Abstract
In seed plants, shoot branching is initiated by the formation of new meristems in the axils of leaves, which subsequently develop into new axes of growth. This study describes the genetic control of axillary meristem formation by the LATERAL SUPPRESSOR (LAS) gene in Arabidopsis thaliana. las mutants show a novel phenotype that is characterized by the inability to form lateral shoots during vegetative development. The analysis shows that axillary meristem formation is differently regulated during different phases of development. During reproductive development, axillary meristems initiate in close proximity to the shoot apical meristem and do not require LAS function. In contrast, during the vegetative phase, axillary meristems initiate at a distance to the SAM and require LAS function. This control mechanism is conserved between the distantly related species tomato and Arabidopsis. Monitoring the patterns of LAS and SHOOT MERISTEMLESS transcript accumulation allowed us to identify early steps in the development of leaf axil identity, which seem to be a prerequisite for axillary meristem initiation. Other regulators of shoot branching, like REVOLUTA and AUXIN RESISTANT 1, act downstream of LAS. The results are discussed in the context of the “detached meristem” and the “de novo formation” concepts of axillary meristem formation.
Keywords: Shoot branching, axillary meristem, LATERAL SUPPRESSOR, Arabidopsis thaliana
In flowering plants, elaboration of the primary shoot axis can be traced back to the activity of the shoot apical meristem (SAM), a small group of mitotic cells initiated during embryogenesis. During the postembryonic phase of development, secondary axes of growth are established, which determine to a large extent the architectural form of plants. These shoot branches originate from secondary meristems initiated in the axils of leaf primordia. Axillary meristems function like the SAM of the primary shoot initiating the development of lateral organs, a process that results in the formation of an axillary bud. In many plant species, further development of axillary buds into shoots is blocked to a different extent by the influence of the primary shoot. This phenomenon is known as apical dominance and seems to be mediated by plant hormones: auxin was found to inhibit lateral bud outgrowth, whereas cytokinin promotes it (Cline 1997; Napoli et al. 1999).
With respect to timing of meristem initiation and further development, axillary shoot formation seems to be differentially regulated during different phases of development. During prolonged vegetative development in Arabidopsis thaliana, axillary meristems are initiated at a distance from the SAM, develop into buds, and finally into side shoots, thereby establishing acropetal gradients of axillary meristem initiation and axillary bud outgrowth (Hempel and Feldman 1994; Stirnberg et al. 1999, 2002; Grbic and Bleecker 2000). In the reproductive phase, axillary meristems initiate evenly (Stirnberg et al. 1999, 2002) or in a basipetal sequence (Hempel and Feldman 1994; Grbic and Bleecker 2000) at close proximity to the SAM in the axils of the youngest leaf primordia. Outgrowth of the resulting axillary buds starts in the youngest leaf axil and progresses toward older leaves, resulting in a basipetal gradient of axillary shoot outgrowth (Hempel and Feldman 1994; Grbic and Bleecker 1996; Stirnberg et al. 1999, 2002). In tomato (Lycopersicon esculentum), axillary shoot development is comparable to that in Arabidopsis thaliana with two differences: during the vegetative phase, axillary meristems are initiated in closer proximity to the SAM, and only two vigorously developing axillary branches are formed during the reproductive phase (Schmitz and Theres 1999).
The origin of axillary meristems is presently unclear. For several plant species it has been suggested that axillary meristems are initiated from cell groups detached from the primary SAM and retain their meristematic identity (Steeves and Sussex 1989; Long and Barton 2000). Alternatively, axillary meristems may originate de novo later in development from partially or fully differentiated cells. Studies on the Arabidopsis phabulosa-1d mutant have demonstrated that the adaxial leaf base plays an important role in axillary meristem initiation leading to a new concept for axillary meristem formation (McConnell and Barton 1998). The homeodomain-containing protein SHOOT MERISTEMLESS (STM) was found to be expressed in all types of shoot meristems. STM function is required for initiation and maintenance of the SAM (Long et al. 1996). When leaf primordia are formed, STM expression is down-regulated. Using STM transcript accumulation as a marker for SAM fate, Long and Barton (2000) were able to define four stages of axillary meristem development. However, the interprimordial expression of STM made it difficult to distinguish between the “detached meristem” and “de novo formation” concepts of lateral meristem initiation.
So far, the genetic control of axillary shoot development is only poorly understood. Mutants with altered patterns of shoot branching have been described in several plant species including tomato, pea, maize, and Arabidopsis. Whereas many mutants show either an increase or a decrease in the outgrowth potential of lateral shoots [e.g., max mutants (Stirnberg et al. 2002); sps/bus (Reintanz et al. 2001; Tantikanjana et al. 2001); tb1 (Doebley et al. 1997); axr1 (Stirnberg et al. 1999); and dad mutants (Napoli 1996)], only a few mutants are characterized by defects in lateral meristem initiation. Besides the Arabidopsis mutants revoluta (rev; Talbert et al. 1995) and pinhead (pnh; McConnell and Barton 1995), the tomato mutants lateral suppressor (ls; Williams 1960) and blind (bl; Rick and Butler 1956) are particularly interesting, because their inability to form lateral meristems is accompanied by only moderate pleiotropic effects on other aspects of plant development. In the tomato ls mutant, axillary meristem formation is blocked during the vegetative growth phase (Malayer and Guard 1964). In addition, ls flowers fail to develop petals and display a reduced male and female fertility (Groot et al. 1994). Characterization of the Lateral suppressor (Ls) gene (Schumacher et al. 1999) revealed that the encoded protein is a putative transcription factor that belongs to the GRAS family of regulatory proteins.
In this study, we have analyzed the role of the Arabidopsis LATERAL SUPPRESSOR (LAS) gene in the process of axillary meristem initiation. Characterization of knockout mutants has demonstrated that this process is differently regulated during vegetative versus reproductive development. Furthermore, comparison of the phenotypes of tomato and Arabidopsis lateral suppressor mutants, in conjunction with the result of a trans-complementation experiment, revealed that the described control mechanism is conserved during evolution. Monitoring the patterns of LAS and STM transcript accumulation allowed us to identify early steps in the development of leaf axil identity, which seem to be a prerequisite for axillary meristem initiation. Double-mutant analysis and expression studies have been instrumental to position LAS function relative to the activities of the SHOOT MERISTEMLESS and REVOLUTA genes.
Results
Isolation and structure of the Arabidopsis LATERAL SUPPRESSOR gene
Initially, a PCR-based strategy was used to isolate a Lateral suppressor (Ls)-homologous gene from Arabidopsis thaliana (see Materials and Methods). Sequence analysis revealed that the Ls-homologous gene contains an open reading frame (ORF) encoding a protein with 50.5% sequence identity to the tomato Ls protein. The next most closely related protein encoded by the Arabidopsis genome shows only 34.8% identity. After completion of the Arabidopsis genome sequence, BLAST analysis resulted in the identification of two BAC clones, F20N2 (GenBank accession no. AC002328) and T5A14 (GenBank accession no. AC005223), which both contain the most homologous LATERAL SUPPRESSOR gene (At1g55580). Microsynteny studies between the tomato Ls region and the identified BAC clones demonstrated that the gene repertoire within this region is completely conserved, but two inversions distinguish the arrangements of genes in both species (Rossberg et al. 2001). These results demonstrate that the isolated DNA segment contains the Ls-orthologous gene from Arabidopsis (LAS).
The sequence of the LAS cDNA (GenBank accession no. AY196482) was deduced from PCR products amplified on cDNA that was prepared from shoot tip RNA. The 5′ end (position 1) and the 3′ end (position 1742) of the transcript have been determined in rapid amplification of cDNA ends (RACE) experiments. Comparison of the cDNA to its genomic counterpart revealed that the LAS gene contains an uninterrupted ORF starting with an ATG at position 96 and ending with a stop codon at position 1431, followed by an untranslated 3′ region of 309 bp. The ORF has a coding capacity for 445 amino acids.
The LAS protein is a member of the GRAS family of putative transcriptional regulators (Pysh et al. 1999). LAS, which was previously named SCL18, represents an independent branch of the GRAS protein family in Arabidopsis. The characteristic sequence motifs of this protein family, like the VHIID domain, the two leucine heptad repeats, and a so-called SAW domain (Pysh et al. 1999), were also identified in LAS. In contrast to GAI, RGA, and a few other members of the GRAS family, the LAS protein has a very short N terminus characterized by repetitions of the amino acids serine and threonine and does not contain a DELLA domain.
Identification of las mutants
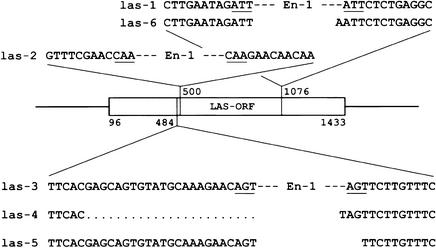
las mutants were identified by screening a collection of Arabidopsis plants mutagenized by the maize transposable element Enhancer-1 (En-1; Baumann et al. 1998; Wisman et al. 1998). Using a PCR-based screening strategy, three independent En-1 insertions within the LAS ORF were identified (las-1, las-2, las-3; Fig. 1). Insertion points were determined by sequencing diagnostic PCR products.
Figure 1.
Structure of different LAS alleles. Transcription is from left to right. Numbers are positions in the LAS cDNA. Enhancer-1 (En-1) insertions in independent mutant alleles are indicated at their positions in LAS. The underlined bases represent the target site duplications produced by the insertions of the transposable element En-1.
To establish a causal relationship between the mutant phenotype and a particular En-1 insertion, several derivatives of the insertion alleles that have excised the En-1 element from the LAS locus were analyzed. For this purpose, progeny of outcrosses to the Columbia (Col) wild type were screened for loss of En-1 from the LAS locus using a PCR-based approach. From the insertion alleles las-1 and las-3, four independent derivatives were obtained. Two stable mutants, las-4 and las-5, as well as a wild-type revertant, were derived from las-3. An additional stable mutant, las-6, was isolated from the F1 progeny of las-1 (Fig. 1). Plants homozygous for the different mutant alleles exhibited a very similar phenotype (see below; Fig. 2), whereas the revertant was indistinguishable from the wild type. Sequence analysis revealed that las-4 is characterized by a deletion of 20 nucleotides together with the insertion of 1 nucleotide, whereas the wild-type revertant had the complete wild-type sequence. In the stable mutant las-5, an insertion of a T nucleotide was found at the site of the former En-1 insertion. The las-6 allele is characterized by an insertion of the tetranucleotide AATT. The sequence alterations observed in the stable mutant alleles las-4, las-5, and las-6 gave rise to frameshifts, which result in a premature stop of protein translation. Overall, these data demonstrate a causal relationship between the mutations at the LAS locus and the observed phenotypic alterations (Fig. 2).
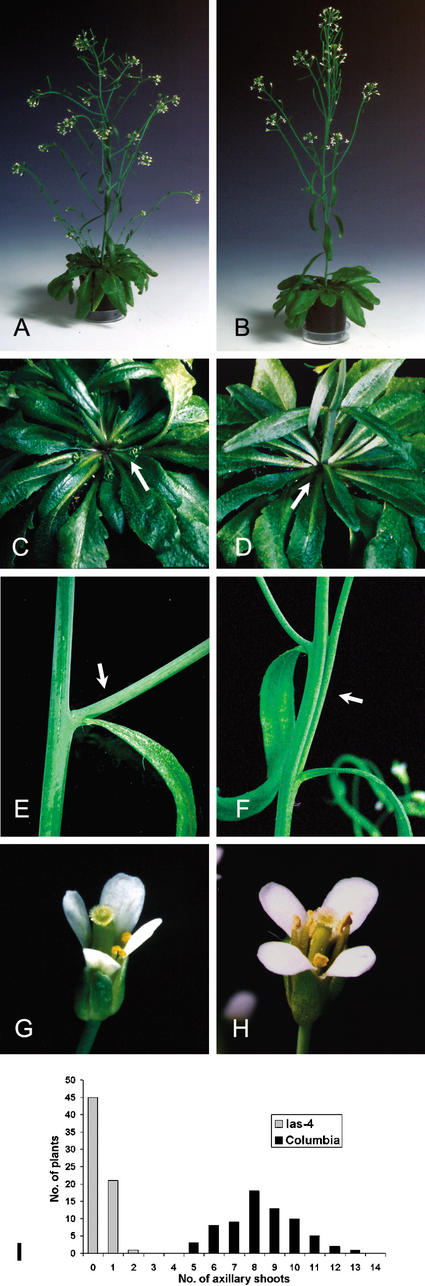
Figure 2.
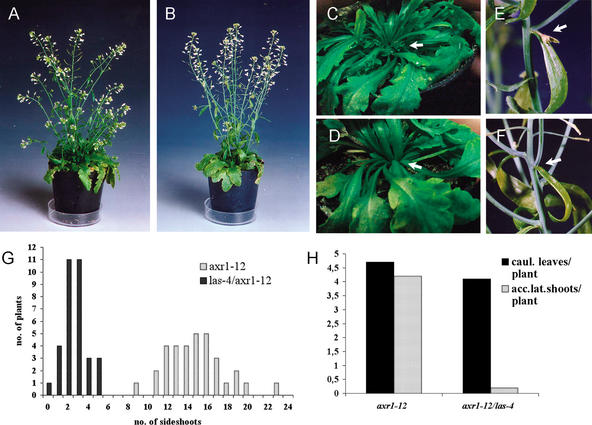
Comparison of wild-type (A,C,E,G) and las-4 (B,D,F,H) plants during vegetative and inflorescence development. In Columbia wild-type plants (A,C) multiple axillary inflorescences have developed from the axils of rosette leaves (C, arrow), whereas axillary shoot development from rosette leaves is blocked in las-4 mutant plants (B,D). Arrows indicate the formation of axillary shoots in Columbia (C) and their absence in las-4 (D) plants, respectively. (E–H) Comparisons of cauline leaf axils (E,F) and flowers (G,H) from the wild-type (E,G) and the las-4 mutant (F,H) plants. The arrows in E and F point to the axes of lateral shoots, which are separated from (Columbia) or attached to the primary axis (las-4) of the plant. (I) A comparison of side-shoot development after decapitation. Plants were first grown under short-day conditions for 28 d and then shifted to long days to induce flowering (Col., n = 76; las-4, n = 67). Primary bolts were removed when the plants reached a height of ∼10–15 cm. Side shoots were counted 10 d after the decapitation.
las mutants are characterized by an almost complete suppression of axillary shoot formation during vegetative development
Six independent lines were established, in which the LAS ORF is disrupted by mutations at different positions. Because all these lines displayed a very similar phenotype, only plants homozygous for the las-4 deletion allele were used in a detailed characterization of the mutant phenotype (Fig. 2). In comparison with Columbia wild-type plants, las mutants showed an almost complete suppression of secondary bolt formation from the axils of rosette leaves (Fig. 2A–D,I). However, side-shoot formation from the axils of cauline leaves of the primary bolt was not inhibited. In some las-4 plants, also the topmost rosette leaves formed axillary shoots. Similar changes in branching patterns were also observed when the las-4 allele was introduced into the Landsberg erecta and Wassilewskija genetic backgrounds (data not shown). The branching phenotype of las mutants was more easily recognized when plants were grown first under short-day conditions to allow the formation of many rosette leaves, and then shifted to long days to induce flowering. Secondary branching was also affected: whereas in the wild type, tertiary shoots developed from the axils of all leaves of secondary shoots, branching from the first two or three leaves of secondary shoots was blocked in the las-4 mutant.
Furthermore, positioning of lateral shoots originating from the axils of cauline leaves was frequently affected in the las-4 mutant. Different from the wild type, in which lateral shoots are positioned in the axils of their subtending leaves, the las-4 mutant showed a tendency toward concaulescence, that is, the point of separation between lateral shoots and the main axis was often displaced acropetally (Fig. 2E,F).
In addition to its defect in side-shoot development, the tomato ls mutant is characterized by the absence of petals from the flower (Williams 1960; Schumacher et al. 1995). Different from tomato, the Arabidopsis las-4 mutant developed a complete whorl of petals (Fig. 2G,H). However, in the mutant, petal abscission seems to be delayed. Root development was analyzed on plantlets grown on vertically oriented agar plates. In contrast to shoot branching, the pattern of root branching did not reveal any significant difference between the las-4 mutant and the Columbia wild type (data not shown).
The Lateral suppressor gene function is conserved between tomato and Arabidopsis
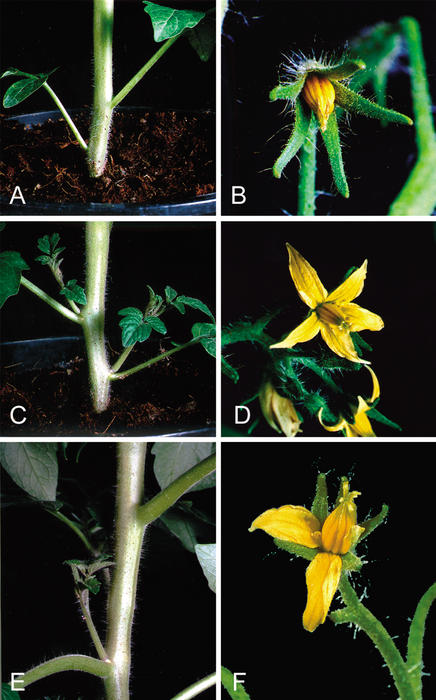
Comparison of the phenotypes of lateral suppressor mutants of tomato and Arabidopsis revealed both similarities and differences. Whereas both mutants showed a severe reduction in the number of axillary shoots, only the tomato mutant was characterized by a failure to develop petals (Williams 1960). In addition, a reduction in male and female fertility was observed in the tomato ls mutant (Groot et al. 1994), but not in the Arabidopsis las-4 mutant. To test for functional complementation between these two distantly related plant species, a 6.3-kb genomic DNA fragment, harboring the Arabidopsis LAS ORF, as well as ∼2.9 kb of 5′ sequence and 2.1 kb of 3′ sequence, was introduced into the tomato lateral suppressor (ls1/ls1) mutant. Four independent transgenic lines harboring at least one complete copy of the LAS gene were established. Two transgenic lines developed side shoots in almost every leaf axil and a whorl of petals on all flowers (Fig. 3C,D). The two additional lines developed side shoots in only a fraction of their leaf axils and also showed an incomplete restoration of the wild-type flower phenotype (Fig. 3E,F). Inheritance of the complementation phenotype was analyzed in transgenic line 258 harboring a single-copy T-DNA insertion. Among 19 plants of the self-pollinated progeny of 258, 14 plants showed complementation and 5 plants had the ls phenotype. By Southern analysis, the T-DNA was detected only in those plants showing complementation. This result is consistent with the assumption that a single-copy T-DNA insertion, segregating in a Mendelian fashion, rescues the ls phenotype.
Figure 3.
Functional complementation of the tomato ls1 mutant with the Arabidopsis LAS gene. Comparison of phenotypes of Antimold B-ls1 (A,B) and Antimold B-ls1 transformed with construct C28.1 (C–F). The pictures show close-ups of leaf axils (A,C,E) and flowers (B,D,F). Note the absence of an axillary shoot in one leaf axil in E and the incomplete whorl of petals in F.
LAS transcripts accumulate in the axils of all primordia derived from the SAM
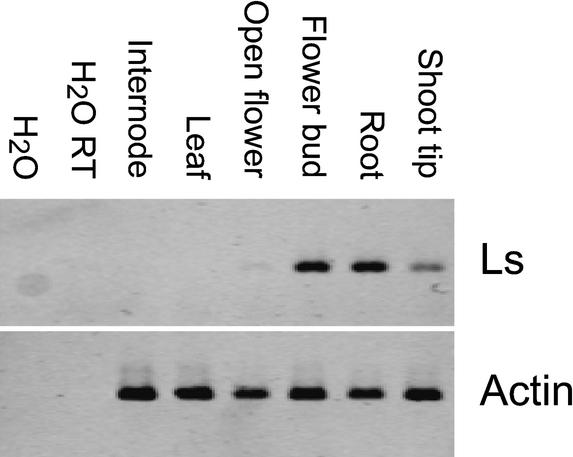
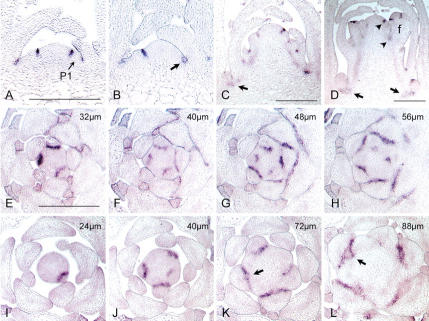
LAS mRNA distribution in different plant tissues was first monitored by reverse transcriptase PCR (RT–PCR) analysis. The LAS transcript was detected in shoot tips, flowers buds, roots, and open flowers, but not in internodes and leaves (Fig. 4). To analyze the distribution of LAS mRNA at the cellular level, RNA in situ hybridization experiments on tissue sections were performed. Columbia and las-4 Arabidopsis plants were grown under short-day conditions and fixed 28 d after sowing. During the vegetative phase of development, LAS transcripts accumulated in the axils of leaf primordia and leaves from P1 to P20/22 (Fig. 5A,E–H). In the axil of P1, signals were observed when the primordium was barely recognizable as a bulge protruding from the SAM. The LAS expression domain was initially about 3–5 cell layers deep, including the L1–L3 layers of the SAM, and extended 1 or 2 cell layers in the adaxial–abaxial dimension. Transverse sections demonstrated that LAS mRNA accumulated in a band-shaped domain at the adaxial side of the leaf primordium, marking the inner boundary of the primordium (Fig. 5E–H). From P5 to P22, LAS expression was restricted to a 1–2-cell-layer-deep domain located within the cleft between the leaf primordium and the inner part of the shoot apex, slightly shifted toward the base of the primordium. In the axils of older leaf primordia, LAS expression faded out until it was no longer detectable in the axils of primordia older than P20/P22. A similar distribution of LAS transcript was observed in vegetative apices of the las-4 mutant, although the intensity of the signal was reduced in comparison to wild-type plants (data not shown).
Figure 4.
RT–PCR analysis of LAS transcript accumulation. Total RNA from different plant tissues was analyzed by RT–PCR, and the PCR products were hybridized to a probe from the LAS gene. Amplification of actin cDNA was used to ensure that equal amounts of cDNA were added to each PCR reaction.
Figure 5.
Patterns of LAS mRNA accumulation during vegetative and reproductive development. Longitudinal and transverse sections through shoot tips of Columbia (A–H) and Landsberg erecta (I–L) plants were hybridized with a probe from the LAS gene. (A) Longitudinal section through the shoot apex of a 28-day-old plant grown under short-day conditions. The arrow points to the P1 primordium. (B–D) Longitudinal sections through shoot tips of Columbia plants grown for 28 d in short days and subsequently for 2 d (B), 4 d (C), or 6 d (D) under long-day conditions. The arrows in B, K, and L point to LAS signals that show a down-regulation in their central region. The arrows in C and D indicate residual amounts of LAS transcripts at the base of developing axillary meristems. The arrowheads in D indicate LAS expression domains in the axils of flower primordia. (E–L) Successive transverse sections from 28-day-old Columbia (E–H) and Landsberg erecta (I–L) plants grown in short days. (E–L) The approximate distance from the top of the meristem to the middle of the section is given in the upper right corner of the image. As Landsberg erecta plants are more elongated at this time point than Columbia plants, not all serial sections are depicted in this case. Bars: A (for A,B), C,D,E (for E–L), 200 μm; f, flower primordium; P1, P1 primordium.
Because histological studies revealed differences in the timing of axillary meristem formation between Arabidopsis ecotypes (Long and Barton 2000; Stirnberg et al. 2002), LAS expression during the vegetative phase was also analyzed in Landsberg erecta. Different from Columbia, in Landsberg erecta plants a down-regulation of the LAS signal was observed in a small oval area in the center of the LAS expression domain (Fig. 5K,L, arrows) from P6/P7 on, which coincides with the position of the presumptive axillary meristem. Sections of flowering shoots were obtained from plants grown under short-day conditions for 28 d and then induced to flower by two, four, or six successive long days. After the shift to long days, LAS mRNA was found in the axils of all leaf primordia originating from the elongating stem (Fig. 5B,C). Some of the LAS expression domains in cauline leaf axils showed a similar down-regulation in their center (Fig. 5B, arrow) to those observed during Landsberg erecta vegetative development. At the base of outgrowing axillary shoots, residual amounts of LAS transcripts have been detected (Fig. 5C,D, arrows). After the transition to flower formation, LAS transcripts were detected at the adaxial base of flower primordia (Fig. 5D, arrowheads). This LAS expression domain was first detectable at the adaxial border of stage 1 flower primordia with cellular dimensions comparable to those domains observed in the axils of leaf primordia. During flower development, LAS expression was observed in the axils of sepal primordia of stage 3–5 flowers (Fig. 5D). This signal faded out at later stages of flower development.
In contrast to the results of the RT–PCR analyses, LAS transcripts could not be detected in the root by RNA in situ hybridization. These experiments were controlled by using a SCARECROW (SCR; Di Laurenzio et al. 1996) antisense probe on parallel sections, which successfully detected the SCR transcript (data not shown).
las mutants fail to initiate axillary meristems during vegetative development
To test whether the inability to form lateral shoots from the axils of rosette leaves was due to a defect in axillary meristem initiation or to an inhibition of lateral bud outgrowth, axillary shoot formation was analyzed in Columbia wild-type plants and las-4 mutants after removal of the primary bolt. Plants were first grown under short-day conditions for 28 d and then shifted to long days to induce flowering. Primary bolts were removed when the plants reached a height of 10–15 cm. After decapitation, the wild-type plants developed between 5 and 13 axillary shoots, whereas none, one, or two axillary shoots were recorded for the las-4 mutant (Fig. 2I). If the las-4 mutant developed secondary bolts, these always originated from the axils of the topmost rosette leaves. These data suggested that the las-4 mutant phenotype was not a consequence of extreme apical dominance, but more likely the result of a defect in axillary meristem initiation during vegetative development.
Long and Barton (2000) have demonstrated that STM expression can be used as an early marker for axillary meristem development. In this study, STM expression was monitored in the wild-type and the las-4 mutant plants to test for initiation of axillary meristems. Transverse sections were prepared from vegetative plants grown under short-day conditions for 28 d and hybridized to an antisense STM probe. These experiments revealed STM transcript accumulation in the SAM and interprimordial regions of both wild-type and las-4 plants (Fig. 6A–D, arrowheads). As reported by other groups (e.g., Long and Barton 2000), leaf primordia and incipient leaf primordia (from P0) were found to be free of STM signal. In the axils of older leaf primordia of the wild type (from P16), STM expression was found to be focused to a group of small, densely cytoplasmic cells localized next to the adaxial center of the primordium border (Fig. 6A,C, arrows, E). These cell groups developed into new meristems, which could first be recognized at the morphological level in axils of P21. However, older leaf axils of the las-4 mutant did not show this focused STM expression (Fig. 6B,D, arrows, F), suggesting that the focusing of STM expression indicative of the onset of meristem organization is dependent on LAS function. Comparison of the patterns of LAS and STM transcript accumulation indicated that the LAS expression domain in the axils of leaf primordia was contained within the STM-positive region (Figs. 5, 6). The developmental timing of STM focusing to the small cell group next to the center of the adaxial primordium boundary seems to be correlated with a down-regulation of the LAS signal, as observed in the Landsberg erecta ecotype (Fig. 5L). Taken together, the results of the decapitation experiments and the analyses of STM expression demonstrate that in the las-4 mutant, axillary meristem initiation is blocked during vegetative development.
Figure 6.
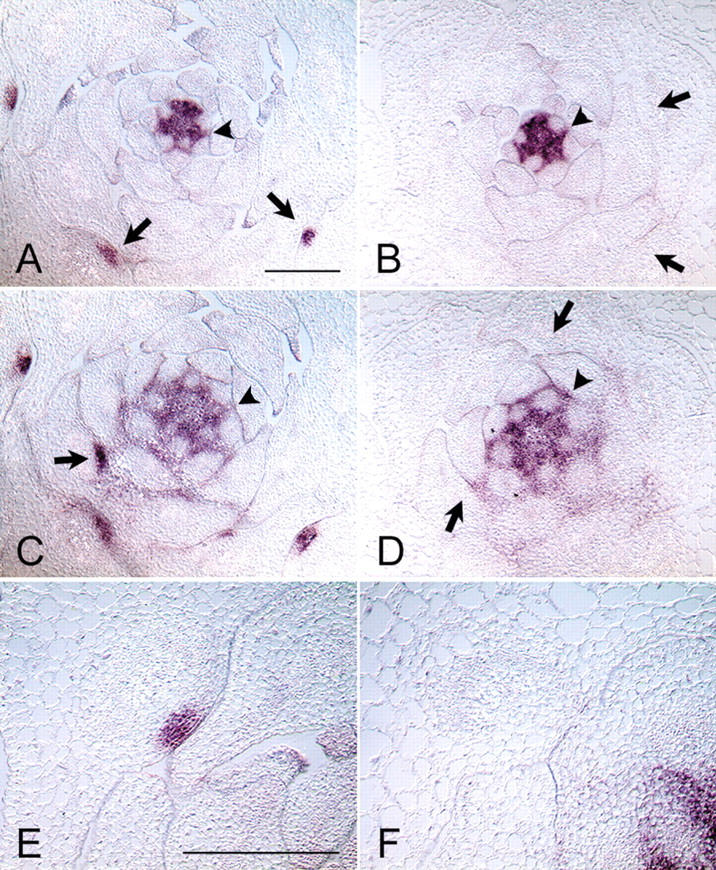
Comparison of STM mRNA accumulation in vegetative shoot apices of wild-type (A,C,E) and las-4 (B,D,F) plants. Consecutive transverse sections through shoot apices of Columbia wild-type (A,C,E) and las-4 (B,D,F) plants were hybridized with an STM antisense probe. Sections were prepared from plants grown under short-day conditions for 28 d before fixation. In wild-type and las-4 plants, STM mRNA is detected in the SAM and in interprimordial regions (A–D, arrowheads). In addition, a focused STM expression domain was observed close to the adaxial center of older leaf primordia of the wild type (E; A,C, arrows) but not in the las-4 mutant (F; B,D, arrows). Bars, 200 μm. Same magnifications in A–D and E and F.
LATERAL SUPPRESSOR acts upstream of REVOLUTA in axillary meristem development
In the Arabidopsis revoluta (rev) mutant, shoot branches in the axils of rosette and cauline leaves frequently fail to develop (Talbert et al. 1995). Double-mutant analysis and expression studies suggested that REV is an activator of STM (Otsuga et al. 2001). To unravel the relationship between REV and LAS, RNA in situ hybridization experiments were performed on tissue sections from wild-type (Columbia) and las-4 plants using REV sense and antisense probes. In Columbia, the REV transcript was detected in vascular bundles of leaves, in the vascular traces of leaf primordia, and in an inverse cup-shaped domain in the SAM itself (Fig. 7A,C). From P17 on, REV mRNA was also found in a cell group located next to the center of the adaxial base of leaf primordia (Fig. 7C,E,G, arrows). This expression domain marks the presumptive axillary meristem and shows an extended overlap with the STM expression domain. These findings support the results of expression studies reported by Otsuga et al. (2001). Comparison of the patterns of REV transcript accumulation in wild-type and las-4 plants revealed no differences in the SAM and in the vascular elements of leaf primordia and leaves (Fig. 7C,D). In contrast, the REV expression domain observed in the cell group located at the central adaxial base of older leaf primordia of wild-type plants was absent in las-4 (Fig. 7E–H, arrows).
Figure 7.
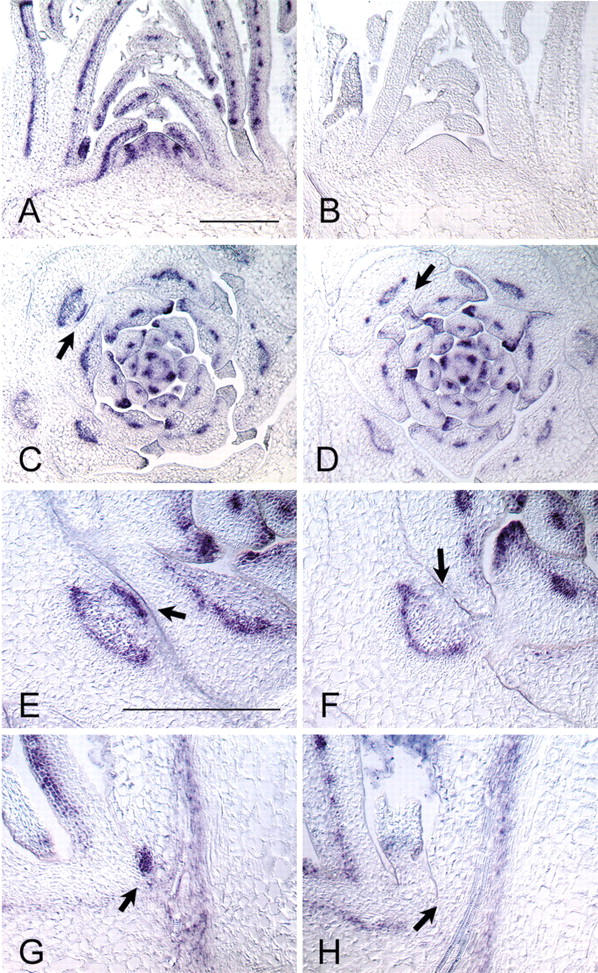
Patterns of REV transcript accumulation in wild-type and las-4 plants. Longitudinal (A,B,G,H) and transverse (C–F) sections through shoot tips of Columbia (A,B,C,E,G) and las-4 (D,F,H) plants were hybridized with REV antisense (A,C–H) and sense (B) probes, respectively. REV transcripts were detected in vascular bundles and provascular tissues of leaf primordia in Columbia (A,C) and las-4 (D) plants. In addition, REV is expressed in an inverse cup-shaped domain in the SAM (A). In leaf primordia older than P16 of Columbia wild-type plants, REV transcript accumulation was also observed in a cell group close to the adaxial center of the primordium (C,E,G, arrows). This expression domain was not found in las-4 plants (D,F,H, arrows). Hybridization with the sense probe (B) showed that the observed signals were specific for REV mRNA.
To test whether LAS expression is dependent on REV function, we have also analyzed LAS transcript accumulation in the rev-5 mutant. Comparison of vegetatively growing wild-type (Columbia) and rev-5 plants did not reveal any difference in the LAS expression pattern (data not shown). Taken together, the results of these expression studies demonstrate that during vegetative development, REV expression in developing axillary meristems is dependent on LAS function.
las is epistatic to axr1 with respect to lateral shoot development during the vegetative phase
The recessive Arabidopsis axr1 mutant shows a pleiotropic phenotype. Beside defects in root, hypocotyl, stamen, and stem elongation, one prominent aspect of its phenotype is an increased branching at maturity (Estelle and Somerville 1987; Lincoln et al. 1990; Stirnberg et al. 1999). All axr1 tissues examined showed reduced auxin sensitivity, suggesting that the AXR1 protein is required for auxin signaling (Abel et al. 1995; Timpte et al. 1995). Phenotypic analyses of loss-of-function mutants suggested that axr1 enhances shoot branching during both the vegetative and the reproductive phases of development (Stirnberg et al. 1999).
To test whether the enhanced shoot-branching potential conferred by the axr1 mutation was able to override the defect caused by las, we have constructed an axr1-12 las-4 double mutant. Homozygous axr1-12 las-4 plants showed the characteristic features of axr1 loss-of-function mutants: for example, reduction in plant height and alterations in leaf morphology (Fig. 8B). The potential for shoot branching was analyzed in a comparison of axr1-12 and axr1-12 las-4 plants. To enhance the outgrowth of lateral buds, primary bolts were removed when the plants reached a height of 10–15 cm. The experiment revealed that, with respect to shoot branching, the axr1-12 las-4 double mutant was similar to las-4: axillary shoot development was blocked in the older rosette leaf axils; however, lateral shoots developed from the axils of cauline leaves and at the topmost rosette leaves (Fig. 8A–D,G). This finding demonstrates that, with respect to lateral shoot development during the vegetative phase, las-4 is epistatic to axr1-12. Different from las-4, the zone of axillary shoot formation extended basipetally into the rosette. axr1-12 las-4 double mutants developed between zero and five shoots from the axils of the topmost rosette leaves (Fig. 8G), whereas only zero to two side shoots were observed in the rosette leaf axils of las-4 mutants (Fig. 2I). The above experiments were controlled by examining las-4 and wild-type populations grown in parallel under the same conditions.
Figure 8.
Phenotypic analysis of axr1-12 and axr1-12 las-4 plants. In axr1-12 (n = 33), multiple axillary shoots developed from the axils of rosette leaves (A,C,G), whereas axillary shoot development from rosette leaf axils was strongly reduced in axr1-12 las-4 double mutants (n = 33; B,D,G). The arrows point to the rosette leaf axils of axr1-12 (C) and axr1-12 las-4 (D) plants. In addition, axr1-12 plants developed accessory side shoots in almost every cauline leaf axil (arrow in E,H). This process was found to be blocked in most cauline leaf axils of the axr1-12 las-4 double mutants (arrow in F,H).
The enhanced shoot-branching potential of axr1 plants leads also to an increase in the formation of accessory side shoots from the axils of cauline leaves (Stirnberg et al. 1999). Comparing axr1-12 with axr1-12 las-4 plants, we observed a strong reduction in the development of accessory side shoots in the double mutant (Fig. 8E,F,H). This finding indicates that, despite the formation of lateral shoots observed in the cauline leaf axils of the las-4 mutant, the LAS gene has an influence on the shoot-branching potential during reproductive development. This conclusion is also supported by the pattern of LAS transcript accumulation.
Discussion
The Lateral suppressor gene (Ls) is a key regulator of shoot branching in tomato. In this study, we have identified the Ls-orthologous gene from Arabidopsis thaliana (LAS), based on its sequence identity with the tomato Ls gene and on microsynteny studies with the corresponding chromosome segments from tomato and Arabidopsis. The LAS gene is more closely related to the tomato Ls gene (50.5% at amino acid level) than to its closest relative from Arabidopsis (At3g03450, 32%). The LAS protein contains all the sequence motifs described for members of the GRAS family (Pysh et al. 1999), except for the DELLA domain that was found in GAI, RGA, and a few other members of this gene family.
To investigate the role of LAS during Arabidopsis development, mutants were identified that harbor En-1 transposable element insertions or stable mutations at different positions of the LAS ORF. las mutants exhibit a new phenotype that is characterized by the inability to form lateral shoots during vegetative development. Double-mutant analysis has shown that las-4 is epistatic to axr1 with respect to axillary shoot development during the vegetative phase.
LAS function is required for axillary shoot formation during vegetative development
In Arabidopsis, shoot branching is differently controlled during the vegetative and reproductive phases of development. Under prolonged short-day conditions, initiation of axillary meristems starts in the axils of the oldest leaves and then progresses toward younger leaf axils. Subsequently, axillary shoot development progresses in parallel with development of the subtending leaf, resulting in an acropetal gradient of lateral shoot formation (Hempel and Feldman 1994; Grbic and Bleecker 1996; Stirnberg et al. 1999, 2002). After the transition to flowering, axillary meristems are initiated evenly (Stirnberg et al. 1999, 2002) or in a basipetal sequence (Hempel and Feldman 1994; Grbic and Bleecker 2000) along the flowering stem. During this phase, bud outgrowth follows a basipetal gradient starting with the youngest leaf axils and progressing toward older leaf axils (Hempel and Feldman 1994; Grbic and Bleecker 1996; Stirnberg et al. 1999, 2002). Under ambient light conditions, both gradients may overlap with each other. Characterization of las mutants has shown that axillary meristems do not form during vegetative development. After the floral transition, however, axillary shoots develop from the axils of cauline leaves and sometimes from the topmost rosette leaves. This phenotype suggests that in Arabidopsis, the LAS gene is an essential component of the genetic mechanism governing the acropetal gradient of axillary meristem formation. On the other hand LAS function is not required for the initiation of primary axillary meristems during the reproductive phase of development. In this period, either its function is not needed or it is provided by a different gene, maybe a different member of the GRAS gene family.
LAS function is, however, not completely dispensable in the axils of cauline leaves. When comparing the number of accessory shoots in cauline leaf axils of axr1-12 and axr1-12 las-4 plants, we observed a strong reduction in the double mutant. In addition, in situ hybridization experiments revealed that LAS mRNA accumulates also in the axils of cauline leaves. After outgrowth of axillary buds, residual amounts of LAS transcripts have been detected at the base of axillary shoots. These findings suggest that LAS has an influence on the potential of cauline leaf axils to form lateral shoots, which is not reflected in the pattern of primary side-shoot development.
LAS is a marker for axil identity
LAS transcripts accumulate in the axils of all primordia derived from the SAM. During vegetative development, LAS transcripts can be detected in the axils of leaf primordia from P1 to P21 in a narrow domain extending 3–5 cell layers in the apical–basal and 1–2 cell layers in the adaxial–abaxial dimension. After floral transition, LAS mRNA was found in the axils of flower primordia and sepals, where the expression domains show similar dimensions. The pattern of LAS transcript accumulation in the axils of leaf primordia is similar to those of the genes LATERAL ORGAN BOUNDARIES (LOB), CUP-SHAPED COTYLEDON1 (CUC1), and CUP-SHAPED COTYLEDON2 (CUC2). Transcripts of all these genes are detected at the boundary of two tissues, the SAM and the new leaf primordium (Aida et al. 1999; Takada et al. 2001; Shuai et al. 2002). It is tempting to speculate that these expression domains are established at this boundary as a result of the influence of transcriptional regulators from both tissue types. The finding that transcripts of different genes accumulate in the axils of leaf primordia in very similar patterns suggests that the axillary tissue has a special identity. Based on the pattern of LAS transcript accumulation, one can assume that axil identity is established together with, or immediately after, the formation of the corresponding leaf primordium. Future experiments have to show whether or not LAS, LOB, CUC1, and CUC2 are related to each other in a hierarchical order of gene interactions.
In situ hybridization experiments with transverse sections demonstrated that LAS transcripts accumulate along the whole adaxial border of leaf primordia. However, axillary meristems are only initiated in the middle of this expression domain close to the center of the adaxial leaf base. This observation suggests that additional positional information is required to determine the specific position of lateral meristem initiation. The close proximity between the developing vascular elements and developing axillary meristems, combined with the observation that ectopic meristems form preferentially over underlying vascular bundles (Chuck et al. 1996; Takada et al. 2001), suggests that vascular bundles contribute to the induction of meristems.
LAS function is required to establish axillary meristems at a distance to the SAM
In Arabidopsis, the first lateral organ that is formed after the shift from short-day to long-day conditions is a flower primordium; cauline leaves develop from the youngest leaf primordia initiated before the floral transition (Hempel and Feldman 1994). With respect to the las mutant phenotype, this observation implies that the youngest leaf axils, which will develop axillary shoots in the absence of LAS function, are either predisposed for axillary meristem formation, or acquire this ability as a consequence of the floral transition. The fact that las-4 plants at different stages of shoot development respond in the same manner when shifted from short-day to long-day conditions indicates that lateral meristem induction is not affected by the number of leaves initiated before the transition. The zone of axillary meristem formation in las mutants may be defined by the range of a signal from the apical meristem or a region of competence of axillary cells extending several nodes from the SAM. Formally, the signal could either be an activator released from the inflorescence meristem or the loss of a repressor of lateral meristem initiation produced by the vegetative SAM. The latter would be very much reminiscent of the phenomenon of correlative inhibition of axillary bud outgrowth known to be exerted by a repressing signal from the shoot apex. In this context, it is interesting to note that in las-4 axr1-12 plants, the zone of side-shoot development is extended basipetally into the rosette. In this double mutant, the zone of axillary meristem formation may be larger because of a modified signal from the SAM or of an alteration in competence of axillary cells. The above observation also indicates that the AXR1 gene does not only repress lateral bud outgrowth but may also negatively control the initiation of axillary meristems. In summary, the youngest leaf axils of vegetatively growing Arabidopsis plants have always the potential to form axillary shoots independent of the LAS function. This potential is realized when the vegetative SAM undergoes the transition into an inflorescence meristem.
It is still a matter of debate, whether axillary meristems are detached from the SAM of the main shoot or develop de novo from fully or partially differentiated cells (Steeves and Sussex 1989; Long and Barton 2000). Because this study demonstrates that axillary meristems develop from LAS-positive cell groups, LAS transcript accumulation in leaf axils can be considered as a marker for meristem formation competence. Comparisons of the patterns of LAS and STM transcript accumulation in leaf axils revealed that LAS-positive cells are also positive for STM mRNA, until the time point when the STM signal is focused to the center of the leaf axil (from P16 in Columbia) and REV expression is turned on. The coexpression of LAS and STM in the axils of P1 to P15 primordia suggests that the identity of axillary meristem progenitor cells is determined together with or shortly after the initiation of the corresponding leaf primordium and that the meristematic character of these cells is retained, whereas neighboring cells lose their meristematic identity. These findings support the “detached meristem” concept. However, meristematic identity by itself, as revealed by STM mRNA accumulation, is not always sufficient to allow axillary meristem initiation, because in the las-4 mutant many STM-positive axils do not develop axillary meristems. During the vegetative phase of development, an additional function, which requires the LAS protein, is needed to initiate axillary meristems. The view that an additional function is required to trigger the formation of axillary meristems at a distance to the SAM supports the argument in favor of a de novo formation of axillary meristems. Taken together, these results suggest that the process of axillary meristem formation during Arabidopsis vegetative development shows characteristics of both, the “detached meristem” and the “de novo formation” concepts.
LATERAL SUPPRESSOR function is conserved during evolution
Microsynteny analysis between the chromosomal segments harboring the LATERAL SUPPRESSOR genes in tomato and Arabidopsis has demonstrated that both genes are orthologous (Rossberg et al. 2001). However, the LATERAL SUPPRESSOR proteins of both species show only 50.5% identity. The defects observed in the corresponding loss-of-function mutants display both similarities and differences. In both cases, side-shoot development is abolished during the vegetative phase of development and not affected during the reproductive phase. Tomato ls mutants develop lateral shoots only in the two leaf axils preceding an inflorescence, whereas the number of leaf axils with lateral shoots in Arabidopsis las-4 plants is ∼3–13, depending on growing conditions. This difference in the length of the zone, where axillary shoots are formed, may be explained by the more elongated stature of the tomato stem during vegetative development or by the slower leaf initiation rate in tomato. In addition, tomato ls mutants fail to develop petals on the flower and are characterized by a reduced male and female fertility. Such defects have not been observed in Arabidopsis plants homozygous for LAS loss-of-function alleles. In this study, we have demonstrated that the Arabidopsis LAS gene can fully complement the tomato lateral suppressor mutant. This result demonstrates that both genes are functionally equivalent and that an essential control mechanism in the process of side-shoot development is conserved between two distantly related species, tomato and Arabidopsis. Differences in the mutant phenotypes of both species may be explained by redundant gene activities during flower development in Arabidopsis leading to a masking of a possible mutant phenotype.
Materials and methods
Plant materials
Tomato seed material of Lycopersicon esculentum L. cv. Antimold B-ls1 (LA0329) was obtained from the Tomato Genetics Resource Center, Davis, CA. Seeds of Arabidopsis thaliana (Heynh.), ecotypes Columbia (Col-O) and Landsberg erecta, were obtained from R. Schmitt, Max-Delbrück-Laboratorium, Köln, Germany. Tomato plants were grown under standard glasshouse conditions with additional artificial light (16-h photoperiod) during the winter period. For cultivation under short-day conditions, Arabidopsis plants were grown in a controlled environment room with 8 h of light (16 h of dark), 20°C–25°C day temperature, and 10°C–15°C night temperature. Short-day-grown plants were induced to flower after 24 or 26 d by successive 16-h photoperiods. Cultivation under long-day conditions was done in the greenhouse with additional artificial light when needed.
Isolation of las mutants
LAS∷En insertion lines were identified in an En-1-mutagenized population of Arabidopsis plants (Wisman et al. 1998) following the protocol of Baumann et al. (1998). PCR screens were performed using the gene-specific primers LSEN2 (5′-GTAC GAATCAACCGTCAGCAACAAG-3′) and LSEN3 (5′-GCCA CAACATCCAAAATCTCCTTACC-3′) in combination with the En-1-specific primer En205 (Baumann et al. 1998). Candidate lines were grown first under short-day conditions (e.g., 26 d), then shifted to long days and inspected for alterations in their shoot-branching pattern. To obtain stable mutant alleles, the En-1 insertion lines were outcrossed to the Columbia (Col) wild type, and progeny plants were screened by PCR for loss of the En-1 element. Candidates that had lost the En-1 element were self-pollinated, and their progenies were analyzed for segregation of the mutant phenotype and sequence alterations within the LAS gene. Mutant alleles were back-crossed six times into the Columbia genetic background, without noticeable changes in phenotypic expression. For phenotypic analyses we used las-4 lines selected after at least three back-crosses into the Columbia background.
DNA isolation
Plant DNA preparation and Southern blot analysis have been described (Brandstädter et al. 1993). Standard techniques were carried out according to Sambrook and Russell (2001).
Isolation of the Arabidopsis LAS gene
Alignment of the tomato Ls amino acid sequence with the related A. thaliana proteins GAI (Peng et al. 1997), RGA (Silverstone et al. 1998), SCARECROW (Di Laurenzio et al. 1996), and SCL13 (Pysh et al. 1999) revealed regions of high and low sequence conservation. Degenerated primers deduced from different regions were tested in PCR experiments under reduced stringency conditions on Arabidopsis genomic DNA. A fragment of ∼700 bp could be amplified with the primer combination CD61-38 (5′-CARTGGCCNCCNYTNATGCA-3′) and CD61-41 (5′-TGRTTYTGCCANCCNARRAA-3′). The PCR reaction was performed using the following regime: 95°C for 30 sec, 50°C for 1 min, 72°C for 1 min (35 times). This PCR product was used to isolate a clone from a genomic Arabidopsis cosmid library by colony hybridization.
RNA isolation, reverse transcription (RT)-PCR Analysis, and rapid amplification of cDNA ends (RACE)
Total RNA was isolated using the RNeasy plant mini kit (QIAGEN) following the manufacturer's instructions. For RT–PCR analysis, 1 μg of total RNA was reverse transcribed using the Superscript II polymerase (Life Technologies) according to the manufacturer's instructions. The product of the first-strand cDNA synthesis reaction was amplified by PCR using the LAS-specific primers AtLs2949F (5′-GTCTCCAATTCCAGTTTCA CAC-3′) and AtLs3532R (5′-TTGCCAGCCAAGAAACAAAG-3′). Amplification of actin cDNA by using the primers ActinATfor (5′-TGGTGTCATGGTTGGGATG-3′) and ActinATrev (5′-CACCACTGAGCACAATGTTAC-3′) was performed as a control to ensure that equal amounts of cDNA were added to each PCR reaction.
RACE experiments were performed using the RACE system of Life Technologies according to the manufacturer's instructions. For 5′ RACE, oligo(dT)-primed cDNA was tailed and amplified using the general amplification primers and the LAS-specific primers AtLs2717R (5′-CCGTTAAATGACCGAAC CGA-3′), AtLs2618R (5′-ACTGTTCTTTGCATACACTGCT-3′), and AtLs2535R (5′-GCCGTCTGATCTTGTTGCT-3′).
For 3′ RACE, first-strand cDNA was amplified using the general amplification primer and the LAS-specific primers AtLs2858F (5′-CCTCCATCTCTCCGCATAAC-3′), AtLs2949F (5′-GTCTCCAATTCCAGTTTCACAC-3′), and AtLs3339F (5′-CGGAAGAGACGGAGAGAAAG-3′). Amplified fragments were cloned into the pGEM-T (Promega) plasmid vector and sequenced.
Tomato transformation
For complementation experiments, a 6.3-kb NheI–BanI fragment of the genomic cosmid clone harboring the Arabidopsis LAS gene was cloned into the binary vector pGPTV-Kan (C28.1; Becker et al. 1992) and transferred into the Agrobacterium tumefaciens strain GV3101 (Koncz and Schell 1986). Transformation of tomato leaf explants was done as described previously (Knapp et al. 1994).
RNA in situ hybridizations
Sample preparations and in situ hybridizations of 8-μm sections were done as described by Coen et al. (1990) with slight modifications. Tween 20 (0.03%) was added to the fixative, and dewatering of the fixed material was done without NaCl. LAS probes contained the nucleotides 2–1348, REV probes the nucleotides 682–2611, in both cases relative to the ATG. STM probes included base pairs 78–1122 of the ORF. The sequences were cloned into pGEM vectors in sense and antisense orientation, respectively, relative to the T7 promoter. Linearized plasmids were used as templates for probe synthesis with T7 RNA polymerase. The time for probe hydrolysis was calculated for an average length of 150 nucleotides as described (Jackson 1992). After the color reaction, slides were mounted in 30% glycerol and photographed using differential interference contrast microscopy.
DNA sequencing and analysis
DNA sequences were determined by the DNAcore facility of the Max-Planck-Institut für Züchtungsforschung (Cologne, Germany) on Applied Biosystems (Weiterstadt, Germany) Abi Prism 377 and 3700 sequencers using BigDye-terminator chemistry. Premixed reagents were from Applied Biosystems. Oligonucleotides were purchased from Life Technologies or Metabion. Analysis of sequences was performed using the Wisconsin Package (version 10.0-UNIX; Genetics Computer Group). Database searches were performed using BLAST programs (Altschul et al. 1990) against NCBI sequence databases.
Acknowledgments
The authors thank Luca Comai, the Nottingham Arabidopsis Stock Center, and the Tomato Genetics Resource Center for providing seed stocks. We thank E. Wissmann for the En lines, R. Schmidt for the cosmid clone, and the ADIS group for DNA sequencing. We are grateful to E. Tillmann for excellent technical assistance. We thank G. Coupland, H.M.O. Leyser, and F. Salamini for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 572 of the University of Cologne and the Schwerpunktprogramm 1005.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL theres@mpiz-koeln.mpg.de; FAX 49-221-5062 413.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.260703.
References
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baumann E, Lewald J, Saedler H, Schulz B, Wisman E. Successful PCR-based reverse genetic screen using an En-1-mutagenised Arabidopsis thaliana population generated via single-seed descent. Theor Appl Genet. 1998;97:729–734. [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Brandstädter J, Rossbach C, Theres K. The pattern of histone H4 expression in the tomato shoot apex changes during development. Planta. 1993;192:69–74. doi: 10.1007/BF00198694. [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG. Concepts and terminology of apical dominance. Amer J Bot. 1997;84:1064–1069. [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Fledmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- Grbic B, Bleecker AB. An altered body plan is conferred on Arabidopsis plants carrying dominant alleles of two genes. Development. 1996;122:2395–2403. doi: 10.1242/dev.122.8.2395. [DOI] [PubMed] [Google Scholar]
- ————— Axillary meristem development in Arabidopsis thaliana. Plant J. 2000;21:215–223. doi: 10.1046/j.1365-313x.2000.00670.x. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Keizer LCP, De Ruiter W, Dons JJM. Seed and fruit set of the lateral suppressor mutant of tomato. Sci Horti. 1994;59:157–162. [Google Scholar]
- Hempel FD, Feldman LJ. Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta. 1994;192:276–286. [Google Scholar]
- Jackson DP. In situ hybridisation in plants. In: Bowles DJ, et al., editors. Molecular plant pathology: A practical approach. Oxford, UK: Oxford University Press; 1992. pp. 163–174. [Google Scholar]
- Knapp S, Larondelle Y, Rossberg M, Furtek D, Theres K. Transgenic tomato lines containing Ds elements at defined genomic positions as tools for targeted transposon tagging. Mol Gen Genet. 1994;243:666–673. doi: 10.1007/BF00279576. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promotor of T-L DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Dev Biol. 2000;218:341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Malayer JC, Guard AT. A comparative developmental study of the mutant sideshootless and normal tomato plants. Amer J Bot. 1964;51:140–143. [Google Scholar]
- McConnell JR, Barton MK. Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristem. Dev Genet. 1995;16:358–366. [Google Scholar]
- ————— Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Napoli C. Highly branched phenotype of the Petunia dad1-1 mutant is reversed by grafting. Plant Physiol. 1996;111:27–37. doi: 10.1104/pp.111.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC. Reevaluation concepts of apical dominance and the control of axillary bud outgrowth. Curr Top Dev Biol. 1999;4:127–169. doi: 10.1016/s0070-2153(08)60469-x. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes & Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM, Butler L. Cytogenetics of the tomato. Adv Genet. 1956;8:267–382. [Google Scholar]
- Rossberg M, Theres K, Acarkan A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R. Comparative sequence analysis reveals extensive microcolinearity in the LATERAL SUPPRESSOR regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell. 2001;13:979–988. doi: 10.1105/tpc.13.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schmitz G, Theres K. Genetic control of branching in Arabidopsis and tomato. Curr Opin Plant Biol. 1999;2:51–55. doi: 10.1016/s1369-5266(99)80010-7. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Ganal M, Theres K. Genetic and physical mapping of the Lateral suppressor (Ls) locus in tomato. Mol Gen Genet. 1995;246:761–766. doi: 10.1007/BF00290724. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. 2nd ed. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Stirnberg P, Chatfield SP, Leyser HMO. AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 1999;121:839–847. doi: 10.1104/pp.121.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HMO. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129:1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON 1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Paris DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JWH, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes & Dev. 2001;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Williams W. The effect of selection on the manifold expression of the ‘suppressed lateral’ gene in the tomato. Heredity. 1960;14:285–296. [Google Scholar]
- Wisman E, Cardon GH, Fransz P, Saedler H. The behaviour of the autonomous maize transposable element En/Spm in Arabidopsis thaliana allows efficient mutagenesis. Plant Mol Biol. 1998;37:989–999. doi: 10.1023/a:1006082009151. [DOI] [PubMed] [Google Scholar]