, have the disadvantage of being relatively time consuming and laborious, requiring large amounts of purified protein. On the other hand, mass spectrometry-(MS)based methods have emerged as powerful tools for the analysis of post-translational modifications due to higher sensitivity, selectively and speed. Over the past few years MS, combined with enrichment strategies for phosphorylated proteins e.g. isotope-coded affinity tags (ICAT) (3), stable isotopic amino acids in cell culture (SILAC) (4) and isobaric reagent iTRAQ (5), has been increasingly employed to identify novel phosphorylation sites. One consequence of this change in phosphorylation research is that bioinformatics resources need to be adapted and expanded to accommodate the new data.
For the thousands of phosphorylation sites identified by phosphoproteomic MS the information on which kinase phosphorylates them, and consequently the pathway in which they act, is still missing. To improve the link between experimentally identified phosphorylation sites and protein kinases, Linding and collaborators (6) have recently used the Phospho.ELM data set to develop and train a method, NetworKIN (http://networkin.info/) that combines computational methods for predicting which group of kinases are likely to phosphorylate a given site with information about signaling pathways and protein interaction data.
The analysis of protein phosphorylation by MS will clearly prove to be an invaluable source of information for understanding cellular signaling. For this reason, we consider it increasingly important to create and maintain publicly available phospho-protein databases, where the exponentially increasing number of known phosphorylation sites (7–12) can be easily accessed by the research community.
MATERIALS AND METHODS
The Phospho.ELM database
The content and the format of Phospho.ELM have been previously described in Diella et al. (13). While the general format of the database has remained essentially unchanged, some additions have been implemented to improve the data retrieval and presentation. The updated version also contains a much larger number of phosphorylation sites (see Figure 1), a new search tool based on sequence comparison and a Web Services interface.
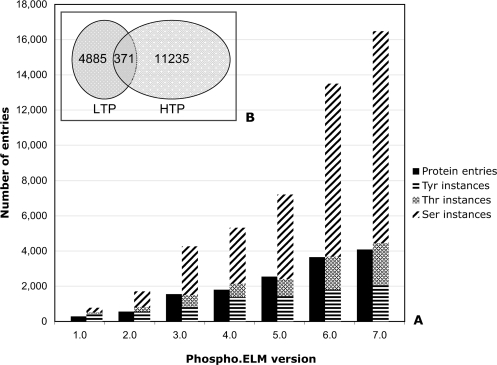
Figure 1.
The plot shows the growth of the Phospho.ELM data set beginning with version 1.0 in December 2003 (panel A). The exponential growth of the phosphorylation instances from Version 5.0 is mainly due to incorporation of the high-throughput data sets. The overlapping of the instances derived from low-throughput (LTP) and high-throughput (HTP) experiments is also shown (panel B).
The user can query the database by protein name, UniProt accession number/identifier, kinase name or binding motif to get a list of all known phosphorylation sites (instances) in a specific protein. The main results page summarizes information about the substrate protein (e.g. a brief description of the protein, protein type, the UniProt protein identification number), the phosphorylation sites contained within it and its surrounding amino acids (+/−10). The annotations to each instance include (where available) the PubMed reference, the kinase(s) phosphorylating the given site, the phospho-peptide binding domain(s) and a link to the ELM server (14) to retrieve further information about the kinase. Also where available, hyperlinks are provided to protein structures containing phosphorylated residues (15). Recently, Zanzoni and collaborators (16) have developed Phospho3D, a database of three-dimensional structures of phosphorylation sites, which stores data derived from the Phospho.ELM database and is focused on the annotation of structural information at the residue level.
Additional information for each protein kinase substrate includes the subcellular compartment [annotated with the Gene Ontology terms (17)], the tissue distribution and a list of interaction partners derived from the MINT (18) and STRING databases (19). The STRING interactors are shown in a summary graphic (network) that opens in a pop-up window. The network views provide links to the STRING database, where the information relative to the interactors is described in detail.
Data set
The current release of the Phospho.ELM data set (version 7.0, July 2007) contains 4078 phospho-protein sequences covering 12 025 phospho-serine, 2362 phospho-threonine and 2083 phospho-tyrosine sites with a total of 16 470 sites. The dataset is currently limited to metazoan species. This is partly due to our annotation capacity and partly because the kinases and nomenclature are so different in other lineages that they should be placed in separate databases. Although no animal species is purposely excluded from the data, currently human (11 197 phospho-sites) and mouse (2073 phospho-sites) are the most representative species due to the prevalence of their use as model organisms in biological research e.g. phosphoproteomic MS analyses have been mainly performed on human/mouse cell lines/tissues.
For each phospho-site we report if the phosphorylation evidence has been identified by small-scale analysis (low throughput; LTP) that typically focus on one or a few proteins at a time or by large-scale experiments (high throughput; HTP), which mainly apply MS techniques. It is noteworthy that in our data set there is a small overlap between instances identified by LTP and HTP experiments (Figure 1). This implies that most of the human phosphoproteome remains to be discovered. Figure 1 also shows that the rate of identification of additional phosphorylation sites on proteins has been increasing at a much faster rate than identification of novel phosphoproteins (e.g. see the srmm2 protein, UniProt accession Q9UQ35). While revealing that many more proteins are heavily phosphorylated than was previously known, it may be worth investigating whether the data also imply a strong bias in the proteins retrieved in the MS experiments.
The kinase responsible for the phosphorylation is known for ∼21% of the Phospho.ELM instances. Currently, more than 250 kinases are annotated in the database (for a detailed list of the kinases see the related information at the Phospho.ELM home page).
The PhosphoBLAST search tool
A BLAST search has been implemented which is complementary to the retrieval by keyword or UniProt accession/identifier. This tool identifies phospho-peptides contained in the query sequence that match those stored in Phospho.ELM (Figure 2). It consists of a two-step process: a BLAST (20) search and a parsing of the BLAST output. The BLAST program performs a sequence-similarity search against the Phospho.ELM data set of peptides (16 471), which have been experimentally proven to contain phospho-residues. It returns a set of local gapped alignments between the query sequence peptides and the phospho-peptides. In the parsing stage, those matches that present more than 70% sequence similarity and that conserve the phospho-residue in the same position as the corresponding phospho-peptide are selected. The final output shows the list of chosen matches, with their alignments and links to database records.
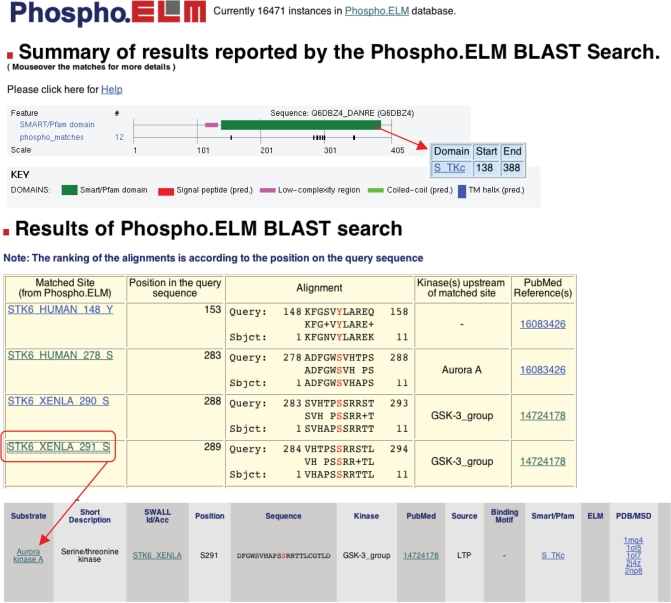
Figure 2.
Output example of a PhosphoBLAST Search using as query the Danio rerio Aurora A kinase sequence. The summary graphic shows the phospho-hits on the query sequence and features from SMART. Details about the matches are shown below in the results table. Clicking on the ‘subject name’ the users can retrieve additional information about the matched Phospho.ELM phosphorylated sites, including the flanking sequence, the PubMed reference, the kinase responsible for the phosphorylation (where known) and links to additional information for the substrate and other relevant databases.
The PhosphoBLAST tool does not aim at predicting phosphorylation motifs in the query protein and is primarily useful for retrieving phosphorylation sites that are conserved in related proteins (whether orthologs or paralogs). Nevertheless, unrelated query proteins occasionally yield matching phosphorylation sites in Phospho.ELM that can be equally interesting: it will be up to the user to consider carefully the possible biological meaning (e.g. shared kinase and/or phospho-peptide-binding domain specificities) associated with these match(es).
Web service
In order to facilitate remote tool integration, a Web Service to access the phospho.ELM database programmatically has been implemented and is available at: http://phospho.elm.eu.org/webservice/phosphoELMdb.wsdl.
The WSDL (Web Service Description Language) (21) file is WS-I compatible. The WS-Interoperability Basic Profile (22) proposes a set of rules to achieve interoperability of web services between different platforms. The WSDL file implements an XML wrapped document/literal style (23). The backend code is implemented in Java and runs on Axis2 (24) inside a Tomcat servlet container (25).
The functionality provided by the Web Service encompasses the current interface functionality with some additional filters. The extra options implemented in the Web Service are to search by PubMed ID and to retrieve all instances with a PDB entry assigned to them.
Database access
Phospho.ELM is developed and deployed with open source software (26). Software is developed in Python including some modules from the BioPython project (27) to retrieve information from UniProt and PubMed. The web interface software uses the CGImodel framework (28).
The data set is publicly available for academic users. Phospho.ELM can be accessed on the public Apache2 powered website at: http://phospho.elm.eu.org.
SUMMARY
Since its inception in 2004, the Phospho.ELM data set has been adopted for numerous bioinformatics tools and pipelines e.g. the protein kinase-specific prediction server GPS (group-based phosphorylation scoring method) (29), the RLIMS-P, a rule-based text-mining program designed to extract information on phosphorylation sites from abstracts (30), PhosphoregDB, a database of tissue and sub-cellular distribution of mammalian protein kinases and phosphatases (31), and NetworKin, a computational approach which combines consensus sequence motifs and contextual data to predict which kinases phosphorylate experimentally identified phosphorylation sites (6).
While anticipating that the size of the Phospho.ELM data set will constantly grow, we consider that the resource should be kept relatively lean in terms of the categories of data to be incorporated. On the other hand, links to external resources are under regular review and likely to be augmented from time to time. For example, resources such as KEGG (32) and Reactome (33) that annotate cell signaling networks are increasing their pathway coverage and it will clearly become essential to provide links to such resources. In the near future we intend to equip Phospho.ELM with links to the predicted kinase-substrate relations from the NetworKIN database (R.Linding, et al., submitted for publication).
ACKNOWLEDGEMENTS
We would like to acknowledge all the Phospho.ELM users who, by reporting missing sites or sending us their data sets, have contributed to improve the database. We wish to thank the EU EMBRACE grant (LHSG-CT-2004-512092) for funding. Many thanks to Ivica Letunic and Arnaud Ceol for technical support. We are grateful to Lars Juhl-Jensen and Rune Linding for their insightful comments and suggestions. We are thankful to Niall Haslam for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat. Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling–2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal. Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 9.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl Acad. Sci. USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 12.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutselakis H, Dimitropoulos D, Fillon J, Golovin A, Henrick K, Hussain A, Ionides J, John M, Keller PA, Krissinel E, et al. E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res. 2003;31:458–462. doi: 10.1093/nar/gkg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35:D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Web Services Description Language. http://www.w3.org/TR/wsdl (30 August 2007, date last accessed).
- 22.Web Services Interoperability Basic Profile Version 1.0. http://www.ws-i.org/Profiles/BasicProfile-1.0-2004-04-16.html (30 August 2007, date last accessed).
- 23.Document/Literal Wrapped style. http://www-128.ibm.com/developerworks/webservices/library/ws-whichwsdl/ (30 August 2007, date last accessed).
- 24.Axis 2. http://ws.apache.org/axis2 (30 August 2007, date last accessed).
- 25.Tomcat. http://tomcat.apache.org/ (30 August 2007, date last accessed).
- 26.PostgreSQL. http://www.postgresql.org (30 August 2007, date last accessed).
- 27.BioPython. http://BioPython.org (30 August 2007, date last accessed).
- 28.Ramu C, Gemund C, Gibson TJ. Object-oriented parsing of biological databases with Python. Bioinformatics. 2000;16:628–638. doi: 10.1093/bioinformatics/16.7.628. [DOI] [PubMed] [Google Scholar]
- 29.Zhou FF, Xue Y, Chen GL, Yao X. GPS: a novel group-based phosphorylation predicting and scoring method. Biochem. Biophys. Res. Commun. 2004;325:1443–1448. doi: 10.1016/j.bbrc.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Hu ZZ, Narayanaswamy M, Ravikumar KE, Vijay-Shanker K, Wu CH. Literature mining and database annotation of protein phosphorylation using a rule-based system. Bioinformatics. 2005;21:2759–2765. doi: 10.1093/bioinformatics/bti390. [DOI] [PubMed] [Google Scholar]
- 31.Forrest AR, Taylor DF, Fink JL, Gongora MM, Flegg C, Teasdale RD, Suzuki H, Kanamori M, Kai C, et al. PhosphoregDB: the tissue and sub-cellular distribution of mammalian protein kinases and phosphatases. BMC Bioinformatics. 2006;7:82. doi: 10.1186/1471-2105-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]