Abstract
Fibroblast growth factor 2 (FGF-2) is a powerful mitogen involved in proliferation, differentiation, and survival of various cells including neurons. FGF-2 expression is translationally regulated; in particular, the FGF-2 mRNA contains an internal ribosome entry site (IRES) allowing cap-independent translation. Here, we have analyzed FGF-2 IRES tissue specificity ex vivo and in vivo by using a dual luciferase bicistronic vector. This IRES was active in most transiently transfected human and nonhuman cell types, with a higher activity in p53 −/− osteosarcoma and neuroblastoma cell lines. Transgenic mice were generated using bicistronic transgenes with FGF-2 IRES or encephalomyocarditis virus (EMCV) IRES. Measurements of luciferase activity revealed high FGF-2 IRES activity in 11-d-old embryos (E11) but not in the placenta; activity was high in the heart and brain of E16. FGF-2 IRES activity was low in most organs of the adult, but exceptionally high in the brain. Such spatiotemporal variations were not observed with the EMCV IRES. These data, demonstrating the strong tissue specificity of a mammalian IRES in vivo, suggest a pivotal role of translational IRES- dependent activation of FGF-2 expression during embryogenesis and in adult brain. FGF-2 IRES could constitute, thus, a powerful tool for gene transfer in the central nervous system.
Keywords: central nervous system, development, FGF, in vivo, translational control
Introduction
FGF-2 or bFGF is a prototype member of the ever increasing FGF family that comprises, to date, 22 genes involved in the control of cell proliferation and differentiation (Nishimura et al. 1999). FGF-2 is a pleiotropic factor that induces the proliferation of most mesoderm- and neuroectoderm-derived cells (Slack and Isaacs 1994; Bikfalvi et al. 1997). In vitro, it stimulates endothelial cell growth and migration; in vivo, it promotes angiogenesis and is a major tumor angiogenic factor (Kandel et al. 1991). FGF-2 is also implicated in wound-healing processes (Bikfalvi et al. 1997; Ortega et al. 1998). It regulates gap junction communications in astroglial cells (Reuss et al. 1998). Furthermore, FGF-2 is a neurotrophic factor able to promote nerve survival and regeneration after central nervous system (CNS) injury (Eckenstein 1994; Houle and Ye 1999). Finally, invalidation of the FGF-2 gene in mice has established its role in controlling the fate, migration, and differentiation of neuronal cells (Dono et al. 1998; Ortega et al. 1998; Zhou et al. 1998). The homeostatic process of vascular tone control by the autonomous nervous system is impaired in these knockout mice (Dono et al. 1998; Zhou et al. 1998).
Five FGF-2 isoforms are expressed by the FGF-2 mRNA, resulting from a process of alternative initiation of translation from five different initiation codons, including four noncanonical CUGs (Florkiewicz and Sommer 1989; Prats et al. 1989; Arnaud et al. 1999). These isoforms have different cellular localizations and functions (Bugler et al. 1991; Couderc et al. 1991; Quarto et al. 1991; Bikfalvi et al. 1995; Arnaud et al. 1999). Their relative abundance varies according to cells and tissues (Coffin et al. 1995; Vagner et al. 1996; Bikfalvi et al. 1997). Interestingly, the murine and human FGF-2 isoforms undergo the same tissue-specific control in mice overexpressing a human FGF-2 transgene (Coffin et al. 1995), suggesting conservation of the regulatory mechanisms between species.
Several cis-acting elements located within the FGF-2 mRNA leader sequence are involved in the control of FGF-2 translation (Prats et al. 1992). The FGF-2 mRNA, in particular, contains an internal ribosome entry site (IRES) that allows translation via a nonclassical cap-independent mechanism (Vagner et al. 1995). This IRES is constitutively active in transformed cells and could account for the deregulated expression of CUG1, -2, and -3–initiated FGF-2 isoforms (HMW FGF-2; Galy et al. 1999). In contrast, HMW FGF-2 expression in normal human cultured cells is translationally controlled by cell density and stress (Vagner et al. 1996). Given the involvement of FGF-2 in cancer and injury repair, such data suggest a pivotal role of the IRES in the subtle regulation of FGF-2 expression in vivo.
To test this hypothesis, we analyzed the regulation of the FGF-2 mRNA IRES ex vivo in transfected cells and in vivo using transgenic mice. Our strategy was to express a bicistronic mRNA encoding two different luciferase reporter genes, the second being under the control of the human FGF-2 IRES. IRES activity was determined from the ratio of second cistron to first cistron expression. Our results from transfection of various mammalian cell types, including mouse, human, hamster, bovine, and monkey cells, showed that the human FGF-2 IRES functions in different species and in all the tested cell types, with greater efficiency in neuroblastoma and p53 minus osteosarcoma cells. IRES activity was elevated in the embryo of transgenic mice, particularly in the heart and brain. In contrast, IRES activity in the adult was low in most organs but remained very high in brain. The in vivo specificity of this activity contrasted strikingly with the relatively ubiquitous expression of viral EMCV IRES.
Materials and Methods
Plasmid Construction
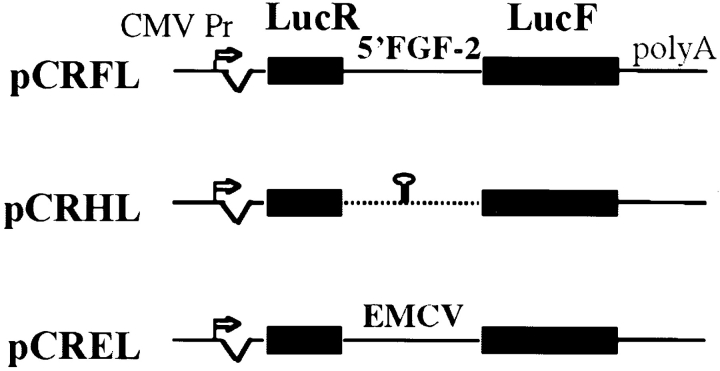
The two plasmids pCRHL and pCREL have been described already (Huez et al. 1998). The two luciferase genes, Renilla luciferase (LucR) and firefly luciferase (LucF), are controlled by the cytomegalovirus promoter (CMV) and separated by either a hairpin or the encephalomyocarditis virus (EMCV) IRES, for pCRHL and pCREL, respectively (see Fig. 1; Huez et al. 1998). The plasmid pCRFL was constructed by replacing the EMCV IRES with the FGF-2 IRES between the two LUC genes. In this plasmid, the 5′ 539 nucleotides of the FGF-2 cDNA are fused to the LucF coding sequence.
Figure 1.
Schematic representation of the bicistronic LucR-I-LucF vectors. Vector construction is described in Materials and Methods. They contain the CMV promoter controlling the expression of a bicistronic LucR-LucF mRNA. A synthetic intron is present 5′ of LucR, and a poly(A) site is present 3′ of LucF (Huez et al. 1998). In the pCRFL construct, the complete FGF-2 5′ leader and beginning of coding sequence (up to nucleotide 539) has been fused to the firefly luciferase open reading frame. In the pCRHL construct, a hairpin has been introduced between the two luciferase genes. The pCREL construct contains the EMCV IRES between the two LUC genes.
Cell Transfection
Cell lines were obtained from American Type Culture Collection (ATCC European Collection of Cell Cultures [ECACC]). NIH 3T3 is a mouse immortalized fibroblast cell line. C2C12 (accession number for ECACC, 91031101) is a mouse muscle myoblast. ABAE is an adult bovine aortic endothelial primary cell (Couderc et al. 1991). CHO is a Chinese hamster ovary carcinoma. HeLa (accession number for ATCC, CCL2) is a human uterus carcinoma of epithelial origin. COS-7 (accession number for ATCC, CRL 1654) is a monkey kidney cell transformed by SV-40 large T antigen. Jurkat (accession number for ATCC, TIB-152) is a human acute T cell leukemia. Skin fibroblast is a primary culture cell from human skin (Touriol et al. 1999). ECV304 (accession number for ECACC, 92091712) is a spontaneously transformed human endothelial cell. 293 (accession number for ATCC, CRL-1573) is a human kidney epithelial cell, transformed with adenovirus 5. SK-Hep-1 (accession number for ATCC, HTB 52) is a human liver adenocarcinoma of endothelial origin; SK-N-AS (accession number for ECACC, 94092302) and SK-N-BE (accession number for EACC, 95011815) are human neuroblastoma. Saos2 (accession number for ATCC, HTB-85) is a p53 −/− human osteosarcoma of epithelial origin. Cells were cultivated according to ATCC or ECACC instructions.
The different cell types were transfected with 1 μg of plasmid and Fugene 6 reagent (Boehringer-Roche) in 12-well petri dishes. 48 h after transfection, cell lysates were prepared for luminescence activity as previously described (Huez et al. 1998).
Generation of Transgenic Mice
Transgenic mice were obtained by injecting fragments containing either the CMV-LucR-EMCV IRES-LucF or the CMV-LucR-FGF2IRES-LucF sequences into one of the pronuclei of (C57Bl/6×CBA)2 fertilized eggs (Brinster et al. 1985). Transgenic embryos and adult mice were identified by PCR and Southern blot analysis of placenta or tail DNA using a LucF 500-bp-long probe amplified with the LucF-specific oligonucleotides 5′-CAGTATGAACATTTCGCAGCC-3′ and 5′- CTGAAGGGACTGTAAAAACAGC-3′. Stable lines were maintained by successive crosses with (C57Bl/6×CBA) F1 mice. The transgene copy number was determined by PhosphorImager scanning of Southern blots.
Reverse Transcription and PCR Analysis
The cDNAs were synthesized as previously described, using 5 μg of DNase-treated total RNA (Touriol et al. 1999). The PCR was performed with the primer couples, 5′-GATTACCAGGGATTTCAGTCG-3′ and 5′-CTGAAGGGACTGTAAAAACAGC-3′, 5′-CCACATATTGAGCCAGTAGC-3′ and 5′-CCATGATAATGTTGGACGAC-3′, to amplify the LucF or LucR DNA sequences, respectively. The PCR reactions were carried out using 0.3 U of Goldstar Taq DNA polymerase (Eurogentec), in a final volume of 30 μl, with 1 μl of cDNA. The reaction was performed on a Perkin Elmer apparatus, under the following conditions: 94°C for 3 min, then 25 cycles of 94°C for 30 s, 58°C for 45 s, 72°C for 45 s, and finally 72°C for 5 min. Amplification results (one third of the reactions) were analyzed on 2% agarose gels (Tris-borate/EDTA), followed by ethidium bromide staining.
Luciferase Activity Analysis
The two luciferase activities were measured in cell or tissue extracts and the IRES activity was determined by calculating the LucR/LucF ratios. The IRES efficiency between different cell types was compared ex vivo by calibrating the pCRFL ratio (FGF IRES activity) to pCRHL (background activity without an IRES). For the in vivo analysis, E11 embryos, heart, limb bud, tail, brain, liver, and placenta of the E16 embryos, and tissue fragments of the adults were frozen in liquid nitrogen and stored at −80°C. They were homogenized in 200 μl of passive lysis buffer (Promega) using thurax and centrifuged for 20 min at 4,500 rpm at 4°C. The supernatant was centrifuged for 15 min at 13,000 rpm at 4°C, and the last supernatant was used for luminescence dosage (30 μl). LucR and LucF activities were measured using the Dual Luciferase kit from Promega.
Results
FGF-2 IRES Activity Is Optimal in Neuroblastoma and Osteosarcoma Cell Lines
The bicistronic vector strategy, which was previously used to demonstrate the existence of the two vascular endothelial growth factor IRESes (Huez et al. 1998), was used here to test the activity of the FGF-2 IRES in different cell types. This bicistronic vector expresses two highly sensitive luciferases, LucR and LucF, which are encoded by the same mRNA under the control of the cytomegalovirus promoter (Fig. 1). This promoter was chosen because it had been shown previously to control the strong expression of reporter genes in tissue-cultured cells and the quasi-ubiquitous (although variable) expression of transgenes in transgenic mice (Furth et al. 1991). The FGF-2 mRNA leader sequence (nucleotides 1–539), including the IRES, was introduced between the LucR and LucF genes (Fig. 1, pCRFL). LucR, the first cistron, provides the level of cap-dependent translation. It is expected to reflect the activity of CMV sequences in the tissues or cells and, thus, to be proportional to the amount of bicistronic mRNA. LucF, the second cistron, is expressed in proportion to the IRES activity. The use of such an assay, based on dual highly sensitive reporters, allows rapid and concomitant quantitation of reporter activity. The IRES activity between different cell types and independently of transfection efficiency was calibrated by using the vector pCRHL, which contains a hairpin but no IRES between the two cistrons as a control (Fig. 1, pCRHL).
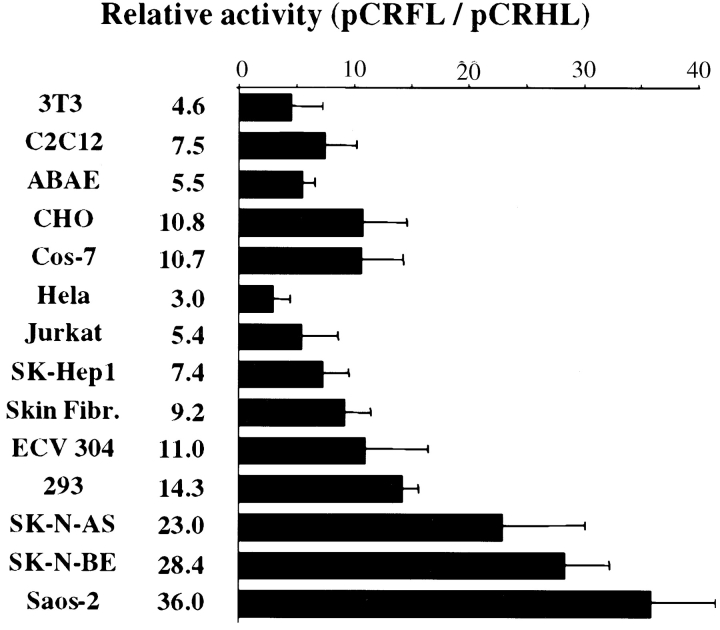
The FGF-2 IRES activity was measured in 14 different cell types from human, monkey, bovine, hamster, and mice that were transiently transfected with either pCRFL or pCRHL vector. As indicated in Fig. 2, the FGF-2 IRES showed a large spectrum of activity since it was active in all the different mammalian cell types tested. However, it was subjected to strong cell type–specific regulation. For example, a 12-fold factor was observed between the lowest (HeLa) and the highest values (Saos-2) within the human cells. Although IRES activation was not directly correlated to cell transformation, the activity in the four noncancerous cell lines (3T3, C2C12, ABAE, and skin fibroblasts) was low or very low, whereas it was medium or high in 6/10 transformed cell lines. Within these latter, low activity was observed in the Jurkat cell lines of lymphocyte origin, whereas high activity was observed in SK-N-BE and SK-N-AS, the two neuroblastoma cell lines. This suggested that the tissue origin of the cell line might be one parameter conditioning FGF-2 IRES activity.
Figure 2.
Analysis of the FGF-2 IRES activity in transiently transfected cells. 14 different cell types were transiently transfected with pCRFL or pCRHL DNA. Cells were harvested 48 h after transfection, and the luciferase activities present in cell extracts were measured. FGF-2 IRES activity was determined by calculating the LucF/LucR ratio. To calibrate the data from the different cell types, the LucF/LucR ratio of pCRFL was divided by the ratio of the pCRHL negative control. Experiments were repeated 5–10 times, and the results are expressed as means ± SEM. The names of the different cell types (described in Materials and Methods) and the histograms values are indicated on the left.
Generation of Transgenic Mice Expressing Bicistronic LucR-IRES-LucF Constructs
To determine the tissue specificity of the FGF-2 IRES in the absence of any bias because of cell transformation or cell culture conditions, we decided to study the expression of the LucR-FGF2-IRES-LucF construct in vivo. Therefore, transgenic mice were produced that contained the LucR-IRES-LucF DNA fragment (see Materials and Methods). Two independent lines, RFL12 and RFLD, were established that contained 28 and 1 copies of the transgene, respectively. Two other transgenic lines that contained a similar construct with the viral EMCV IRES in place of FGF-2 IRES (Fig. 1, pCREL) were also produced. Both of these lines, RELA and RELB, contained 12–15 copies of the construct (data not shown).
FGF-2 IRES Activity Is High in the Embryo, Especially in the Heart and Brain
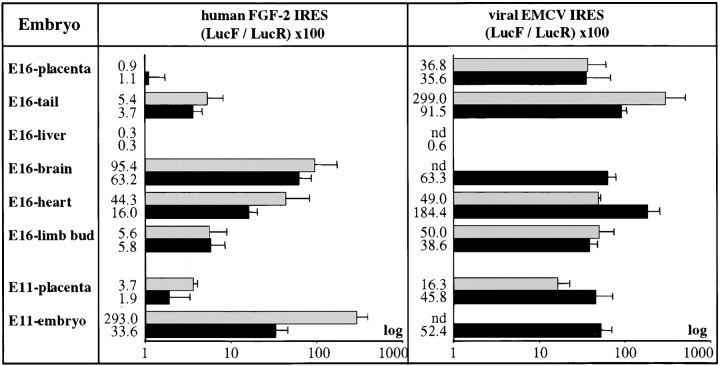
Luciferase activities in the transgenic lines were measured in the whole embryo and placenta for E11 and in various tissues of the E16 embryo. As shown in Fig. 3 (left), the activity of FGF-2 IRES was high in the E11 embryo, whereas it was low in the placenta of the two RFL transgenic lines. Such a difference was also observed in the E16 embryo, where the placenta showed no significant FGF-2 IRES activity, whereas IRES was active in several tissues exhibiting the same CMV transcriptional efficiency, such as the heart and brain (Fig. 3 and Table ). The level of IRES activity varied according to the organ: it was hardly detectable in the liver; and low in limb bud and tail, regardless of the high activity of the CMV promoter in these tissues; and was high in the heart and brain. In contrast, EMCV IRES activity was high in all the tissues of the two REL transgenic lines embryos tested, except in the liver of E16 embryos (Fig. 3, right). These data show that FGF-2 IRES is active in the early embryo, but is strongly regulated in a tissue-specific manner.
Figure 3.
FGF-2 and EMCV IRESes activities in transgenic mice embryos. Embryos from the transgenic mouse lines were recovered at 11 and 16 d postcoitum, and luciferase activities were measured in different organs (indicated on the left) as described in Materials and Methods. IRES activities are represented by histograms and were measured by calculating the LucF/LucR ratio (Table ). The results are shown for each of the two independent transgenic lines expressing one or other bicistronic construct pCRFL (FGF-2) or pCREL (EMCV), as means ± SEM of experiments that were performed 3–10 times. The lines expressing pCRFL (left) are RFLD (gray boxes) and RFL12 (black boxes). The lines expressing pCREL (right) are RELB (gray boxes) and RELA (black boxes). nd corresponds to experiments giving no detectable LucR activity.
Table 1.
Measurement of Renilla and firefly luciferase Activities in Whole Embryo (E11) and in Embryonic (E16) and Adult Tissues of Transgenic Mice Lines RFL12 and RELA
| RFL:human FGF-2 | REL:EMCV | |||
|---|---|---|---|---|
| Embryo | lucR×10−3 | lucF×10−2 | lucR×10−3 | lucF×10−2 |
| E11-embryo | 39.5 | 130 | 12 | 62 |
| E11-placenta | 11.5 | 1 | 22.4 | 102 |
| E16-limb bud | 10,400 | 6,240 | 22.5 | 100 |
| E16-heart | 56 | 170 | 8.6 | 80 |
| E16-brain | 45 | 270 | 32.5 | 225 |
| E16-liver | 43 | 1.3 | 1.6 | 0.1 |
| E16-tail | 970 | 450 | 119 | 470 |
| E16-placenta | 54 | 8.1 | 55 | 190 |
| Adult | ||||
| Testis | 164 | 340 | 1,787 | 20,550 |
| Ovary | 28.5 | 13.4 | 13 | 52 |
| Intestine | 81 | 4.1 | 45 | 77.8 |
| Kidney | 122 | 18.3 | 64 | 96 |
| Stomach | 168 | 10 | 300 | 360 |
| Heart | 96 | 11.5 | 141 | 141 |
| Thymus | 3.4 | 1.7 | 10 | 50 |
| Tongue | 326 | 65 | 2,800 | 19,600 |
| Brain | 14 | 550 | 3.2 | 9.6 |
| Muscle | 29,000 | 5,800 | 109 | 54.5 |
| Skin | 202 | 33 | 640 | 5,880 |
| Liver | 20 | 1.8 | nd | nd |
| Lung | 2 | 0.8 | 13 | 220 |
This corresponds to representative values of experiments repeated at least three times. nd, not detected.
In Adult Mice, the FGF-2 IRES Shows Very High Brain-specific Activity
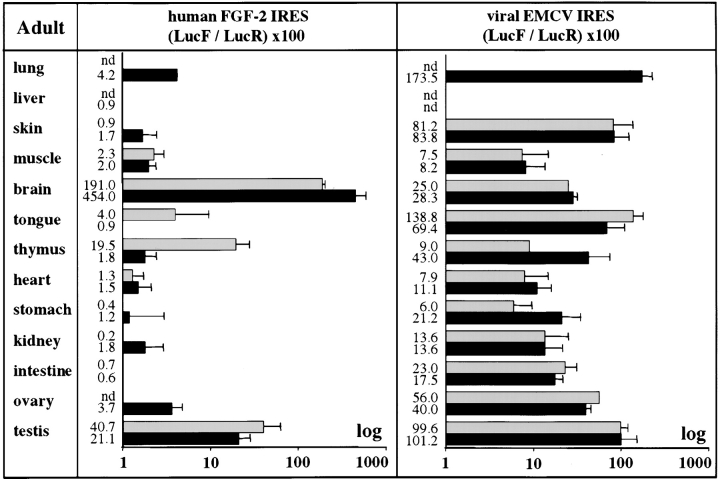
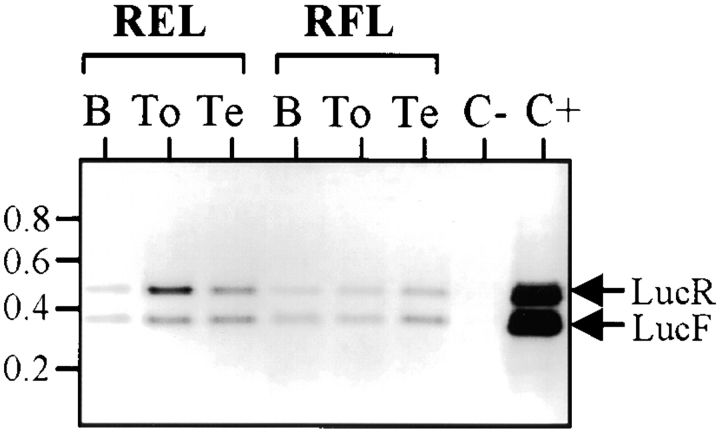
The activity of FGF-2 IRES was also measured in different adult organs. As shown in Fig. 4 and Table , the IRES activity variations were exacerbated in comparison with the data obtained from cell cultures and embryos. The variation in FGF-2 IRES activity between the different organs attained 400-fold, as illustrated by the activities found in the stomach (0.4 and 1.2 for the lines RFLD and RFL12, respectively) and in the brain (191 and 454, respectively; Fig. 4, left). FGF-2 IRES was weakly expressed in most organs, except the testis (up to 40.7) and the brain, where it presented exceptionally high activity (up to 454). In contrast, except for the liver where neither LucR nor LucF activity was significantly detected (this could result from proteolytic activities known to be present in this organ), the EMCV IRES activity was high in most adult organs (Fig. 4, right, and Table ). The high activity of FGF2 IRES observed in the brain strongly suggests that this cellular IRES is upregulated in this tissue. To check that the strong signal observed in the brain was due to IRES activity and not to any aberrant event, comparative reverse transcriptase (RT)–PCR was used to measure the relative levels of LucF and LucR cistrons. We show that the ratio of LucF RNA/LucR RNA cistrons was similar in all the organs tested, including the brain, as expected for such bicistronic mRNAs (Fig. 5). In summary, these data indicate that the FGF-2 IRES is subjected to stringent regulation in the adult, and that its activity is strongly upregulated in the brain.
Figure 4.
FGF-2 and EMCV IRESes activities in transgenic adult mice. Different organs (indicated on the left) were prepared from adult transgenic mice and luciferase activities were measured as in Fig. 3. IRES activities are represented by histograms corresponding to the LucF/LucR activities (see Table ). The results are shown for each of the two independent transgenic lines, expressing one or other bicistronic construct pCRFL (FGF-2) or pCREL (EMCV), as means ± SEM of experiments that were performed 3–10 times. The lines expressing pCRFL (left) are RFLD (gray boxes) and RFL12 (black boxes). The lines expressing pCREL (on the right) are RELB (gray boxes) and RELA (black boxes). nd corresponds to experiments giving no detectable LucR activity.
Figure 5.
Analysis of the bicistronic mRNA in transgenic tissues by RT-PCR. Total RNA was extracted from RFL12 or RELA transgenic mice brain (B), tongue (To), or testis (Te), and the level of LucF and LucR cistrons was analyzed by RT-PCR (as described in Materials and Methods). The negative control (C−) corresponds to a PCR experiment performed using RNA from the brain of a nontransgenic mouse. The positive control (C+) was performed with an in vitro–transcribed bicistronic RFL mRNA. The bands corresponding to amplification of LucR (465 nucleotide) or LucF (366 nucleotide) cDNAs are indicated by arrows. This experiment enabled us to check the LucF/LucR ratios at the RNA level in the different organs.
Discussion
IRESes, which allow cap-independent translation initiation, have been discovered in several cellular capped mRNAs during the last few years. The presence of such elements in capped mRNAs has raised the question of their function in the regulation of gene expression. This report shows for the first time that in contrast to the broad-range activity of a picornavirus IRES, a mammalian IRES, (i.e, human FGF-2) is highly tissue-specific in vivo. The tissue specificity that we observed is strongly reminiscent of the proposed role of FGF-2 in embryogenesis and the adult CNS (Eckenstein 1994; Slack and Isaacs 1994; Houle and Ye 1999). These data constitute a new step in our understanding of the physiological relevance of cellular mRNA IRESes.
The analysis of our transgenic mice showed that the human FGF-2 IRES is particularly active in the embryo proper at midgestation. At a later stage, its activity is particularly high in the brain and persists in the adult. In contrast, as observed from transfection experiments, FGF-2 IRES functions in most tissue culture cell types ex vivo. All this suggests that the IRES-dependent translation of FGF-2 might be correlated with cell status, as it is functional in actively proliferating cells, embryonic or in vitro cultured, and extinguished in fully differentiated and quiescent cells, except for the CNS. Interestingly, among the different cell lines tested, the strongest IRES activity is observed in Saos2, a p53 −/− osteosarcoma cell line. As p53 has been shown to have an inhibitory effect on FGF-2 translation (B. Galy, L. Créancier, C. Zanibellato, A.C. Prats, and H. Prats, unpublished results), this could indicate that the FGF-2 IRES is not only subjected to activation, but also to silencing by trans-acting factors. This silencing could be temporarily removed in response to specific stimuli, such as stress or tissue lesion (Vagner et al. 1996). An aberrant activation of the FGF-2 IRES could participate in acquisition and/or maintenance of the transformed cell status, in concordance with our previous observations that the HMW FGF-2 isoforms are constitutively expressed in transformed cells while they are regulated as a function of cell density in subprimary skin fibroblasts (Galy et al. 1999).
These data also show that the human FGF-2 IRES activity is more tissue-specific than species-specific. Indeed, it is not only active in human cultured cells, but also in hamster, monkey, and mouse cells or tissues, suggesting that specific cellular trans-acting factors are able to recognize FGF-2 IRES independently of their origin. This corroborates previous observations in transgenic mice that expressed a human FGF-2 cDNA transgene encoding the different FGF-2 isoforms (Coffin et al. 1995). In those transgenic animals, the pattern of expression of the different human isoforms was highly tissue-specific and corresponded to that of the endogenous FGF-2 isoforms. Interestingly, FGF-2 proteins were observed in tissues such as the brain where the FGF-2 mRNA could not be detected, even by RT-PCR (Coffin et al. 1995). This suggests that the level of translation of the endogenous FGF-2 mRNA could be upregulated in a few tissues, a hypothesis which is consistent with our observations of very high IRES activity in the brain of the IRES FGF-2 transgenic mice.
While the transgenic mice that overexpress FGF-2 (Coffin et al. 1995) provide important information concerning the pattern of expression of the various FGF-2 isoforms and its conservation between human and mouse, they cannot be used to distinguish between cap-dependent and IRES-dependent translation. The bicistronic vector approach that we describe in the present study allows us to enlighten the involvement of the FGF-2 IRES in vivo. Strikingly, this mechanism of cap-independent translation is more generally active during embryogenesis than in the adult, where it is restricted to a few organs. Indeed, we find high IRES activity in the brain and weaker activity in the testes, ovaries and lung, whereas all the other tested organs show no significant IRES activity, even those in which the CMV promoter is active. In particular, the IRES is silent in the heart where HMW murine and human FGF-2 proteins have been detected in FGF-2 transgenic mice (Coffin et al. 1995), suggesting that a majority of these isoforms could result from cap-dependent translation of FGF-2 mRNAs in this organ. The physiological activation of FGF-2 IRES observed in the brain, correlated with the high IRES efficiency in two neuroblastoma cell lines, suggests the existence of a specific CNS internal entry activating factor. The location of the IRES just upstream from the CUG1 codon and the observation of higher HMW FGF-2 synthesis in nervous system cells, in particular after nerve injury, strongly suggest that IRES could have an essential role in the brain (Vagner et al. 1995; Huber et al. 1997; Meisinger and Grothe 1997).
IRESes constitute powerful tools as they allow the expression of several genes from the same transcription unit. Two groups have previously reported that a bicistronic retroviral vector with the EMCV IRES works stably in chicken and mouse embryos (Ghattas et al. 1991; Kim et al. 1992). We show in this study that EMCV IRES functions in a ubiquitous manner not only in the embryos, but also in most organs of adult transgenic mice. This is in agreement with a recent report that demonstrates a quasi-ubiquitous activity of the IRES of another picornavirus, Theiler's murine encephalomyelitis virus, in young and adult transgenic mice (Shaw-Jackson and Michiels 1999). In contrast, we demonstrate that the FGF-2 IRES, a cellular mRNA IRES, shows strong tissue-specific activity. These observations may be related to another study performed in transgenic flies indicating that the IRES contained in the 5′ untranslated regions of antennapedia and ultrabithorax exhibited a high degree of developmental regulation and were not functionally equivalent (Ye et al. 1997). Furthermore, the IRES of an avian retrovirus shows considerable activity in multipotent neural cells and in their glial and neuronal progeny (Derrington et al. 1999). We hypothesize from these results that retroviral IRESes and cellular IRESes, which both belong to capped and polyadenylated transcripts, would present stringent tissue specificity, whereas the activity spectrum of picornavirus IRESes, which in most cases belong to uncapped mRNA, would be broader.
Finally, the stringent tissue specificity of a given cellular IRES might be very useful as a biotechnological tool to drive specific gene expression in a particular tissue. As illustrated in this study, the FGF-2 IRES could be powerfully used to express neurotrophic factors from multicistronic vectors in CNS regeneration programs.
Acknowledgments
We are grateful to J. Auriol for technical assistance. We thank A. Mahfoudi, C. Orsini, and H. Prats, for helpful discussions, and D. Warwick for English proofreading.
This work was supported by grants from the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, the Conseil Régional Midi-Pyrénées, the European Commission BIOTECH program (contrat 94199-181), and Rhone-Poulenc-Rorer (now Aventis). L. Créancier was financed first by the European Commission BIOTECH program, and then by Retina France.
Footnotes
Abbreviations used in this paper: CMV, cytomegalovirus; CNS, central nervous system; EMV, encephalomyocarditis virus; IRES, internal ribosome entry site; RT, reverse transcriptase.
References
- Arnaud E., Touriol C., Boutonnet C., Gensac M.C., Vagner S., Prats H., Prats A.C. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol. Cell. Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikfalvi A., Klein S., Pintucci G., Quarto N., Mignatti P., Rifkin D.B. Differential modulation of cell phenotype by different molecular weight forms of basic fibroblast growth factorpossible intracellular signaling by the high molecular weight forms. J. Cell Biol. 1995;129:233–243. doi: 10.1083/jcb.129.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikfalvi A., Klein S., Pintucci G., Rifkin D.B. Biological roles of fibroblast growth factor-2. Endocr. Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- Brinster R.L., Chen H.Y., Trumbauer M.E., Yagle M.K., Palmiter R.D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc. Natl. Acad. Sci. USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugler B., Amalric F., Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol. Cell. Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J.D., Florkiewicz R.Z., Neumann J., Mort-Hopkins T., Dorn G.W., II, Lightfoot P., German R., Howles P.N., Kier A., O'Toole B.A. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol. Biol. Cell. 1995;6:1861–1973. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc B., Prats H., Bayard F., Amalric F. Potential oncogenic effects of basic fibroblast growth factor requires cooperation between CUG and AUG-initiated forms. Cell Regul. 1991;2:709–718. doi: 10.1091/mbc.2.9.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrington E.A., Lopez-Lastra M., Chapel-Fernandez S., Cosset F.L., Belin M.F., Rudkin B.B., Darlix J.L. Retroviral vectors for the expression of two genes in human multipotent neural precursors and their differentiated neuronal and glial progeny. Hum. Gene Ther. 1999;10:1129–1138. doi: 10.1089/10430349950018120. [DOI] [PubMed] [Google Scholar]
- Dono R., Texido G., Dussel R., Ehmke H., Zeller R. Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenstein F.P. Fibroblast growth factors in the nervous system. J. Neurobiol. 1994;25:1467–1480. doi: 10.1002/neu.480251112. [DOI] [PubMed] [Google Scholar]
- Florkiewicz R.Z., Sommer A. Human basic fibroblast growth factor gene encodes four polypeptidesthree initiate translation from non-AUG codons. Proc. Natl. Acad. Sci. USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth P.A., Hennighaussen L., Baker C., Beatty B., Woychick R. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 1991;19:6205–6208. doi: 10.1093/nar/19.22.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy B., Maret A., Prats A.C., Prats H. Cell transformation results in the loss of the density-dependent translational regulation of the expression of fibroblast growth factor 2 isoforms. Cancer Res. 1999;59:165–171. [PubMed] [Google Scholar]
- Ghattas I.R., Sanes J.R., Majors J.E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol. Cell. Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle J.D., Ye J.H. Survival of chronically-injured neurons can be prolonged by treatment with neurotrophic factors. Neuroscience. 1999;94:929–936. doi: 10.1016/s0306-4522(99)00359-0. [DOI] [PubMed] [Google Scholar]
- Huber K., Meisinger C., Grothe C. Expression of fibroblast growth factor-2 in hypoglossal motoneurons is stimulated by peripheral nerve injury. J. Comp. Neurol. 1997;382:189–198. [PubMed] [Google Scholar]
- Huez I., Creancier L., Audigier S., Gensac M.C., Prats A.C., Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel J., Bossy-Wetzel E., Radvanyi F., Klagsbrun M., Folkman J., Hanahan D. Neovascularisation is associated with a switch to the export of bFGF in the multistep development of fibrosarcoma. Cell. 1991;66:1095–1104. doi: 10.1016/0092-8674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- Kim D.G., Kang H.M., Jang S.K., Shin H.S. Construction of a bifunctional mRNA in the mouse by using the internal ribosomal entry site of the encephalomyocarditis virus [published erratum appears in Mol. Cell. Biol. 1992. 12:4807] Mol. Cell. Biol. 1992;12:3636–3643. doi: 10.1128/mcb.12.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Grothe C. Differential expression of FGF-2 isoforms in the rat adrenal medulla during postnatal development in vivo. Brain Res. 1997;757:291–294. doi: 10.1016/s0006-8993(97)00341-7. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Utsonomiya Y., Hoshikawa M., Ohuchi H., Itoh N. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim. Biophys. Acta. 1999;1444:148–151. doi: 10.1016/s0167-4781(98)00255-3. [DOI] [PubMed] [Google Scholar]
- Ortega S., Ittmann M., Tsang S.H., Ehrlich M., Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc. Natl. Acad. Sci. USA. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A.C., Vagner S., Prats H., Amalric F. Cis-acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor. Mol. Cell. Biol. 1992;12:4796–4805. doi: 10.1128/mcb.12.10.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats H., Kaghad M., Prats A.C., Klagsbrun M., Lélias J.M., Liauzun P., Chalon P., Tauber J.P., Amalric F., Smith J.A., Caput D. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc. Natl. Acad. Sci. USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarto N., Talarico D., Florkiewicz R., Rifkin D.B. Selective expression of high molecular weight basic fibroblast growth factor confers a unique phenotype to NIH 3T3 cells. Cell Reg. 1991;2:699–708. doi: 10.1091/mbc.2.9.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss B., Heinke A., Unsicker K. Gap junction uncoupling of neonatal rat astroglial cultures does not affect their expression of fibroblast growth factor 2. Neurosci. Lett. 1998;258:45–48. doi: 10.1016/s0304-3940(98)00847-7. [DOI] [PubMed] [Google Scholar]
- Shaw-Jackson C., Michiels T. Absence of internal ribosome entry site-mediated tissue specificity in the translation of a bicistronic transgene. J. Virol. 1999;73:2729–2738. doi: 10.1128/jvi.73.4.2729-2738.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack J.M., Isaacs H.V. The role of fibroblast growth factors in early Xenopus development. Biochem. Soc. Trans. 1994;22:585–589. doi: 10.1042/bst0220585. [DOI] [PubMed] [Google Scholar]
- Touriol C., Morillon A., Gensac M.C., Prats H., Prats A.C. Expression of human FGF-2 is post-transcriptionally controlled by a unique destabilization element present in the mRNA 3′ untranslated region between alternative polyadenylation sites. J. Biol. Chem. 1999;274:21402–21408. doi: 10.1074/jbc.274.30.21402. [DOI] [PubMed] [Google Scholar]
- Vagner S., Gensac M.C., Maret A., Bayard F., Amalric F., Prats H., Prats A.C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S., Touriol C., Galy B., Audigier S., Gensac M.C., Amalric F., Bayard F., Prats H., Prats A.C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J. Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Fong P., Iizuka N., Choate D., Cavener D.R. Ultrabithorax and antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Sutliff R.L., Paul R.J., Lorenz J.N., Hoying J.B., Haudenschild C.C., Yin M., Coffin J.D., Kong L., Kranias E.G. Fibroblast growth factor 2 control of vascular tone. Nat. Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]