Abstract
Translation of the open reading frame 2 (ORF-2) of the human respiratory syncytial virus M2 gene initiates at one of the three initiation codons located upstream of the termination codon for the first ORF. Replacement of ORF-2 with the major ORF of the chloramphenicol acetyltransferase reporter gene followed by systematic mutagenesis of the putative initiation codons demonstrated the usage of these codons as the translational initiators for ORF-2 expression both in vitro and in vivo. While the efficiency of translation was maintained when only the first and second AUG codons were preserved in vivo, there was no apparent preference in vitro for any of the three codons when only one was present. Mutagenesis studies showed that the location of the termination codon of ORF-1 protein plays a crucial role in directing translation of ORF-2 from the upstream initiation codons in vivo. This indicates that the second ORF is accessed by the ribosomes that are departing from the first ORF and that these ribosomes reinitiate on AUG codons 5′ to the point of translation termination.
Keywords: M2 gene/pneumovirus/respiratory syncytial virus/termination–reinitiation/translation
Introduction
Viruses have developed a variety of strategies to maximize the number of proteins encoded by their size-limited genomes. While many of these strategies can involve control of gene expression at the transcriptional level, several result from alternatives to conventional translational processes. Viruses depend on the host cell to provide ribosomes to synthesize proteins, and analyses of translation in virus-infected cells have revealed several unexpected features of the translation process. Additional proteins may arise from the use of secondary translation initiation sites within an mRNA to access another open reading frame (ORF). An example of this is seen for the phosphoprotein gene of the paramyxoviruses Sendai virus and pneumonia virus of mice (PVM; Giorgi et al., 1983; Curran et al., 1986; Curran and Kolakofsky, 1987, 1990; Barr et al., 1994; Gupta et al., 1996). In an alternative strategy, the genomes/mRNA of picornaviruses contain large RNA structures (internal ribosomal entry sites, IRES; Pelletier and Sonenberg, 1988) that allow the efficient translation of the non-capped mRNA. The IRES direct ribosomes to bind and initiate translation at a pre-defined initiation codon.
With the exception of IRES-directed initiation of transcription, all initiation events conform to elements of the ribosomal scanning mechanism proposed by Kozak (1987, 1989). In this model, a ribosome initiates translation from the first available initiation codon. Following initiation, the ribosomes move along the template in a 5′→3′ direction until they encounter a termination codon, at which point they depart from the mRNA. Kozak (1987) suggested a consensus sequence (GCC)GCC(A/G)CCAUGG as an optimal sequence for translation initiation by vertebrate ribosomes, where the underlined AUG is the invariant initiation codon. In higher eukaryotes, sequences flanking the AUG modulate its ability to halt the scanning 40S subunit. Mutation in the consensus motif GCCACC in position –6 to –1, especially substitution of a pyrimidine for the A at the –3 position, weakens this adherence and causes some 40S subunits to bypass the first AUG allowing initiation instead at the next AUG. This context-dependent leaky scanning is also seen when the highly conserved G in position +4 immediately following the AUG is mutated, but it is generally not affected by nucleotides at positions +5 and +6. Consequently, while translation initiation preferentially occurs at the AUG codon nearest the cap site, an AUG further downstream could also initiate translation if the first does not fully conform to the consensus.
Respiratory syncytial (RS) virus is the major causative agent for hospitalization of children <1 year of age with respiratory infection worldwide. RS virus is a paramyxovirus and the genome is a negative sense RNA molecule of ∼15 kb in length. Replication and transcription of the virus genome occur in the cytoplasm of the infected cell using virus-encoded proteins. The minimal RNA polymerase requirements of RS virus are the ribonucleoprotein (RNA complexed with nucleoprotein, N), the phosphoprotein, P, and the large protein, L (Grosfeld et al., 1995; Yu et al., 1995). However, transcription is greatly enhanced by the presence of a further viral protein, M2 (also known as the 22K protein; Collins et al., 1995, 1996; Hardy and Wertz, 1998). The M2 gene is unique to the genus Pneumovirus, and no similarity to any known transcription factor has been described, but the functions of the N, P and L proteins are believed to be equivalent to those of the other Paramyxoviridae, and of the Rhabdoviridae. The L protein contains the sequence motifs for RNA-dependent RNA polymerase activity, and is presumed to be responsible for capping and polyadenylation of mRNAs (Stec et al., 1991).
The M2 mRNAs of all pneumoviruses contain two ORFs, conserved in location though not in sequence (Collins and Wertz, 1985; Elango et al., 1985; Baybutt and Pringle, 1987; Collins et al., 1990; Ling et al., 1992; Zamora and Samal, 1992; Alansari and Potgeiter, 1994; Ahmadian et al., 1999). The first major ORF (ORF-1) of the M2 gene of these viruses utilizes 60–75% of the entire coding capacity of the M2 mRNA, and the protein product from this ORF, which is readily detected in virus-infected cells, is the transcriptional activator. The second major ORF (ORF-2) is located towards the 3′ end of the mRNA, overlaps ORF-1 and utilizes the remainder of the unassigned available nucleotide sequence. For human RS virus strain A2, the M2 gene is 956 nucleotides in length with the first major ORF starting at nucleotide 10 and terminating at nucleotide 592, and encodes a polypeptide of 194 amino acids with a predicted mol. wt of 22.1 kDa. The M2 ORF-1 polypeptide was reported to have an electrophoretic mobility corresponding to a mol. wt of ∼24 kDa (Collins and Wertz, 1985). The protein products from the second ORF of the RS virus and PVM M2 genes have recently been detected in virus-infected cells, and the RS virus M2-2 protein has been proposed to be involved in control of the switch between virus RNA replication and transcription (Ahmadian et al., 1999; Bermingham and Collins, 1999). This hypothesis is reinforced by data showing reduced amounts of genomic and anti-genomic viral RNA but equal amounts of viral mRNA during infection with a recombinant RS virus lacking the M2-2 ORF when compared with wild-type RS virus (Jin et al., 2000). Translation of the second ORF (M2-2) of RS virus is thought to initiate at one of the three AUGs located at nucleotides 563, 569 and 581, terminating at residue 832. The largest possible polypeptide, with 90 residues, has a predicted mol. wt of ∼9.5 kDa. A third protein of 19 kDa is also seen in RS virus-infected cells and has been suggested to arise by internal initiation within ORF-1. Collins and Wertz (1985) suggested that ribosomes that terminate translation at nucleotides 592–594 at the end of the first ORF of the RS virus M2 gene may reinitiate at the downstream AUG at nucleotides 650–652 and direct synthesis of two-thirds of the second ORF to generate a polypeptide of 61 amino acids. Western blot analysis of protein from virus-infected cells detected an ORF-2 polypeptide with a mol. wt of ∼9.3 kDa (Ahmadian et al., 1999), which is consistent with a protein generated from the full-length of the second ORF and also with the observed mobility of the in vitro translated product of the full-length RS virus M2 gene ORF-2.
The mechanism by which the RS virus M2 gene second ORF is accessed by ribosomes is unknown, but several possibilities exist. First, it is possible that the second ORF is translated from initiation codons located downstream of the ORF-1 termination signal as originally suggested (Collins and Wertz, 1985). This does not appear to be the case since the protein products from such an event would be considerably smaller than those seen in virus-infected cells (Ahmadian et al., 1999). In addition, the in vitro translation product from the entire M2 ORF-2 migrates with the same mobility as the proteins detected in vivo. Secondly, ribosomes that fail to initiate translation at the first available AUG may scan along the mRNA until they reach the initiation codon for ORF-2. This seems unlikely since there are a further 10 AUG codons located upstream of the first possible initiation codon for ORF-2 and each would have to be ignored by the ribosomes. However, this mechanism has been implicated in the generation of the additional 19 kDa protein presumed to be from ORF-1. The M2 mRNA may contain RNA secondary structures that direct the ribosomes to the second ORF initiation codons, much like the IRES sequences found for picornaviruses and that are thought to be responsible for expression of the crucifer-infecting tobomovirus (crTMV) coat protein (CP) in vitro (Ivanov et al., 1997). Such a large structure would impose considerable evolutionary constraints should they arise in a coding region such as would be the case for the M2 mRNA. To date, such IRES structures have not been observed in any of the mononegavirales. Finally, a less likely possibility is the recently described phenomenon of ribosomal shunting or jumping for adenovirus late mRNAs, and 35S RNA of cauliflower mosaic virus (CaMV) and rice tungro bacilliform virus (RTBV; Fütterer et al., 1993; Yueh and Schneider, 1996; Hemmings-Mieszczak and Hohn, 1999). In this mechanism, a cis-acting element at the 5′ terminus of the mRNA, known as a donor site, directs the ribosomes via a shunt to a predefined acceptor site located further downstream in the RNA. This mechanism would allow the ribosomes to bypass any internal initiation codons and any strong mRNA secondary structures. Latorre et al. (1998) recently attributed the expression of the Y proteins of Sendai virus, a paramyxovirus, to the presence of a shunting mechanism within the Sendai virus P/C mRNA. However, they were unable to demonstrate the presence of donor sites within the 5′ region of the P/C mRNA.
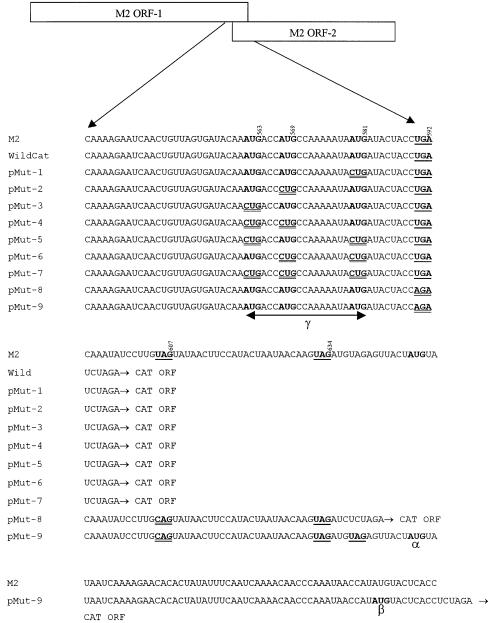
Alignment of the sequence of the overlapping regions of both ORFs 1 and 2 of the M2 genes of various RS virus strains, two avian pneumovirus (APV) strains and PVM fail to show any conservation of the translation initiation and termination codons in this region (Figure 1). The only common feature found is that the potential initiation codon(s) for ORF-2 are located upstream of the termination codon of ORF-1. We have investigated the initiation of translation of the RS virus M2 ORF-2 and have shown that translation of both the first and second ORFs is coupled in a way previously unknown in a eukaryotic system.
Fig. 1. Alignment of the sequence of the overlapping region of ORF-1 and ORF-2 of the M2 genes of pneumoviruses. Stop codons for ORF-1 are italicized and underlined. Potential initiation codons for ORF-2 are shown in bold. DDBJ/EMBL/GenBank accession numbers for the sequences shown are: human respiratory syncytial virus (HRSV) strains A2 (M74568), S2 (U39662) and B1 (AF013254); bovine respiratory syncytial virus (M82816); ovine respiratory syncytial virus (U02510); avian pneumovirus (APV, X63408); and pneumonia virus of mice (PVM; Ahmadian et al., 1999).
Results
To gain insight into the mechanism by which the second ORF of the RS virus M2 gene is accessed by ribosomes, and also to find the most likely translation initiation site(s) of this ORF, mutational analysis of the three possible initiation sites of ORF-2 and the stop codons of ORF-1 of the RS virus M2 gene was undertaken. The majority of the second ORF of the RS strain A2 M2 gene was replaced with a reporter gene encoding the chloramphenicol acetyltransferase (CAT) gene to allow quantitative analysis of expression of the second ORF. The authentic CAT gene ORF initiation codon was deleted and the gene positioned such that the coding region of the CAT gene was located downstream of the termination codon of the M2 first ORF but still in-frame with the three potential ORF-2 initiation codons. The putative ORF-2 initiation codons and ORF-1 termination codons were removed in a systematic manner to determine which were utilized to initiate translation of ORF-2. These constructs were cloned under the control of a T7 promoter and expression directed by T7 RNA polymerase in vitro or provided by a recombinant vaccinia vTF7-3 for in vivo analysis (Fuerst et al., 1986).
Quantitative analysis of CAT expression was determined using the CAT enzyme-linked immunosorbent assay (ELISA) system (Boehringer Mannheim, UK). Figure 2 shows the nucleotide sequence surrounding the overlapping region of the ORFs 1 and 2 of the A2 strain of the RS virus M2 gene in which mutations in the start codons of ORF-2 and stop codons of ORF-1 were introduced. In this study, these AUGs were designated AUG1 (at position 561–563), AUG2 (at position 567–569) and AUG3 (at position 579–581). Three stop codons, at positions 590–592, 605–607 and 632–634, designated Stop 1, 2 and 3, respectively, are located at the end of the RS virus M2 ORF-1 and follow the three closely spaced AUG codons at the start of ORF-2. Plasmid pWildCat retained all three ORF-2 initiation codons whereas plasmids pMut-1, pMut-2 and pMut-3 each contained a single mutation in one of the three AUGs at the start of the second ORF of the M2 gene (Figure 2). Recombinant plasmids pMut-4, pMut-5 and pMut-6 contained two mutations to knock out two of the three AUGs at the start of ORF-2. In these plasmids, AUGs 1 and 2 in plasmid pMut-4, AUGs 1 and 3 in plasmid pMut-5, and AUGs 2 and 3 in plasmid pMut-6 were mutated by changing the AUG to CUG. Recombinant plasmid pMut-7 contains mutations in all three AUGs (1, 2 and 3) of the ORF-2 by changing them to CUG.
Fig. 2. Sequences of mutations made in the overlapping region of ORFs 1 and 2 of the RS virus M2 mRNA. The positions of start codons and mutations are shown in bold, whereas those of the stop codons are shown in bold and single underlined. The nucleotide positions of each of these in the RS virus M2 mRNA are indicated. Mutated codons are shown in bold and double underlined. M2 indicates the original RS virus strain A2 M2 mRNA sequence and Wild indicates the sequence in pWildCat. AUG-α, AUG-β and AUGs in region γ (encompassing AUGs located at positions 561–563, 567–569 and 579–581) represent potential initiation codons for the ORF-2 polypeptide.
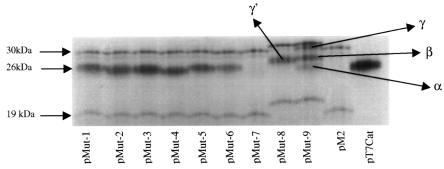
To assess the pattern of protein expression of these constructs in vitro, the plasmid constructs were transcribed and translated in vitro using a coupled rabbit reticulocyte lysate transcription–translation system and the resultant protein products subjected to SDS–PAGE analysis (Figure 3). For comparison, expression of the RS virus M2 and the native CAT protein was also determined using plasmids pM2 and pT7CAT, respectively. Because the CAT gene was cloned after the stop codon of M2 ORF-1 in the recombinant M2–CAT constructs, expression of the CAT gene from the first AUG of ORF-2 resulted in the addition of 10 amino acids, adding ∼1 kDa to the 25 kDa CAT protein. In our hands, the RS virus M2 ORF-1 directs the synthesis of two closely migrating polypeptides of ∼30–31 kDa rather than the predicted 22.1 kDa molecular weight. Following in vitro translation, the smaller of the two polypeptides is considerably fainter than the larger polypeptide (Figure 3). The two polypeptides are more clearly visible in the western blot analysis seen in Figure 5A. An additional polypeptide of ∼19 kDa is observed routinely, as has been described by others (Routledge et al., 1987; Collins et al., 1996).
Fig. 3. Autoradiographs showing in vitro translation protein products of recombinant plasmid constructs. Transcription and translation were directed in a cell-free coupled reaction system using T7 RNA polymerase and rabbit reticulocyte lysate. The polypeptides were analysed by SDS–PAGE on a 17% polyacrylamide gel. The positions of the molecular weight markers are indicated on the left.
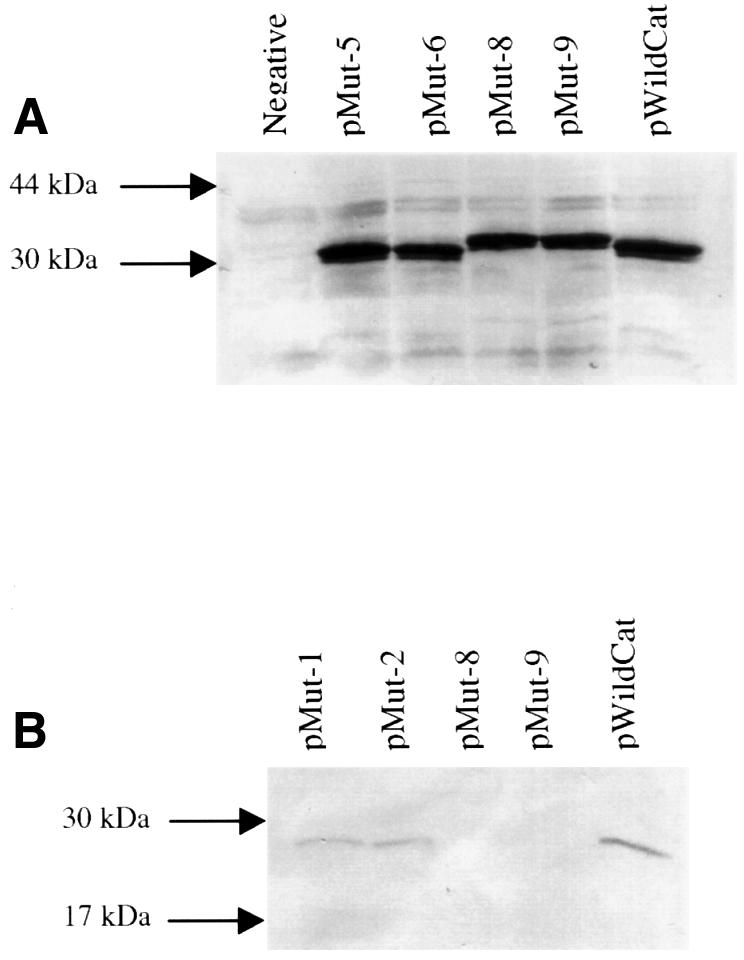
Fig. 5. Western blot analysis of polypeptides expressed in vivo. (A) Detection of M2 ORF-1 polypeptide products from the recombinant plasmid constructs pRSmut-5, pRSmut-6, pRSmut-8, pRSmut-9 and pWildCat. The lane labelled Negative is the analysis of cell lysates following transfection with plasmid pBluescribe. (B) Detection of expressed CAT protein (from ORF-2) of recombinant constructs pRSmut-1, pRSmut-2, pRSmut-8, pRSmut-9 and pWildCat.
Translation of the pWildCat mRNA in vitro directed the synthesis of the 30–31 kDa proteins from ORF-1 together with a 26 kDa protein corresponding to the CAT protein, with a slightly retarded mobility compared with that of the native CAT protein, as expected (data not shown). Translation of mRNA from plasmids pMUT-1 to pMUT-6 generated the same 30–31 and 26 kDa polypeptides corresponding to expression from ORF-1 and the recombinant CAT ORF, respectively (Figure 3). mRNA from plasmid pMut-7, in which all of the AUGs located at the start of the ORF-2 were mutated, directed synthesis of the ORF-1 polypeptides to the same levels as constructs pMut-1 to pMut-6; however, expression of the recombinant CAT protein was totally abrogated. These results indicate that all of the three AUG codons of ORF-2 are, or can be, used in expression of the M2 gene second ORF in vitro.
Plasmids pMut-8 and pMut-9 have the Stops 1 and 2 mutated. In plasmid pMut-8, the CAT ORF is located immediately after Stop 3, whereas in pMut-9 the CAT ORF is located ∼80 nucleotides downstream from Stop 3. In plasmid pMut-9, initiation codons AUG labelled α and β in Figure 2 are in-frame with the CAT ORF and these are located 13 and 67 nucleotides downstream of Stop 3, respectively. In both of these constructs, the ORF-1 polypeptides migrated with slightly retarded mobilities, indicating an increase in the molecular weight of these polypeptides when compared with those of pMut-1 to pMut-6. Additionally, the 19 kDa M2-related polypeptide expressed in constructs pWildCat and pMut-1 to pMut-6 showed an increase in its molecular weight when expressed from plasmids pMut-8 and pMut-9. In these latter constructs, the C-terminus of the M2 ORF-1 protein was extended by moving the translational stop codon further downstream toward the 3′ end of the mRNA, and the concomitant increase in size of the 19 kDa polypeptide confirms that it is generated by internal initiation within ORF-1, as previously proposed (Collins et al., 1990). Polypeptides α, β and γ from pMut-9 (Figure 3) may correspond to expression of ORF-2 initiating at AUG codons α, β and the region labelled γ, containing the three ORF-2 initiation codons, respectively (Figure 2). Similarly, polypeptide γ′ produced from pMut-8 (Figure 3) corresponds to expression of ORF-2 from the AUGs located in region γ. Further analysis is required to assign initiation codons unambiguously to these specific polypeptides that are expressed in vitro. The results suggest that translation of M2 ORF-2 in vitro may be by a ribosomal scanning mechanism.
The experiments described above were performed in vitro, which leaves open the possibility that the results were not representative of the in vivo situation. For this reason, the mechanism of the expression of the CAT gene as a model of expression of RS virus M2 in vivo was investigated. The vaccinia vTF7-3 expression system was selected to drive expression of these constructs from the T7 promoter. Expression of the CAT gene was determined using a CAT ELISA, which allowed accurate estimation of the yield of protein. Cells transfected with plasmid pT7CAT were used as a positive control and those transfected with plasmid pBluescribe without any insert as a negative control.
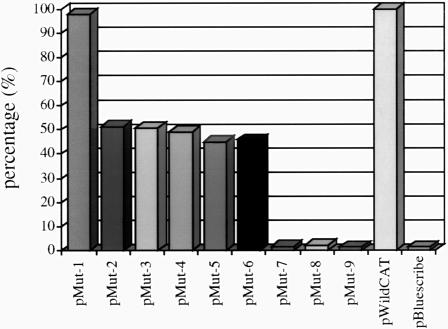
Expression from plasmid pWildCat (Figure 4) produced readily detectable amounts of CAT protein, and the levels of CAT protein from this construct were taken as 100%. Expression levels of CAT from plasmid pT7CAT were several fold higher than that of plasmid pWildCat (data not shown). Plasmids pMut-2 to pMut-6 (Figure 4) produced CAT protein to levels ∼45–52% of those seen with pWildCat, which could be attributed to the mutations in one or more of the AUGs of ORF-2. Interestingly, expression of CAT levels from pMut-1 were similar to those seen for pWildCat in vivo although no similar preference was observed in vitro. In this construct, AUGs 1 and 2 were left intact, with only AUG 3 being mutated. This may indicate that AUGs 1 and 2 are preferred to AUG 3 for initiation of translation of ORF-2 in vivo. In other RS virus strains (e.g. S2, ovine and bovine), only two AUG codons are located upstream of the ORF-1 termination codon (Figure 1). Plasmid pMut-7, in which AUGs 1, 2 and 3 at the start of ORF-2 were removed (Figure 4), as expected from the in vitro analysis, produced no detectable CAT protein. This confirmed that translation of ORF-2 must be initiated from one or more of these AUG codons. Plasmids pMut-8 and pMut-9 (Figure 4), which retain all three ORF-2 potential initiation codons but are mutated in the Stop 1 and 2 of ORF-1, did not produce any detectable CAT protein in vivo. This contrasts with the result obtained from the in vitro translations and may be a reflection of the artificial nature of the in vitro system. We have noted that different reticulocyte lysate preparations, in uncoupled transcription–translation systems, display considerable batch-dependent variation in whether a product is produced from mutants 7, 8 and 9 (not shown). The reasons for this variability are not known, but may reflect variations in the concentration of a critical factor.
Fig. 4. Diagram showing the level of expression of the CAT protein detected from recombinant plasmid constructs which had the CAT ORF fused in-frame with the second ORF of the RS virus strain A2 M2 gene. Levels of expression of CAT protein are shown as a percentage of the expression observed from plasmid pWildCat after normalization. The data are averages of the results of five experiments.
Since constructs pMut-8 and pMut-9 failed to express any CAT protein in vivo, it was plausible that the addition of a large number of extra amino acids to the N-terminus of the CAT protein resulted in a conformational change that prevented any expressed recombinant CAT protein being detected using the CAT ELISA assay. Also, it was necessary to determine that ORF-1 from these constructs was expressed in vivo. Polypeptides from cell lysates were separated by SDS–PAGE and the ORF-1 products detected by western blotting (Figure 5A). ORF-1 products were detected from constructs pMut-5, pMut-6, pMut-8, pMut-9 and pWildCat. Also noted was the expected slight increase in the molecular weight of the ORF-1 products of pMut-8 and pMut-9 resulting from the increase in length of ORF-1. Furthermore, the ORF-1 products of all of the mutants were expressed at the same levels as those of pWildCat. Western blot analysis was performed on pMut-1, pMut-2, pMut-8, pMut-9 and pWildCat cell lysates to detect CAT protein. This detected low levels of CAT protein for pMut-1, pMut-2 and pWildCat (Figure 5B). However, no CAT protein was detected for pMut-8 and pMut-9. The latter results demonstrate that recombinant CAT protein, in its denatured form, was readily detectable but was not expressed in vivo.
Discussion
In this study, we demonstrate not only that expression of the RS virus M2 ORF-2 protein is initiated at AUGs located upstream of the ORF-1 termination codon, but also that expression from these initiation codons requires the termination of M2 ORF-1 translation in vivo. A series of constructs was prepared containing mutations in one or more of the potential start codons of the RS virus strain A2 M2 gene ORF-2 and the stop codons of ORF-1. These were used to find the mode of expression of the RS virus M2 ORF-2 protein. In these constructs, a CAT gene, in which the authentic AUG codon was deleted, was cloned in-frame with the coding sequence of ORF-2. Translation of the CAT gene could only be initiated from one or more of the 5′-proximal AUG codons at the beginning of ORF-2.
Translation of mRNAs in vitro confirmed that the initiation of translation of the M2 ORF-2 could, in principle, occur at any of the three potential initiation sites. Deletion of any one or two of these sites did not significantly affect the synthesis of the 26 kDa chimeric CAT protein, whereas deletion of all three AUG codons in plasmid pMut-7 prevented production of the CAT-related protein. Mutation of the ORF-1 stop codon did not alter translation of ORF-2 in vitro (Figure 3). In the mutants pMut-8 and pMut-9, in which the first two ORF-1 stop codons were mutated thereby moving termination of translation towards the 3′ end of the mRNA, the ORF-1 protein was increased in size by the expected amount. Translation from ORF-2 in vitro was generally unaffected by mutation of the ORF-1 stop codon, but it was noted that with different translation lysate preparations, mRNAs from plasmids pMut-8 and pMut-9 failed to synthesize an ORF-2 protein.
In addition to the effects on translation described above, a protein of ∼19 kDa, which is observed consistently when RS virus M2 mRNA is translated in vitro, was also increased in size. This observation, taken together with that of pMut-7 (Figure 3), where expression of the chimeric CAT protein is abrogated, but not expression of the 29 and 19 kDa polypeptides, indicates that the 19 kDa polypeptide is generated from ORF-1 and most probably arises by initiation at an internal AUG codon. The absence of suitable antibodies previously has prevented unambiguous assignment of this protein to ORF-1. The role of this polypeptide in vivo is not known, but the complete ORF-1 polypeptide has been shown to enhance RNA synthesis in RS virus. The presence of an N-terminally deleted form of this transcriptional enhancer may be important, and further investigation is required.
The in vitro translation analysis suggested that there was no preference for a particular ORF-2 AUG initiation codon as the CAT-related polypeptides were of similar intensities on autoradiographs. Since it is known that the optimized in vitro translation systems do not always reflect accurately the situation in vivo, we decided to confirm the data using the recombinant vaccinia virus T7 RNA polymerase system to direct transcription of the plasmids in transfected tissue culture cells.
This analysis showed that all of the recombinant constructs of the RS virus M2 gene, except for plasmids pMut-7, pMut-8 and pMut-9, expressed a polypeptide of ∼26 kDa, representing the chimeric CAT product, translation of which was initiated from one of the three AUGs of ORF-2 that are located upstream of the termination codon for ORF-1. The data from plasmids pMut-1 to pMut-7 confirmed that the presence of one of the AUG codons at the beginning of ORF-2 was essential for translation of M2 ORF-2 both in vitro and in vivo. In contrast to the data generated in vitro, the deletion of ORF-2 AUG 3 in pMut-1 had little effect on translation and this may indicate a preference for AUG codons 1 and 2 in vivo.
Unlike the situation frequently observed in vitro, plasmids pMut-8 and pMut-9 did not direct the synthesis of a detectable CAT-related polypeptide in vivo. In these two plasmids, the first and second stop codons of ORF-1 were removed and, in the case of pMut-8, an AUG codon adjacent to the third stop was also removed. This increased the size of ORF-1, which now terminated at stop codons further downstream in the mRNA. In plasmid pMut-9, an AUG codon in-frame with ORF-2 and, consequently, with the CAT gene, is located 13 nucleotides downstream of the Stop 3. This codon was not used to express the CAT gene in vivo; however, it may have been used in the in vitro translation system since a 25 kDa polypeptide that corresponded to the predicted molecular weight of CAT protein was detected (polypeptide α; Figures 2 and 3).
The data generated in transfected cells indicate that the initiation of translation of the RS virus M2 gene ORF-2 is coupled to the termination of translation of ORF-1 such that the initiation of translation depends on prior termination of translation of ORF-1. Thus, by moving the stop codon of ORF-1 further down the mRNA, without affecting the environment of the upstream AUG codons at the start of ORF-2, translation from ORF-2 was completely ablated. In the case of the pneumovirus M2 ORF-2, the proteins are not abundant and presumably reinitiation is not common. Increasing the distance between the stop codon of ORF-1 and the initiation site(s) of ORF-2 prevented ribosomes from reinitiating at the upstream AUG(s). This indicates that the normal distance of up to 29 nucleotides between the stop codon of ORF-1 and the 5′-proximal start codon of ORF-2 is sufficiently short to allow ribosomes to reinitiate before they detach completely from the mRNA. Reinitiation of translation at AUG codons downstream of a 5′-proximal ORF is uncommon in eukaryotic mRNA but it can occur in entirely artificially made mRNAs (Kozak, 1989) and has been shown to occur for translation of the hepatitis B virus P protein (Hwang and Su, 1998). Reinitiation of translation has also been reported for the expression of influenza B virus BM2 protein from segment 7 (M1) bicistronic mRNA where the initiation and termination codons overlap, coupling both processes. (Horvath et al., 1990). A translation reinitiation process has been described for the bacteriophage MS2 and fr in which ribosomes were envisaged to move bi-directionally along the mRNA (Adhin and Duin, 1990). In the case of the hepatitis B virus P protein, it has been suggested that a small proportion of translation may arise by ribosomes that terminate at a downstream stop codon, but this has not been proven directly. To date, no example of the exclusive initiation of translation of an ORF in a eukaryotic mRNA similar to that described here for the RSV M2 gene has been identified.
The mechanism by which the reverse translocation of the ribosome occurs is not known, and a number of possibilities exist. A specific sequence may be present in the region of the M2 mRNA overlap between ORF-1 and ORF-2, which positively directs a proportion of ribosomes to reinitiate translation at the upstream start codon. However, the absence of this phenomenon in many, but not all, reticulocyte lysate extracts may indicate that the process requires the presence, or less probably the absence, of a cellular factor(s), whose concentration is affected by preparation. It is also possible that a specific feature of the M2 mRNA secondary structure allows translocation of ribosomes in a 3′→5′ direction on the same mRNA strand. The conformation may bring the stop and start codons of ORF-1 and ORF-2, respectively, even closer, such that after an 80S ribosome has translated the upstream ORF and the 60S subunit has dissociated, the 40S ribosomal subunit holds on to the mRNA and is able to reinitiate at an upstream AUG. The frequent absence of the process in vitro would then be due to a failure of the mRNA to adopt this conformation. This appears unlikely unless the conformation is dependent on components of the lysate that may be limiting. Alternatively, in the overoptimized and enriched ribosomal fraction of the lysate, the high concentration of ribosomes may result in a polysome structure that prevents translocation of the ribosome terminating at the end of ORF-1 through steric hindrance. Finally, the process may be a common one in which, after termination of ORF-1, a proportion of the ribosomes retained on mRNA adopt a scanning mechanism and are able to scan the mRNA in both the 5′→3′ and 3′→5′ direction. The relatively rare occurrence of overlapping ORFs would mean that the reinitiation process described here is not common. With artificial mRNAs, the extent of ribosomal backwards scanning was suggested to be dependent on the distance between the first ORF termination codon and the upstream initiation codon and this would also reduce the frequency of such translation initiation events (Peabody and Berg, 1986; Peabody et al., 1986). Any of these mechanisms would be unlikely to occur frequently and this would result in low levels of expression of the second ORF protein. This process may represent a novel mechanism for controlling the expression of certain gene products.
Materials and methods
Construction of recombinant plasmids
A CAT gene ORF lacking the AUG initiation codon was generated by PCR using Vent DNA polymerase (New England Biolabs) and oligonucleotide primers 5′-CAT (GGGGTCTAGAGAAAAAAATCACTGGA) and 3′-CAT (GGGGTCTAGATTACGCCCCGCCCTGCCACTC). The 5′-CAT primer for this PCR was designed such that the first AUG codon of the CAT gene was deleted. Both primers were designed to have an XbaI recognition site at their 5′ end (underlined). Plasmid pT7CAT containing a copy of the CAT gene was use as a template to amplify the CAT gene but missing its authentic initiation codon.
Recombinant plasmids pMut-1 to pMut-7 were designed to have mutations in one or more of the AUG codons of ORF-2, whereas plasmid pWildCat was designed to have all of the putative RS virus strain A2 gene initiation codons for ORF-2 left intact. The overall strategy to make all of these recombinant plasmids was similar. To make these plasmid constructs, inverse PCR using Vent DNA polymerase and primers that contained one or more mutations at the desired points were employed. The plasmid pM2, in which the full-length copy of the RS virus M2 gene was cloned, was used as a template to delete the region of the second ORF located downstream of the first stop codon of the ORF-1 (overlapping region). This was replaced with the complete ORF of the CAT gene that lacked its own initiation codon. Mutagenesis of the specific AUG codons was achieved using inverse PCR with synthetic oligonucleotides that contained one or more mutations at the required points. The PCR primers were designed in such a way that the resulting constructs retained the first stop codon of the major ORF (ORF-1) at position 592 while all three possible initiation codons of ORF-2 could be mutated. All of the primers contained an XbaI site at their 5′ end so that the ORF of the CAT gene was in-frame with the second ORF. In all of these constructs, the primer M2END (GGTCTAGACAATTCAAGTTGTGGGACAAAATGGATCCC) was common, and primers M2WILD (GGTCTAGATCAGGTAGTATCATTATTTTTGG), M2MUT1 (GGTCTAGATCAGGTAGTATCAGTATTTTTGGCATGGTGGTCAT), M2MUT2 (GGTCTAGATCAGGTAGTATCATTATTTTTGGCAGGGTGGTCATTTGTATCACTAA), M2MUT3 (GGTCTAGATCAGGTAGTATCATTATTTTTGGCATGGTGGTCAGTTGTATCACTAA), M2MUT4 (GGTCTAGATCAGGTAGTATCATTATTTTTGGCAGGGTGGTCAGTTGTATCACTAA), M2MUT5 (GGTCTAGATCAGGTAGTATCAGTATTTTTGGCATGGTGGTCAGTTGTATCACTAA), M2MUT6 (GGTCTAGATCAGGTAGTATCAGTATTTTTGGCAGGGTGGTCATTTGTATCACTAA) and M2MUT7 (GGTCTAGATCAGGTAGTATCAGTATTTTTGGCAGGGTGGTCAGTTGTATCACTAA) were used to make plasmid constructs pWildCat and pMut-1 to pMut-7, respectively.
The three stop codons at the end of the M2 gene ORF-1 are designated Stop 1 (UGA at position 592), Stop 2 (UAG at position 607) and Stop 3 (UAG at position 634). Recombinant plasmids pSTOPM2, pMut-8 and pMut-9 were designed to have mutations in the stop codons of ORF-1. In all of these plasmids, Stops 1 and 2 were mutated. Plasmid pSTOPM2 was generated to be used as a template in the PCRs to make plasmids pMut-8 and pMut-9, which contained the AUG-minus CAT gene in-frame with the ORF-2. Plasmid pSTOPM2 was constructed such that although it contained the whole of the RS virus M2 gene (both ORFs 1 and 2), Stops 1 and 2 were mutated. Primer Stopmut (ATACTGCAAGGATATTTGTCTGGTAGTATCATT) contained two altered nucleotides (shown in bold) that mutated Stops 1 and 2 by changing them to AGA and CAG, respectively, but kept Stop 3 intact. PCR amplification of this primer in conjunction with primer Stopend (AACTTCCATACTAATAACAAGTAGATG) was performed using plasmid pM2. This plasmid was used as a template to make plasmids pMut-8 and pMut-9.
Inverse PCR using primers M2MUT8 (GGCTCTAGAGATCTACTTGTTATTAGTATGGAAG) and M2END and plasmid pSTOPM2 as template was used to delete the region of ORF-2 after the Stop 3 of ORF-1 and replace it with the AUG-minus CAT gene. Using primer M2MUT8 in this PCR, a BglII recognition site was introduced before the XbaI site to facilitate screening of the desired plasmid by restriction enzyme digestion. The resulting recombinant plasmid was designated pMut-8. In this plasmid, an AUG that was located immediately after Stop 3 was removed by mutation to ensure that it did not interfere with the interpretation of the results.
PCR using primers Stopwild (GGGGTCTAGAGGTGAGTACATATGGTT) and M2END with plasmid pSTOPM2 as a template was used to delete the region of the M2 gene after nucleotide 714 and replace it with the CAT gene. In this plasmid, Stops 1 and 2 were removed and Stop 3 was kept. This plasmid contains an AUG codon 13 nucleotides downstream of the Stop 3 that is in-frame with the ORF-2 coding sequence. The CAT gene with the deleted AUG, described previously, was cloned in such a way that it was in-frame with the coding sequence of ORF-2.
In vitro transcription and translation
In vitro expression of the recombinant constructs was analysed using the Coupled TNT expression system (Promega Corporation, Southampton, UK). A 1 µg aliquot of supercoiled plasmid DNA was incubated in the presence of T7 RNA polymerase, [35S]methionine and rabbit reticulocyte lysate according to the manufacturer’s recommendations.
Transfection and measurement of CAT reporter protein
For the transfection procedure, Hep-2 cells were grown overnight, in wells of 2 cm diameter, in Glasgow modified Eagle’s medium (GMEM) supplemented with 5% fetal calf serum to ∼75% confluency, and infected with vaccinia virus vTF7-3 (Fuerst et al., 1986) at a multiplicity of infection of 3–5. At 1 h post-infection, cells were transfected with a mixture of 2 µg of DNA and 4 µl of Lipofectace™ (Gibco-BRL Life Technologies Ltd, UK), as previously described (Randhawa et al., 1997). Of the 2 µg of total DNA used in the transfections, 1.8 µg were that of the recombinant plasmids with the remaining 0.2 µg being made up of plasmid pT7-β-gal, a construct that contains the β-gal reporter gene under the control of the T7 promoter. This plasmid was used to standardize the results of the CAT protein detected from each recombinant plasmid. At 48 h post-transfection, the cells were washed with 1× phosphate-buffered saline and lysed in 150 µl of CAT lysis buffer (1% Triton X-100, 10 mM MOPS pH 6.5, 10 mM NaCl and 1 mM EGTA). Cellular debris was removed by centrifugation and CAT reporter protein quantified using a CAT ELISA kit (Boehringer Mannheim, UK), by comparison with purified CAT protein standards.
Western blot analysis
Aliquots of the same cell lysates obtained above were subjected to SDS–PAGE separation and the proteins transferred to nitrocellulose using standard blotting techniques. The nitrocellulose filters were then blocked in a solution of a 10% (w/v) mixture of dried milk (Marvel). For the detection of the ORF-1 polypeptides of RS virus strain A2 M2 gene, nitrocellulose filters were incubated with a 1:500 dilution of antibody 5H5 (raised against RS virus M2 protein) in PBS–0.1% Tween for 1 h followed by three washes with PBS–Tween. Following a further incubation period of 1 h in 1:400 dilution of a goat anti-mouse IgG (H+L)–horseradish peroxidase (HRP, Bio-Rad) conjugate, the filters were developed in Sigma Fast™ 3,3′-diaminobenzidine peroxidase substrate (DAB; Sigma) according to the manufacturer’s recommendations. For the detection of CAT protein, nitrocellulose filters were incubated for 1 h sequentially with digoxigenin (DIG)-labelled anti-CAT antibody (Boehringer Mannheim, UK) and antibody anti-DIG–HRP conjugate (Boehringer Mannheim, UK). Filters were developed in fast-acting DAB (Sigma).
Acknowledgments
Acknowledgements
We would like to thank Dr G.Toms, University of Newcastle for providing the M2-specific monoclonal antibody 5H5. This work was supported by the Medical Research Council and the Wellcome Trust.
References
- Adhin M.R. and van Duin,J. (1990) Scanning model for translational reinitiation in Eubacteria. J. Mol. Biol., 213, 811–818. [DOI] [PubMed] [Google Scholar]
- Ahmadian G., Chambers,P. and Easton,A.J. (1999) Detection and characterization of proteins encoded by the second ORF of the M2 gene of pneumoviruses. J. Gen. Virol., 80, 2011–2016. [DOI] [PubMed] [Google Scholar]
- Alansari H. and Potgieter,L.N.D. (1994) Molecular cloning and sequence analysis of the phosphoprotein, nucleocapsid protein, matrix protein and 22K (M2) protein of the ovine respiratory syncytial virus. J. Gen. Virol., 75, 3597–3601. [DOI] [PubMed] [Google Scholar]
- Barr J., Chambers,P., Harriot,P., Pringle,C.R. and Easton,A.J. (1994) Sequence of the phosphoprotein gene of pneumonia virus of mice: expression of multiple proteins from two overlapping reading frames. J. Virol., 68, 5330–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baybutt H.N. and Pringle,C.R. (1987) Molecular cloning and sequencing of the F and 22K membrane protein genes of the RSS-2 strain of respiratory syncytial virus. J. Gen. Virol., 68, 2789–2796. [DOI] [PubMed] [Google Scholar]
- Bermingham A. and Collins,P.L. (1999) The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl Acad. Sci. USA, 96, 11259–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L. and Wertz,G.W. (1985) The envelope-associated 22K protein of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polytranscript. J. Virol., 54, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Hill,M.G. and Johnson,P.R. (1990) The two open reading frames of the 22K mRNA of human respiratory syncytial virus: sequence comparison of antigenic subgroups A and B and expression in vitro. J. Gen. Virol., 71, 3015–3020. [DOI] [PubMed] [Google Scholar]
- Collins P.L., Hill,M.G., Camargo,E., Grosfeld,H., Chanock,R.M. and Murphy,B.R. (1995) Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl Acad. Sci. USA, 92, 11563–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Hill,M.G., Cristina,J. and Grosfeld,H. (1996) Transcription elongation factor of respiratory syncytial virus, a non-segmented negative strand RNA virus. Proc. Natl Acad. Sci. USA, 93, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. and Kolakofsky,D. (1987) Identification of an additional Sendai virus non-structural protein encoded by the P/C mRNA. J. Gen. Virol., 68, 2515–2519. [DOI] [PubMed] [Google Scholar]
- Curran J. and Kolakofsky,D. (1990) Sendai virus P gene produces multiple proteins from overlapping open reading frames. Enzyme, 44, 244–249. [DOI] [PubMed] [Google Scholar]
- Curran J., Richardson,C.D. and Kolakofsky,D. (1986) Ribosomal initiation at alternate AUGs on the Sendai virus P/C mRNA. J. Virol., 57, 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N., Satake,M. and Vankatesan,S. (1985) mRNA sequence of three respiratory syncytial virus genes encoding two nonstructural proteins and a 22K structural protein. J. Virol., 55, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Kiss-László,Z. and Hohn,T. (1993) Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell, 73, 789–802. [DOI] [PubMed] [Google Scholar]
- Giorgi C., Blumberg,B.M. and Kolakofsky,D. (1983) Sendai virus contains overlapping genes expressed from a single mRNA. Cell, 35, 829–836. [DOI] [PubMed] [Google Scholar]
- Grosfeld H., Hill,M.G. and Collins,P.L. (1995) RNA replication by respiratory syncytial virus (RSV) is directed by the N, P and L proteins: transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol., 69, 5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K.C., Ono,E. and Xu,X. (1996) Lack of correlation between Sendai virus P/C mRNA structure and its utilization of two AUG start sites from alternate reading frames: implications for viral bicistronic mRNAs. Biochemistry, 35, 1223–1231. [DOI] [PubMed] [Google Scholar]
- Hardy R.W. and Wertz,G.W. (1998) The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol., 72, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M. and Hohn,T. (1999) A stable hairpin preceded by a short open reading frame promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA, 5, 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M., Williams,M.A. and Lamb,R.A. (1990) Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J., 9, 2639–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.L. and Su,T.S. (1998) Translational regulation of hepatitis B virus polymerase gene by termination–reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol., 79, 2181–2189. [DOI] [PubMed] [Google Scholar]
- Ivanov P.A., Karpova,O.V., Skulachev,M.V., Tomashevskaya,O.L., Rodionova,N.P., Dorokhov,Y.L. and Atabekov,J.G. (1997) A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology, 232, 32–43. [DOI] [PubMed] [Google Scholar]
- Jin H., Cheng,X., Chou,H.Z.Y., Li,S. and Seddiqui,A. (2000) Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J. Virol., 74, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre P., Kolakofsky,D. and Curran,J. (1998) Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol., 18, 5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling R., Easton,A.J. and Pringle,C.R. (1992) Sequence analysis of the 22K, SH and G genes of turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J. Gen. Virol., 73, 1709–1715. [DOI] [PubMed] [Google Scholar]
- Peabody D.S. and Berg,P. (1986) Termination–reinitiation occurs in the translation of mammalian cell mRNAs. Mol. Cell. Biol., 6, 2695–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D.S., Subramani,S. and Berg,P. (1986) Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol., 6, 2703–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J. and Sonenberg,N. (1988) Internal initiation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- Randhawa J.S., Marriott,A.C., Pringle,C.R. and Easton,A.J. (1997) Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J. Virol., 71, 9849–9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge E.G., Willcocks,M.M., Morgan,L., Samson,A.C.R., Scott,R. and Toms,G.L. (1987) Heterogeneity of the RSV 22K protein revealed by western blotting with monoclonal antibodies. J. Gen. Virol., 68, 1209–1215. [DOI] [PubMed] [Google Scholar]
- Stec D.S., Hill,M.G.,III and Collins,P.L. (1991) Sequence analysis of the polymerase L gene of human respiratory syncytial virus and the predicted phylogeny of nonsegmented negative-strand viruses. Virology, 183, 273–287. [DOI] [PubMed] [Google Scholar]
- Yu Q., Hardy,R.W., Wertz,G.W. (1995) Functional cDNA clones of the human respiratory syncytial (RS) virus N, P and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol., 69, 2412–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A. and Schneider,R.J. (1996) Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev., 10, 1557–1567. [DOI] [PubMed] [Google Scholar]
- Zamora M. and Samal,S.K. (1992) Sequence analysis of M2 mRNA of bovine respiratory syncytial virus obtained from an F-M2 dicistronic mRNA suggest structural homology with that of human respiratory syncytial virus. J. Gen. Virol., 73, 737–741. [DOI] [PubMed] [Google Scholar]