Abstract
The MED1/TRAP220 subunit of the Mediator plays a key role in facilitating ligand-dependent interactions of this multisubunit coactivator complex with nuclear receptors through their ligand binding domains. The isolated MED1/TRAP220 protein previously was shown to interact with glucocorticoid receptor (GR) in a ligand-dependent manner. However, the functional role of MED1/TRAP220, within the context of the entire Mediator, is not well studied in GR-mediated transcription. In this study, we show that GR binds directly to the Mediator complex and that both LXXLL motifs of MED1/TRAP220 contribute to its binding to GR. Furthermore, using a Med1/Trap220−/− mouse embryonic fibroblast (MEF) line that lacks entirely MED1/TRAP220, we show that MED1/TRAP220 enhances GR-mediated transcription from an MMTV promoter based-reporter gene and that mutations in the MED1/TRAP220 LXXLL motifs reduce, but do not eliminate, GR-dependent transcription. An analysis of endogenous genes in Med1/Trap220−/− cells has confirmed a variable MED1/TRAP220 requirement for different GR target genes. Taken together, these findings support the idea that Mediator, at least in part through MED1/TRAP220, plays a coregulatory role in ligand-dependent GR-mediated gene expression.
INTRODUCTION
Nuclear receptors are a large family of transcription factors that generally regulate transcription in a ligand-dependent manner. Typical nuclear receptor domains include a less conserved N-terminal activation domain (AF1), a highly conserved central DNA-binding domain (DBD), a hinge region and a conserved C-terminal ligand binding domain (LBD) that contains activation domain 2 (AF2). Nuclear receptors form homo- or heterodimers and selectively bind to specific hormone responsive elements (HREs) on target gene promoters (1). Receptor dimers regulate gene expression through recruitment of various transcription coregulators. These include factors, such as the p160 family, that directly or indirectly facilitate chromatin remodeling, and other factors, such as Mediator, that act more directly on the general transcription machinery through ligand-independent (AF1 function) and ligand-dependent (AF2 function) mechanisms (2–4).
The Mediator is a well-known nuclear receptor coactivator complex that was first identified in mammalian cells through intracellular ligand-dependent interactions with thyroid hormone receptor (TR) and shown to mediate TR function on DNA templates containing T3 response elements (TREs) in a cell-free system reconstituted with purified general transcription factors (5). Subsequent studies in this and other laboratories identified similar or identical complexes and coactivation functions for a variety of transcription activators (6–11). The MED1/TRAP220 subunit of Mediator has been implicated in nuclear receptor functions by demonstrations of (i) ligand-dependent interactions of isolated MED1/TRAP220 (or derived fragments) with nuclear receptors (12,13), (ii) stimulatory effects of ectopically expressed MED1/TRAP220 on nuclear receptor functions (12,13), (iii) joint ligand- and MED1/TRAP220-dependent interactions of complete Mediator complexes with nuclear receptors (14–16), (iv) dramatically decreased ability of Mediator complexes lacking MED1/TRAP220 to coactivate nuclear receptors in vitro (16) and, most convincingly, (v) compromised nuclear receptor functions in Med1/Trap220−/− cells (15,17,18) or in MED1/TRAP220 siRNA-treated cells (19). Consistent with demonstrations of the role of LXXLL domains in mediating interactions of a variety of coactivators with nuclear receptors (2), the LXXLL domains in MED1/TRAP220 have been shown to be important for ligand-dependent interactions of isolated MED1/TRAP220 (or derived fragments) with nuclear receptors (13,20,21), for interactions of complete Mediator with nuclear receptors (14,16) and for optimal nuclear receptor coactivator functions in vitro (16).
Glucocorticoids, through the action of the glucocorticoid receptor (GR), play important roles in regulating many biological processes that include hematopoiesis, immune responses and metabolism. Following ligand binding, dissociation of chaperones, and transport to the nucleus, dimerized GR binds to glucocorticoid response element (GRE) sequences in target gene promoters and positively or negatively regulates gene expression through recruitment of various transcription coregulators and chromatin remodeling factors (22–24). Besides studies implicating p160 family members in GR function (25–27), a role for the Mediator has also been demonstrated (28,29). Thus, isolated MED1/TRAP220 and MED14/TRAP170 proteins were shown to interact with GR through AF1 and AF2, respectively, and their ectopic expression was shown to synergistically increase GR-dependent transcription in transient transfection assays. Although ectopic MED1/TRAP220 alone failed to increase GR-mediated transcription (28), this likely reflected the presence of endogenous MED1/TRAP220. Recent studies using MED14/TRAP170 and MED1/TRAP220 siRNAs showed that expression of some GR-target genes was specifically affected upon reduction of MED14/TRAP170 or MED1/TRAP220 (29). MED1/TRAP220 was shown to be recruited to the promoter of endogenous GR-target genes and to the transfected MMTV promoter in a ligand-dependent manner (29,30). These results strongly suggest that MED1/TRAP220 plays an important role in GR-mediated transcription.
The present study confirms a ligand-dependent interaction between GR-LBD and the LXXLL-containing central domain of MED1/TRAP220 and, importantly, shows for the first time that GR interacts directly with the complete Mediator complex. It further evaluates contributions of the two MED1/TRAP220 LXXLL domains both to GR-MED1/TRAP220 interactions and to the enhancement of GR-dependent transcription by MED1/TRAP220. In vitro binding assays show that both LXXLL domains contribute to the interaction between MED1/TRAP220 and GR, and functional assays with a Med1/Trap220 null mouse embryonic fibroblast (MEF) cell line show that MED1/TRAP220, in part through its LXXLL domains, enhances GR-mediated transcription. These findings support our proposal that the Mediator, through MED1/TRAP220, plays a stimulatory role in GR-mediated gene expression.
MATERIALS AND METHODS
Plasmid construction
For GST-fusion protein expression, cDNAs were amplified from corresponding sequences and subcloned into pGEX vector. A mammalian expression vector for GR was kindly provided by Dr Kai Ge. cDNAs for MED1/TRAP220 and mutants were subcloned and expressed using the pIRESneo vector (Clontech).
Cell culture
HeLa S cells were cultured in suspension in DMEM with 10% bovine calf serum. Wild-type and Med1/Trap220−/− MEF cells were cultured in monolayer in DMEM with 10% fetal bovine serum. For preparation of HeLa nuclear extract under dexamethasone (Dex) conditions, HeLa S cells were cultured in suspension in DMEM with 10% charcoal-stripped serum for 24 h before harvesting. For preparation of HeLa nuclear extract under +Dex conditions, HeLa S cells were cultured in suspension in DMEM with 10% fetal bovine serum and Dex was added to the culture at a final concentration of 10 µM 24 h before cells were harvested. To prepare charcoal-stripped serum, 500 ml of serum was incubated with 25 g of AG1-X resin (BioRad) at room temperature overnight with stirring. After the resin was allowed to settle, fresh resin (25 g) was added with 5 g of activated charcoal powder and stirred again at room temperature for 4–6 h. The charcoal and resin were removed by centrifugation and filtration through a 0.8 µm filter.
Protein purification
GST and GST-fusion proteins were expressed in bacteria and purified as described (14). Briefly, the bacterial lysate was incubated with Glutathione-agarose beads for 4 h at 4°C. The beads were washed five times with BC500-0.1% NP40. The amounts of GST and GST-fusion proteins on beads were normalized by SDS-PAGE. Mediator was purified as described (16).
In vitro protein binding assay
The assay was performed with GST-fusion proteins (5 µg) immobilized on glutathione-Sepharose beads (Amersham Pharmacia) and in vitro-translated 35S-labeled proteins (TNT kit, Promega). After incubating at 4°C for 4 h and washing with buffer A (20 mM Tris, pH 7.9, 10% glycerol, 0.2 mM EDTA, 300 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, 0.1% NP-40), bound proteins were eluted by boiling in SDS sample buffer, resolved by SDS-PAGE and visualized by autoradiography.
Transient transfection assay
Wild-type and Med1/Trap220−/− MEF cells were maintained in DMEM with 10% fetal bovine serum. The MMTV-luciferase reporter contains the MMTV promoter with GR response elements. Cells were transfected with the aid of Fugen 6 (Roche) on 24-well plates and total levels of transfected DNA were kept constant by adjusting levels of empty vectors. A β-gal plasmid was used as an internal control for normalizing transfection efficiency. Cells were cultured in DMEM with 10% charcoal-stripped fetal bovine serum after transfection. One day after transfection, Dex was added to +Dex wells to a final concentration of 100 nM and luciferase activity was measured 24 h after ligand addition (2 days after transfection). Luciferase activities are shown as relative luciferase units (RLU) after normalization by β-gal activity.
Statistical analysis
Figures show the mean and SD of double or triple samples in representative experiments. Statistical differences between treated samples were determined using unpaired Student's t-test. P-values are stated in the figure legends.
Real-time PCR
For preparation of cells for real-time PCR studies, wild-type and Med1/Trap220−/− MEFs were seeded in six-well plates at 105 cells/well. The cells were transfected with 0.5 µg GR expression vector with the aid of Fugen 6 in MEM containing charcoal-stripped serum. Two days after transfection, Dex or vehicle (100% ethanol) was added to the cells at a final concentration of 100 nM. Total RNA was collected 10 h after Dex or vehicle treatment using RNeasy Mini kit (Qiagen). Equal amounts of total RNA were used for first strand cDNA synthesis using SuperScriptIII First Strand cDNA Synthesis for RT-PCR kit (Invitrogen) following the manufacturer's instructions. Gene expression was analyzed by quantitative real-time PCR in 25 µl reactions using SYBR Green (Applied Biosystems). 18S RNA was used as an internal control for normalization. Fold inductions are shown relative to vehicle-treated control cells. The figures show mean and SD of two independent experiments. Primers used in real-time PCR were: 18S, Forward, 5′-AGTCCCTGCCCTTTGTACACA-3′ Reverse, 5′-CGATCCGAGGGCCTCACTA-3′; GILZ, Forward, 5′-GGCCCTAGACAACAAGATTG-3′ Reverse, 5′-TCAAGCAGCTCACGAATCTG-3′; IRF8, Forward, 5′-TCCCAGCTGGACATTTCTGA-3′ Reverse, 5′-CCACAGAAGGTTCCTTGATC-3′.
RESULTS
GR interacts directly with the Mediator complex in vitro
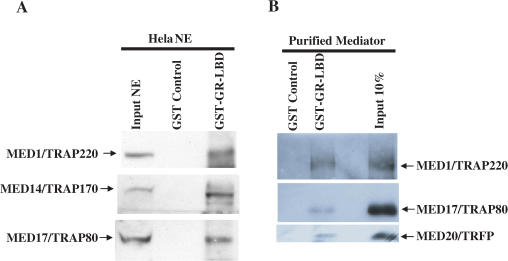
To investigate the role of the Mediator in GR-mediated transcription, we first set out to determine whether GR shows a direct interaction with the complete Mediator complex. A GST-fusion protein (GST-GR-LBD) containing the GR ligand binding domain (residues 439–795) was expressed in bacteria, purified and immobilized on glutathione-Sepharose beads. Immobilized GST-GR-LBD and control GST proteins were incubated with either HeLa S nuclear extract or with purified Mediator complex in the presence of 10 µM Dex. After washing, bound proteins were eluted by boiling in SDS sample buffer and analyzed by immunoblot. This analysis (Figure 1) shows specific binding to the GR-LBD of representative Mediator components (MED1/TRAP220, MED14/TRAP170, MED17/TRAP80 and MED20/TRFP) both from HeLa nuclear extract and from the purified Mediator preparations. These results demonstrate that the Mediator complex associates directly with GR through the LBD.
Figure 1.
GR interacts with the Mediator complex. (A) Binding of Mediator in nuclear extract. (B) Binding of purified Mediator. Beads containing bound GST or GST-GR-LBD were incubated with HeLa nuclear extract (A) or purified Mediator complex (B) in the presence of 10 µM Dex and processed as described in Materials and Methods. Associated components were identified by immunoblot with antibodies (all generated in this lab) to MED1/TRAP220, MED14/TRAP170, MED17/TRAP80 and MED20/TRFP.
The LXXLL-containing domain of MED1/TRAP220 interacts with the GR-LBD in a ligand-dependent manner
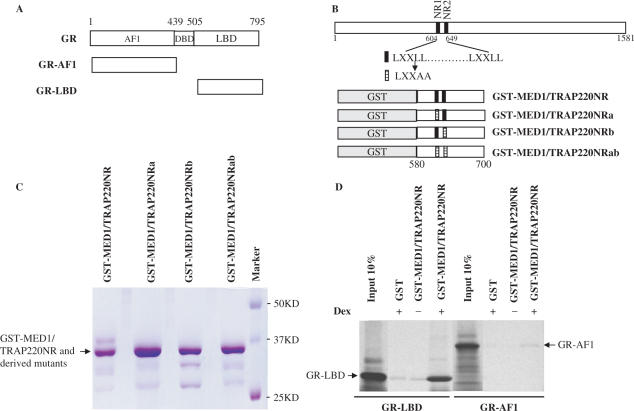
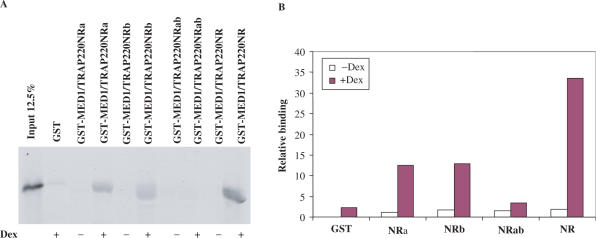
To determine whether MED1/TRAP220 can interact with GR, GST was fused to a MED1/TRAP220 fragment (residues 580–701) that contains the two LXXLL domains (Figure 2B), expressed in and purified from bacteria (Figure 2C), and immobilized on GST beads. The immobilized GST-MED1/TRAP220(580-701) was incubated with in vitro translated, 35S-Met-labeled GR-AF1-DBD (residues 1–505) or GR-DBD-LBD (residues 439–795) fragments (Figure 2A) in the presence or absence of Dex and, after washing, bound proteins were eluted and detected by autoradiography. The analysis in Figure 2D shows that MED1/TRAP220(580-701) binds strongly to GR-DBD-LBD in a ligand-dependent manner, but not to GR-AF1-DBD, thus confirming a similar earlier-reported result (28). MED1/TRAP220 contains two of the LXXLL motifs (within NR boxes) typically associated with nuclear receptor coactivators (Figure 2B, NR1 and NR2). To determine the relative contributions of these motifs to MED1/TRAP220 interactions with GR-LBD, mutations were introduced as shown in Figure 2B (NRa for the NR1 mutation, NRb for the NR2 mutation and NRab for the NR1 and NR2 double mutation) and corresponding proteins were expressed and purified (Figure 2C). As shown in Figure 3A, GST-MED1/TRAP220NRa and GST-MED1/TRAP220NRb retain ligand-dependent interactions with GR-LBD, although the interactions are significantly weaker than for wild-type GST-MED1/TRAP220NR. Mutation of both NR1 and NR2 completely abolishes the interaction with GR-LBD. These results demonstrate that MED1/TRAP220 NR1 and NR2 are both essential for optimal binding of MED1/TRAP220 to GR and that they contribute equally to this interaction.
Figure 2.
The NR boxes of MED1/TRAP220 interact with GR-LBD in a ligand-dependent manner. (A) Schematic representation of GR. (B) Schematic representation of MED1/TRAP220 and MED1/TRAP220 mutant proteins fused with GST. Numbers indicate positions of amino acid residues, black bars represent NR (LXXLL) boxes, shaded bars represent NR mutants (LXXAA). (C) Comassie staining of purified GST-MED1/TRAP220 fusion proteins. (D) Binding of MED1/TRAP220 NR to GR domains. Glutathione-Sepharose beads containing immobilized GST or GST-MED1/TRAP220NR (amino acids 580–701) proteins were incubated with in vitro translated, 35S-labelled GR-AF1 or GR-LBD proteins (shown in A) as indicated and bound proteins were resolved by SDS-PAGE and visualized by autoradiography.
Figure 3.
Both NR boxes contribute to the interaction between MED1/TRAP220 and GR. (A) Binding assays with the indicated fusion proteins (Figure 2B) and 35S-labeled GR-LBD were performed as described in Figure 2D. (B) Quantitation of the binding results in A. Values are from the representative experiment shown in A. Similar results were obtained from an independent experiment.
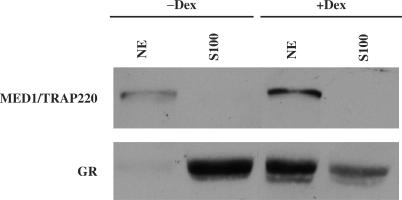
MED1/TRAP220 and GR co-localize in the nucleus after ligand treatment
To further explore the intracellular relationships of GR and MED1/TRAP220 inside a cell in response to ligand treatment, cellular localizations of GR and MED1/TRAP220 were monitored in the absence and presence of Dex. As shown in Figure 4, and as expected, GR is predominantly cytoplasmic in HeLa S cells cultured in medium with hormone-depleted serum and predominantly nuclear following treatment with Dex for 24 h. In contrast, MED1/TRAP220 is found exclusively in the nuclear fraction regardless of Dex treatment. This suggests that, in vivo, MED1/TRAP220 and GR may interact with each other only in the presence of ligand due to their localization in different compartments in the absence of ligand.
Figure 4.
MED1/TRAP220 and GR co-localize in the nucleus after ligand treatment. Nuclear (NE) and cytoplasmic (S100) fractions were prepared from HeLa S cells grown in the absence or presence of Dex. MED1/TRAP220 and GR were detected by immunoblot with corresponding anti-MED1/TRAP220 and anti-GR antibodies.
MED1/TRAP220 is required for optimal GR-mediated transcription activation
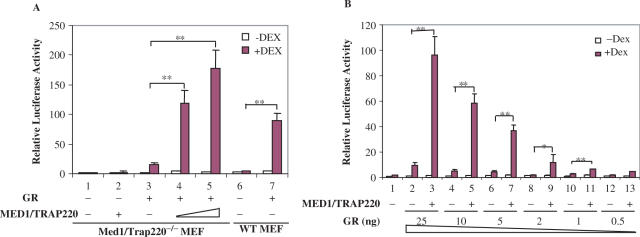
The results of the GST-GR-LBD pull down assay strongly suggest that the Mediator complex is recruited to GR target gene promoters through direct interactions between GR- and Mediator-associated MED1/TRAP220. Thus, the Mediator could play a functional role in GR-mediated transcription activation. To test this, Med1/Trap220−/− and wild-type MEF cells were transiently transfected with a GR expression vector and a MMTV-luc reporter. A β-gal vector was cotransfected in order to monitor transfection efficiency. The activation of MMTV-luc was measured as relative luciferase units (RLU) after normalization to the β-gal activity. As a control, transfection of MMTV-Luc alone into Med1/Trap220−/− and wild-type MEFs gave only background luciferase activity in the presence of Dex, indicating that endogenous GR is present at only a low level and/or inactive in the transfection assay (Figure 5A, lanes 1 and 6). Cotransfection of GR increased luciferase activity in a ligand-dependent manner in both wild-type and Med1/Trap220−/− MEFs. However, the induction level in Med1/Trap220−/− MEFs was very low compared to the level in wild-type MEFs (relative luciferase units of 16 and 90 in Med1/Trap220−/− and wild-type MEFs, respectively) (Figure 5A, lane 3 versus lane 7). This indicates that MED1/TRAP220 plays an important role in enhancing GR-mediated MMTV-luc activation. To test if the reduced reporter activity is due to lack of MED1/TRAP220 in Med1/Trap220−/− MEFs, these cells were cotransfected with GR and MED1/TRAP220 expression vectors. As shown in Figure 5A (lanes 4 and 5) the MMTV-luc reporter was activated, in a dose-dependent manner, to a level exceeding that observed in wild-type cells. Transfection of MED1/TRAP220 alone (Figure 5A, lane 2) did not activate the reporters, demonstrating that the activation of MMTV-luc is dependent on transfected GR. When Med1/Trap220−/− MEFs were transfected with a constant amount of a MED1/TRAP220 vector and increasing amounts of the GR vector, a clear dose-dependent response to GR was observed (Figure 5B). These results clearly demonstrate that MED1/TRAP220 enhances GR-mediated transcription activation on the MMTV-promoter.
Figure 5.
MED1/TRAP220 enhances GR-mediated transcription. (A) MED1/TRAP220 requirement for optimum GR-mediated transcription activation. Med1/Trap220−/− and WT MEFs were co-transfected with MMTV-luc reporter (150 ng) and vectors expressing GR (25 ng) or MED1/TRAP220 as indicated and cultured in the presence or absence of Dex. Conditions were as described in Materials and Methods. (B) Effect of GR dosage on MED1/TRAP220 enhanced transcription. Luciferase activities are shown as relative luciferase units (RLU) after normalization by β-gal activity. Values are the mean and SD of triple (A) or duplicate (B) samples in a representative experiment of at least three. * indicates p < 0.05. ** indicates p < 0.01 with unpaired Student's t-test.
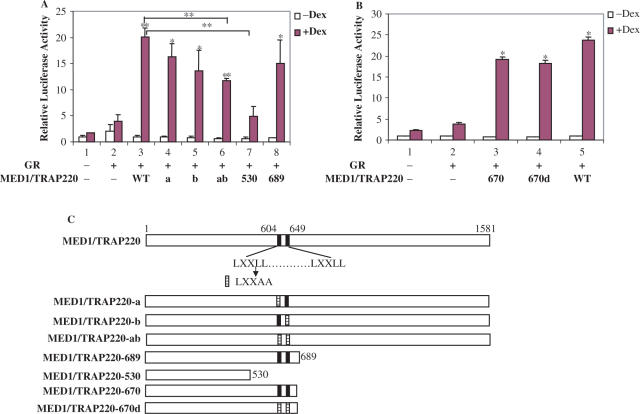
Since the NR boxes of MED1/TRAP220 interact with GR-LBD in a ligand-dependent manner, we next tested whether the NR boxes are essential for GR-mediated transcription. To address this question, we introduced point mutations that changed the LXXLL motif to LXXAA in the first NR box (MED1/TRAP220-a), in the second NR box (MED1/TRAP220-b) or in both NR boxes (MED1/TRAP220-ab) (Figure 6B). Wild-type and mutated MED1/TRAP220 vectors were cotransfected with MMTV-luc and the GR vector into Med1/Trap220−/− MEFs and relative luciferase units were measured. Compared to the high level of reporter induction observed with wild-type MED1/TRAP220, MED1/TRAP220-a and MED1/TRAP220-b showed slightly (but reproducible) lower levels of induction while MED1/TRAP220-ab showed an even lower level of induction (Figure 6A, lane 1 versus lanes 3–6). However, significant activation of MMTV-luc was still observed with the double mutant (Figure 6A). These results indicate that, while not essential for a significant level of MED1/TRAP220 enhancement of GR-mediated transcription, the two NR boxes of MED1/TRAP220 do contribute to the optimal MED1/TRAP220 enhancement of GR-mediated transcription activation.
Figure 6.
Effect of MED1/TRAP220 mutations on enhancement of GR-mediated transcription. (A) Functional analysis. Med1/Trap220−/− MEFs were transfected with MMTV-luc reporter (150 ng) and vectors expressing GR (10 ng) or MED1/TRAP220 (300 ng) mutants (described in C) as indicated, grown in the presence or absence of Dex as indicated, and processed as described in Materials and Methods. Luciferase activities are shown as relative luciferase units (RLU) after normalization to β-gal activity. Values are the mean and SD of duplicate samples in a representative experiment of at least three. Lanes 3-8 were compared with lane 2, lanes 4–8 were compared with lane 3. * indicates p < 0.05, ** indicates p < 0.01 with unpaired Student's t-test. (B) Mutation of the two LXXLL motives does not affect the function of the N-terminal domain of TRAP220 in enhancing GR-mediated transcription. The experiments were performed and analyzed as described in (A). Lanes 3–5 were compared to lane 2. * indicates p < 0.05. (C) Schematic representation of MED1/TRAP220 and MED1/TRAP220 mutants.
The function of the large (∼800 amino acids) region of MED1/TRAP220 C-terminal to the LXXLL domain is poorly characterized. However, recent studies have shown that this region is phosphorylated by several kinases (31) and that phosphorylation by the extracellular signal-regulated kinase (ERK) of the mitogen-activated protein kinase (MAPK) family stabilizes and increases the intrinsic activity of MED1/TRAP220 (32). To determine whether the C-terminal domain of MED1/TRAP220 is required for GR-mediated transcription, Med1/Trap220−/− MEFs were cotransfected with GR and MED1/TRAP220-689 (Figure 6C) expression vectors. In this assay, MED1/TRAP220-689 enhanced MMTV-luc activity to a level equivalent to ∼75% of that observed with full length MED1/TRAP220 (Figure 6A, lane 8 versus lane 3). This indicates that the C-terminus of MED1/TRAP220 is also dispensable for a substantial level of GR-mediated transcription activation. As shown in Figure 6A (lane 7 versus lane 2), further deletion of a region (amino acid 531–689) containing two LXXLL motifs and the surrounding sequences severely reduced the level of induction. However, point mutations of the two NR boxes (LXXLL→LXXAA) did not significantly affect the luciferase reporter activation by this N-terminal fragment (1–670) (Figure 6B). MED1/TRAP220-530, which contains only the conserved region of MED1/TRAP220, did not significantly enhance the level of activity seen with GR alone. All these results strongly suggest that although the two NR boxes contribute to the optimal MED1/TRAP220 activity, sequences surrounding the NR boxes play a more important role for MED1/TRAP220 enhancement of GR-mediated transcription activation.
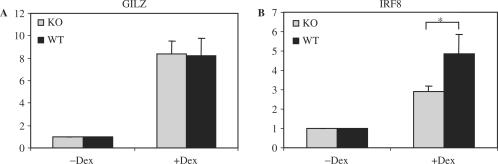
In order to examine the role of MED1/TRAP220 in endogenous GR target gene expression, we analyzed the expression of GR target genes GILZ and IRF8 in wild-type and Med1/Trap220−/− MEFs. Wild-type and Med1/Trap220−/− MEFs were transfected with GR vector. Two days after transfection, Dex or vehicle was added to the transfected cells at a final concentration of 100 nM to induce GILZ and IRF8 expression. RNA was extracted 10 h after treatment and real-time PCR was performed with the RNA. As shown in Figure 7, GILZ and IRF8 were both induced after Dex treatment in wild-type and Med1/Trap220−/− MEFs. GILZ was induced equally well in Med1/Trap220−/− and wild-type MEFs while the induction of IRF8 was reduced in Med1/Trap220−/− MEFs. These results indicate that TRAP220/MED1 contributes to the IRF8 induction but not to GILZ induction in response to Dex. These results with MEFs that are completely devoid of MED1/TRAP220 parallel those reported by Garabedian and colleagues using siRNA-mediated knockdown of MED1/TRAP220 (29).
Figure 7.
Expression profile of endogenous GR target genes in wild-type and Med1/TRAP220−/− MEFs. Cells were treated as described in the text, and real-time PCR was performed as described in Materials and Methods. (A) GILZ is induced to similar levels in wild-type and Med1/TRAP−/− MEFs. (B) Induction of IRF8 is reduced in Med1/TRAP220−/− MEFs relative to wild-type MEFs. * indicates p < 0.05.
DISCUSSION
Through interactions both with transcriptional activators and with RNA polymerase II, the Mediator plays a key role, as a bridging complex, in transmitting signals from activators, including a broad range of nuclear receptors, to the general transcription machinery (11). In the case of nuclear receptors, the MED1/TRAP220 subunit of Mediator plays a key role in this process through ligand-dependent interactions with the AF2 domain of many nuclear receptors. Despite a reported interaction between GR and the isolated MED1/TRAP220 subunit (28), it has not been established that GR can interact with the more physiologically relevant Mediator complex and the role of the Mediator complex in GR-mediated transcription has not been well defined.
In this study, we further investigated the functional role of MED1/TRAP220 in GR-mediated transcription activation. We first showed that a purified GR-LBD could bind the intact Mediator complex in a HeLa nuclear extract. We then demonstrated that the interaction is direct since the purified GR-LBD also showed a similarly strong interaction with a purified Mediator complex. Given the reported role of MED1/TRAP220 in facilitating Mediator recruitment by nuclear receptors, we then confirmed that MED1/TRAP220 interacts with GR-LBD in a ligand-dependent manner. These results strongly suggested that the Mediator complex functions as a GR coactivator. In support of this hypothesis, Med1/Trap220 null MEFs that are devoid of MED1/TRAP220 showed a near complete loss of GR-mediated transcription compared to wild-type MEFs. That this was due directly to the loss of MED1/TRAP220, rather than an indirect effect resulting from establishment of the null cell line, was shown by the ability of an ectopic MED1/TRAP220 to fully restore GR function in Med1/Trap220 null MEFs (Figure 5A lanes 4 and 5). Of note, and as shown here for GR and elsewhere for PPARγ and ER (15,19), Med1/Trap220 null MEFs are useful for establishing cellular functions of MED1/TRAP220 because the ubiquitous and abundant expression of MED1/TRAP220 may obscure visualization of significant effects of ectopically MED1/TRAP220 in common transfection assays. Furthermore, whereas siRNA-mediated knockdown of MED1/TRAP220 offers an alternative approach, the presence of residual levels of MED1/TRAP220 and potential off target effects could complicate such analyses.
A role of MED1/TRAP220 in GR-mediated transcription was also studied by Garabedian and colleagues (28). An initial study showed physical interactions of GR with two isolated Mediator components, MED14/TRAP170/DRIP150 and MED1/TRAP220/DRIP205, whereas transfected reporter assays showed enhancement of GR function by MED14/TRAP170/DRIP150, but not by MED1/TRAP220/DRIP205 alone. The later results likely reflected saturating levels of endogenous MED1/TRAP220 in HeLa cells (Chen,W. and Roeder,R.G., unpublished data), a complication avoided in the present assays. A more recent study from Garabedian and colleagues (29) showed that siRNA-mediated knockdown of MED1/TRAP220 reduced the expression of some GR-target genes such as LAD1 and IRF8 and that MED1/TRAP220 is recruited to the regulatory region of GR-target genes (IRF8 and IGFBP1) in a Dex-dependent manner. Although MED1/TRAP220 was also recruited to the GR target gene GILZ in response to Dex, siRNA-mediated knockdown of MED1/TRAP220 had no effect on GILZ expression (29). That this reflects a true MED1/TRAP220-independent expression of GILZ, and not the presence of residual MED1/TRAP220, was established by our demonstration of normal Dex-induced GILZ expression in Med1/Trap220 null cells. In combination, the previous (29) and current results strongly implicate MED1/TRAP220 as an important, albeit gene selective, coregulator of GR transactivation.
MED1/TRAP220 binding to nuclear receptors involves interactions of MED1/TRAP220 NR box/LXXLL motifs with the LBD of nuclear receptors (12,13). We also demonstrated a role for NR boxes in ligand-dependent interactions between MED1/TRAP220 and the LBD of GR. However, in contrast to other studies showing preferential interactions of other nuclear receptors with one or other of the two MED1/TRAP220 NR boxes (13,20,21), our analysis indicates that the two NR boxes contribute equally to MED1/TRAP220 interactions with the GR-LBD and that, whereas individual NR boxes mediate a low level of binding, both boxes are required for optimal interactions of MED1/TRAP220 to the GR-LBD. In functional studies of NR box-mutated MED1/TRAP220 in Med1/Trap220 null cells, and somewhat surprisingly, it was found, first, that single NR box mutations only slightly reduced MED1/TRAP220 enhancement of GR function, and second, that while mutations of both NR boxes effected a greater reduction of activity, there was still a substantial residual activity. Similar observations were reported for aryl hydrocarbon receptor (33) and for the estrogen receptor (34,38). One possibility is that ectopic reporters, as used in these analyses, are somewhat permissive because of relaxed chromatin structure (35,36). Another possibility is that although the LXXLL motif is important for optimal MED1/TRAP220/Mediator binding to nuclear receptors, the sequences surrounding or even distal to the NR boxes also contribute to GR interactions. In this regard, flanking sequences have been shown to be important for MED1/TRAP220 interactions with other nuclear receptors (20). In the case of the GR coactivator GRIP1, structure-functional studies have revealed that at least two GRIP1 components, the LXXLL motif and the sequences flanking the motif, contribute to the specific interactions between GR and GRIP1 (37). On the other hand, as discussed above, GR also was shown to interact physically and functionally, through the AF1 domains, with the MED14/TRAP170/DRIP150, a subunit of the Mediator complex. Hence, GR may efficiently recruit the Mediator complex through association with MED14/TRAP170/DRIP150 and MED1/TRAP220/DRIP205 (28).
In our previous study of the role of MED1/TRAP220 in adipogenesis, we found that the inability of Med1/Trap220−/− MEFs to undergo adipogenesis and to fully express PPARγ target genes could be restored by full length MED1/TRAP220 (15). More recently, we have found that this deficiency can be rescued not only by the full length MED1/TRAP220, but also by MED1/TRAP220(1-689), containing the NR box region but lacking the C-terminus, and, most surprisingly, by MED1/TRAP220(1-530), lacking both the NR box region and the C-terminus (Ge,K. and Roeder,R.G., unpublished data). These results, along with previous reports of NR box requirements for optimal nuclear receptor functions, indicate conditional requirements for NR boxes for MED1/TRAP220 functions. The present results, indicating significant MED1/TRAP220 enhancement of GR function in the absence of the MED1/TRAP220 C-terminus and NR boxes, are consistent with these findings. Nonetheless, none of these studies eliminate the possibility that the NR box and C-terminal regions of MED1/TRAP220 are required for efficient function of nuclear receptors, including GR, on at least some of the many nuclear receptor target genes that have been identified.
ACKNOWLEDGEMENTS
We thank Dr Ge K. for providing MED1/TRAP220 and mutant expression vectors. This work was supported by a National Institutes of Health National Research Service Award to W.C. and by a National Institutes of Health grant (DK071900) to R.G.R. Funding to pay the Open Access publication charges for this article was provided by Rockefeller University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 3.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 4.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 5.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, et al. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 10.Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 11.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 13.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang YK, Guermah M, Yuan CX, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl Acad. Sci. USA. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 16.Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, Le Roith D, Chambon P, Gonzalez FJ, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J. Biol. Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell JD. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 2000;20:5433–5446. doi: 10.1128/mcb.20.15.5433-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachez C, Gamble M, Chang CP, Atkins GB, Lazar MA, Freedman LP. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol. Cell. Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 23.Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim. Biophys. Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Reichardt HM, Tronche F, Berger S, Kellendonk C, Schutz G. New insights into glucocorticoid and mineralocorticoid signaling: lessons from gene targeting. Adv. Pharmacol. 2000;47:1–21. doi: 10.1016/s1054-3589(08)60108-8. [DOI] [PubMed] [Google Scholar]
- 25.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grenier J, Trousson A, Chauchereau A, Amazit L, Lamirand A, Leclerc P, Guiochon-Mantel A, Schumacher M, Massaad C. Selective recruitment of p160 coactivators on glucocorticoid-regulated promoters in Schwann cells. Mol. Endocrinol. 2004;18:2866–2879. doi: 10.1210/me.2004-0241. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa H, Yu RT, Haraguchi T, Hiraoka Y, Nakatani Y, Morohashi K, Umesono K. Nuclear structure-associated TIF2 recruits glucocorticoid receptor and its target DNA. Biochem. Biophys. Res. Commun. 2004;320:218–225. doi: 10.1016/j.bbrc.2004.05.161. [DOI] [PubMed] [Google Scholar]
- 28.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol. 2006;20:560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misra P, Owuor ED, Li W, Yu S, Qi C, Meyer K, Zhu Y-J, Rao MS, Kong A-NT, et al. Phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. J. Biol. Chem. 2002;277:48745–48754. doi: 10.1074/jbc.M208829200. [DOI] [PubMed] [Google Scholar]
- 32.Pandey PK, Udayakumar TS, Lin X, Sharma D, Shapiro PS, Fondell JD. Activation of TRAP/mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell. Biol. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Ge K, Roeder RG, Hankinson O. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J. Biol. Chem. 2004;279:13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- 34.Wu Q, Burghardt R, Safe S. Vitamin D-interacting protein 205 (DRIP205) coactivation of estrogen receptor alpha (ERalpha) involves multiple domains of both proteins. J. Biol. Chem. 2004;279:53602–53612. doi: 10.1074/jbc.M409778200. [DOI] [PubMed] [Google Scholar]
- 35.Sheldon LA, Smith CL, Bodwell JE, Munck AU, Hager GL. A ligand binding domain mutation in the mouse glucocorticoid receptor functionally links chromatin remodeling and transcription initiation. Mol. Cell. Biol. 1999;19:8146–8157. doi: 10.1128/mcb.19.12.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CL, Htun H, Wolford RG, Hager GL. Differential activity of progesterone and glucocorticoid receptors on mouse mammary tumor virus templates differing in chromatin structure. J. Biol. Chem. 1997;272:14227–14235. doi: 10.1074/jbc.272.22.14227. [DOI] [PubMed] [Google Scholar]
- 37.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JE, Kim K, Sacchettini JC, Smith CV, Safe S. DRIP150 coactivation of estrogen receptor α in ZR-75 breast cancer cells is independent of LXXLL motifs. J. Biol. Chem. 2005;280:8819–8830. doi: 10.1074/jbc.M413184200. [DOI] [PubMed] [Google Scholar]