Abstract
Initiation factor 3 (eIF3) forms a multifactor complex (MFC) with eIF1, eIF2, and eIF5 that stimulates Met-tRNAiMet binding to 40S ribosomes and promotes scanning or AUG recognition. We have previously characterized MFC subcomplexes produced in vivo from affinity-tagged eIF3 subunits lacking discrete binding domains for other MFC components. Here we asked whether these subcomplexes can bind to 40S ribosomes in vivo. We found that the N- and C-terminal domains of NIP1/eIF3c, the N- and C-terminal domains of TIF32/eIF3a, and eIF5 have critical functions in 40S binding, with eIF5 and the TIF32-CTD performing redundant functions. The TIF32-CTD interacted in vitro with helices 16–18 of domain I in 18S rRNA, and the TIF32-NTD and NIP1 interacted with 40S protein RPS0A. These results suggest that eIF3 binds to the solvent side of the 40S subunit in a way that provides access to the interface side for the two eIF3 segments (NIP1-NTD and TIF32-CTD) that interact with eIF1, eIF5, and the eIF2/GTP/Met-tRNAiMet ternary complex.
Keywords: Eukaryotic translation initiation factor (eIF), multifactor complex (MFC), translational control, protein synthesis, 40S ribosome binding, TIF32/NIP1
Supplemental material is available at http://www.genesdev.org.
Formation of the 80S translation initiation complex is a multiple-step process involving many soluble eukaryotic initiation factors (eIFs). According to present models, the ternary complex (TC), comprised of eIF2, GTP, and Met-tRNAiMet, binds to the 40S ribosome with the help of eIF1, eIF1A, and eIF3. The 43S preinitiation complex thus formed interacts with mRNA in a manner stimulated by eIF4F, poly(A)-binding protein, and eIF3, and the resulting 48S complex scans the mRNA until the Met-tRNAiMet base pairs with the AUG start codon. On AUG recognition, eIF5 stimulates GTP hydrolysis by eIF2, the eIFs are ejected, and the resulting 40S–Met-tRNAiMet–mRNA complex joins with the 60S subunit. For a new round of initiation, the ejected eIF2–GDP complex must be recycled to eIF2–GTP by the exchange factor eIF2B (for reviews, see Hershey and Merrick 2000; Hinnebusch 2000).
Biochemical data obtained in the 1970s suggested that mammalian eIF3 binds first to the 40S ribosome independently of the other factors and subsequently promotes TC and mRNA binding to the small ribosomal subunit (Hershey and Merrick 2000; Hinnebusch 2000). Whereas mammalian eIF3 contains 11 different subunits (eIF3a–eIF3k, or 3a–3k), the yeast factor has only five core subunits (TIF32/3a, PRT1/3b, NIP1/3c, TIF34/3i, and TIF35/3g) and one substoichiometric component (HCR1/3j). The five-subunit complex purified from yeast can restore binding of Met-tRNAiMet (Danaie et al. 1995; Phan et al. 1998) and mRNA (Phan et al. 2001) to 40S ribosomes in heat-inactivated prt1-1 (eIF3b) mutant extracts. Thus, yeast eIF3 possesses two critical functions ascribed to the more complex mammalian factor. However, our finding that yeast eIF3 forms a multifactor complex (MFC) with eIFs 1, 2, and 5 and Met-tRNAiMet that can exist free of ribosomes in vivo suggested that all of these factors may bind to the 40S subunit as a preformed unit (Asano et al. 2000).
Our laboratory and others recently completed a comprehensive analysis of subunit interactions in the MFC using a combination of two-hybrid analysis, in vitro binding assays, and the purification of MFC subcomplexes formed in vivo by affinity-tagged eIF3 subunits lacking various predicted binding domains (Verlhac et al. 1997; Asano et al. 1998, 1999, 2000; Phan et al. 1998; Vornlocher et al. 1999; Valášek et al. 2001b, 2002). The results of these studies (summarized in Fig. 1H) suggest that each of the three largest subunits of eIF3 (TIF32/3a, NIP1/3c, and PRT1/3b) has a binding site for the other two subunits, and that the extreme C-terminal domain (CTD) of PRT1 additionally interacts with eIF3 subunits TIF34/3i and TIF35/3g. HCR1/3j binds simultaneously to both the N-terminal domain (NTD) of PRT1 and the TIF32-CTD. eIF1 is tethered to the MFC through interactions with the TIF32-CTD and NIP1-NTD. In addition to eIF1, the NIP1-NTD also binds to the CTD of eIF5. The β-subunit of eIF2 makes two critical contacts with eIF3, a direct interaction with the extreme CTD of TIF32 and an indirect association with the NIP1-NTD via the eIF5-CTD (Fig. 1H).
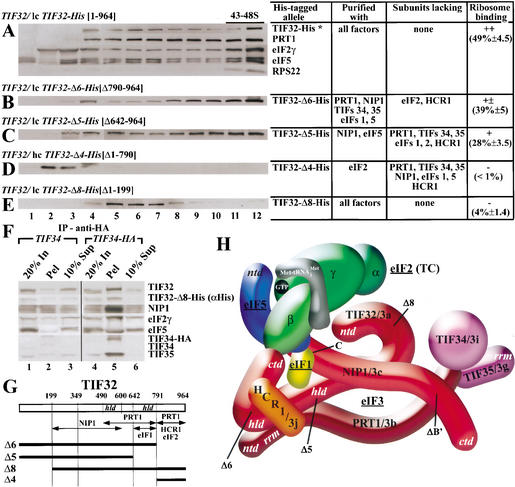
Figure 1.
The TIF32-NTD is required for association of the MFC with 40S ribosomes in vivo and identification of the MBU. (A–E) Transformants of wild-type strain W303 containing low-copy (lc) plasmid pRSTIF32-His (A), lc pRSTIF32-Δ6-His (B), lc pRSTIF32-Δ5-His (C), high-copy (hc) YEpTIF32-Δ4-His (D), and lc pRSTIF32-Δ8-His (E) were grown in SD medium containing minimal requirements to an OD600 of ∼1.5, and 50 μg/mL cycloheximide was added 5 min prior to harvesting. Whole-cell extracts (WCEs) were prepared and separated by velocity sedimentation on 7.5%–30% sucrose gradients for 5 h at 41,000 rpm. Nineteen gradient fractions were collected, and the first 12, which contained more than one-half of the total amount of eIF3 (data not shown), were resolved by SDS-PAGE, followed by immunoblot analysis using antibodies against the proteins indicated immediately to the right of each panel. The presence of 40S ribosomes was detected by the A254 profile and by probing for RPS22. For each construct, the deleted TIF32 amino acids are indicated in brackets next to the construct name. The table to the right of panels A–E indicates which components of the MFC copurified (column 2), and which did not (column 3), with the relevant His8-tagged proteins (column 1; Valášek et al. 2002). Column 4 lists the mean proportions (with standard deviations) of the total tagged proteins found in fractions 11 and 12 of the gradients calculated from quantification of Western data from two independent experiments. High-copy plasmids bearing TIF32-His alleles were used when the mutant proteins were expressed at low levels relative to full-length TIF32-His expressed from a single-copy plasmid. (F) Coimmunoprecipitation of TIF32-Δ8-His with other MFC components. WCEs were prepared from TIF34 strain H1485 and TIF34-HA strain H2768 transformed with single-copy plasmid YCpTIF32-Δ8-His-U, and aliquots containing 500 μg of protein were immunoprecipitated with mouse monoclonal anti-HA antibodies (12CA5). Immune complexes were analyzed by Western blotting using antibodies against the proteins indicated to the right. (Lanes 1,4) Twenty percent of the input WCE used for immunoprecipitation (20% In). (Lanes 2,5) The entire immunoprecipitated fraction (Pel). (Lanes 3,6) Ten percent of the supernatant fractions from the immunoprecipitations (10% Sup). (G) Schematic of TIF32 with arrows delimiting the minimal binding domains for the indicated proteins identified previously. The lines beneath the schematic depict the various truncated TIF32-His proteins (designated on the left) that were analyzed in panels A–F. (H) A 3D model of the MFC based on a comprehensive analysis of subunit interactions (Valášek et al. 2002). The labeled protein subunits are shown roughly in proportion to their molecular weights. The degree of overlap between two different subunits depicts the extent of their interacting surfaces. The boundaries of the relevant truncations (summarized in Figs. 1G, 2F) are indicated at the appropriate positions in TIF32 and NIP1. See text for further details. ntd, N-terminal domain; ctd, C-terminal domain; hld, HCR1-like domain; rrm, RNA recognition motif.
The stable association of eIFs 1, 3, and 5 and TC in the MFC suggests that the different functions of these factors are physically linked. eIF1, eIF2, and eIF5 were implicated in selecting AUG as the start codon (Yoon and Donahue 1992; Huang et al. 1997; Pestova et al. 1998; Pestova and Kolupaeva 2002), and their related functions in this process may be coordinated by mutual association with the NIP1-NTD and TIF32-CTD of eIF3 (Asano et al. 2000; Valášek et al. 2002). Association between eIF2 and eIF3 in the MFC could underlie the ability of eIF3 to stimulate TC binding to the 40S ribosome. Indeed, we found that sequestering eIF2 in a defective subcomplex comprised of NIP1-NTD, eIF1, and eIF5 produced a Gcd− phenotype in yeast, indicating a reduced rate of TC binding to 40S subunits during reinitiation on GCN4 mRNA (Valášek et al. 2002). These data suggested that incorporation of TC into the MFC facilitates its binding to 40S subunits in vivo (Valášek et al. 2002). Consistently, a cluster of alanine substitutions in the eIF5-CTD (tif5–7A) that disrupts the indirect contact between eIF2 and eIF3 in the MFC reduced the binding of TC to 40S subunits in vitro (Asano et al. 2001). In tif5–7A cells, however, the diminished rate of translation initiation was accompanied by accumulation of 48S complexes containing eIF1, eIF2, and eIF3 and lacking only eIF5 (Asano et al. 2000). Even disrupting the direct eIF2–eIF3 contact in the MFC by overexpressing CTD-less TIF32 protein (TIF32-Δ6) did not decrease the amount of TC bound to 40S subunits, although it exacerbated the translation defect in tif5–7A cells (Valášek et al. 2002). Hence, the multiple contacts linking eIF2 and eIF3 in the MFC seem to be most critical for a step(s) downstream of TC binding, including scanning, AUG recognition, or GTP hydrolysis by eIF2.
Having demonstrated that several steps in the initiation pathway are stimulated by formation of the MFC, we sought to identify the minimal requirements for association of the MFC with the 40S ribosome. We show here that NIP1, the TIF32-NTD, and eIF5 comprise a minimal 40S-ribosome-binding unit (MBU) that is sufficient for 40S binding in vivo. Consistently, the TIF32-NTD and NIP1 interacted in two-hybrid and in vitro binding assays with the small ribosomal subunit protein RPS0A located on the solvent side of the 40S subunit (Spahn et al. 2001a). We also detected specific interaction in vitro between the TIF32-CTD and a short segment from domain I of 18S rRNA that may provide eIF3 with access to the 60S-interface side of the 40S subunit. We conclude that TIF32, NIP1, and eIF5 have key functions not only in connecting eIF3 to TC in the MFC, but also in anchoring the MFC to the 40S ribosome.
Results
The extreme N terminus of TIF32 is essential for association of the MFC with 40S ribosomes
We recently established a protein linkage map between subunits of eIF3 and eIFs 1, 2, and 5 in the MFC (Fig. 1H; Valášek et al. 2002). By individually His8-tagging the three largest subunits of eIF3 and deleting predicted binding domains in the tagged subunits, we could affinity-purify subcomplexes containing only the MFC components that were associated with the mutant tagged subunits. Here we investigated whether these subcomplexes can interact with 40S ribosomes in vivo as a means of identifying the minimal requirements for MFC binding to the 40S ribosome. Using this approach, we showed previously that expression of a truncated form of PRT1 lacking the N-terminal RRM sequestered TIF34 and TIF35 in a defective subcomplex that could not stably associate with 40S ribosomes (Valášek et al. 2001b). This finding suggested that TIF32 and NIP1 play crucial roles in 40S binding.
We tested this prediction by first analyzing subcomplexes formed by His8-tagged TIF32 proteins truncated at the N or C terminus (Fig. 1G). Whole-cell extracts (WCEs) from wild-type strains expressing the mutant TIF32 proteins from low- or high-copy plasmids were resolved on sucrose density gradients and probed by Western blotting for the tagged proteins and other MFC components. In extracts from all strains, a significant fraction of PRT1, eIF2γ, and eIF5 were found in the fractions (11–12) that contain the 43–48S initiation complexes, as judged by the presence of 40S subunit protein RPS22 (Fig. 1A; data not shown). As expected, full-length TIF32-His also peaked in fraction 12, ensuring that the His8 tag does not interfere with association of TIF32 with 40S ribosomes (Fig. 1A). Based on quantification of the Western signals, we calculated that ∼49% of the TIF32-His in these gradient fractions was associated with 40S ribosomes (Fig. 1A).
We showed previously that removal of the extreme C terminus from TIF32 (in the mutant allele TIF32-Δ6-His; Fig. 1G) results in dissociation of eIF2 and HCR1 from the MFC (Supplementary Fig. 1A; Valášek et al. 2002). However, this CTD-less TIF32 mutant showed strong 40S binding, suggesting that the TIF32-CTD, eIF2, and HCR1 are largely dispensable for ribosome binding by the remaining MFC components associated with TIF32-Δ6-His (Fig. 1B). A high level of 40S binding also was observed for the larger C-terminal truncation in TIF32-Δ5-His (Fig. 1C), which forms a subcomplex containing only NIP1 and eIF5 (Supplementary Fig. 1A; Fig. 3F, lanes 3,4 below; Valášek et al. 2002). However, a reduced proportion of the TIF32-Δ5-His protein cosedimented with the 40S ribosomes compared with that seen for TIF32-His (28% vs. 49%; Fig. 1C vs. 1A). Thus, removing residues 642–964 from the C terminus of TIF32, thereby disrupting its association with PRT1, TIF34, TIF35, eIF1, and eIF2, seems to weaken but does not abolish 40S binding by the remaining subcomplex comprised of TIF32-Δ5-His, NIP1, and eIF5. Based on results shown below, this subcomplex will be referred to as the minimal 40S-binding unit (MBU).
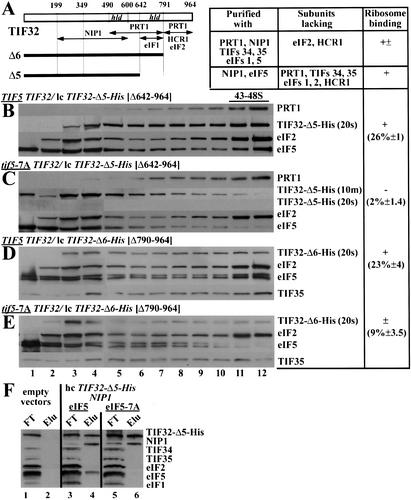
Figure 3.
Evidence that eIF5 and the TIF32-CTD have redundant functions in association of the MFC with 40S ribosomes in vivo. (A) Same as Figure 1G. (B–E) Western blot analysis of sucrose gradient fractions, conducted and evaluated as described in Figure 1 except that WCEs were prepared from the wild-type TIF5 (H2898; B,D) and tif5-7A (H2899; C,E) strains bearing lc pRSTIF32-Δ5-His (B,C) and lc pRSTIF32-Δ6-His (D,E). The membrane probed with anti-His antibodies is shown with different exposure times in C. (F) The eIF5-7A mutation does not affect the interaction between the TIF32-NTD and NIP1. WCEs were prepared from strains H2896 (lanes 1,2), YLVMBU (lanes 3,4), and YLVMBU-7A (lanes 5,6), incubated overnight with Ni2+-NTA-silica resin, and the bound proteins were eluted and subjected to Western blot analysis. Lanes 1, 3, and 5 contained 3% of the flow-through fractions (FT); lanes 2, 4, and 6 contained 3% of the total volume of the corresponding eluted fractions (Elu).
Deleting the extreme N terminus of TIF32 in TIF32-Δ8-His did not greatly reduce the yield of any copurifying MFC components (Supplementary Fig. 1A; Valášek et al. 2002). As shown in Figure 1E, however, the majority of TIF32-Δ8-His was found in fractions 5–7, most likely corresponding to intact MFC, and only a trace of the mutant protein cosedimented with the 40S ribosome. To verify that most of the TIF32-Δ8-His protein is associated with other MFC components, WCE prepared from a TIF34-HA strain harboring TIF32-Δ8-His was immunoprecipitated with anti-HA antibodies, and the immune complexes were probed with antibodies against eIF3 subunits, eIF5 and eIF2γ. As expected, the anti-HA antibodies immunoprecipitated a large fraction (∼90%) of TIF34-HA, TIF35, and NIP1 (all eIF3 subunits) and ∼30% of eIF5 and eIF2γ (Fig. 1F, lanes 4–6). Importantly, similar amounts (∼50%) of the native TIF32 and TIF32-Δ8-His proteins, which must compete for incorporation into eIF3, coimmunoprecipitated with TIF34-HA. In a control experiment, only a small fraction of all factors were immunoprecipitated nonspecifically with anti-HA antibodies from WCE containing untagged TIF34 (Fig. 1F, lanes 1–3). Taken together, the data in Figures 1E and F strongly suggest that the TIF32-NTD plays a crucial role in association of the MFC with the 40S ribosomes. Because its removal had a minimal impact on MFC composition, the TIF32-NTD may interact directly with the 40S ribosome.
The extreme C-terminal fragment of TIF32 (in TIF32-Δ4-His), previously shown to bind only eIF2 in vivo (Supplementary Fig. 1B; Valášek et al. 2002), accumulated at the top of the gradient in fractions 2–4 (Fig. 1D). The fact that none of the TIF32-Δ4-His protein was associated with ribosomes may indicate that interaction with eIF2 alone is not sufficient for 40S binding in vivo. Alternatively, it is possible that the TIF32-Δ4-His association with eIF2 is too weak to withstand centrifugation through a sucrose gradient.
NIP1/3c plays a critical role in 40S ribosome association
We proceeded next to examine the role of NIP1 in 40S ribosome binding by the MFC. As expected, full-length NIP1-His peaked in fractions 11 and 12 containing the 43–48S initiation complexes (Fig. 2A). In sharp contrast, removal of N-terminal residues 1–156 led to accumulation of nearly all of the resulting NIP1-C-His protein in fraction 4, and only a trace amount in fractions 11 and 12 (Fig. 2B). We have demonstrated that this deletion of the NIP1-NTD abolishes association of eIF3 with eIFs 1, 2, and 5 without significantly destabilizing the eIF3 complex (Supplementary Fig. 1C; Valášek et al. 2002). Consistent with these findings, we observed an approximately threefold increase in the proportion of eIF3 subunits, but not eIFs 1, 2, and 5, that cosedimented with NIP1-C-His in fraction 4 compared with that seen in the same fraction with full-length NIP1-His (Fig. 2, cf. B and A). Thus, the NIP1-C-His protein sequesters each of the eIF3 subunits in a nonribosomal complex lacking eIFs 1, 2, and 5. These results indicate that the NIP1-NTD, and possibly one or more of its binding partners eIFs 1, 5, and 2, is essential for MFC association with 40S ribosomes.
Figure 2.
NIP1 plays a critical role in 40S binding by the MFC. (A–E) Western blot analysis of the sucrose gradient fractions conducted and evaluated as described in Figure 1, except that WCEs were prepared from W303 transformants bearing lc YCpNIP1-His (A), lc YCpNIP1-C-His (B), lc YCpNIP1-N‘-His (C), hc YEpNIP1-ΔA-His (D), and hc YEpNIP1-ΔB‘-His (E). Black rectangles enclose gradient fraction 4 in which the distributions of NIP1-C-His, TIF32, PRT1, TIF34, and TIF35 differ substantially between the gradients in B versus A, as quantified in the last column of the table to the right of the blots. (F) Same as Figure 1G except that truncations in NIP1-His are under study.
Truncating NIP1-His from the C terminus after residue 370 (in NIP1-ΔA-His) was shown previously to dissociate eIF3 subunits PRT1, TIF34, and TIF35 from the purified MFC (Supplementary Fig. 1C; Valášek et al. 2002). As shown in Figure 2D, the residual MFC complex formed by NIP1-ΔA-His sedimented almost exclusively in fractions 4 and 5, free of 40S ribosomes. As would be expected from the absence of PRT1, TIF34, and TIF35, the sedimentation rate of the NIP1-ΔA-His subcomplex is slightly less than that of the intact MFC formed by TIF32-Δ8-His (Fig. 1E). A smaller truncation of NIP1-His after residue 571 (in NIP1-ΔB‘-His) did not destabilize the MFC (Supplementary Fig. 1C; Valášek et al. 2002), but it substantially reduced 40S binding of the resulting mutant protein, with the majority sedimenting at the same rate as the intact MFC formed by TIF32-Δ8-His (cf. Figs. 2E and 1E). Thus, the C-terminal one-third of NIP1 (lacking in NIP1-ΔB‘-His) may directly interact with the 40S subunit.
The His8-tagged segment containing only the N-terminal 205 residues of NIP1 (in NIP1-N‘-His) was shown previously to copurify with only eIFs 1, 2, and 5 (Supplementary Fig. 1D; Valášek et al. 2002). Its overexpression conferred a Gcd− phenotype that was suppressed by overproducing the TC, and strongly exacerbated by simultaneous overexpression of eIF1 and eIF5. Hence, we concluded that the NIP1-NTD–eIF1–eIF2–eIF5 subcomplex is defective for 40S binding. Consistent with this interpretation, we found that NIP1-N‘-His sedimented at the top of the gradient in fractions 2–4 (Fig. 2C).
Evidence that eIF5 plays an important role in association of the MFC with the 40S subunit
The MBU consists of TIF32-Δ5-His, NIP1, and eIF5 (Fig. 1C). To determine whether eIF5 actively contributes to 40S binding by the MBU, we took advantage of the tif5–7A allele containing a cluster of alanine substitutions in the eIF5-CTD. These mutations disrupted eIF5 interactions with NIP1 and eIF2β in vitro and destabilized the MFC in vivo (Asano et al. 2000). We introduced TIF32-Δ5-His into isogenic TIF5 and tif5–7A strains and measured the amount of TIF32-Δ5-His that bound to 40S subunits, as described above. TIF32-Δ5-His showed substantial 40S binding in the TIF5 extract (Fig. 3B, lanes 11,12), in accordance with the findings in Figure 1C for a different wild-type strain. In contrast, TIF32-Δ5-His showed little 40S binding in the tif5–7A mutant, with the majority of the mutant protein detected in fractions 3–4 (Fig. 3C). In agreement with previous results (Asano et al. 2001), the tif5–7A mutation eliminated 40S binding of eIF5 itself (Fig. 3C), indicating that interaction of the eIF5-CTD with eIF2β and NIP1 is required to anchor eIF5 to 43–48S complexes. Whereas the loss of mutant eIF5 from 43–48S complexes does not lead to dissociation of wild-type eIF3 from 40S ribosomes (Fig. 3C, PRT1 panel), wild-type eIF5 is clearly required for 40S binding by the TIF32-Δ5-His–NIP1–eIF5 subcomplex (Fig. 3C, TIF32-Δ5-His panel).
To eliminate the possibility that dissociation of eIF5–7A from NIP1 would also dissociate NIP1 from TIF32-Δ5-His and thereby destroy 40S binding by TIF32-Δ5-His, we overexpressed TIF32-Δ5-His and NIP1 in combination with either eIF5 or eIF5–7A and purified TIF32-Δ5-His-containing subcomplexes on nickel silica resin. As expected, both NIP1 and wild-type eIF5 copurified with TIF32-Δ5-His independently of other MFC components. The tif5–7A mutation dissociated eIF5 from TIF32-Δ5-His, but did not reduce binding of the latter to NIP1 (Fig. 3F, lanes 4,6).
It should be noted that the TIF32-Δ5-His protein is much less stable in tif5–7A versus TIF5 cells (Fig. 3B,C). Thus, the failure of TIF32-Δ5-His to bind to 40S ribosomes might result from low-level expression, rather than loss of a direct contribution of eIF5 to 40S binding. At odds with this last possibility, we found that 40S binding of TIF32-Δ6-His also was diminished by the tif5–7A mutation, even though TIF32-Δ6-His expression was not reduced in tif5–7A cells (Fig. 3D,E). In addition, simultaneous overexpression of TIF32-Δ5-His, NIP1, and eIF5–7A substantially increased the stability of TIF32-Δ5-His, yet the mutant protein showed no binding to 40S ribosomes (data not shown). Thus, it seems clear that eIF5 promotes ribosome binding by eIF3 but that its contribution is redundant with the 40S-binding function of TIF32-CTD, being required only when the TIF32-CTD is lacking.
The MBU but not eIF3 requires eIF5 for 40S ribosome binding in vitro
To confirm our conclusion that eIF5 is crucial for 40S binding by the MBU but dispensable for binding of intact eIF3 to 40S subunits, we purified the MBU (TIF32-Δ5-His–NIP1–eIF5) and the MBU-7A (TIF32-Δ5-His–NIP1) complexes described in Figure 3F. As expected, the MBU contained eIF5, whereas MBU-7A did not, whereas both were devoid of detectable eIF2 and eIF1 (Fig. 4A). We also purified the five-subunit eIF3 holoprotein (PRT1-His–TIF32–NIP1–TIF34-HA–TIF35-Flag) from a strain overexpressing all five core eIF3 subunits. This eIF3 preparation was devoid of eIF2, eIF1, and eIF5 (Fig. 4A). It also contained a substantial amount of the PRT1-His–TIF34-HA–TIF35-Flag (P45) subcomplex described previously (Phan et al. 2001), as these three subunits are overproduced to higher levels than are TIF32 and NIP1 (Fig. 4A). (The complete absence of eIF5 from this eIF3 preparation was unexpected and may reflect the presence of multiple tags on the different eIF3 subunits.) Each complex was incubated with purified 40S ribosomes (Fig. 4A), and the bound and unbound fractions were separated by centrifugation on 10% sucrose cushions and analyzed by Western blotting. As a control, reactions lacking 40S ribosomes were carried out in parallel. Wild-type eIF3 interacted strongly with 40S ribosomes in vitro, as we observed approximately fivefold higher amounts of all five eIF3 subunits in the pellet fractions when ribosomes were present in the reaction (Fig. 4B, lanes 1,3). The stoichiometries of bound versus unbound eIF3 subunits indicated that the five-subunit eIF3 holoprotein bound to 40S subunits much more effectively than did the P45 subcomplex (Fig. 4B, lanes 1,2). This agrees with our previous finding that the PRT1-His–ΔRRM–TIF34–TIF35 subcomplex cannot bind to ribosomes in vivo (Valášek et al. 2001b). The fact that at least 60%–70% of the TIF32 and NIP1 in the eIF3 preparation bound to 40S subunits eliminates the possibility that binding was dependent on trace amounts of eIF5 or any other bridging factor that might exist in the preparation. These data provide the first evidence that yeast eIF3 can interact with the 40S subunit in the absence of other initiation factors. They also support our conclusion that eIF5 is dispensable for eIF3 binding to 40S subunits when the TIF32-CTD is intact (Fig. 3; Asano et al. 2000).
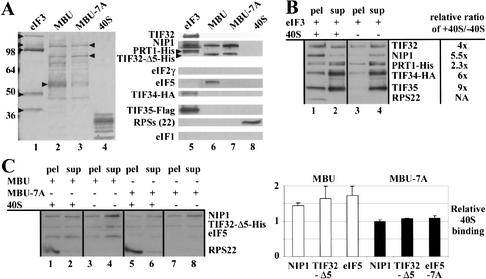
Figure 4.
Purified MBU binds to 40S ribosomes in vitro dependent on eIF5. (A, lanes 1–4) SDS-PAGE separation and Coomassie staining of the following components used in the binding assays of B and C: eIF3, MBU, and MBU-7A purified from LPY87 (Phan et al. 2001), YLVMBU, and YLVMBU-7A strains, respectively, as described in Figure 3F, and purified 40S ribosomal subunits. (Lanes 5–8) Western blot analysis of the purified samples in lanes 1–4, respectively, probed with antibodies against the proteins indicated on the left. (B) Purified eIF3 binds to 40S ribosomes in vitro. The affinity-purified eIF3 shown in A was incubated with (lanes 1,2) or without (lanes 3,4) purified 40S ribosomes and the bound (pel, lanes 1,3) and unbound (sup, lanes 2,4) fractions were separated by sedimentation onto 10% sucrose cushions at 90,000 rpm. Aliquots of the supernatant and pellet fractions were fractionated by SDS-PAGE, analyzed by Western blotting, and the ratio of the Western signals in the pellet fractions measured in the presence or absence of 40S subunits was calculated for each eIF3 subunit and listed to the right. The Western signals were quantified by video densitometry using NIH Image software. (C) The MBU subcomplex containing the TIF32-NTD, NIP1, and eIF5 is sufficient for 40S binding in vitro. Same as in B except that the MBU was incubated with (lanes 1,2) or without (lanes 3,4) 40S ribosomes, and the MBU-7A subcomplex lacking eIF5 was incubated with (lanes 5,6) or without (lanes 7,8) 40S ribosomes. The average ratios and standard errors of the Western signals in the pellet fractions measured in the presence versus the absence of 40S subunits were calculated from four independent experiments and plotted in the histogram on the right. White and black bars show the values obtained for the reactions containing MBU or MBU-7A, respectively.
In the in vivo experiments described above, the MBU bound to 40S subunits less tightly than did the intact MFC (Fig. 1A,C). Consistently, we found that purified MBU bound less tightly than purified eIF3 to 40S ribosomes in the in vitro binding assay (Fig. 4C). Although MBU binding was relatively weak, it occurred reproducibly in four independent experiments; moreover, the MBU-7A complex lacking eIF5 showed no binding at all in the same assays (Fig. 4C). Thus, we conclude that the N-terminal half of TIF32, NIP1, and eIF5, comprising the MBU, are sufficient for weak 40S binding in vitro that is wholly dependent on eIF5.
The TIF32-CTD specifically interacts with 18S rRNA via helices 16–18
Having established that TIF32, NIP1, and eIF5 play critical roles in 40S binding in vivo, we investigated whether these proteins can interact specifically with 18S rRNA in vitro. [35S]-labeled subunits of the MFC were incubated with biotinylated full-length 18S rRNA, and the rRNA–protein complexes were precipitated with Streptavidin agarose. As shown in Figure 5B, [35S]-TIF32 (lanes 5,6) and to a lesser extent [35S]-NIP1 (lanes 8,9) bound specifically to biotinylated 18S rRNA, whereas [35S]-PRT1 (lanes 2,3) and all remaining components of the MFC did not (data not shown). Neither TIF32 nor NIP1 interacted specifically with an equivalent amount of biotinylated β-globin mRNA or 25S rRNA under similar conditions (Fig. 5B). We then mapped the 18S rRNA-binding domain in TIF32 by testing purified GST fusions containing full-length or truncated versions of TIF32 (Fig. 5A) for binding to biotinylated 18S rRNA in a Northwestern assay (Fig. 5C). Biotinylated 18S rRNA interacted with full-length TIF32 (Fig. 5C, lane 2), the C-terminal half of TIF32 (Fig. 5C, lane 4), and with the extreme CTD of TIF32 (Fig. 5C, lane 6). In sharp contrast, none of the GST–TIF32 fusions lacking the extreme CTD, nor GST alone, bound to 18S rRNA (Fig. 5C, lanes 1,3,5,7).
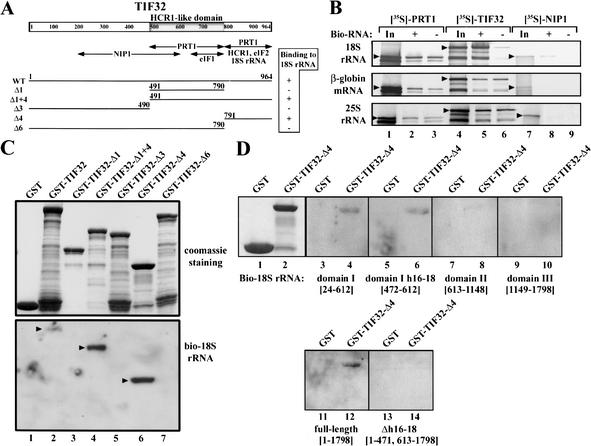
Figure 5.
The TIF32 CTD directly interacts with helices 16–18 of domain I of 18S rRNA in vitro. (A) Same as Figure 1G, with the HCR1-like domain (HLD) shaded. Arrows beneath the schematic depict the minimal segments required for interactions with the indicated molecules, based on results shown in C and reported previously (Valášek et al. 2002). The lines beneath the arrows depict the segments of TIF32 used in the Northwestern blot analysis of C with amino acid endpoints and clone designations indicated. The binding of each segment to full-length biotinylated 18S rRNA measured in C is summarized on the right. (B) Full-length TIF32 and NIP1 interact specifically with biotinylated 18S rRNA in Streptavidin pull-down assays. Biotinylated 18S rRNA, 25S rRNA, and β-globin mRNA, respectively, were incubated with [35S]-labeled PRT1 (lane 2), TIF32 (lane 5), or NIP1 (lane 8), and the RNA–protein complexes were precipitated with Streptavidin agarose (GIBCO-BRL), washed, and separated by SDS-PAGE. Gels were dried and subjected to autoradiography. The corresponding nonbiotinylated RNAs were used in lanes 3, 6, and 9. Lanes 1, 4, and 7 show 50% of the input amounts of [35S]-labeled proteins added to each reaction (In). Arrowheads indicate the full-length proteins. (C) The TIF32-CTD binds to biotinylated 18S rRNA in Northwestern assays. GST fusions to full-length TIF32 (lane 2), TIF32-Δ1 (lane 3), TIF32-Δ1 + 4 (lane 4), TIF32-Δ3 (lane 5), TIF32-Δ4 (lane 6), and TIF32-Δ6 (lane 7), or GST alone (lane 1), were expressed in Escherichia coli, immobilized on glutathione-Sepharose beads, and separated by SDS-PAGE. The resolved GST proteins were transferred to a membrane, renatured overnight, and incubated with biotinylated full-length 18S rRNA synthesized in vitro. The RNA–protein complexes were visualized by the Brightstar biodetect kit from Ambion. The amounts of fusion proteins used in the assay are shown in the upper panel stained with Gelcode Blue Stain Reagent (Pierce). (D) Same as in C except that various domains of 18S rRNA containing the nucleotides indicated in brackets were biotinylated and tested for binding to GST–TIF34-Δ4 or GST alone in Northwestern assays. Lanes 1 and 2 show the stained fusion proteins used in the assays and lanes 3–14 show the results of Northwestern assays for the indicated fusion proteins and biotinylated probes.
The yeast 18S rRNA can be divided into three domains based on its tertiary structure, with domain I forming the body (b) and shoulder (sh) of the 40S ribosome, domain II forming the protuberance (pt), and domain III forming the head (h), beak (bk), and common core (helix 44) of the 40S subunit (Spahn et al. 2001a). To investigate which domain mediates the interaction between 18S rRNA and the TIF32-CTD, three RNA transcripts corresponding to isolated domains I, II, and III were biotinylated in vitro and tested for binding to GST–TIF32-Δ4 (TIF32-CTD) in Northwestern assays. As shown in Figure 5D, only domain I (lanes 3,4) and, in particular, a short segment spanning helices 16–18 (lanes 5,6) showed specific interaction with GST–TIF32-Δ4 (lanes 4,6 vs. lanes 8,10). Importantly, removal of helices 16–18 from full-length 18S rRNA completely eliminated interaction of 18S rRNA with GST–TIF32-Δ4 (Fig. 5D, lane 14 vs. lane 12). Thus, we conclude that the extreme CTD of TIF32 can directly interact with the segment of 18S rRNA containing helices 16–18. The results discussed above showed that the TIF32-CTD is required for 40S binding of the eIF3 when the eIF5-CTD harbors the tif5–7A mutations. Hence, we propose that the TIF32-CTD stimulates 40S binding at least partly through interaction with helices 16–18 of 18S rRNA.
The TIF32-NTD strongly interacts with the CTD of 40S ribosomal protein RPS0A
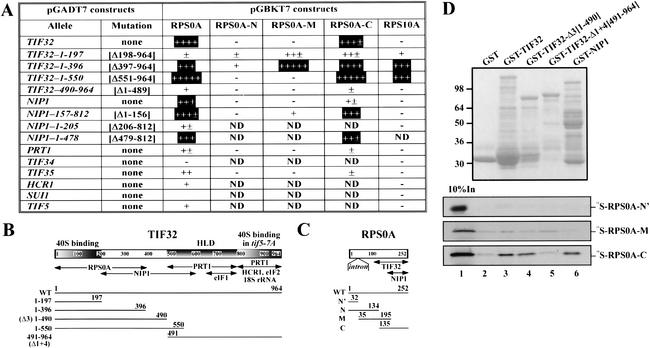
We next analyzed interactions between the subunits of the MFC and 40S subunit proteins using the yeast two-hybrid assay. [Note that the nomenclature for ribosomal proteins of Planta and Mager (1998) was adopted in this study.] The complete coding sequences for the subunits of eIF3, eIF1, and eIF5, and all 32 40S ribosomal proteins (RPSs) were fused to the GAL4-activation and DNA-binding domains, respectively. The resulting constructs were tested in all combinations for two-hybrid interactions in yeast. Because two-hybrid analysis conducted with large proteins has many limitations (Phizicky and Fields 1995; Asano et al. 1998), we also screened various segments of TIF32, NIP1, and PRT1 for two-hybrid interactions with all RPSs. As shown in Figure 6A, the N-terminal 396 residues of TIF32 are sufficient for strong interactions with RPS0A and RPS10A, whereas residues 1–197 interacted weakly with both proteins (constructs in rows 1–5, columns 3 and 7). Consistently, the TIF32 segment 490–964 failed to interact with RPS0A and RPS10A. It is intriguing that the TIF32-NTD interacts with RPS0A and RPS10A and also is critical for MFC association with 40S ribosomes in vivo (Fig. 1E). In addition, it appears that the internal segment of NIP1 between residues 157 and 478 contains a binding site for RPS0A but does not interact with RPS10A (Fig. 6A, rows 6–9). None of the other eIF3 subunits, eIF1, or eIF5 showed strong binding to RPS0A or RPS10A in the two-hybrid assay (Fig. 6A, rows 10–15). None of the other RPSs showed two-hybrid interactions with any of the MFC components tested.
Figure 6.
The N-terminal one-third of TIF32 and an internal segment of NIP1 can interact with the C-terminal half of the 40S protein RPS0A in vivo and in vitro. (A) Two-hybrid interactions between subunits of the MFC and 40S ribosomal proteins (RPSs). Transformants of strain AH109 containing the pGADT7 derivates in the first column encoding subunits of eIF3, eIF1, or eIF5 were transformed with the pGBKT7 constructs listed in columns 3–7 encoding full-length RPS0A, N-terminal (N), middle (M), or C-terminal (C) fragments of RPS0A, or full-length RPS10A. The resulting strains were tested for growth on SC-Leu-Trp-His plates. The amount of growth was judged after incubation at 30°C for 3 d and compared with that given by positive control constructs encoding T-antigen and p53 (Clontech). Boxed interactions were judged to be highly specific; ND, not determined. (B) Same as Figure 5A, with the 40S-binding domains shaded and labeled above. Arrows beneath the schematic depict the minimal segments required for interactions with the indicated molecules, based on results shown in A and D and reported previously (Valášek et al. 2002). The lines beneath the arrows depict the segments of TIF32 used in the two-hybrid assay (A) and in the binding assays of D with amino acid endpoints and clone designations indicated. (C) Schematic of RPS0A. Arrows beneath the schematic depict the minimal segments required for interactions with the TIF32-NTD and NIP1 based on results shown in A and D. The lines beneath the arrows depict the segments of RPS0A used in the two-hybrid experiments (A) and binding assays of D with amino acid endpoints and clone designations indicated. (D) The C-terminal half of RPS0A interacts with the TIF32-NTD in vitro. GST fusions to full-length TIF32 (lane 3), TIF32-Δ3 (lane 4), TIF32-Δ1 + 4 (lane 5), and full-length NIP1 (lane 6), or GST alone (lane 2), were expressed in Escherichia coli, immobilized on glutathione-Sepharose beads, and incubated with 10 μL of [35S]-labeled RPS0A-N‘, PRS0A-M, and RPS0A-C at 4°C for 2 h. The beads were washed three times with 1 mL of phosphate-buffered saline, and bound proteins were separated by SDS-PAGE. Gels were first stained with Gelcode Blue Stain Reagent (Pierce; top panel) followed by autoradiography (bottom panels). Lane 1 shows 10% of the input amounts of in vitro translated proteins added to each reaction (10% In).
We mapped the binding sites in RPS0A for TIF32 and NIP1 by testing each of the latter for two-hybrid interactions with three overlapping segments of RPS0A derived from the N terminus (N), middle (M), or C terminus (C) of the protein. The results in Figure 6A suggest that RPS0A contains a TIF32-binding site in the C-terminal half of the protein and an NIP1-binding site at the extreme C terminus (columns 4–6).
Finally, we verified the two-hybrid interactions just described by testing GST fusions made to full-length TIF32, TIF32-NTD, or TIF32-CTD for binding to [35S]-labeled segments of RPS0A synthesized in vitro. The C-terminal segment of RPS0A (RPS0A-C) bound specifically to GST fusions bearing full-length TIF32 and the TIF32-NTD (Δ3), but not to the TIF32-CTD (Δ1 + 4) or GST alone (Fig. 6D, lanes 3,4 vs. lanes 2,5). Full-length GST-NIP1 also interacted with RPS0A-C (Fig. 6D, lane 6). In contrast, the middle segment (RPS0A-M), which overlaps RPS0A-C by 60 residues, and the extreme N terminus (RPS0A-N‘) of RPS0A showed little or no interaction with all GST fusions tested (Fig. 6D). It is unlikely that these interactions were mediated by RNA, because RNAse A treatment did not alter the results just described (data not shown). These findings confirm that the TIF32-NTD and NIP1 both interact with the C-terminal half of RPS0A.
Discussion
The TIF32-NTD, NIP1, and eIF5 comprise a minimal 40S-binding unit
We investigated here whether various MFC subcomplexes formed in vivo by His8-tagged versions of TIF32 and NIP1 lacking binding domains for other MFC components can compete with native MFC for stable 40S binding in vivo. We discovered that the N-terminal half of TIF32, NIP1, and eIF5 comprise a minimal 40S-binding unit (MBU) that is sufficient for 40S binding in vivo and in vitro. The N and C termini of NIP1 and the TIF32-NTD were found to be required for 40S binding by the MFC in vivo, whereas eIF5 was necessary for binding only when the TIF32-CTD was absent. Because deletion of the TIF32 N terminus by the Δ8 mutation eliminated 40S binding by the MFC (Fig. 1E) but had only a minimal effect on MFC integrity (Valášek et al. 2002), it seems likely that the TIF32-NTD makes a critical contact with the 40S ribosome itself. Consistent with this idea, we uncovered specific interactions between the TIF32-NTD and the 40S subunit proteins RPS0A and RPS10A.
We found that the TIF32-Δ8 allele partially complemented the Ts− lethal phenotype of the rpg1-1 allele of TIF32 (Valášek et al. 2002). This finding seems at odds with the nearly complete inability of the MFC containing TIF32-Δ8-His to bind to 40S subunits in vivo (Fig. 1E). However, in the present study, the MFC harboring TIF32-Δ8-His must compete with wild-type MFC for 40S binding. If rpg1-1 also impairs 40S binding, then the MFC with TIF32-Δ8-His may be capable of low-level 40S binding in rpg1-1 cells because of the absence of competition. Interestingly, NIP1 also interacts with RPS0A, which might help to explain how the mutant MFC with TIF32-Δ8-His could interact with the 40S ribosome in rpg1-1 cells.
Deletion of the C-terminal segment of NIP1 after residue 571 (ΔB‘) abolished 40S binding (Fig. 2E) and impaired NIP1 function in vivo, even though it did not disrupt interaction of the truncated protein with any MFC components (Valášek et al. 2002). Hence, the NIP1-CTD, like the TIF32-NTD, may contact the 40S ribosome directly. Although the NIP1-CTD was dispensable for interaction with RPS0A (Fig. 6A), it could be required for the interaction we observed between NIP1 and 18S rRNA.
Deleting the NTD of NIP1 does not reduce interaction of the truncated protein (NIP1-C-His) with other core eIF3 subunits or HCR1, but it dissociates the eIF3 complex from eIF1, eIF2, and eIF5 (Valášek et al. 2002) and is lethal in vivo (L. Valášek and A.G. Hinnebush, unpubl.). We provided strong evidence here that the intact eIF3 complex formed by NIP1-C-His binds poorly to 40S subunits in vivo (Fig. 2B). This 40S-binding defect cannot result merely from dissociation of eIF1 and eIF2 from eIF3 because these factors are not present in the MBU formed by TIF32-Δ5-His. It also does not result simply from the absence of eIF5 because removing eIF5 from the MFC by the tif5–7A mutation does not decrease 40S binding by the MFC in vivo (Asano et al. 2001). In addition, we showed that purified eIF3 devoid of eIF5 and other MFC components can interact strongly with 40S ribosomes in vitro (Fig. 4B). Accordingly, we propose that the NIP1-NTD makes a critical contact with a 40S component in addition to its role in tethering eIF1, eIF5, and TC to eIF3. It remains to be seen whether disrupting this putative 40S contact without affecting association with eIF5 would impair 40S binding by an otherwise intact MFC.
Although elimination of eIF5 from the MFC by the tif5–7A mutation did not diminish 40S binding by other MFC components, it did reduce ribosome binding by the mutant subcomplex formed by TIF32-Δ6-His, which lacks eIF2 and HCR1. An even stronger defect was observed when eIF5 was eliminated from the eIF5–NIP1–TIF32-Δ5-His subcomplex (the MBU) by tif5–7A (Fig. 3). These last findings indicate that eIF5 is critically required for 40S binding in vivo when the TIF32-CTD is missing. Consistent with these in vivo results, we found that the purified MBU showed weak binding to 40S subunits in vitro, whereas the TIF32-Δ5-His–NIP1 binary complex lacking eIF5 (MBU-7A) failed to bind under the same conditions (Fig. 4C). The weaker binding of the MBU versus eIF3 holoprotein in vitro is consistent with the idea that the TIF32-CTD enhances 40S binding by intact eIF3.
We found that a 140-nt segment of domain I of rRNA encompassing helices 16–18 is necessary and sufficient for specific binding of 18S rRNA to the TIF32-CTD in vitro. Hence, the 40S-binding activity of the TIF32-CTD may involve a direct interaction with domain I of rRNA. Because this segment of TIF32 also interacts directly with eIF2 and promotes HCR1 binding to the MFC (Valášek et al. 2002), the reduced 40S binding of the complex formed by TIF32-Δ6-His in tif5–7A cells might derive partly from the absence of eIF2 or HCR1.
It was not surprising that deletion of all NIP1 residues C-terminal to residue 206 (NIP1-N‘-His) abolished 40S binding by the residual NTD of NIP1 (Fig. 2), because the NIP1-NTD cannot interact with TIF32 even though it forms a subcomplex with eIF5, eIF1, and eIF2. Overexpressing the NIP1-NTD together with eIF1 and eIF5 sequestered a fraction of eIF2 in an NIP1-NTD–eIF1–eIF5–eIF2 subcomplex and produced a Gcd− phenotype, signifying a reduced rate of TC binding to 40S subunits in vivo (Valášek et al. 2002). We found recently that the NIP1-NTD cannot associate with 40S ribosomes even when eIF1 and eIF5 are overexpressed (data not shown), supporting our contention that the NIP1-NTD–eIF1–eIF5–eIF2 subcomplex cannot compete effectively with native MFC for 40S binding in vivo. Thus, it appears that association of eIFs 1, 2, and 5 with eIF3 in the MFC enhances the binding of these factors to 40S subunits in vivo.
Evidence that eIF3 binds to the solvent side but has access to the 60S-interface side of the 40S ribosome
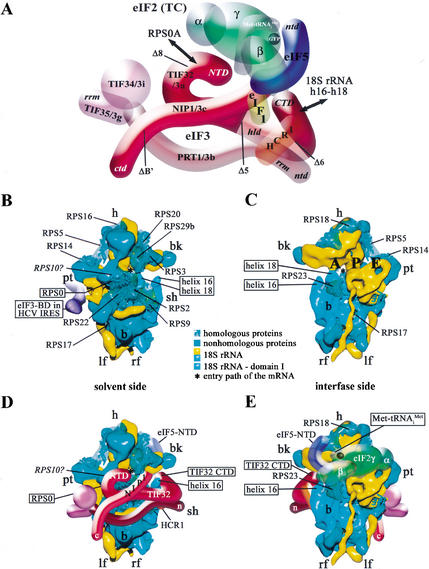
The results in Figure 6 suggest that both the N-terminal one-third of TIF32 and the internal segment of NIP1 spanning residues 157–478 can interact with the C-terminal half of the 40S protein RPS0A. RPS0A has a homolog (S2) in the Escherichia coli 30S ribosomal subunit and, thus, its location in the 3D structure of the yeast 40S ribosome has been predicted (Spahn et al. 2001a). RPS0A is expected to reside on the solvent side of the 40S subunit (i.e., opposite to the 60S-interface side), between the protuberance (pt) and beak (bk; Fig. 7B). Hence, binding of the TIF32-NTD and NIP1 to RPS0A would place this portion of eIF3 on the solvent side of the 40S ribosome (Fig. 7D). The TIF32-NTD also interacted with RPS10A, but the location of the latter in the 40S ribosome cannot be predicted. There is, however, unidentified protein mass surrounding RPS0A in the 40S 3D structure (Fig. 7B; Spahn et al. 2001a), and we found that RPS10A and RPS0A interact in the two-hybrid assay (L. Valášek, A. Mathew, and A.G. Hinnebusch, unpubl.). Thus, it is possible that RPS10A lies adjacent to RPS0A in the 40S ribosome, allowing it to interact with the TIF32-NTD (Fig. 7D).
Figure 7.
Model predicting the interaction of eIF3 with the Saccharomyces cerevisiae 40S ribosomal subunit. (A) Same as Figure 1H except that the model was rotated 180° about the vertical axis. The segments with solid shading represent the domains shown here to play a critical role in association of the MFC with the 40S ribosome. Interactions of the TIF32-NTD with RPS0A and of the TIF32-CTD with helices 16–18 of 18S rRNA are indicated by arrows. See text for further details. (B,C) Cryo-EM reconstruction of the S. cerevisiae 40S subunit docked with modified atomic models of 18S rRNA and 40S ribosomal proteins, adapted from Spahn et al. (2001a). The 40S subunit is shown from the solvent (B) or interface (C) sides, with RNA segments in yellow or turquoise and proteins in green. The ribbon models of the proteins with homology to Escherichia coli small ribosomal proteins are shown in a transparent envelope and labeled and boxed for RPS0. The solid protein segments are predicted to be composed of proteins without prokaryotic homologs. Domain I of 18S rRNA is shown in turquoise, with the positions of helices 16 and 18 indicated. The predicted position of RPS10A based on the data in Figure 6A is indicated with a question mark. (B) The position of the eIF3-binding domain in the HCV IRES (violet) bound to the small subunit is indicated [adapted from Spahn et al. (2001b)]. (D,E) Hypothetical location of the eIF3 complex on the 3D model of the 40S subunit based on the results of this study, including the requirements for the N- and C-terminal domains in both NIP1 and TIF32 for 40S binding, interaction between the TIF32-NTD and RPS0A, binding of the TIF32-CTD to helices 16–18 of 18S rRNA, and binding of NIP1 to RPS0A and 18S rRNA. The other MFC components, eIF1, eIF2, and eIF5, which are tethered to eIF3 via the NIP1-NTD and TIF32-CTD, are predicted to interact with the interface side of the 40S subunit, as shown for a small portion of eIF5 visible from the solvent side in D and for eIF5 and the TC (eIF2–GTP–Met-tRNAiMet) in E. The dimensions of the MFC model relative to the dimensions of the 40S subunit were determined from the molecular weights of both macromolecules, from published EM reconstitutions of the eIF3–40S complex (Lutsch et al. 1986; Srivastava et al. 1992), and from the relative dimensions of the 3D structures of the 30S ribosomal subunit (Thermus thermophilus) and eIF2γ (Pyrococcus abyssi) obtained from the Protein Data Bank (http://www.rcsb.org/pdb) and visualized by Swiss-PdbViewer 3.7.
Although its interaction with RPS0A would place eIF3 on the solvent side of the 40S subunit, the interaction we observed between the TIF32-CTD and 18S rRNA would provide eIF3 with access to the 60S-interface side. Helices 16 and 18 are accessible from both sides of the 40S subunit, and helix 16 protrudes into the solvent (Fig. 7B,C). If the TIF32-CTD wraps around helix 16, as shown in Figure 7D, it would be exposed on the interface side of the 40S subunit (Fig. 7E). The NTD of NIP1 might then penetrate the cleft between the beak (bk) and shoulder (sh) and also gain access to the interface side (Fig. 7E). We showed recently that the N-terminal tail of eIF1A can physically interact with eIF3 and eIF5 when these factors are all bound to the same 40S subunits (Olsen et al. 2002). As an ortholog of bacterial IF1, eIF1A is expected to bind to the A-site of the 40S subunit in the vicinity of helices 16 and 18 (Carter et al. 2001). In this location, eIF1A would be in close proximity to the binding sites for TIF32-CTD, NIP1-NTD, and eIF5 predicted by our model (Fig. 7C,E).
The binding of mammalian eIF3 to the 40S subunit has been visualized by two groups using 3D reconstructions from electron micrographs of negatively stained native 40S subunits (Lutsch et al. 1986; Srivastava et al. 1992). Both studies described contacts between eIF3 and the platform side of the 40S subunit in the vicinity of the protuberance, relatively close to the locations of the TIF32-NTD, PRT1-CTD, and TIF34/TIF35 depicted in our model (Fig. 7D). This binding site is also consistent with the location of the eIF3-binding domain in the HCV IRES (Sizova et al. 1998; Kieft et al. 2001) and the recent cryo-EM map of the IRES–40S complex (Spahn et al. 2001b). The binding domain for eIF3 in domains IIIa–b of the HCV IRES extends from the platform side of the 40S subunit just below the midline of the particle (Fig. 7B). It should be noted, however, that our model is more consistent with the eIF3–40S complex visualized by Lutsch et al. (1986), in which eIF3 appears to make extensive contact with most of the solvent side of the 40S subunit.
The TIF32-CTD and NIP1-NTD–eIF5 subassembly make independent contacts with eIF2β in the MFC that have additive stimulatory effects on translation initiation in vivo (Asano et al. 2000; Valášek et al. 2002). The available data suggest that these contacts are required not only for efficient recruitment of MFC components to the ribosome but also during later steps in the initiation process, such as scanning, AUG recognition, or GTP hydrolysis (Asano et al. 2001; Valášek et al. 2002). Hence, it is appropriate to consider whether all of the contacts identified in the MFC free in solution can persist on the surface of the 40S ribosome. The eIF2 is expected to interact at least partly with the 60S-interface side of the 40S subunit in the vicinity of the P-site, where Met-tRNAiMet binds. Based on the results of immunoelectron microscopy of TC bound to the 40S subunit, cross-linking of eIF2 subunits to 40S proteins, and the ability of antibodies against ribosomal proteins to inhibit eIF2 binding, it was proposed that eIF2 binds partly to the interface side of the 40S above the protrusion in the vicinity of the P-site and partly to the platform side in proximity to the bound eIF3 (for review, see Bommer et al. 1991). The P-site is located between the beak (bk) and protuberance (pt) on the interface side of the 40S ribosome (Fig. 7C), ∼50–55 Å from the binding sites for the TIF32-CTD and NIP1-NTD predicted in our model. This separation is comparable to the dimensions of the γ-subunit of eIF2 (Schmitt et al. 2002). Thus, it is reasonable to propose that the NTD of eIF2β can remain connected to the TIF32-CTD and the NIP1-NTD–eIF5 assembly, and still allow Met-tRNAiMet binding to the P-site (Fig. 7E). Interestingly, we showed previously that the TIF32-CTD–eIF2β interaction is dispensable for MFC integrity when the TIF32-NTD is deleted, indicating a potential negative role for the TIF32-NTD in regulating eIF2–eIF3 interactions (Valášek et al. 2002). It would be intriguing if this negative function was neutralized when the TIF32-NTD interacts with RPS0 on the solvent side of the 40S subunit, strengthening the connections between eIF2 and eIF3 on the interface side of the ribosome.
The eIF1 functions in conjunction with eIF1A in scanning and formation of a stable 48S complex positioned at the AUG start codon (Pestova et al. 1998). Both in vivo and in vitro data indicate that eIF1 is required to reject pairing between Met-tRNAiMet and near-cognate start codons in the P-site to ensure stringent selection of AUG as the start site (Yoon and Donahue 1992; Pestova and Kolupaeva 2002). It was proposed that eIF1 binds near the P-site and produces a conformational change that is instrumental to the rejection of near-cognate triplets (Pestova and Kolupaeva 2002). The protrusion of the NIP1-NTD into the 60S-interface side of the 40S subunit predicted by our model (Fig. 7E) might not be extensive enough to permit eIF1 to bind near the P-site without severing its connections to the TIF32-CTD and NIP1-NTD. Thus, the interactions between eIF3 and eIF1 may be dissolved on binding of the MFC to the 40S ribosome. Perhaps interaction of the TIF32-CTD with helices 16–18 of 18S rRNA triggers the release of eIF1 from eIF3 for its interaction with the P-site. Although many aspects of our model are speculative, we think it should provide a useful framework for examining the importance of different contacts made by eIF3 with the 40S subunit and the role of eIF3 in delivering eIF1, eIF5, and the TC to their binding sites on the interface side of the ribosome.
Materials and methods
Yeast strains and plasmids
The following strains were used in the work reported here: F556 (W303) MATa ade2-1 trp1-1 can1-100 his3-11,15 ura3 (A. Hopper); AH109 MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3∷MEL1UAS-MEL1TATA-lacZ (Clontech); H2898 MATa ura3-52 leu2-3,112 trp1-Δ63 gcn2Δ tif5Δ∷hisG tif34Δ∷hisG p[TIF5-FL TRP1] p[TIF34-HA LEU2]; H2899 MATa ura3-52 leu2-3,112 trp1-Δ63 gcn2Δ tif5Δ∷hisG tif34Δ∷hisG p[tif5-FL-7A TRP1] p[TIF34-HA LEU2]; H2768 MATa ura3-52 leu2-3,112 his1-29 gcn2-508 tif34Δ-1 〈HIS4-lacZ ura3-52〉 p[TIF34-HA LEU2]; H1485 MATa ura3-52 leu2-3 leu2-112 his1-29 gcn2-508 〈HIS4-lacZ ura3-52〉; H2896 MATa ura3-52 leu2-3,112 trp1-Δ63 gcn2Δ tif5Δ∷hisG p[TIF5-FL LEU2 2μ]; YLVMBU, H2896 + p[TIF32-Δ5-His URA3 2μ] p[NIP1 TRP1 2μ] (this study); H2897 MATa ura3-52 leu2-3,112 trp1-Δ63 gcn2Δ tif5Δ∷hisG p[tif5-FL-7A LEU2 2μ]; YLVMBU-7A, H2897 + p[TIF32-Δ5-His URA3 2μ] p[NIP1 TRP1 2μ] (this study). Plasmids used in this study and details of their construction are available as Supplemental Material.
Northwestern blot analysis and biotin pull-down assay
Northwestern blot analysis and in vitro synthesis of biotinylated 18S rRNA were carried out as described previously (Valášek et al. 2001a). We used a modification of a published protocol (Scherly et al. 1990) for the biotin pull-down assay, described in Supplemental Material.
40S-ribosome-binding assay
Binding of MBU and eIF3 to 40S ribosomes was measured using a method described previously (Moreno et al. 1998) with minor modifications described in Supplemental Material.
Other biochemical techniques
Sucrose gradient separations and Western blot analysis of gradient fractions, preparation of whole-cell extracts, coimmunoprecipitations, and Ni2+ chelation chromatography of eIF3 complexes containing His-tagged proteins were all conducted as described previously (Valášek et al. 2002).
Acknowledgments
We are thankful to Věra Valáškova for her help with figures, and Nilsa Rivera del Valle and Mark J. Swanson for technical assistance. We are thankful to Thomas Dever for critical reading of the manuscript and to members of the Hinnebusch and Dever laboratories for helpful discussions and suggestions during the course of this work. B.S. received a Ph.D. fellowship grant from the Bay Zoltan Foundation for Applied Research, Hungary.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ahinnebusch@nih.gov; FAX (301) 496-6828.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1065403.
References
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP–GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes & Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valášek L, Donahue TF, Hinnebusch AG. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer UA, Lutsch G, Stahl J, Bielka H. Eukaryotic initiation factors eIF-2 and eIF-3: Interactions, structure and localization in ribosomal initiation complexes. Biochimie. 1991;73:1007–1019. doi: 10.1016/0300-9084(91)90142-n. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Danaie P, Wittmer B, Altmann M, Trachsel H. Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- Hinnebusch AG. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 185–243. [Google Scholar]
- Huang H, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsch G, Benndorf R, Westermann P, Bommer UA, Bielka H. Structure and location of initiation factor eIF-3 within native small ribosomal subunits from eukaryotes. Eur J Cell Biol. 1986;40:257–265. [PubMed] [Google Scholar]
- Moreno JM, Kildsgaard J, Siwanowicz I, Mortensen KK, Sperling-Petersen HU. Binding of Escherichia coli initiation factor IF2 to 30S ribosomal subunits: A functional role for the N-terminus of the factor. Biochem Biophys Res Commun. 1998;252:465–471. doi: 10.1006/bbrc.1998.9664. [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2002;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valášek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein–protein interactions: Methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B" and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever RNAs. J Virology. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell. 2001a;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001b;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit–subunit interface. J Mol Biol. 1992;220:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- Valášek L, Hašek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem. 2001a;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- Valášek L, Phan L, Schoenfeld LW, Valášková V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001b;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Nielsen KH, Hinnebusch AG. Direct eIF2–eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac M-H, Chen R-H, Hanachi P, Hershey JWB, Derynck R. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornlocher HP, Hanachi P, Ribeiro S, Hershey JWB. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem. 1999;274:16802–16812. doi: 10.1074/jbc.274.24.16802. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Donahue TF. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]