Abstract
ATP-dependent chromatin remodeling has an important role in the regulation of cellular differentiation and development. For the first time, a topological view of one of these complexes has been revealed, by mapping the interactions of the catalytic subunit Isw2 with nucleosomal and extranucleosomal DNA in the complex with all four subunits of ISW2 bound to nucleosomes. Different domains of Isw2 were shown to interact with the nucleosome near the dyad axis, another near the entry site of the nucleosome, and another with extranucleosomal DNA. The conserved DEXD or ATPase domain was found to contact the superhelical location 2 (SHL2) of the nucleosome, providing a direct physical connection of ATP hydrolysis with this region of nucleosomes. The C terminus of Isw2, comprising the SLIDE (SANT-like domain) and HAND domains, was found to be associated with extranucleosomal DNA and the entry site of nucleosomes. It is thus proposed that the C-terminal domains of Isw2 are involved in anchoring the complex to nucleosomes through their interactions with linker DNA and that they facilitate the movement of DNA along the surface of nucleosomes.
The paradox of ATP-dependent chromatin remodeling is the ability of these complexes to mobilize nucleosomes and overcome more than 100 protein-DNA contacts in the nucleosome without completely displacing the nucleosome from DNA (7, 9, 19). Models of how movement of the nucleosome could occur, either by the generation of DNA bulges that propagate around the surface of the nucleosome or by the diffusion of DNA twist through the nucleosome in a propeller-like fashion, have been proposed (11, 17, 25). Nucleosome mobilization appears to be done through the translocation of the catalytic subunit along DNA, presumably by the SNF2, a DNA helicase-like domain that has lost its helicase activity but retained its translocase activity (4). The situation has become more complex with recent discoveries that the site of DNA translocation is far inside the nucleosome, two helical turns from the dyad axis or superhelical location 2 (SHL2), for four different chromatin remodeling complexes (23, 26, 34). Translocation along DNA at SHL2 would be expected to be extremely difficult due to the steric constraints of the surrounding DNA being extensively bound on either side of the surface of the histone octamer.
The data suggest that, in order for ISW2 to execute the movement of DNA around the surface of the nucleosome, there are three functional domains of ISW2. The first is a translocation domain that binds to nucleosomal DNA at SHL2 and translocates along DNA (23, 26, 34). The second is a regulatory domain of ISW2 that contributes to the unidirectional movement of DNA into the nucleosome (33). The third is an anchor domain to tether ISW2 to the core nucleosome by way of extranucleosomal DNA and to orient ISW2 on the nucleosome surface so that it has a preferred direction for moving nucleosomes (5). Previously, DNA footprinting had shown this tripartite architecture as suggested, in that ISW2 contacts (i) a small, 10-bp region spanning the minor groove of DNA at SHL2, (ii) a 10- to 20-bp region just inside the nucleosome from the entry/exit site, and (iii) 50 to 60 bp of extranucleosomal DNA (15). Photochemical cross-linking has shown that the catalytic subunit Isw2 contacts SHL2, the entry/exit site, and extranucleosomal DNA as far as 20 bp from the edge of the nucleosome, while Itc1 contacts similar regions but contacts extranucleosomal DNA more extensively and is bound as far as 53 bp from the entry/exit site (15). The importance of the anchoring domain for ISW2 is clearly seen as the affinity of ISW2 is dramatically reduced when there is less than 20 bp of extranucleosomal DNA (15). The anchoring of ISW2 not only is important for efficient binding but also properly orients ISW2 because, at higher concentrations, ISW2 can bind nucleosomes and yet is still unable to mobilize nucleosomes with 20 bp or less of extranucleosomal DNA (33). The structural domains of ISW2 involved in the translocation, regulation, and anchoring of the complex have not been previously identified.
Only a few structures of subregions of the ISWI type of remodeling complexes or related proteins have been solved. The ATPase subunit of these enzymes contains seven superfamily 2 (SF2) helicase motifs (22) and four additional SNF2-specific motifs (29). The core catalytic domain of ISWI has not been solved, but structures have been obtained for similar motifs in the Rad54 protein from zebrafish and Sulfolobus solfataricus (8, 29). Two lobes were observed in these structures, which are referred to as the DEXD and HELIC domains, and each contains a central beta-sheet flanked by alpha helices. The Rad54 protein structure from Sulfolobus was solved with it bound to DNA, and it was found that the DEXD domain interacts with the minor groove of double-stranded DNA (8). The structure solved for the C terminus of Drosophila melanogaster ISWI revealed three structural domains, referred to as the SANT, SLIDE (SANT-like domain), and HAND domains, that form an extended dumbbell-like structure with two globular domains connected by a spacer helix (14). There has been speculation about how the C-terminal domain of ISWI interacts with nucleosomes, but there have been no data showing directly how this occurs. What has been lacking is information on how these domains are utilized in the complete chromatin remodeling complex, not only with the catalytic subunit intact but also with the other auxiliary or regulatory subunits properly assembled into the complex, and how this holoenzyme complex then interacts with the nucleosomal substrate.
In this study, we determined the domain architecture of the catalytic subunit bound to nucleosomes as part of the four-subunit ISW2 complex comprising Isw2, Itc1, Dpb4, and Dls1 by peptide mapping of Isw2 cross-linked at different nucleosomal and extranucleosomal sites. We have used site-directed modification of nucleosomal and extranucleosomal DNA to photochemically cross-link the Isw2 subunit to different sites and peptide mapping to determine the domains of Isw2 contacting these regions.
MATERIALS AND METHODS
Isolation of Isw2 subunits cross-linked to DNA for peptide mapping.
Synthesis of radiolabeled DNA photoaffinity probes with cross-linkers 17 to 18, 60 to 62, and 92 bp from the dyad axis and photoaffinity-labeling reactions with FLAG-purified yeast ISW2 complex were performed as described previously (15). Cross-linked Isw2 was isolated by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), visualized by phosphorimaging, excised, electroeluted with a buffer containing 50 mM NH4HCO3 and 10% SDS, dried, and stored at −20°C.
Peptide mapping with NTCB digestion.
Photoaffinity-labeled Isw2 was resuspended in a freshly prepared denaturing buffer containing 8 M urea, 10 mM 2-mercaptoethanol, and 0.2 M Tris-acetate (pH 8.5) and incubated at 37°C for 2 h prior to the addition of 12.5 to 25 mM NTCB (2-nitro-5-thiocyanobenzoic acid in ethanol) and incubation for 15 min. The pH was adjusted to approximately 9.0 to 9.5 with NaOH, followed by incubation at 37°C for 1 to 16 h. The reaction was stopped by the addition of SDS sample buffer containing 250 mM Tris-HCl (pH 8.8), 10% glycerol, 2% SDS, 1 mM EDTA, 0.5 M 2-mercaptoethanol, and 0.088% Coomassie blue G250 and analyzed by 4 to 20% or 8 to 20% Tris-glycine SDS-PAGE and phosphorimaging. The apparent molecular masses of the released fragments were estimated by comparing them to the 125I-labeled Mark12 protein standard and Isw2-NTCB markers prepared by in vitro transcription/translation.
Peptide mapping with CNBr digestion.
Photoaffinity-labeled Isw2 was resuspended in 70% formic acid, mixed with an equal volume of freshly prepared CNBr solution (125 to 500 mM cyanogen bromide in 70% formic acid), and incubated at 25°C in the dark for 5 min to 2 h. The cleavage reaction was stopped by diluting samples fivefold with water and drying by vacuum centrifugation. The dried CNBr cleavage samples were resuspended in SDS sample buffer and analyzed by 10% bis(2-hydroxyethyl)imino (BIS)-Tris SDS-PAGE (13) and phosphorimaging. The apparent molecular masses of the released fragments were estimated by comparing them to the 125I-labeled Mark12 protein standard and Isw2-CNBr markers were prepared by in vitro transcription/translation.
Preparation and analysis of peptide-mapping markers.
Salt and detergent were removed from 50 μl (∼44 μg) of Mark12 unstained protein standard (Invitrogen) by using a Sephadex G-10 (GE Healthcare) spin column equilibrated in 200 mM sodium borate (pH 9.0). The protein standard was incubated on ice for 3 h with 0.25 mg/ml sulfo-SHPP [sulfosuccinimidyl-3-(4-hydroxyphenyl)propionate; Pierce]. Excess reagents were removed by using Sephadex G-10 spin columns equilibrated with phosphate-buffered saline (100 mM sodium phosphate, 150 mM NaCl, pH 7.2). The derivatized protein standard was transferred to an Iodo-Gen (1,3,4,6-tetrachloro-3a-6a-diphenylglycouril; Pierce)-coated tube containing 100 μCi of Na125I (Perkin Elmer) and incubated at 25°C for 15 min. The reaction was terminated by removing the sample from the tube and adding NaI to 1 mM. Free Na125I was removed by using Sephadex G-10 spin columns, and the radiolabeled Mark12 protein standard was mixed with SDS sample buffer (65 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.09% bromophenol blue) and stored at −20°C.
The Isw2 fragments corresponding to both the single-hit and the complete cleavage peptides digested by using NTCB or CNBr were prepared in vitro by PCR amplification from the plasmid pET-ISW2-FLAG encoding ISW2 with a C-terminal double FLAG tag and by using a TNT quick coupled transcription/translation system (Promega) with [35S]methionine (Perkin Elmer). Transcription/translation reactions were carried out at 30°C for 90 min, followed by the addition of 20 μg/ml leupeptin, and the reaction products were stored at −80°C.
The synthesized Isw2 markers were analyzed by 8 to 20% Tris-glycine SDS-PAGE and/or 4 to 12% or 10% BIS-Tris SDS-PAGE and phosphorimaging (see Fig. 3 and Table 1). The apparent molecular masses were calculated by comparing the electrophoretic mobilities of the markers to those of the 125I-labeled Mark12 protein standard. Table 1 lists all Isw2 peptide-mapping markers with their calculated and apparent molecular masses on Tris-glycine SDS-PAGE or BIS-Tris SDS-PAGE.
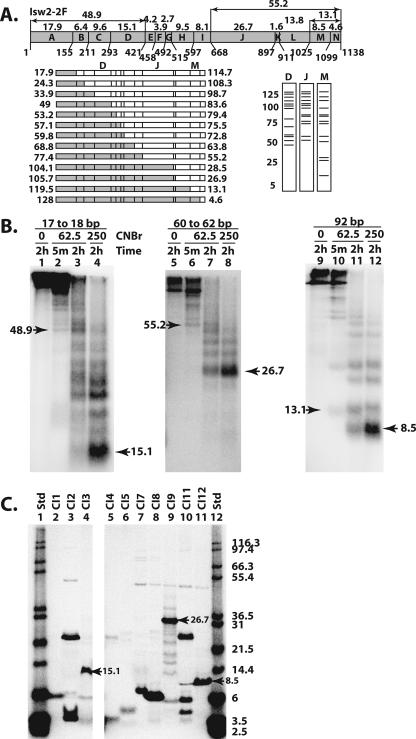
FIG. 3.
The DEXD, HAND, and SLIDE domains were cross-linked to nucleosomal and extranucleosomal DNA. (A) The methionine sites (numbers below the schematic) and predicted molecular masses (numbers above the schematic) of the proteolytic fragments are shown for Isw2. The set of fragments obtained with single-hit CNBr digestion are shown below the Isw2 schematic on the left, with two fragments created from each single hit depicted as either a shaded or an open box. Below on the right are shown the predicted radiolabeled fragments as resolved by SDS-PAGE and obtained from cross-linking in three different regions of Isw2, labeled D, J, and M. (B) Photoaffinity-labeled Isw2 was digested with CNBr at the indicated concentrations (mM) and incubation times, followed by analysis by 10% BIS-Tris SDS-PAGE. The apparent molecular masses, in kDa, of the released bands were calculated as described in Materials and Methods. The molecular masses for the smallest bands released by limited and extensive CNBr digestion are indicated by arrowheads. (C) Peptides corresponding to fragments of Isw2 that could be expected from complete CNBr digestion (Table 1) were synthesized and were analyzed by 10% BIS-Tris SDS-PAGE. Arrowheads highlight the markers that show migrations similar to those of the bands observed in CNBr peptide-mapping experiments. The molecular masses in kDa of 125I-labeled Mark12 protein standards (Std) are indicated.
TABLE 1.
A list of Isw2 markers synthesized by in vitro coupled transcription/translationa
| Marker | Fragment (residues)b | No. of amino acids | Calculated molecular mass (kDa) | Apparent molecular mass (kDa) in:
|
|
|---|---|---|---|---|---|
| Tris-glycine SDS-PAGE | BIS-Tris SDS-PAGE | ||||
| NN1 | Isw2 (1-462) | 462 | 53.7 | 57.86 | 54.10 |
| NN2 | Isw2 (1-525) | 525 | 61.0 | 63.74 | 59.27 |
| NN3 | Isw2 (1-902) | 902 | 104.5 | 108.04 | 113.71 |
| NN4 | Isw2 (1-973) | 973 | 113.1 | 116.15 | 121.49 |
| NC1 | Isw2 (464-C) | 675 | 78.7 | 81.11 | 80.35 |
| NC2 | Isw2 (534-C) | 605 | 70.4 | 71.18 | 72.34 |
| NC3 | Isw2 (903-C) | 236 | 28.0 | 32.78 | 31.80 |
| NC4 | Isw2 (974-C) | 165 | 19.4 | 25.63 | 20.79 |
| NI1 | Isw2 (464-525) | 62 | 7.2 | NA | NA |
| NI2 | Isw2 (534-902) | 369 | 42.4 | 50.01 | 47.63 |
| NI3 | Isw2 (903-973) | 71 | 8.6 | NA | 5.45 |
| CN1 | Isw2 (1-155) | 115 | 17.9 | 27.62 | 35.75 |
| CN2 | Isw2 (1-211) | 211 | 24.3 | 34.49 | NA |
| CN3 | Isw2 (1-293) | 293 | 33.9 | 44.93 | 46.82 |
| CN4 | Isw2 (1-421) | 421 | 48.9 | 53.99 | 53.77 |
| CN5 | Isw2 (1-458) | 458 | 53.2 | NA | NA |
| CN6 | Isw2 (1-492) | 492 | 57.0 | NA | NA |
| CN7 | Isw2 (1-515) | 515 | 59.7 | 62.63 | 61.00 |
| CN8 | Isw2 (1-597) | 597 | 69.2 | NA | NA |
| CN9 | Isw2 (1-668) | 668 | 77.3 | 79.39 | 79.29 |
| CN10 | Isw2 (1-897) | 897 | 103.9 | 108.63 | 106.16 |
| CN11 | Isw2 (1-911) | 911 | 105.5 | NA | NA |
| CN12 | Isw2 (1-1025) | 1025 | 119.3 | NA | 120.00 |
| CN13 | Isw2 (1-1099) | 1099 | 127.9 | NA | NA |
| CC1 | Isw2 (156-C) | 983 | 114.6 | 109.39 | 107.05 |
| CC2 | Isw2 (212-C) | 927 | 108.2 | NA | NA |
| CC3 | Isw2 (294-C) | 845 | 98.6 | 94.82 | 96.45 |
| CC4 | Isw2 (422-C) | 717 | 83.6 | 83.18 | 81.95 |
| CC5 | Isw2 (459-C) | 680 | 79.3 | 78.88 | NA |
| CC6 | Isw2 (493-C) | 646 | 75.5 | NA | NA |
| CC7 | Isw2 (516-C) | 623 | 72.7 | 70.97 | 70.70 |
| CC8 | Isw2 (598-C) | 541 | 63.3 | NA | NA |
| CC9 | Isw2 (669-C) | 470 | 55.2 | 57.18 | 57.51 |
| CC10 | Isw2 (898-C) | 241 | 28.6 | NA | NA |
| CC11 | Isw2 (912-C) | 227 | 27.0 | 30.07 | 33.47 |
| CC12 | Isw2 (1027-C) | 112 | 13.1 | 16.74 | 18.74 |
| CC13 | Isw2 (1100-C) | 39 | 4.6 | NA | NA |
| CI1 | Isw2 (156-211) | 56 | 6.4 | NA | 5.98 |
| CI2 | Isw2 (212-293) | 82 | 9.6 | NA | 23.43 |
| CI3 | Isw2 (294-421) | 128 | 15.1 | NA | 12.96 |
| CI4 | Isw2 (422-458) | 37 | 4.2 | NA | 3.82 |
| CI5 | Isw2 (459-492) | 34 | 3.9 | NA | 4.60 |
| CI6 | Isw2 (493-515) | 23 | 2.7 | NA | NA |
| CI7 | Isw2 (516-597) | 82 | 9.5 | NA | 7.68 |
| CI8 | Isw2 (598-668) | 71 | 8.1 | NA | 5.70 |
| CI9 | Isw2 (669-897) | 229 | 26.7 | NA | 32.23 |
| CI10 | Isw2 (898-911) | 14 | 1.6 | NA | NA |
| CI11 | Isw2 (912-1025) | 114 | 13.8 | NA | 25.05 |
| CI12 | Isw2 (1027-1099) | 73 | 8.5 | NA | 10.44 |
A marker number is given to each peptide. The first letter denotes the cutting agent (N for NTCB and C for CNBr). The second letter denotes whether the marker contains the N or C terminus of the protein; “I” indicates a marker containing neither terminus. The numbers are in order of the markers' positions relative to the full-length protein from N to C terminus. NA, not analyzed.
C, C terminus.
Structure modeling of Isw2 ATPase and C-terminal domains.
The modeling of the Isw2 ATPase and C-terminal domains was performed with Accelrys DS modeling 1.1 with the MODELER module based on a comparative homology modeling algorithm (24). The structure of Sulfolobus solfataricus Rad54 ATPase domain in complex with 25-bp double-stranded DNA (PDB ID 1Z63) (8) and the Drosophila ISWI-C domain structure (PDB ID 1OFC) (14) were used as structural templates for the modeling of Isw2 ATPase in complex with DNA and the Isw2 C-terminal domains, respectively. The sequence alignments of Isw2 ATPase with the Sulfolobus Rad54 ATPase structure and Isw2-C with the Drosophila ISWI-C domain structure were performed with the “Align2D” command with default settings. For each structure, 200 models were built with the “refined” setting. All of the nucleotide residues in DNA strands “C” and “D” in the 1Z63 structure were selected as the ligand in building the structures for the Isw2 ATPase structure in complex with DNA. The 20 models with the lowest PDF (probability density functions) values were exported as Protein Data Bank files, and the “combined energy” values were compared by using ProSa2003 (http://www.came.sbg.ac.at/came-frames/prosa.html) (27). The structural model with the lowest “combined energy” was selected as the final model for Isw2 ATPase in complex with DNA. Since yeast Isw2-C contains two regions that are not aligned with Drosophila ISWI-C, the Isw2-C model with the lowest “combined energy” was subject to loop refinement. A total of 100 loop models were built using the “Refine Loop” command with the “refined” setting. The 10 loop models with the lowest PDF values were exported, and the model with the lowest “combined energy” was selected as the final model for Isw2-C.
RESULTS
Strategy for identification of Isw2 domains associated with nucleosomal and extranucleosomal DNA regions.
Previously, DNA footprinting and photochemical cross-linking had shown a tripartite architecture for Isw2. Isw2 was shown to be proximal to DNA (i) two helical turns from the dyad, (ii) near the entry/exit site, and (iii) up to 20 bp from the edge of nucleosomes with extranucleosomal DNA (15). Itc1 also contacts the same regions as Isw2, except that it binds a larger stretch of the extranucleosomal DNA from the entry/exit site, to at least 53 bp from the edge of the nucleosome. In this study, the same photo-cross-linking technique was used, in combination with peptide mapping by chemical proteolysis, in order to identify the domains of Isw2 that interact with these regions of the nucleosome. Photoreactive DNA was used to cross-link Isw2 17 to 18, 60 to 62, and 92 bp from the dyad axis (Fig. 1), corresponding to the three distinct regions protected by ISW2 binding (15). DNA was made photoreactive by the targeted incorporation of nucleotides conjugated through a short tether to a photoreactive aryl azide, as shown in Fig. 1 (18, 32). Adjacent to the photoreactive nucleotide, a 32P-labeled nucleotide in DNA was incorporated such that, after cross-linking and DNA digestion, the radiolabel was transferred to the cross-linked protein (15). The region of Isw2 cross-linked to DNA was mapped by chemical proteolysis of the cross-linked Isw2, separation of the peptide fragments by gel electrophoresis, and visualization of the labeled peptide fragment(s) by phosphorimaging. The cleavage conditions were adjusted to examine both single-hit cutting (i.e., one cleavage per polypeptide) and extensive digestion of the labeled protein in order to determine the location of the radiolabel (i.e., the cross-linking site) based on the electrophoretic mobilities of the radiolabeled fragments. The single-hit conditions created two sets of proteolytic fragments, containing either the N or C terminus of the protein. Appropriate size markers of the different truncated forms of Isw2 containing the N or C terminus were generated by using an in vitro coupled transcription/translation system and labeled [35S]methionine for comparison with the photoaffinity-labeled peptide fragments (Table 1 and Fig. 2C and 3C). Particular internal fragments were also synthesized in this way for comparison with cross-linked products that were obtained by exhaustive proteolysis.
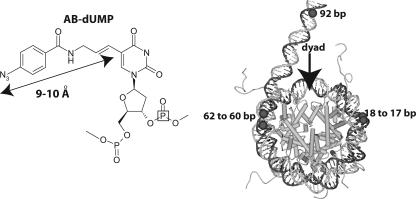
FIG. 1.
Site-specific cross-linking of DNA to Isw2. In the left panel, the structure of the photoreactive nucleotide AB-dUMP is shown as incorporated into DNA. The distance from the cross-linking site (N3) to the C-5 position of deoxyuridine is 9 to 10 Å, as indicated. The location of the 32P radiolabel is boxed in the structure to illustrate its general position. In the right panel, a model of the nucleosome is shown with the sites that are targeted for modification and photo-cross-linking to Isw2 highlighted in black; the numbering refers to the number of base pairs from the dyad axis.
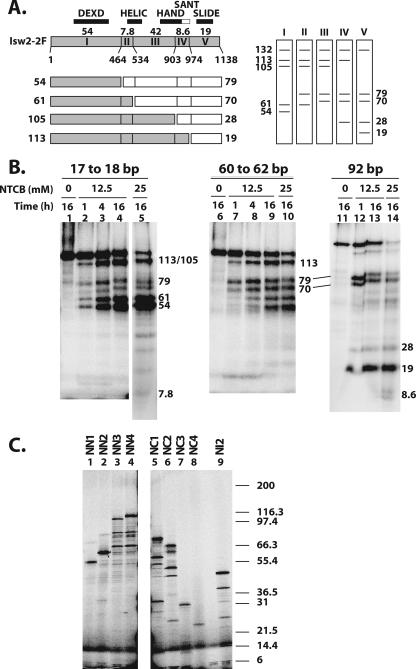
FIG. 2.
Mapping the regions of Isw2 cross-linked to three sites in the nucleosome by NTCB digestion. (A) A schematic diagram of Isw2 is shown, with residue numbers for the different cysteine cut sites indicated. The locations of the conserved Isw2 domains are indicated, along with the size of each proteolytic fragment (kDa). The set of fragments obtained with single-hit NTCB digestion are shown below the Isw2 schematic, with two fragments created from each single hit depicted as either a shaded or open box. The predicted sets of radiolabeled single-hit proteolytic fragments that can be obtained depending on the site of Isw2 that is cross-linked to DNA are shown to the right. (B) Photoaffinity-labeled Isw2 was digested with the indicated concentrations of NTCB and incubation times, followed by analysis by 4 to 20% Tris-glycine SDS-PAGE. The apparent molecular masses of the radiolabeled bands were calculated as described in Materials and Methods and are shown in kDa. (C) Fragments of Isw2 were synthesized in vitro that correspond to fragments that would be obtained by NTCB digestion (Table 1) and contain either the N terminus (NN1, NN2, NN3, and NN4), the C terminus (NC1, NC2, NC3, and NC4), or an internal fragment (NI2). These truncated proteins were analyzed by 4 to 20% Tris-glycine SDS-PAGE. The molecular masses of the 125I-labeled Mark12 protein standards are indicated to the right. The slowest-migrating major bands were fully translated products and were used in calculating the apparent molecular masses.
Figure 2A shows a schematic diagram of the NTCB cleavage sites at cysteines in Isw2 and the fragment sizes of labeled peptides predicted for single-hit NTCB digestion of Isw2 (12). There are six cysteines in Isw2, with two pairs of cysteines either immediately adjacent to each other (Cys 463 and Cys 464 or separated by 7 amino acids (Cys 526 and Cys 534), such that NTCB cleavage creates only one of five distinct sets of radiolabeled peptide fragments, depending on the location of the radiolabeled tag. This particular type of proteolysis is done first in order to simplify the interpretation of the proteolysis because of the reduced number of cleavage sites. These results then provide an initial view of the location of the cross-link, which can be refined by using more extensive proteolysis conditions.
The DEXD domain of Isw2 was bound to DNA at 17 and 18 bp from the dyad axis.
NTCB digestion of Isw2 cross-linked to DNA 17 and 18 bp from the dyad axis generated primarily 113-, 105-, 62-, and 54-kDa radiolabeled peptides (Fig. 2B, lanes 2 to 3), consistent with cross-linking to region I (residues 1 to 463). Lesser amounts of the labeled 70- and 79-kDa proteolytic fragments were detected, consistent with minor cross-linking to regions II and III. The major cross-linking site was to region I, as shown by extensive NTCB digestion, primarily producing a 54-kDa radiolabeled proteolytic fragment, with very little of a detectable 7.8-kDa fragment (Fig. 2B, lane 5). The relative mobilities of these proteolytic fragments were confirmed by comparison with truncated versions of Isw2 that were synthesized by coupled transcription/translation production (Table 1 and Fig. 2C). The region of Isw2 cross-linked near the dyad axis includes a region having homology with the DEXD domain of the conserved ATPase domain and an N-terminal flanking region. Delineating whether the N terminus or the DEXD domain was associated with DNA was done by CNBr digestion of cross-linked Isw2 (12). There are 13 methionine, or CNBr cleavage, sites in Isw2 (Fig. 3A). Extensive CNBr digestion produced a labeled proteolytic fragment with an electrophoretic mobility corresponding to a 15.1-kDa fragment spanning residues 294 to 421 (Fig. 3A, region D, and B, lane 4). This peptide was synthesized to determine the electrophoretic mobility of this peptide region (Fig. 3C, lane 4, and Table 1). The electrophoretic mobility of this fragment was measured and compared to the predicted mobility and the mobilities of the nearby peptide fragments that were synthesized in vitro with a coupled transcription/translation system with [35S]methionine. As can be seen in Table 1, there are no other proteolytic fragments generated by cleavage with CNBr from the N-terminal two-thirds of Isw2 that have an electrophoretic mobility comparable to that of this region. Consistent with this conclusion, the limited digestion generated peptide fragments with the smallest fragment corresponding to the 48.9-kDa peptide, or residues 1 to 421 of Isw2 (Fig. 3A, lane D, and B, lane 2). These results showed that the DEXD domain, and not its N-terminal flanking region, is close to SHL2 inside the nucleosome (Fig. 4A). Isw2, as part of the four-subunit ISW2 complex, has its DEXD domain close to the internal contact site at SHL2. Translocation along DNA from this same DNA site has recently been found to be essential for ISW2 remodeling, consistent with the DEXD domain of the conserved ATPase region contacting this region (34).
FIG. 4.
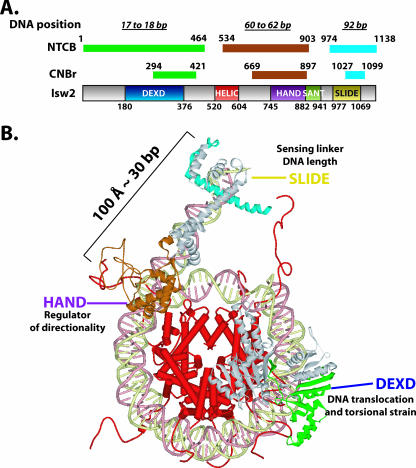
Model of Isw2 docking to the nucleosome. (A) A summary of the mapping results for regions of Isw2 cross-linked to the nucleosomal and extranucleosomal DNA. Isw2 is schematically shown as a gray bar with conserved domains boxed in various colors. The fragments cross-linked to indicated DNA sites (17 to 18, 60 to 62, and 92 bp from the dyad axis of the nucleosome) are shown as color-coded narrow bars above the Isw2 schematic and are denoted by mapping agent, NTCB or CNBr. Beginning and ending amino acid positions for each domain and mapped fragment are given. (B) Structural model of Isw2 interacting with the nucleosomal core particle. The regions of Isw2 that interact with the SHL2 site, the entry/exit site, and extranucleosomal DNA are colored green, brown, and cyan, respectively, as in panel A and are modeled based on homology and peptide mapping data. The domains of Isw2 are labeled, and their proposed functional roles in chromatin remodeling by ISW2 are given.
The HAND domain of Isw2 was bound near the entry/exit of the nucleosome.
Isw2 cross-linked 60 to 62 bp from the dyad axis was digested with NTCB and generated primarily 113-, 105-, 79-, and 70-kDa radiolabeled proteolytic fragments that corresponded to cross-linking to region III—residues 534 to 903 (Fig. 2B, lane 7). More extensive digestion with NTCB enriched for a 42-kDa peptide, with no significant amounts of smaller radiolabeled peptides, consistent with cross-linking to region III (Fig. 2B, lane 10). The 42-kDa peptide has an electrophoretic mobility that is close to that of the polypeptide from residue 1 to 464 created by extensive NTCB digestion, but under our gel conditions, they are separated from each other (Table 1 and data not shown). Further confirmation of cross-linking at region III was also obtained by CNBr digestion (see below). Region III has sequence homology to a portion of the C terminus of ISWI from Drosophila and corresponds to the HAND domain. Region III also contains a part of the conserved ATPase domain observed in other Snf2 subfamily members and corresponds to the HELIC domain. Further mapping of the region of Isw2 cross-linked to DNA 60 and 62 bp from the dyad axis was done by digestion of photoaffinity-labeled Isw2 with CNBr. The smallest fragment from extensive CNBr digestion of Isw2 cross-linked to DNA had an electrophoretic mobility corresponding to that of the 26.7-kDa fragment of Isw2 from residues 669 to 897 (Fig. 3A, region J, and B, lane 8) and correlates to the region sharing homology with the HAND domain. These results demonstrated more clearly that the HAND domain of Isw2 was cross-linked to DNA near the entry/exit site and not the HELIC domain. The 26.7-kDa CNBr proteolytic fragment is distinctive because it is the largest CNBr fragment that can be obtained under exhaustive digestion conditions (Fig. 3A and C, lane 9, and Table 1). Limited CNBr digestion was consistent with this observation as the smallest radiolabeled proteolytic fragment observed, of 55.2 kDa, corresponds to a single cut at residue 668 and the radiolabel residing on the C-terminal half of this cut site (Fig. 3A and B, lane 6).
The SLIDE domain of Isw2 was associated with extranucleosomal DNA.
The peptide fragments generated by limited NTCB digestion of Isw2 cross-linked 92 bp from the dyad axis were 113, 79, 70, 28, and 19 kDa and corresponded to cross-linking to region V, residues 974 to 1120 (Fig. 2B, lane 12, and C, lane 8). Under more-exhaustive NTCB digestion conditions, less than 10% of the cross-linking was found to be at region IV (residues 903 to 973), consistent with the amount of the labeled 8.6-kDa peptide compared to the amount of 19-kDa peptide detected in lane 14. Further refinement of the region of Isw2 cross-linked to extranucleosomal DNA was done by CNBr cleavage. After extensive digestion with CNBr, Isw2 cross-linked to DNA 92 bp from the dyad axis produced a radiolabeled 8.5-kDa peptide corresponding to residues 1025 to 1099 (Fig. 3B, lanes 11 to 12, and C, lane 11), consistent with the 13.1-kDa fragment (residues 1027 to 1138) of Isw2 obtained under limited digestion conditions (Fig. 3A, region M, and B, lane 10). Peptide mapping of cross-linked Isw2 with cyanogen bromide narrowed down the region of Isw2 cross-linked to extranucleosomal DNA from residues 974 to 1138 to residues 1025 to 1099, corresponding to a region that has sequence homology with the SLIDE domain observed in the C terminus of ISWI from Drosophila. These data show that the SLIDE domain of Isw2 is directly associated with extranucleosomal DNA 92 bp from the dyad axis or 19 bp from the edge of the nucleosome.
Structural model for Isw2-nucleosome interaction.
A complete map of the conserved domains of ISW2 that were cross-linked to the three nucleosomal and extranucleosomal DNA sites is summarized in Fig. 4A. The regions of Isw2 cross-linked to nucleosomal and extranucleosomal DNA correspond to conserved regions in which crystal structure information is available from homologous proteins. In order to better visualize the domain interactions between Isw2 and the nucleosome, a structural model of Isw2-nucleosome interactions was made based on the mapping data. This overall model, for the first time, demonstrates structurally how the catalytic subunit of an ISWI chromatin remodeling complex interacts with its nucleosomal and extranucleosomal substrate and likely catalyzes the remodeling reaction as part of the ISW2 holoenzyme activity (Fig. 4B).
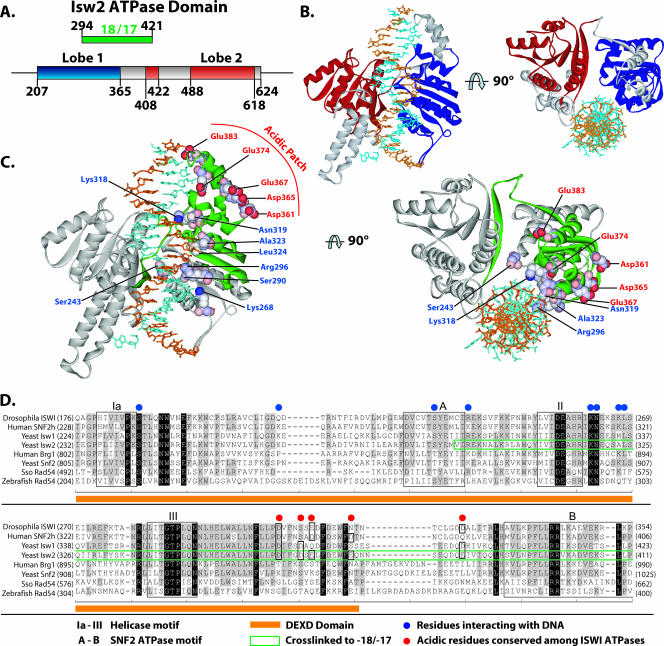
The DEXD domain is the most-conserved region in the ATPase domain of Isw2 compared to other chromatin-remodeling ATPase domains and, based on sequence homology, is located in the first lobe of the ATPase domain (8, 29). The ATP-binding site is also believed to be located within this conserved region (30). The stretch of nucleosomal DNA about 20 bp from the dyad axis was shown by two independent laboratories to be a critical point for chromatin remodelers to translocate along DNA and to generate localized torsional strains (23, 34). Our mapping results, for the first time, have demonstrated the physical interaction between the ATPase domain and this critical point on nucleosomal DNA and suggest that chromatin-remodeling events indeed are powered by the direct contact of the ATPase domain inside the nucleosome.
A model of the binding of the ATPase domain of Isw2 to nucleosomal DNA was constructed by modeling the structure based on homology to the Rad54 ATPase domain structure from Sulfolobus. The ATPase domain obtained by protein modeling was docked onto the nucleosome structure at the region found to be cross-linked with nucleosomal DNA and oriented to the nucleosomal DNA similarly to that observed in the cocrystal of the Rad54 ATPase domain with DNA. In this model, it can be clearly seen that the DEXD domain makes more-extensive contact with DNA, consistent with it being the most likely to be cross-linked to nucleosomal DNA (Fig. 5B and C, regions highlighted in green and blue).
FIG. 5.
Model for the binding of the ATPase domain of Isw2 to nucleosomal DNA. (A) The locations of the α/β helicase lobes (Lobe 1 and Lobe 2) in the primary amino acid structure of the Isw2 ATPase domain, based on sequence alignment with Sulfolobus Rad54, are shown, along with the relationship to the region of Isw2 cross-linked to DNA 17 and 18 bp from the dyad axis. (B) The Isw2 ATPase domain model was generated as described in Materials and Methods, based on homology modeling and the structure of Sulfolobus Rad54 bound to DNA. The first helicase lobe is depicted in blue and the second in red, with the region between the lobes shown in gray. (C) The region of Isw2 cross-linked to the SHL2 site is highlighted in green in the same model that is shown in panel B. The amino acids contacting DNA as suggested by the structure of Rad54 bound to DNA are highlighted in blue and displayed in a space fill format. A putative acidic patch is highlighted in red with a space fill format on the surface of the first α/β-helicase lobe. (D) An acidic patch on the surface of Isw2 is moderately conserved in the ISWI subfamily. A sequence alignment of SNF2 family ATPases is shown. Helicase- and SNF2-specific motifs are boxed and labeled as indicated. The fragment identified as cross-linking DNA at SHL2 is boxed in green.
Another outcome from this model is the observation of a moderately conserved acidic patch on the surface of the DEXD domain that would be in the correct location to make contact with the basic N-terminal tail region of histone H4 (Fig. 5C and D). Isw2 contact at SHL2 requires the presence of the basic tail region of histone H4 (6). The histone H4 tail is also required for efficient remodeling by ISW2 and other ISWI complexes (2, 3, 6, 10). These data suggest that the acidic patch may interact with the H4 tail and facilitate its recruitment of the DEXD domain of Isw2 to nucleosomal DNA at SHL2. The acidic patch, comprising residues Asp361, Asp365, Glu367, Glu374, and Glu383, is conserved in other ISWI homologs. The acidic patch seems to have a unique role in chromatin remodeling by the ISWI-type complexes as it is not found in other members of the SF2 family, such as Brg1, Swi2/Snf2, and Rad54.
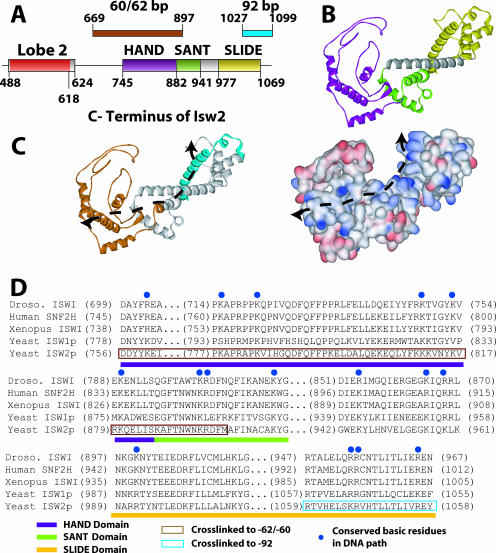
Models of the C terminus of Isw2 docking to the entry site of the nucleosome and linker DNA were constructed based on sequence homology with the C terminus of ISWI from Drosophila and its known structure. Although previously the structure of the C terminus of ISWI from Drosophila was postulated to bind and bridge across two gyres of the nucleosomes, our cross-linking data were not consistent with this model (14). This elongated structure does, however, quite well accommodate having the HAND/SANT domain combination bound at the entry site, while the SLIDE domain, connected by a long, conserved, alpha-helical spacer domain, is bound to linker DNA 19 bp from the edge of the nucleosome (Fig. 4B and 6). The 30-bp distance between the nucleosome entry/exit site bound by the HAND domain and the part of extranucleosomal DNA bound by the SLIDE domain is remarkably consistent with the 100-angstrom distance observed between the SLIDE and HAND/SANT domains (14). The model of the C terminus of Isw2 docked to the nucleosome and linker DNA was such that both regions shown to be cross-linked to DNA could be simultaneously bound to DNA. The path likely traversed by DNA across the surface of the C terminus of Isw2 is strongly suggested by the relative locations of the two regions in the C terminus that are cross-linked to DNA (Fig. 6C, indicated by a dotted line). Docking was done such that the long alpha helix of the SLIDE domain was bound to the major groove of DNA at the appropriate distance from the edge of the nucleosome. Aside from the cross-linking data suggesting that this part of the SLIDE domain is in contact with linker DNA, other data indicate that the alpha helix may be involved in DNA binding and that the basic amino acids conserved in this helix are required (14). A striking feature of the path of DNA as suggested by cross-linking is the highly conserved basic amino acids that are found on this surface (Fig. 6C, shown in blue). These basic amino acids are well conserved throughout the ISWI family of chromatin-remodeling complexes, span the entire length of the postulated DNA path, and provide supporting evidence for the model of the interaction of the C terminus of Isw2 with the nucleosome and linker DNA (Fig. 6D).
FIG. 6.
Model for the C terminus of Isw2 and its interaction with the nucleosome. (A) The positions of the HAND, SANT, and SLIDE domains in Isw2 are shown and correlated with the regions of Isw2 that were cross-linked to nucleosomal and extranucleosomal DNA 60 to 62 and 92 bp from the dyad axis. (B) A model of a portion of the C terminus of Isw2 is shown, with the HAND domain in purple, the SANT domain in light green, the SLIDE domain in yellow, and the spacer helix in gray. (C) A DNA binding path is suggested by the surface charge distribution of the Isw2 C-terminal domain model and the regions found to be cross-linked to DNA. The same model that is shown in panel B is depicted with the regions cross-linked to the entry/exit site in brown and extranucleosomal DNA in cyan (left panel). The distribution of the surface charges (positive in blue, negative in red) of the Isw2 C-terminal domain model suggests a putative path of DNA as indicated by the dashed line (right panel). (D) A sequence alignment of the C-terminal domains of ISWI subfamily ATPases illustrates the conserved, positively charged residues (highlighted by blue dots) that are likely to interact with DNA. Conserved domains are indicated by colored bars below the sequence alignment. The regions that cross-link to the entry/exit site of the nucleosome (positions 60 to 62) and the extranucleosomal DNA (position 92) are boxed in brown and cyan, respectively.
DISCUSSION
Although ATP-dependent chromatin remodeling has been shown to regulate cellular processes, such as transcription, DNA replication and damage repair, heterochromatin formation and maintenance, and cell cycle control, there has not been much structural information regarding how these complexes interact with chromatin. Due to the size and complexity of these complexes and the intrinsic difficulty in crystallizing them, structural information has remained scarce. Yet knowledge of the molecular interactions of these enzymes and their substrates is critical for understanding the mechanism of ATP-dependent mobilization and reorganization of nucleosomes. Towards this goal, we have determined the molecular interactions of the catalytic subunit of one of these complexes with nucleosomes, leading to a structural model of this subunit bound to nucleosomes. The DEXD lobe of the ATPase domain was found to be physically associated near the dyad axis and likely promotes nucleosome movement by translocation along DNA at this location. The SLIDE domain located near the C terminus of Isw2 interacts with linker or extranucleosomal DNA 19 bp from the edge of the nucleosome and, based on its binding site, helps anchor ISW2 to nucleosomes through its interaction with extranucleosomal DNA. For the first time, the HAND domain of an ISWI-type remodeler has been shown to bind near the entry site of nucleosomes. As such, the HAND domain may facilitate the directional preference that ISW2 has for mobilizing nucleosomes.
The DEXD domain consists of several key ATPase motifs conserved among most SF1 and SF2 ATPases and is the most-conserved domain among all ATP-dependent chromatin-remodeling ATPases (22, 29). A mutation of K215R in Isw2 completely eliminates the ATPase and remodeling activities of ISW2 and is located within the ATP-binding pocket of the protein within the DEXD domain (30). The demonstration that the DEXD domain is associated with the internal SHL2 site of the nucleosome when bound by ISW2 coincides well with the recent finding that ISW2 translocation at this site is required for nucleosome mobilization. Translocation by other ATP-dependent remodelers, such as NURF, RSC, and SWI/SNF, also occurs at this same location on nucleosomes and is vital for nucleosome mobilization by their respective complexes (23, 26, 34). The interactions of ISW2 at this region are also affected by the histone H4 N-terminal tail and the length of the extranucleosomal DNA (6).
ISW2 must be efficiently anchored to nucleosomes so it can make the appropriate contacts for regulating the direction of nucleosome movement and provide the impetus for nucleosome movement through its translocation along DNA at the correct position. A separate domain likely facilitates the anchoring and orienting of ISW2 onto the nucleosome. The importance of the anchoring domain of ISW2 is clearly seen as the affinity of ISW2 for nucleosomes is dramatically reduced when there is less than 20 bp of extranucleosomal DNA (15). The anchoring function is important not only for efficient binding but also for properly orienting ISW2. Nucleosomes with 20 bp or less of extranucleosomal DNA can be bound using higher concentrations of ISW2 but cannot be made mobile (33). The inability of ISW2 to mobilize nucleosomes under these conditions is due to improper ISW2 binding, with the translocation domain being unable to bind to the SHL2 site (6). Cross-linking data showed that the SLIDE domain is responsible for binding to extranucleosomal DNA and likely serves as part of the anchor domain of ISW2.
Earlier studies with the C terminus of Drosophila ISWI had suggested that the SLIDE domain might interact with the H4 N-terminal tail and, thus, be bound to the core nucleosome surface (14). At the time of this study, there was no way to know how these structural domains were oriented on the surface of the nucleosome. We would expect or predict that the SLIDE domains in the Drosophila CHRAC and ACF complexes, like that observed for Isw2, are also bound to extranucleosomal DNA. Consistent with these studies, the SANT domain was found not to be directly bound to DNA, due to its absence of cross-linking at the three contact sites tested. Although the SANT domain is structurally similar to the c-Myb DNA binding domain, the particular residues involved in DNA binding in c-Myb were not conserved in the ISWI SANT domain, and the overall negatively charged surface of the SANT domain is incompatible with binding DNA. These structural aspects and earlier biochemical results indicated that the SANT domain was likely not involved in binding DNA. Several studies have suggested instead that similar SANT domains, like those in Isw2 and ISWI, contact the H3 histone tail (1, 20, 28). The location of the SANT domain near the entry/exit site, as deduced by cross-linking of the HAND domain, places the SANT domain proximal to the base of the histone H3 N-terminal tail. However, a potential role for the SANT domain binding the H3 tail region in nucleosome remodeling is not clear, as the H3 tail does not appear to be required for ISWI or ISW2 remodeling (6, 14). It is, however, not unexpected that the histone H3 tail is not critical, since the SANT domain itself does not appear to be essential for ISWI remodeling (14).
The HAND domain consists of four helices, three forming an L-shaped structure while the fourth is located in its concave surface (Fig. 6B, purple), with no other similar structures found in the current databases. The HAND and SANT domains probably function together in Isw2 as one functional module and are not able to move independently of each other due to the extensive interactions observed between these two domains. The loss of ISW2 contacts near the entry/exit site of nucleosomes interferes with the directional preference of nucleosome movement by ISW2, while not affecting the general ability of ISW2 to mobilize nucleosomes (33). The cross-linking data therefore suggest that one functional role for the novel HAND domain is to regulate the ISW2 preferred direction for mobilizing nucleosomes, since the HAND domain bound near the entry/exit site could block the release of DNA in that direction (14).
The HAND, SANT, spacer, and SLIDE domains form a path of highly conserved, surface-accessible, positively charged residues ideal for binding DNA, extending from just inside the nucleosome entry/exit site to 19 bp of the extranucleosomal DNA (Fig. 5C and D). The structure formed by the tight association of the SLIDE, spacer, SANT, and HAND domains appears, based on structural information from the C terminus of ISWI, to be a rigid, elongated scaffold that could readily facilitate pushing DNA into the nucleosome, in conjunction with Itc1 as it clamps onto DNA. The path of DNA would be predicted to make a sharp turn as DNA encounters alpha helix SL3 of the SLIDE domain, based on the predicted structure of the C terminus of Isw2.
Our current model is that the highly conserved catalytic-core DEXD domain interacts with the critical internal site of the nucleosome for translocation along DNA and the generation of localized torsional strain. DNA bound to the protein scaffold formed from the HAND, SANT, spacer, and SLIDE domains is pushed or forced into the nucleosome due to a conformational shift of Isw2. As the conformation of Isw2 is altered by ATP hydrolysis, the extranucleosomal DNA fixed onto Isw2 can be forced into the nucleosome as the two sides of Isw2 come closer together and the second side remains attached to its nucleosomal site. A DNA bulge on the surface of the nucleosome would be formed between the two attachment points of Isw2. The formation of the bulge would be enhanced by the generation of DNA torsional strain due to the translocation of Isw2 on nucleosomal DNA. Nucleosome remodeling by ISW2 with DNA translocation and twisting at SHL2 creating torsional strain in the DNA between the translocation site and the entry/exit site of DNA from the nucleosome has recently been proposed (34). ISW2 accommodates or relieves the torsional strain by promoting the formation of a small DNA bulge on the surface of the nucleosome through coordinated conformational changes near the entry/exit site of the nucleosome. The bulge propagates through the nucleosome in one direction, presumably by the release of contacts near SHL2. The unidirectional movement of the DNA bulge is controlled by ISW2 contacts with the entry/exit site of the nucleosome. Further experiments will be needed to test different aspects of this model and the roles of conserved domains or residues in ISW2 remodeling.
Acknowledgments
We thank Mekonnen Dechassa for technical assistance in radio-iodination. Thanks go to members of the Bartholomew lab for their critical review of and comments on the manuscript.
This work was supported by Public Health Service grant GM 70864 from the National Institutes of Health.
Footnotes
Published ahead of print on 1 October 2007.
REFERENCES
- 1.Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. [DOI] [PubMed] [Google Scholar]
- 2.Clapier, C. R., G. Langst, D. F. Corona, P. B. Becker, and K. P. Nightingale. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapier, C. R., K. P. Nightingale, and P. B. Becker. 2002. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 5.Dang, W., M. N. Kagalwala, and B. Bartholomew. 2007. The Dpb4 subunit of ISW2 is anchored to extranucleosomal DNA. J. Biol. Chem. 282:19418-19425. [DOI] [PubMed] [Google Scholar]
- 6.Dang, W., M. N. Kagalwala, and B. Bartholomew. 2006. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol. Cell. Biol. 26:7388-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319:1097-1113. [DOI] [PubMed] [Google Scholar]
- 8.Durr, H., C. Korner, M. Muller, V. Hickmann, and K. P. Hopfner. 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121:363-373. [DOI] [PubMed] [Google Scholar]
- 9.Edayathumangalam, R. S., P. Weyermann, P. B. Dervan, J. M. Gottesfeld, and K. Luger. 2005. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 345:103-114. [DOI] [PubMed] [Google Scholar]
- 10.Fazzio, T. G., M. E. Gelbart, and T. Tsukiyama. 2005. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 25:9165-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flaus, A., and T. Owen-Hughes. 2003. Mechanisms for nucleosome mobilization. Biopolymers 68:563-578. [DOI] [PubMed] [Google Scholar]
- 12.Fontana, A., and E. Gross. 1986. Fragmentation of polypeptides by chemical methods, p. 67-119. In A. Darbre (ed.), Practical protein chemistry: a handbook. John Wiley & Sons, Inc., New York, NY.
- 13.Graham, D. R., C. P. Garnham, Q. Fu, J. Robbins, and J. E. Van Eyk. 2005. Improvements in two-dimensional gel electrophoresis by utilizing a low cost “in-house” neutral pH sodium dodecyl sulfate-polyacrylamide gel electrophoresis system. Proteomics 5:2309-2314. [DOI] [PubMed] [Google Scholar]
- 14.Grune, T., J. Brzeski, A. Eberharter, C. R. Clapier, D. F. Corona, P. B. Becker, and C. W. Muller. 2003. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12:449-460. [DOI] [PubMed] [Google Scholar]
- 15.Kagalwala, M. N., B. J. Glaus, W. Dang, M. Zofall, and B. Bartholomew. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23:2092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Langst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 18.Lannutti, B. J., J. Persinger, and B. Bartholomew. 1996. Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry 35:9821-9831. [DOI] [PubMed] [Google Scholar]
- 19.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 20.Mo, X., E. Kowenz-Leutz, Y. Laumonnier, H. Xu, and A. Leutz. 2005. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 19:2447-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Richmond, E., and C. L. Peterson. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24:3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha, A., J. Wittmeyer, and B. R. Cairns. 2005. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12:747-755. [DOI] [PubMed] [Google Scholar]
- 24.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 25.Schiessel, H., J. Widom, R. F. Bruinsma, and W. M. Gelbart. 2001. Polymer reptation and nucleosome repositioning. Phys. Rev. Lett. 86:4414-4417. [DOI] [PubMed] [Google Scholar]
- 26.Schwanbeck, R., H. Xiao, and C. Wu. 2004. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J. Biol. Chem. 279:39933-39941. [DOI] [PubMed] [Google Scholar]
- 27.Sippl, M. J. 1993. Recognition of errors in three-dimensional structures of proteins. Proteins 17:355-362. [DOI] [PubMed] [Google Scholar]
- 28.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 29.Thoma, N. H., B. K. Czyzewski, A. A. Alexeev, A. V. Mazin, S. C. Kowalczykowski, and N. P. Pavletich. 2005. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12:350-356. [DOI] [PubMed] [Google Scholar]
- 30.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Zofall, M., and B. Bartholomew. 2000. Two novel dATP analogs for DNA photoaffinity labeling. Nucleic Acids Res. 28:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zofall, M., J. Persinger, and B. Bartholomew. 2004. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol. Cell. Biol. 24:10047-10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zofall, M., J. Persinger, S. R. Kassabov, and B. Bartholomew. 2006. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13:339-346. [DOI] [PubMed] [Google Scholar]