Abstract
The centromere is a complex structure, the components and assembly pathway of which remain inadequately defined. Here, we demonstrate that centromeric α-satellite RNA and proteins CENPC1 and INCENP accumulate in the human interphase nucleolus in an RNA polymerase I–dependent manner. The nucleolar targeting of CENPC1 and INCENP requires α-satellite RNA, as evident from the delocalization of both proteins from the nucleolus in RNase-treated cells, and the nucleolar relocalization of these proteins following α-satellite RNA replenishment in these cells. Using protein truncation and in vitro mutagenesis, we have identified the nucleolar localization sequences on CENPC1 and INCENP. We present evidence that CENPC1 is an RNA-associating protein that binds α-satellite RNA by an in vitro binding assay. Using chromatin immunoprecipitation, RNase treatment, and “RNA replenishment” experiments, we show that α-satellite RNA is a key component in the assembly of CENPC1, INCENP, and survivin (an INCENP-interacting protein) at the metaphase centromere. Our data suggest that centromere satellite RNA directly facilitates the accumulation and assembly of centromere-specific nucleoprotein components at the nucleolus and mitotic centromere, and that the sequestration of these components in the interphase nucleolus provides a regulatory mechanism for their timely release into the nucleoplasm for kinetochore assembly at the onset of mitosis.
The centromere is a specialized structure on chromosomes for microtubule attachment to ensure the equal partitioning of chromosomes during cell division. This structure comprises two defined domains: the central core for the assembly of the kinetochore and the flanking pericentric heterochromatin for centromere cohesion. In Schizosaccharomyces pombe, the outer centromeric repeat sequences give rise to small interfering RNAs (siRNA) that participate in chromatin repression (Volpe et al. 2002). The depletion of Dicer (a nuclease required for the processing of siRNAs) in a chicken cell line leads to the disruption of heterochromatin assembly and cohesion (Fukagawa et al. 2004). However, Dicer depletion has no observable effect on the binding of the core kinetochore proteins CENPA and CENPC1 (Fukagawa et al. 2004), indicating that while the evolutionarily conserved RNA interference (RNAi) machinery is crucial for the establishment of the pericentric heterochromatin, it may not be essential for the core kinetochore region. A recent study in maize has further described the association of single-stranded centromeric transposable element and repeat RNA with the core kinetochore complex that is distinct from those at pericentric heterochromatin (Topp et al. 2004); however, the functional significance of the observed centromere RNA transcripts is unclear. Furthermore, little is known about the subnuclear distribution of centromere RNA, and the pathway and significance, if any, of such RNA in kinetochore formation and function.
The nucleolus is a specialized organelle of the interphase nucleus where ribosomal DNA is transcribed, pre-rRNA is processed, and ribosome subunits are assembled. In addition, this organelle is known to serve a variety of other essential cellular and cell cycle control functions by allowing the timely sequestration of specific trans-acting factors and the biogenesis of many cellular ribonucleoprotein particles (RNPs) including transfer tRNAs, small nuclear snRNAs, different mRNAs, and telomerase RNA (Thiry and Lafontaine 2005). Specifically, numerous studies have directly demonstrated the accumulation of different centromere-associated proteins at the nucleolus, including borealin, PARP1, and PARP2 (Earle et al. 2000; Adams et al. 2001; Saxena et al. 2002; Gassmann et al. 2004; Meder et al. 2005; Rodriguez et al. 2006).
Here, we show the enrichment of centromeric α-satellite RNA and centromere proteins CENPC1 and INCENP in the interphase nucleolus. We also present evidence that the centromere satellite RNA is required for the assembly of centromere-associated nucleoprotein components at the nucleolus and kinetochore.
Results
Centromere RNA accumulates in the transcriptionally active nucleolus
To gain insight into the subnuclear distribution pattern of centromere RNA, we performed preliminary RNA-FISH using various centromere-specific α-satellite probes and observed positive RNA-FISH signals in discrete compartments resembling nucleoli (data not shown). To further investigate this observation, we used antisera specific to two known nucleolar proteins, nucleolin and UBF1 (nucleolar transcription factor), as markers for the nucleolus in immunofluorescence studies. The results with these antisera confirmed that α-satellite RNA was enriched in the nucleolus (Fig. 1A,C,E,G). Competition with tRNA and rRNA did not affect the detection of the α-satellite RNA signals at the nucleoli (examples of chromosome 2- and 4-specific α-satellite RNA are shown in Fig. I,J).
Figure 1.
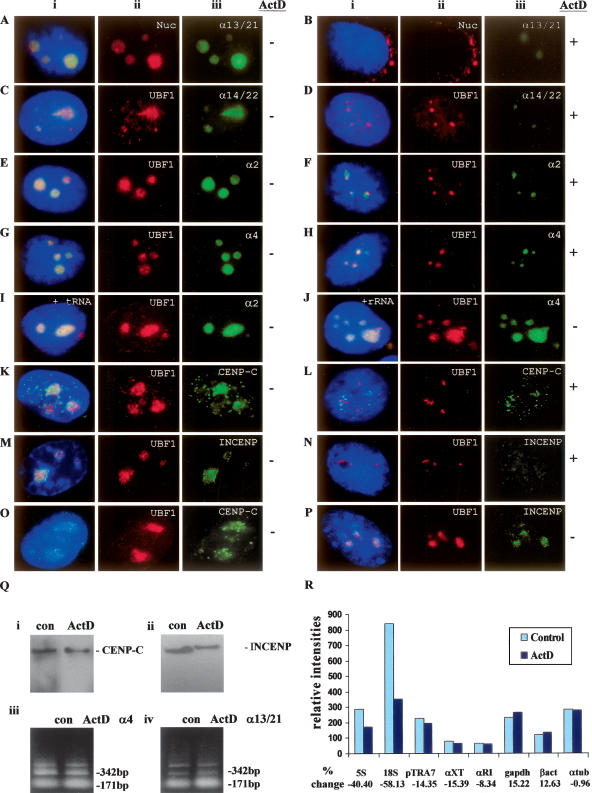
(A–J) Subnuclear distribution of centromeric α-satellite RNA and proteins in HeLa cells. Unless otherwise stated, images were captured on a conventional fluorescence microscope. Simultaneous RNA-FISH detection of α-satellite RNA specific for chromosomes 13/21 (Aiii; green), 14/22 (Ciii; green), 2 (Eiii; green) and 4 (Giii; green); and immunofluorescence detection of nucleolar proteins nucleolin (Nuc) (Aii; red) and UBF1 (Cii,Eii,Gii; red) in human HeLa cells. ActD treatment at 0.05 μg/mL resulted in the delocalization of α13/21- (Biii; green), α14/22- (Diii; green), α2- (Fiii; green), and α4- (Hiii; green) satellite RNA, nucleolin (Bii; red), and UBF1 (Dii,Fii,Hii; red) from the nucleoli. UBF1 displayed a typical relocalization toward the nucleolar periphery to give a concentrated cap-like signal at the tip of the nucleoli (Dii,Fii,Hii; red). Competition with tRNA and rRNA (10× excess amount) did not affect the detection of α-satellite RNA at the nucleoli as exemplified by the chromosome 2- and 4-specific α-satellite RNA signals (Iiii,Jiii; green). (K–N) Nucleolar accumulation of CENPC1 (Kiii; green) and INCENP (Miii; green), as indicated by coimmunostaining with UBF1 (Kii, Mii, respectively; red). CENPC1 and INCENP both delocalized from the nucleoli following ActD treatment (Liii,Niii, respectively; green). (O) Confocal laser scanning microscopy analysis showing nucleolar accumulation of CENPC1 by differential interference contrast images (iii; green) and coimmunostaining with UBF1 (ii; red). (P) Confocal microscopy analysis showing nucleolar accumulation of INCENP (iii; green) and coimmunostaining with UBF1 (ii; red). (i) Merged images of ii and iii. Antibodies used were as described in the Methods, except for the anti-CENPC1 antibody used in Oiii, which was kindly provided by Dr. Bill Earnshaw. (Q) Effect of ActD treatment on the expression level of CENPC1, INCENP, and α-satellite RNA. Total cell lysates were prepared from HeLa cells with (ActD) and without (con) prior treatment with 0.05 μg/mL ActD for 6 h, and subjected to Western blotting analysis. The expression levels of both CENPC1 (i) and INCENP (ii) remained unchanged following ActD treatment. (iii) Total RNA was prepared from untreated and ActD-treated HeLa cells. RT-PCR was performed on the RNA using primer sets corresponding to the α4- and α13/21-specific α-satellite sequences. A 171-bp ladder of monomeric α-satellite sequences was seen. No significant difference in the intensities of PCR products was detected following 6 h ActD treatment. (R) Nuclear run-on transcription assay to assess the effect of ActD treatment on α-satellite transcription. Equal numbers of nuclei were used for each assay, as determined by the DNA concentration. The effect of ActD on the transcription of α-satellite was measured by comparing the hybridization signals of in vitro-synthesized 32P-labeled run-on transcripts with immobilized DNA. The radioactive signals for the ribosomal genes 5S and 18S were decreased following ActD treatment, but the signals for the transcription of RNA polymerase II-driven genes (such as GAPDH, beta-actin, and alpha-tubulin) and those of α-satellite DNA (pTRA7 [chromosomes 13/14/21-specific], αXT [chromosomes 14/22-specific], and αRI [chromosomes 13/21-specific]) (Lo et al. 1999) were not significantly affected.
Next, we investigated whether the observed nucleolar accumulation of α-satellite RNA is dependent on RNA transcription by performing RNA-FISH and immunofluorescence on cells following treatment with actinomycin D (ActD) at 0.05 μg/mL for 6 h. Since ActD is a general transcriptional inhibitor, we have used conditions that have been demonstrated in other studies to be specific for the inhibition of nucleolar RNA transcription by RNA polymerase I (see Methods; Etheridge et al. 2002; Zhang et al. 2004; Meder et al. 2005). As expected, nucleolin displaced to the nuclear periphery following ActD treatment, whereas UBF1 displayed a typical relocalization toward the nucleolar periphery to give a concentrated cap-like signal at the tip of the nucleoli (Zhang et al. 2004) (Fig. 1Bii,Dii,Fii,Hii, respectively). In the presence of the ActD level used, α-satellite RNA also significantly, but not completely, delocalized from the nucleoli, indicating that the nucleolar localization of α-satellite RNA is sensitive to RNA polymerase I inhibition (Fig. 1Biii,Diii,Fiii,Hiii).
As controls, parallel experiments were performed to determine the effect of ActD treatment on the expression levels of both centromere proteins and α-satellite RNA by Western blotting and RT-PCR analysis, respectively (Fig. 1Q). The levels of expression of the centromere proteins CENPC1 and INCENP (Fig. 1Q) were not affected with 6 h of ActD treatment at 0.05 μg/mL, suggesting that the observed reduction in nucleolar signals was not due to inhibition of general transcription (Fig. 1Q). Likewise, the α-satellite transcription was not noticeably reduced in cells subjected to treatment with 3–6 h of ActD, as indicated by RT-PCR performed using primer sets corresponding to the chromosome 4- and 13/21-specific α-satellite (Fig. 1Q). We also performed run-on transcription assays to assess the direct effect of 6 h of 0.05 μg/mL ActD treatment on the transcription of α-satellite. As controls, we showed that the expression levels of the ribosomal genes 5S and 18S were greatly reduced by the ActD treatment, but not those of the RNA polymerase II-dependent genes such as GAPDH, beta-actin, and alpha-tubulin (Fig. 1R). There was no detectable difference with the transcriptional level of the various chromosome-specific α-satellite repeat species analyzed, further indicating that RNA polymerase I was unlikely to be directly involved in the regulation of α-satellite RNA transcription. These data therefore show that the transcription of α-satellite RNA (and centromere proteins CENPC1 and INCENP) is not driven by RNA polymerase I, although the nucleolar localization of the satellite RNA itself is dependent on an active RNA polymerase I state.
Nucleolar localization of centromere proteins CENPC1 and INCENP is sensitive to actinomycin D treatment and RNA depletion
A previous study has demonstrated the interaction of CENPC1 with UBF1 (Pluta and Earnshaw 1996). By immunofluorescence analysis with conventional and confocal laser scanning microscopy (Fig. 1K,O, respectively), we have demonstrated a clear subcellular localization of CENPC1 at the nucleolus. The high specificity of the antibody used in immunofluorescence was evident from the detection of a single CENPC1 protein band in Western blot analysis (Fig. 1Q).
In order to provide independent confirmation, we cloned a CENPC1 full-length cDNA fragment in-frame with the C terminus of the enhanced green fluorescent protein (GFP). To circumvent any potential nonspecific subcellular localization of GFP-CENPC1 due to the fixation procedure, we first examined the distribution of the transiently expressed recombinant GFP-CENPC1 protein in living human HeLa and 293 cells, and observed specific association of GFP-CENPC1 with the centromeres and a significant enrichment at the nucleolus (data not shown). Further experiments involving immunofluorescence staining of GFP confirmed the targeting of GFP-CENPC1 to both the nucleolus (Fig. 2C) and centromere (Fig. 2D,E) by counterstaining with the nucleolus marker nucleophosmin-1 and human anti-centromere antiserum CREST. When the GFP construct alone was transfected into cells, no nucleolar localization was observed (data not shown).
Figure 2.
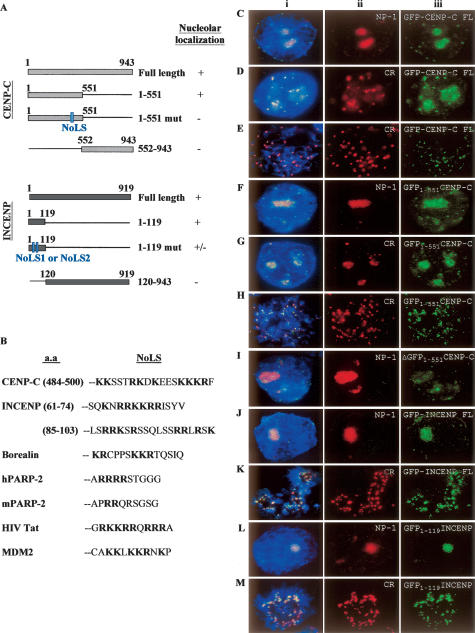
Determination of the nucleolus localization sequence (NoLS) motifs on CENPC1 and INCENP. Full-length and truncated GFP-CENPC1 and GFP-INCENP constructs were transfected into human 293 cells. More than 20 cells from each transfection experiment were scored for their subcellular distribution pattern. (A) Schematic diagrams for GFP fusion constructs for CENPC1 (top panel) and INCENP (bottom panel). Numbers indicate the positions of amino acid residues of the proteins or peptide being expressed. The locations of the NoLS motifs are indicated. Plus and minus signs indicate positive and negative localization to the nucleolus, respectively. (B) Amino acid comparisons between NoLSs of CENPC1, INCENP, and nucleolus proteins borealin, human and mouse PARP2, HIV Tat protein, and MDM2 (Lohrum et al. 2000). Bold letters denote basic amino acids. The amino acid positions of the CENPC1 and INCENP NoLSs are shown. (C–M) Human 293 cells were transfected with either full-length or truncated GFP-CENPC1 and GFP-INCENP constructs (see A). The localization of full-length (FL) GFP-CENPC1 (detected using anti-GFP antibody) (Ciii–Eiii; green) to nucleoli (Ciii), and interphase and metaphase centromeres (Diii,Eiii, respectively), are indicated by coimmunostaining with nucleophosmin-1 (Cii; red/NP-1) and CREST (Dii,Eii; red/CR) antibodies. Some examples of the colocalization of GFP-CENPC1 (Diii,Eiii; green) and CREST at the interphase centromeres (Dii; red) and metaphase centromeres (Eii; red) are presented. Direct visualization of GFP-CENPC1 signals without anti-GFP antibody gave similar results (data not shown). The NoLS at the N-terminal portion of CENPC1 (GFP-CENPC1 amino acids 1–551) efficiently targeted nucleolar (Fiii; green) and interphase and metaphase centromeres (Giii,Hiii, respectively; green) as indicated by costaining with nucleophosmin-1 (Fii; red/NP-1) and CREST (G,Hii; red/CR), respectively. The mutation of Lys residues within NoLS (1–551 mut in A) abrogated the recruitment to both the centromere (not shown) and nucleolus (Iiii; green), as indicated by NP-1 staining (Iii, red). The GFP fusion CENPC1 protein carrying the C-terminal fragment (amino acids 552–943) also fails to target to both the nucleolus and centromere (data not shown). Both full-length (FL) GFP-INCENP (Jiii,Kiii; green) and truncated (amino acids 1–119; containing unmodified NoLS1 and NoLS2) GFP-INCENP (Liii,Miii; green) proteins targeted to the nucleolus and centromere, as indicated by costaining with antibodies against nucleophosmin-1 (Jii,Lii; red/NP-1) and centromere (Kii,Mii; red/CR). Mutating NoLS1 or NoLS2 showed varying effects on nucleolar and centromere localization (see text). The GFP fusion protein carrying the C-terminal portion of INCENP (amino acids 120–943) targeted to neither the nucleolus nor the centromere (data not shown). (i) Merged images of ii and iii.
Recent studies have reported the nucleolar accumulation of centromere proteins including borealin (Rodriguez et al. 2006), PARP1, and PARP2 (Meder et al. 2005). In order to identify other centromere proteins that may localize to the nucleolus, we performed immunofluorescence analysis with a panel of antisera against centromere proteins. We found that INCENP (Fig. 1M,P), but not survivin, CENPA, CENPB, and CENPE (data not shown), were enriched at the nucleolus. Our data are consistent with recent analyses of the nucleolus composition using high-throughput mass-spectrometry-based proteomic techniques (Andersen et al. 2002, 2005; Scherl et al. 2002; Leung et al. 2006; Nucleolar Proteome Database [NOPdb], available at http://lamondlab.com/NOPdb/) that have revealed the presence of endogenous INCENP in the nucleolus. We have also found that treatment with 0.05 μg/mL ActD for 6 h resulted in the delocalization of CENPC1 and INCENP from the nucleolus (Fig. 1Liii,Niii, respectively), suggesting that the nucleolar localization of these proteins, as was the case with the α-satellite RNA, was also dependent on RNA polymerase I activity. Of interest, active nucleolar RNA transcription is required for centromere proteins borealin (Rodriguez et al. 2006), PARP1, and PARP2 (Meder et al. 2005) to reside at the nucleolus.
We were interested to know if the nucleolar localization of CENPC1 and INCENP depended on the presence of RNA. Both HeLa and human 293 cells were permeabilized either in the presence or absence of RNase, followed by immunofluorescence analysis with the relevant antibodies. Treatment of permeabilized cells with RNase prior to antibody incubation resulted in a complete loss of CENPC1 and INCENP signals from the nucleolus (data not shown), suggesting that the nucleolar localization of these proteins was dependent on the presence of RNA.
Identification of nucleolus localization sequences (NoLS) of CENPC1 and INCENP
To identify the region(s) of CENPC1 and INCENP that are required for nucleolar targeting, we generated GFP constructs containing fragments corresponding to various regions of these proteins (Fig. 2A). Each of the construct variants was introduced into human 293 cells, and >20 transfected cells from each experiment were scored for subcellular localization 24 h post-transfection. The N-terminal fragment (amino acids 1–551) of CENPC1 fused to GFP was able to target the recombinant protein to both nucleolus (Fig. 2F) and centromere (Fig. 2G,H), as indicated by colocalization with anti-nucleophosmin-1 and CREST antibodies, respectively. The GFP-CENPC1 fusion protein harboring the C-terminal fragment (amino acids 552–943) gave neither nucleolar nor centromeric staining patterns (data not shown). These data indicated that that the nucleolar targeting motif of CENPC1 is found within the N-terminal fragment (amino acids 1–551).
No precise nucleolus localization sequences (NoLS) consensus amino acid sequences have been reported; however, they usually reside within the nucleus localization sequences (NLS), which contain clusters of basic amino acids that are commonly enriched in Arg (R) and Lys (K) residues. The typical monopartite NLS is a cluster of basic residues starting with Pro (P) followed by five residues, of which at least three are either Lys (K) or Arg (R). The bipartite pattern begins with two basic residues (Lys K or Arg R) followed by 10 spacer residues in which at least three out of five residues are Lys (K) or Arg (R). A bipartite NLS (KKSSTRKD KEESKKKRF, bold letters denote basic residues) is found on the N-terminal portion (amino acids 484–500) of CENPC1 as determined by the PSORT II server (http://psort.ims.u-tokyo.ac.jp/), a program designed for the prediction of protein localization in eukaryotic cells. Of interest, this motif is not only conserved in other species but has also been found to be essential for centromere targeting of CENPC1 (Yang et al. 1996; Politi et al. 2002). Furthermore, it shares significant sequence homology with the NoLSs of other known nucleolar proteins such as the human immunodeficiency virus Tat protein (which contains two stretches of basic amino acids GRKKRRQRRRA), MDM2 (Lohrum et al. 2000), and centromere proteins including human and mouse PARP2 (Meder et al. 2005), and borealin (Rodriguez et al. 2006) (Fig. 2B).
In order to demonstrate that the bipartite NLS found on CENPC1 is also essential for regulating nucleolar localization, we generated a mutated N-terminal GFP-CENPC1 (amino acids 1–551) construct carrying three mutations in K492, K497, and K498 (Fig. 2A). The mutated product failed to localize to the nucleolus (Fig. 2I) and centromere (data not shown) following transfection into 293 cells, indicating that the NLS motif and the specific Lys (K) residues are critical for nucleolar and centromere targeting.
Taken together, the above data indicated the presence of CENPC1 within the nucleolus. However, the data do not allow us to infer whether CENPC1 interacts directly with UBF1 as described earlier (Pluta and Earnshaw 1996), since we have primarily used UBF1 only as a marker for the nucleolus and have not looked directly at the interaction between the two proteins. It is unclear why the previous study (Pluta and Earnshaw 1996) detected only limited localization of CENPC1 at the nucleoli, as we have shown in our own experiments that the anti-CENPC1 antibodies from both laboratories gave similar strong localization within the nucleoli (see Fig. 1K,O). This discrepancy may be due to differences in the underlying immunofluorescence techniques used.
To identify the NoLS on INCENP, we constructed and examined the subcellular localization of GFP-fused full-length and truncated INCENP proteins (Fig. 2A) 24 h following the transfection of the DNA plasmids into 293 cells. For each experiment, >20 cells were analyzed for the subcellular distribution of the GFP fusion proteins. The GFP full-length and N-terminal fragment (amino acids 1–119) of INCENP showed targeting to both the nucleolus (Fig. 2J,L) and centromere (Fig. 2K,M), whereas the C-terminal fragment (amino acids 120–919) distributed throughout the nucleus (data not shown).
Of interest, two bipartite NLSs were also found in INCENP (NLS1-SQKNRRKKRRISYV, amino acids 61–74; and NLS2-LSRRKSRSSQLSSRRLRSK amino acids 85–103, as predicted by PSORT II), and both are conserved between human and mouse (data not shown). Mutating the Arg (R) and Lys (K) residues in the NLS1 (RKK, amino acids 65–67) of the GFP-fused N-terminal (amino acids 1–119) INCENP fragment resulted in a large percentage (>50%) of the cells with diffused nuclear staining, whereas mutations in NLS2 (RRK, amino acids 87–89) disrupted the nucleolar localization of the fragment in ∼20% of the transfected cells. These results indicate that mutating NLS1 or NLS2 alone partially impairs the nucleolar localization of INCENP.
Centromere protein CENPC1 is an RNA-binding protein
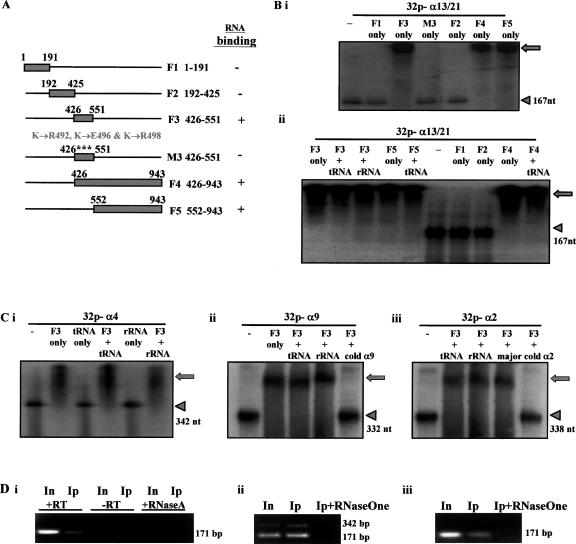
Recent studies have shown the presence of RNA-binding activity in the conserved hinge region of the HP1 family of proteins (HP1 alpha, beta, and gamma); this hinge region connects the chromo and chromoshadow domains and is required for the localization of these proteins to the centromeric heterochromatin (Maison et al. 2002; Muchardt et al. 2002). Homology of this hinge region is also found in CENPC1 (Muchardt et al. 2002). To determine if CENPC1 contains RNA-binding activity, we generated GST-CENPC1 protein fragments corresponding to various regions of CENPC1 (Fig. 3A). These fragments were subjected to RNA-binding/electrophoretic mobility-shift assays using 0.25 μg of radioactive-labeled RNA probes transcribed from various DNA plasmids containing chromosome-specific α-satellite sequences that ranged in size from 167 to 342 nucleotides (Fig. 3B,C). Fragment F3 (amino acids 426–551), which contained the HP1 hinge-homologous sequence, was clearly able to bind α-satellite RNA as evident from the mobility shift of the labeled RNA. The specificity of the binding of F3 to the labeled satellite RNA was shown by the modified fragment M3 containing mutations in three consecutive lysines (K492, K496, and K498) that effectively abolished the RNA-binding property of F3 (Fig. 3B). Both fragments F4 and F5 (corresponding to C-terminal amino acids 426–943 and 552–943, respectively), but not fragments F1 and F2 (amino acids 1–191 and 192–425, respectively), also efficiently bound labeled α-satellite RNA, showing that RNA-binding activity was present on fragments F4 and F5 as well. As a control, the addition of an excess of 2 μg of tRNA to the assay did not result in any significant competition effect on the observed binding of F3, F4, and F5 (Fig. 3B), providing supporting evidence for the binding specificity of these fragments for the α-satellite RNA. The RNA-binding specificity of F3 was also shown with the addition of an excess of rRNA to the assay (Fig. 3B). Furthermore, the RNA-binding specificity of these CENPC1 fragments was shown by positive band shifts when α-satellite RNA probes α9, α4, and α2 that are specific for the centromeres of chromosomes 9, 4, and 2, respectively, were used in the RNA-binding/electrophoretic mobility-shift assays (Fig. 3C). The addition of tRNA, rRNA, and AT-rich mouse pericentric major satellite as a competitor has no significant effects on the binding of these proteins to the labeled α-satellite RNA probe (Fig. 3C).
Figure 3.
RNA-binding properties of CENPC1 and the kinetochore. (A) Schematic representation of the different recombinant GST-CENPC1 constructs. (F1–F5) GST-CENPC1 recombinant fragments corresponding to amino acids 1–191, 192–425, 426–551, 426–943, and 552–943, respectively; (M3) a GST-CENPC1 recombinant fragment corresponding to F3426–551 but containing three mutational substitutions: K492 → R492, K496 → E496, and K498 → R498. Plus and minus signs indicate positive and negative α-satellite RNA binding, respectively (see B). (B) RNA binding assay. (i) Direct incubation of 0.25 μg of radioactively labeled single-stranded α13/21-satellite RNA probes (167 nt in size) with recombinant GST-CENPC1 fragments resulted in a band shift of the labeled RNA (arrowhead) away from its expected position to a significantly larger size (arrow) for fragments F3, F4, and F5 (but not F1, F2, and M3), indicating a positive RNA-binding property for fragments F3, F4, and F5. (ii) The addition of 2 μg of tRNA or rRNA as a competitor has no significant effects on the binding of these proteins to the labeled α-satellite RNA probes. (C) Direct incubation of 0.25 μg of radioactively labeled single-stranded α4 (Ci), α9 (Cii), and α2 (Ciii) satellite RNA probes (342, 332, and 338 nt in size, respectively) with recombinant GST-CENPC1 fragment F3 resulted in a band shift of the labeled RNA (arrowhead) away from its expected position to a significantly larger size (arrow). The addition of 2 μg of tRNA, rRNA, and AT-rich mouse pericentric major satellite as a competitor has no significant effects on the binding of these proteins to the labeled α-satellite RNA probes. However, the addition of 2 μg of cold α9 (Cii) and α2 (Ciii) resulted in loss of band shift. (D) Detection of CENPC1-associated RNA following CENPC1-mediated RNA-ChIP. α-Satellite PCR primers used were specific for chromosome 13/21 (i,ii) or chromosome 4 (iii). RNA was detected as a 171-bp cDNA band (corresponding to an α-satellite monomer) using RT-PCR in the CENPC1-mediated RNA-ChIP sample (Ip) and in the total genomic input DNA (In). In some experiments where the number of PCR cycle was increased from 25 to 30, a 171-bp ladder could be seen (e.g., ii). The absence of products when no reverse transcriptase was added (−RT) or following pretreatment of samples with either RNaseA or RNaseOne for 10 min at room temperature indicated that the RT-PCR products were derived from immunoprecipitated RNA, and not DNA.
Centromere satellite RNA is an integral component of the kinetochore
To further investigate possible in vivo interaction between α-satellite RNA and kinetochore proteins, we performed RT-PCR on RNA isolated following chromatin immunopreciptation (RNA-ChIP) using an anti-CENPC1 antibody. Two different primer sets corresponding to α-satellite sequences on chromosome 13/21 (Fig. 3Di,ii) and chromosome 4 (Fig. 3Diii) were used. We observed RT-PCR products corresponding to the 171-bp α-satellite monomers or multimers, providing evidence for the in vivo binding of α-satellite RNA with the CENPC1-associated kinetochore complex. No PCR product was detected when either RNaseA or RNaseOne (see below) was included in the chromatin immunoprecipitation, suggesting that the PCR products generated were amplified from cDNA following reverse transcription.
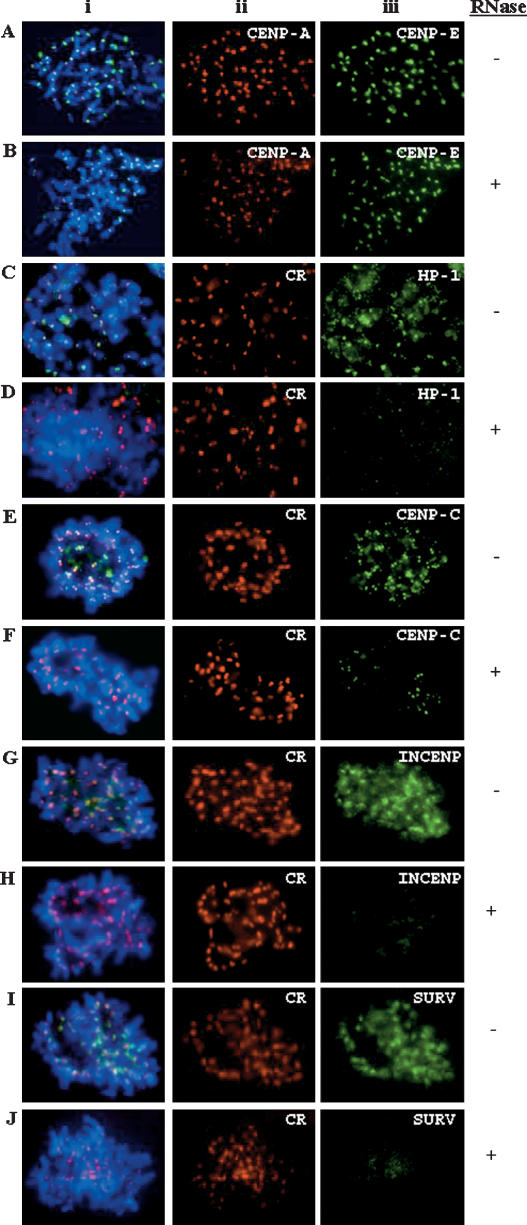
In order to explore the functional significance of RNA components at the kinetochore region, we performed immunofluorescence studies on cells permeabilized either in the presence or absence of RNase, followed by immunofluorescence analysis with relevant antibodies. RNase treatment has relatively little effect on the localization of CENPA, CENPB, and CENPE on the metaphase chromosomes (Fig. 4A,B; data not shown). As a control, we included heterochromatin protein HP1-alpha in our analysis, as RNase treatment has previously been shown to cause dispersion of this protein from the pericentric heterochromatin (Muchardt et al. 2002). As expected, we showed that RNA depletion abolished the localization of HP1-alpha at the metaphase pericentric heterochromatin (Fig. 4C,D). Importantly, we showed that a similar treatment prior to antibody incubation resulted in a significant, but not complete, delocalization of CENPC1 from the metaphase centromere (Fig. 4E,F). In comparison, under the same conditions of RNase treatment, INCENP and survivin showed complete delocalization from the centromere region of the metaphase chromosomes (Fig. 4G–J).
Figure 4.
Immunofluorescence analysis of the role of RNA in the assembly of centromere proteins on HeLa cell metaphase chromosomes. (A,B) Centromere binding of CENPA (Aii,Bii; red) and CENPE (Aiii,Biii; green) was not affected by RNase (which targets both single- and double-stranded RNA) treatment of the cells. (C–J) Centromere localization of human CREST antiserum (CR, which recognizes CENPA and CENPB) was not affected by RNase treatment of the cells (Cii–Jii; red). A similar treatment resulted in the complete delocalization of HP1-alpha (Ciii,Diii; green), INCENP (Giii,Hiii; green), and survivin (SURV) (Iiii,Jiii; green). This treatment caused a significant but not complete delocalization of CENPC1 from the centromere (Eiii,Fiii; green). Plus and minus signs indicate with and without RNase treatment, respectively. (i) Merged images of ii and iii.
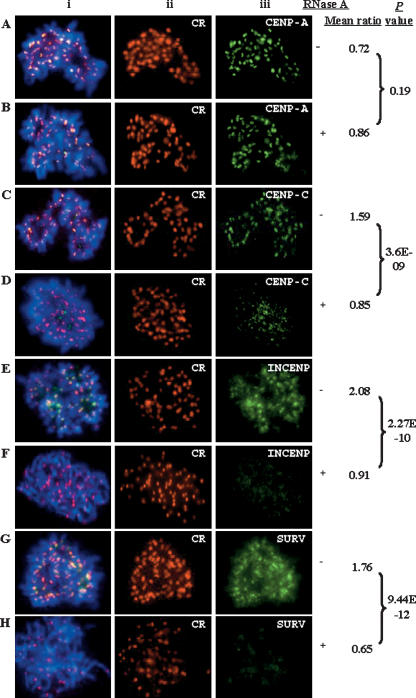
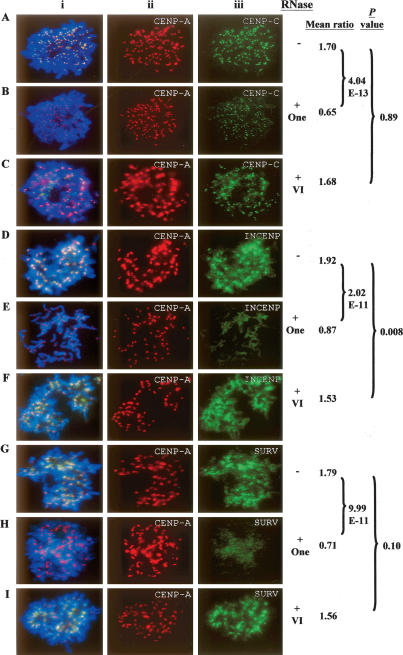
To further investigate the nature of the RNA that was responsible for the above observation, we repeated the experiments using RNases that preferentially cleaved either single-stranded RNA (RNaseA and RNaseOne) or double-stranded RNA (RNaseVI) (Figs. 5 6) (Topp et al. 2004; Martens et al. 2005). A control sample before RNase treatment was included within each experimental set. Treatment with either RNaseA or RNaseOne resulted in a significantly reduced association of CENPC1 (Figs. 5C,D, 6A,B), and the complete removal of INCENP (Figs. 5E,F, 6D,E) and survivin (Figs. 5G,H, 6G,H) at the metaphase centromeres. In a parallel experiment, we showed that CENPA binding at the metaphase centromeres was not significantly affected (Figs. 5A,B, 6A,B,D,E,G,H). We further showed that treatment of cells with double-stranded RNA-specific RNaseVI had no measurable effect on the metaphase centromere-association property of CENPC1, INCENP, survivin, and CENPA (Fig. 6A,C,D,F,G,I). Collectively, these experiments using different RNases suggest that single-stranded RNA, but not double-stranded RNA, was required for the association of CENPC1, INCENP, and survivin at the centromere.
Figure 5.
Effect of single-stranded RNA-specific RNaseA treatment on the localization of centromere proteins on HeLa metaphase chromosomes. In total, 25 cells with (+) or without (−) RNaseA treatments, were scored using the Ip Lab program. For each cell, immunofluorescence intensities for five randomly chosen chromosomes were quantified. Each data set was normalized against the CREST signal (Aii–Hii; red) and expressed as “mean ratio.” The RNaseA treatment resulted in a significant reduction (P-value < 0.001) in the immunofluorescence signals for CENPC1 (Ciii,Diii; green), INCENP (Eiii,Fiii; green), and survivin (SURV) (Giii,Hiii; green), but not for CENPA (Aiii,Biii; green). As with the RNase treatment shown in Fig. 4, signal reduction on the centromere was less complete for CENPC1 compared with CENPA, INCENP, and survivin. A similar reduction in CENPC1 signal at the centromere was observed following RNaseA treatment and immunofluorescence analysis performed using a different anti-CENPC1 antibody provided by Dr. Bill Earnshaw (data not shown). (i) Merged images of ii and iii.
Figure 6.
Effect of single-stranded RNA-specific RNaseOne and double-stranded RNA-specific RNaseVI treatment on the localization of centromere proteins on HeLa metaphase chromosomes. The comparison of the immunofluorescence signals for 25 cells with (+) and without (−) RNaseOne treatments by scoring the signal intensities on five chromosomes for each cell. Each data set was normalized against the CENPA signal (Aiii–Iii; red) and expressed as “mean ratio.” RNase A treatment led to a significant reduction (P-value < 0.001) for immunofluorescence intensities for CENPC1 (Aiii,Biii; green), INCENP (Diii,Eiii; green), and survivin (SURV) (Giii,Hiii; green). As with the RNase or RNaseA treatment shown in Figs. 4 and 5, signal reduction on the centromeres was less complete for CENPC1 compared with CENPA, INCENP, and survivin. Similar pairwise comparisons indicated no significant difference in mean immunofluorescence signals before and after RNaseVI treatment for CENPC1 (Aiii,Ciii; green), INCENP (Diii,Fiii; green), and survivin (SURV) (Giii,Iiii; green). (i) Merged images of ii and iii.
Centromeric α-satellite RNA is capable of partially restoring the relocalization of CENPC1 and INCENP to the nucleolus and centromere following RNase treatment
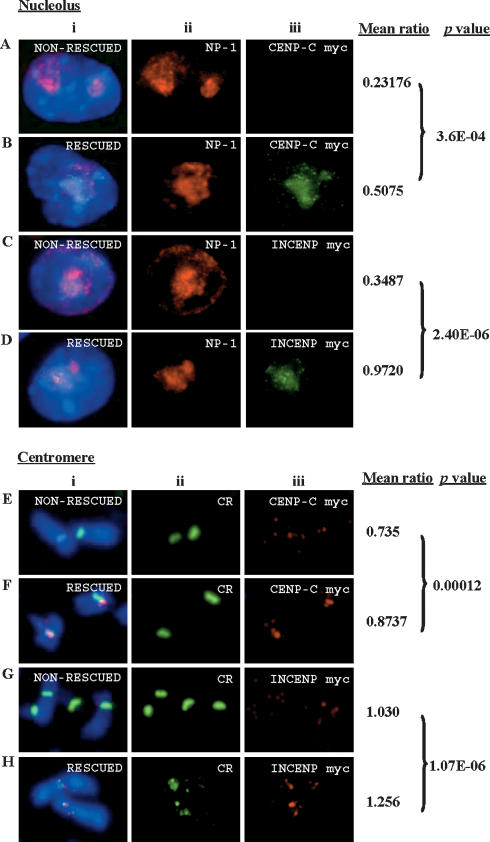
In order to further ascertain the role of α-satellite RNA in centromere protein binding at the nucleolus and centromere, we investigated whether the restoration of this RNA in RNase-treated cells could directly facilitate the in situ recruitment or targeting of CENPC1 and INCENP. For this analysis, we used recombinant myc-tagged CENPC1 and INCENP proteins generated using the T7 RNA polymerase Escherichia coli expression system. After RNase treatment and a brief incubation with α-satellite RNA in the presence of RNase inhibitor, the recombinant proteins were added to the cells. Using this RNA “rescue” assay, the replenishment of α13/21 (Fig. 7) or α14/22 (data not shown) α-satellite RNA was found to give significant restoration of the targeted recruitment of CENPC1 and INCENP to the nucleolus (Fig. 7A–D) and mitotic centromere (Fig. 7E–H).
Figure 7.
RNaseOne treatment and in situ “rescue” assay. Cells cytospun on slides were permeabilized with Triton X-100 and subjected to RNaseOne treatment for 20 min (this treatment resulted in the significant delocalization of both CENPC1 and INCENP from the kinetochores; see Fig. 6) followed by fixation with 4% formaldehyde. Cells were then incubated with RNase inhibitor (RNasin) and in vitro transcribed α-satellite RNA for 1 h at 37°C, before overnight incubation with the relevant myc-tagged CENPC1 and INCENP recombinant proteins. The recruitment of these proteins at the kinetochore (detected with CREST antibody) and nucleolus was detected using antibody against the myc tag. (A–D) Effects on the nucleolus. The “mean ratio” of the immunofluorescence signals of CENPC1-myc (Aiii,Biii; green) and INCENP-myc (Ciii,Diii; green) normalized against nucleophosmin-1 (NP-1) (Aii–Dii; red) was determined from the quantitative immunofluorescence scoring of the nucleoli from 25 cells, with (“rescued”) and without (“non-rescued”) replenishment using in vitro transcribed α-satellite (α13/21) RNA. The RNA replenishment led to a significant enrichment (P-value < 0.001) of CENPC1-myc (Aiii,Biii; green) and INCENP-myc (Ciii,Diii; green) staining in the nucleoli of most cells. (E–H) Effects on the mitotic centromere. The “mean ratio” of the immunofluorescence signals of CENPC1-myc and INCENP-myc normalized against the CREST signals (Eii–Hii; green) for both “rescued” (using α13/21) and “non-rescued” cells was determined from the quantitative immunofluorescence scoring of 50 chromosomes. The α-satellite RNA replenishment resulted in a modest recovery in the binding of CENPC1-myc and INCENP-myc seen only at the kinetochores of some of the chromosomes.
RNAi knockdown of CENPC1 and rescue of function by overexpression of GFP-CENPC1 but not GFP-CENPC NoLS mutant
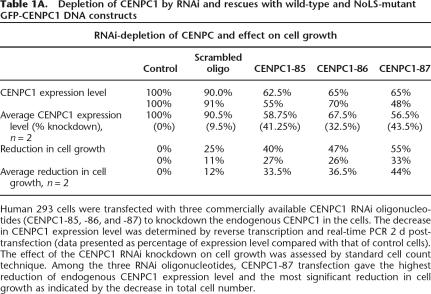
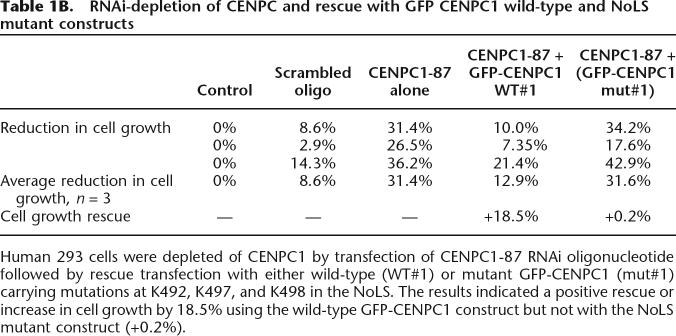
To further understand the functional significance of the interaction between CENPC1 and centromeric α-satellite RNA, we performed RNAi-knockdown experiments to deplete CENPC1 expression level in human 293 cells, followed by transfection with either a GFP-CENPC1 wild-type or mutant plasmid construct as an attempt to restore the function of the protein. Three RNAi oligonucleotides, CENPC-85, -86, and -87, were tested. The efficiency of the RNAi knockdown was determined by reverse transcription and real-time PCR analysis 2 d post-knockdown. As seen in Table 1A, the transfection of CENPC1-85, -86, and -87 RNAi oligonucleotides into cells resulted in a reduction of CENPC1 RNA level by 41.25%, 32.5%, and 43.5%, respectively. Such reductions in endogenous CENPC1 were accompanied by a concomitant decrease in cell growth by 33.5%, 36.5%, and 44.0%, respectively. Among three RNAi knockdowns, CENPC1-87 RNAi knockdown was the most detrimental to cells.
Table 1A.
Depletion of CENPC1 by RNAi and rescues with wild-type and NoLS-mutant GFP-CENPC1 DNA constructs
Human 293 cells were transfected with three commercially available CENPC1 RNAi oligonucleotides (CENPC1-85, -86, and -87) to knockdown the endogenous CENPC1 in the cells. The decrease in CENPC1 expression level was determined by reverse transcription and real-time PCR 2 d post-transfection (data presented as percentage of expression level compared with that of control cells). The effect of the CENPC1 RNAi knockdown on cell growth was assessed by standard cell count technique. Among the three RNAi oligonucleotides, CENPC1-87 transfection gave the highest reduction of endogenous CENPC1 expression level and the most significant reduction in cell growth as indicated by the decrease in total cell number.
Given that CENPC1-87 produced the greatest effect in the depletion of CENPC1 expression level, we chose it for further RNAi depletion of CENPC1 and rescue assays. In order to demonstrate that the NoLS domain on CENPC1 is functionally essential for centromere function, we compared the ability of full-length GFP-CENPC1 and a NoLS mutant GFP-CENPC1 construct in restoring the functionality of the CENPC1 RNA-depleted kinetocore. The NoLS mutant construct carried three mutations (K492, K497, and K498) in the NoLS domain. We transfected the GFP-CENPC1 wild-type and mutant constructs 1 d after RNAi knockdown and determined the extent of rescue from RNAi-induced inhibition of cell growth 2 d later. The GFP-CENPC1 construct increased the cell growth by 18.5%, whereas the mutated construct was incapable of restoring the detrimental effects of the CENPC1 RNAi knockdown (Table 1B). These results provided evidence for the functional significance of the CENPC1 NoLS domain.
Table 1B.
RNAi-depletion of CENPC and rescue with GFP CENPC1 wild-type and NoLS mutant constructs
Human 293 cells were depleted of CENPC1 by transfection of CENPC1-87 RNAi oligonucleotide followed by rescue transfection with either wild-type (WT#1) or mutant GFP-CENPC1 (mut#1) carrying mutations at K492, K497, and K498 in the NoLS. The results indicated a positive rescue or increase in cell growth by 18.5% using the wild-type GFP-CENPC1 construct but not with the NoLS mutant construct (+0.2%).
Discussion
Centromeric α-satellite RNA is essential for the nucleolar localization of CENPC1 and INCENP
In this study, we have demonstrated the nucleolar accumulation of the constitutive centromere protein CENPC1, the centromere-associated chromosomal passenger protein INCENP, and centromeric α-satellite RNA. We have tested and not observed a similar nucleolar accumulation of the constitutive centromere proteins CENPA and CENPB, and the centromeric passenger proteins survivin and CENPE. Using protein truncation and site-specific mutagenesis, we have identified the protein domains required for the localization of CENPC1 and INCENP to the nucleolus. The nucleolus localization sequence (NoLS) motifs for both of these proteins coincide with the nucleus localization sequence (NLS) motifs, and are found at the N-terminal portions of the proteins. These NoLS motifs share significant homology with those of other nucleolus-associated proteins (Fig. 2B). Some of these proteins, including borealin (Gassmann et al. 2004; Rodriguez et al. 2006), PARP1 and PARP2 (Meder et al. 2005), and INCENP (Ainsztein et al. 1998; Rodriguez et al. 2006), also bind directly to the centromere.
We have shown that the presence of centromeric α-satellite RNA is closely associated with the nucleolar localization of CENPC1 and INCENP, since RNase treatment induces a complete disruption of this localization. More specifically, in the RNA “rescue” study, we have shown that the incubation of RNase-treated cells with α-satellite RNA results in the in situ targeting of myc-tagged CENPC1 and INCENP recombinant proteins to the nucleolus. Experiments involving ActD treatment further demonstrate that the nucleolar accumulation of α-satellite RNA, CENPC1, and INCENP is dependent on active RNA polymerase I transcription. Previous studies have similarly shown that the nucleolar localization of centromere proteins borealin (Rodriguez et al. 2006) and PARP1 and PARP2 (Meder et al. 2005) is dependent on active nucleolar transcriptional activity. Recent studies in S. pombe (Djupedal et al. 2005; Kato et al. 2005) and Arabidopsis (Kanno et al. 2005) have shown that RNA polymerase II and IV (a unique RNA polymerase in plants) are essential for the generation of siRNAs. The transcription machinery that modulates the transcription of centromere repeats or siRNA synthesis in vertebrates is still unknown. However, it is unlikely that RNA polymerase I is directly driving the transcription of the nucleolar α-satellite RNA, as indicated by our nuclear run-on transcription data. Furthermore, the depletion of α-satellite RNA-FISH signals is unlikely to be due to the down-regulation of some specific proteins, as the conditions we used for ActD treatment did not affect the expression levels of proteins in general, as exemplified by those of CENPC1 and INCENP. In this context, the mechanism of action for the RNA polymerase I remains undefined, although it may be directly involved in providing a structural network or complex that retains the various centromere components in the nucleolus.
The nucleolus serves the important function of ribosome synthesis. Increasing numbers of studies now show that it is also a multitasking organelle that engages in other important cellular functions (Thiry and Lafontaine 2005). An example of the pluri-functionality of the nucleolus is the cell cycle–dependent nucleolar localization of the telomere components hTERT (reverse transcriptase catalytic subunit of telomerase), hTR (telomerase RNA), and telomeric DNA-binding protein TERF2, where these components have been shown to be released from the nucleolus at late S and G2/M phase at a time that coincides with telomere elongation (Wong et al. 2002; Zhang et al. 2004). Our data suggest that the nucleolus may similarly sequester centromeric components such as centromere RNA and proteins for timely delivery to the chromosomes for kinetochore assembly at mitosis. The centromere proteins, in particular the chromosomal passenger proteins such as borealin (Gassmann et al. 2004; Rodriguez et al. 2006), PARP1 and PARP2 (Meder et al. 2005), and INCENP (Ainsztein et al. 1998; Rodriguez et al. 2006), display dynamic changes in distribution patterns during various stages of the cell cycle. Although these proteins only bind the centromere following mitotic onset (Martineau-Thuillier et al. 1998; Burke and Ellenberg 2002; Gassmann et al. 2004), they are expressed much earlier in mid-S to G2 phase and have been shown to accumulate in the nucleolus. During this period, the nucleolus may serve a repository role for the temporal storage of centromere RNA and proteins, and control the timely release of these components into the nucleoplasm for kinetochore assembly at M phase. It is also possible that the assembly of functional kinetochore nucleoprotein complexes may take place in the nucleolus.
Centromeric α-satellite RNA and CENPC1 associate and facilitate nucleoprotein assembly at the mitotic centromere
We have demonstrated that treatment of metaphase cells with single-stranded RNA-specific RNases (but not with a double-stranded RNA-specific RNase) results in the significant but incomplete delocalization of CENPC1, and the complete delocalization of two centromere-associated chromosomal passenger proteins INCENP and survivin, from the metaphase centromere. Similar treatments have no significant effect on the centromeric localization of CENPA, CENPB, and CENPE. These results indicate that the assembly of INCENP and survivin, and to an extent CENPC1 (discussed below), but not CENPA, CENPB, and CENPE, at the metaphase centromere is dependent on the presence of single-stranded RNA.
Several lines of evidence suggest that at least a component of this single-stranded RNA is directly transcribed from the centromeric α-satellite DNA, and that this α-satellite RNA transcript associates directly with CENPC1. First, we have used recombinant GST-fusion protein truncation constructs and site-specific mutagenesis to show that CENPC1 binds single-stranded α-satellite RNA probes derived from chromosomes 2, 4, 9, or 13/21, and ranging in size from 167–342 nucleotides (nt). This RNA-binding activity resides within the central region (amino acids 426–551) as well as the C-terminal portion of the protein. Second, by performing RT-PCR on RNA isolated following RNA-ChIP using an anti-CENPC1 antibody, we have demonstrated in vivo binding of single-stranded α-satellite RNA with the CENPC1-associated kinetochore protein complex. The RT-PCR results indicate that the kinetochore-associated α-satellite RNA has a size range corresponding to multiples of the 171-nt α-satellite repeating unit. These studies show the presence of single-stranded α-satellite RNA of 171 nt and larger and belonging to the different chromosome-specific α-satellite subfamilies. However, we cannot exclude the possibility that shorter RNA or α-satellite RNA of other sequences may be present, as the present detection may bias against these RNAs.
Third, we have shown that the in situ replenishment of single-stranded α-satellite RNA significantly restores the relocalization of myc-tagged CENPC1 and INCENP to the centromeres of some of the chromosomes following RNase treatment. The incomplete rescue of the recruitment of the myc-tagged recombinant proteins to the kinetochore in our assay could be due to the inappropriate folding or modification of the recombinant proteins or replenishing RNA used. Further investigations are needed to discern these possibilities and define the characteristics and possible heterogeneity associated with these RNA transcripts.
Fourth, we have shown that wild-type GFP-CENPC1, but not the mutant construct with mutated residues within the NoLs domain, was able to partially restore the functionality of the CENPC1-depleted kinetochore in the CENPC1 RNAi-knockdown cells and rescue the cells from the inhibition of cell growth. The failure of the over-expression of the mutated GFP-CENPC1 protein in improving the rate of cell growth suggests that the NoLS domain is essential for the proper function of CENPC1.
Together, the above analyses indicate that, in addition to the important role played by the short, double-stranded siRNA at the pericentric heterochromatin (Fukagawa et al. 2004), a distinct class of longer, single-stranded centromeric α-satellite RNA is an important structural component of the human centromere.
A model for the nucleolar and centromeric assembly of centromere-associated nucleoprotein components
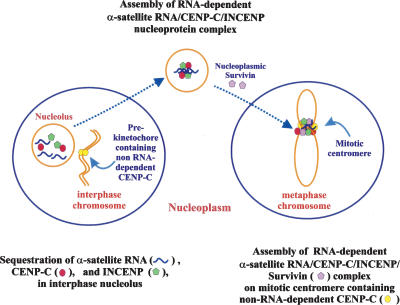
Central to our model (Fig. 8) is the finding that centromeric α-satellite RNA is essential for the enrichment of centromere proteins CENPC1, INCENP, and/or survivin at the nucleolus and mitotic centromere. We propose that the nucleolar sequestration of centromere-specific nucleoprotein components provides a regulatory mechanism for the timely release of these components into the nucleoplasm for kinetochore assembly at the onset of mitosis. Our finding that CENPC1 binds centromere satellite RNA, whereas previous in vitro studies have shown that CENPC1 is also DNA-binding (Yang et al. 1996; Politi et al. 2002), together suggest that this protein has a dual RNA- and DNA-binding function. It is therefore possible that one pool of CENPC1 may play a constitutive centromere DNA-binding role that persists throughout the cell cycle, while a second pool of this protein may act directly as, or be part of, a chaperone mechanism to relocate centromeric α-satellite RNA and centromere proteins, including INCENP, from the nucleolus onto the mitotic centromere. The RNA dependence of the nucleolar localization of both CENPC1 and INCENP suggests that these proteins may associate with the centromeric α-satellite RNA and be assembled into a nucleoprotein complex in the nucleolus. This nucleolar chaperone complex may further include other centromere-associated proteins such as borealin (Rodriguez et al. 2006), PARP1, and PARP2 (Meder et al. 2005), since these components have also been shown to accumulate in the nucleolus. In further support of the proposed dual role of CENPC1, we have consistently observed significantly higher quantitative immunofluorescence signals for CENPC1 (but not other constitutive centromere proteins examined, including CENPA and CENPB) on the metaphase centromere compared with those of the interphase centromere (data not shown). Of interest, a recent study (Kwon et al. 2007) has shown that CENPC1 is targeted to interphase or metaphase centromeres through interactions with different sets of centromere proteins, providing further evidence that CENPC1 is localized to the centromere via two independent pathways during interphase or the mitosis stage. We have shown in the present study that treatment with single-stranded RNA-specific RNases results in a significant, but not complete, delocalization of CENPC1 from the metaphase centromere, consistent with the idea that only the RNA-, but not the DNA-, associated pool of CENPC1 (and components of its complex, such as INCENP) is sensitive to the RNase treatment. Our data demonstrate that survivin does not localize to the nucleolus, although its localization to the metaphase centromere is also RNA-dependent. This can be explained by the incorporation of survivin into the metaphase chromosomes following the breakdown of the nucleolar membrane, via INCENP and/or borealin, as previous work has shown that survivin forms a functional complex with these two proteins for targeting to the centromere (Klein et al. 2006).
Figure 8.
A model for the assembly of human centromere-associated nucleoprotein components at the interphase nucleolus and mitotic centromere. The nucleolar and centromeric accumulation of CENPC1, INCENP, and/or survivin is dependent on the presence of single-stranded α-satellite RNA. The nucleolus may serve as an interphase storehouse for the sequestration of centromere-associated nucleoprotein components and facilitate the assembly of these components into complexes, prior to their release into the nucleoplasm for relocation, together with additional nucleoplasmic centromere-associated protein components such as survivin, onto the mitotic centromere.
Methods
Immunofluorescence, RNA-FISH, RNase treatment, and in situ “rescue” assay
For immunofluorescence, cells on slides were permeabilized in cold PNS buffer (20 mM Pipes at pH 6.8, 200 mM NaCl, 600 mM sucrose, and protease inhibitor), fixed, and incubated with the relevant primary and secondary antibodies (Uren et al. 2000; Saxena et al. 2002; Wong et al. 2005). For RNA-FISH, cells were hybridized using various biotinylated double-stranded α-satellites (Uren et al. 2000; Saxena et al. 2002; Wong et al. 2005) but without subjecting the cells to the DNA denaturation step. For cytospun metaphase spreads, cells were treated with Taxol, cytospun on slides, and incubated in KCM buffer at pH 7.5 (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl, 0.5 mM EDTA, 0.1% [v/v] Triton X-100, Roche protease inhibitor) (Uren et al. 2000). For RNase treatment, cells were incubated with 15 μg/mL RNase, 15 μg/mL RNaseA (Sigma), 25 U RNaseOne (Promega; in 25 μL of KCM buffer), and 10 U RNaseVI (Ambion; in 25 μL of KCM buffer) for 15 min, prior to partial fixation and incubation with the relevant antibodies.
For in situ “rescue” experiments, cells cytospun on slides were permeabilized with Triton X-100 and subjected to RNase treatment as described followed by fixation with KCM buffer with 4% formaldehyde. Cells were then washed, incubated with RNase inhibitor (RNasin) for 10 min at room temperature, and incubated with 1 μg of in vitro unlabeled transcribed α-satellite RNA (produced using T7 or Sp6 RNA polymerase Riboprobe kit from Promega) for 1 h at 37°C, before overnight incubation with 0.5 μg of the relevant myc-tagged CENPC1 and INCENP recombinant proteins (diluted in KCM buffer with RNasin, 0.5% [v/v] Triton X-100 and BSA) generated using the T7 transcription/translation kit (see below). The cells were incubated with anti-myc mouse monoclonal antibody (Invitrogen) and relevant secondary antibodies to detect the recruitment of these proteins at the kinetochore (detected with CREST antibody) and nucleolus (detected with NP-1 antibody). The mean ratios for the fluorescence intensities of CENPC1 and INCENP against CREST and nucleophosmin-1 NP-1, for both rescue and nonrescued cells, respectively, were scored. The templates used for the synthesis of α-satellite RNA were PCR fragments specific to the centromeres of chromosomes 4, 9, 13/21 and mouse major satellite generated using primer sets containing either a T7 or SP6 promoter at the 5′ end of the fragment. The primers used were (underlined sequence indicates either SP6 or T7 promoter sequence, respectively): α2/SP6for-ATTTAGGTGACACTATAGAAGCGCTCT CAGGACTACGTGGAAAAAGG and α-2rev1-GACCTGAAAGC GCTCCAAATGTCCACA; α4/T7for-ATTTAATACGACT CACTATAGGCATTCTCAGAAACTTCTTTGTGATGTG and α4rev-CTTCTGTCTAGTTTTTATGTGAAGATA; α9/SP6for-ATTTAGGTGACACTATAGAAGCCCTATGATGAAAAAGG TAATATCTTC and α-9rev-CCTGAAAGCGCTTAAAACGTCC GCTTG; α13/21/T7for3-AATTTAATACGACTCACTATAG GCAGAAGCATTCTCAGAAACT and α13/21rev4-TCCCGC TTCCAACGAAATCCTCCAAAC; majorT7for-AATTTAATAC GACTCACTATAGGTGGCGAGAAAACTGAAAATCACGGA and majorrev-TCTTGCCATATTCCACGTCCTACAG.
Antibodies and Western blot analysis
Primary antibodies used were rabbit polyclonal antisera against CENPC1 (Kalitsis et al. 1998), CENPE (Uren et al. 2000), INCENP (Abcam), and survivin (Novus Biologicals); mouse monoclonal antisera against CENPA (MBL), HP1-alpha (Chemicon), UBF1 (Santa Cruz Biotechnology), nucleolin (Upstate Biotechnology), nucleophosmin-1 (Sigma); and human CREST6 antiserum (predominantly anti-CENPA and CENPB) (Kalitsis et al. 1998). The secondary antisera used were donkey anti-human Texas Red (Jackson Laboratory, Bar Harbor, ME), and donkey anti-mouse or anti-rabbit Alexa Fluoro 488/594 (Molecular Probes). An additional CENPC1-specific rabbit polyclonal antibody used in some confirmatory experiments was kindly provided by Dr. Bill Earnshaw (Wellcome Trust Centre for Cell Biology, University of Edinburgh, UK).
For Western blotting, cells were lysed in RIPA buffer (150 mM KCl, 20 mM HEPES at pH 7.9, 0.3% NP-40, 0.5 mM EDTA, 0.5 mM PMSF, protease inhibitor) followed by SDS-PAGE and Western analysis.
Plasmids and recombinant GST (glutathione S-transferase) and myc fusion proteins
Full-length and truncated CENPC1 and INCENP cDNAs were generated by PCR amplification and cloned into the C terminus of the enhanced green fluorescent protein GFP (pEGFP-N1; Clonetech) for transfection into cells using lipofectamine 2000 transfection reagent (Invitrogen). The primer sets used in generating the PCR fragments were: GFPCENPC1HindIII 5′-CCGAAGCTTCGATGGCTGCGTCCGGTCTGGATCAT and GFPCENPC1BamHI 3′-CGGGATCCTCTGAGTAAAAAGAAGAA CAC for full-length GFP-CENPC1; GFPCENPC1HindIII 5′ and GFPCENPC1551BamHI 3′-CGGGATCCTGGTAATTCATTCTTAC TGA for N-terminal GFP-CENPC1 1-551; GFPCENPC1552HindIII 5′-CCGAAGCTTCGATGCATCACAATAGTAGCCGA and GFPCENPC1BamHI 3′ for C-terminal GFP-CENPC1 552-943; GFPINCENPHindIII 5′-CCGAAGCTTCGGGGACGACGGCCCC AGGGCCCATT and GFPINCENPBamHI 3′-CGGGATCCGT GCTTCTTCAGGCTGTAGGCCAGGCTGCT for full-length GFP INCENP; GFPINCENPHindIII 5′ and GFPINCENP119BamHI 3′-CGGGATCCGCCGTTCTCCCCGACCACTGT for N-terminal GFP-INCENP1-119; GFPINCENP120HindIII 5′-CCGAAGCTTCG GTTCTGCGGCGTGTGACCCGTGCT and GFPINCENPBamHI 3′ for C-terminal GFP-INCENP120-919.
Mutations of the residues within the NoLS sequences on CENPC1 and INCENP were performed using a site-directed mutagenesis kit (Stratagene) with the following primers: Cenpc NLSmut 5′-CTAGAAAAGATGAGGAAGAATCTAAACAGCAGCG CTT and CenpcNLSmut 3′-AGCGCTGCTGTTTAGATTCTTCCT CATCTTTTCTAG for the N-terminal GFP-1-551 CENPC1 NoLS mutant and full-length GFP CENPC1 NoLS mutant; IncenpNLS1mut 5′-CTTCTCAGAAGAACCAACAGGAGAAGA GACGGATTTCT and IncenpNLS1mut 3′-AGAAATCCGTCTC TTCTCCTGTTGGTTCTTCTGAGAAG for the N-terminal GFP INCENP-1-119 NoLS1 mutant; IncenpNLS2mut 5′-CAGGAG AAGGTTATCCCCCGAAGAGTCTCGGAGCAGCCA and IncenpNLS2mut 3′-TGGCTGCTCCGAGACTCTTCGGGGGA TAACCTTCTCCTG for the N-terminal GFP-INCENP-1-119 NoLS2 mutant.
The myc-tagged CENPC1 and INCENP recombinant proteins were generated using the TNT Quick Coupled Transcription/Translation systems (Promega). The templates used for the generation of the proteins were PCR fragments containing a T7 RNA promoter and myc protein tag at the 5′ and 3′ ends of the product, respectively. The primer sets used for the PCR amplification were (underlined sequence indicates either T7 promoter or myc sequence, respectively): T7-CENPC-TGTAATACGACTCAC TATAGGGAGAATGGCTGCGTCCGGTCTGGATCAT and CENPC1myc-TCACAGATCCTCTTCTGAGATGAGTTTTT GTTCTCTGAGTAAAAAGAAGAACAC for full-length CENPC1-myc tag protein; T7-INCENP-TGTAATACGACTCACTATAGG GAGAATGGGGACGACGGCCCCAGGGCCCATT and INCENP-Cmyc-TCACAGATCCTCTTCTGAGATGAGTTTTT GTTCGTGCTTCTTCAGGCTGTAGGC full-length INCENP-myc tag protein.
For the construction of GST fusion proteins, PCR was used to generate GST:CENPC11–191 (fragment F1), GST:CENPC1192–425 (fragment F2), GST:CENPC1426–551 (fragment F3), GST: CENPC1426–943 (fragment F4), and GST:CENPC1552–943 (fragment F5) for subcloning into the pGex2T vector (Pharmacia). The primers used were: GSTcpc BamHI 5′-CGCGGATCCATGGCTGC GTCCGGTCTGGATCAT and GSTcpc191 EcoRI 3′-CGGAA TTCCATTTACTGAATTTTCAAAAGT for GST:CENPC11–191; GSTcpc 192BamHI5′-CGCGGATCCATGCTGCCTTCAAGTA CAGAG and GSTcpc425EcoRI 3′-CGGAATTCCGAATT CTCTTCTCTGTTTTTG for GST:CENPC1192–425; GSTcpc426 BamHI 5′-CGCGGATCCATGATGGCTAAACCAGCTGAAGAA and GSTcpc551 EcoRI 3′-CGGAATTCCTGGTAATTCATTTCT TACTGA for GST:CENPC1426–551; GSTcpc552 BamHI 5′-CGCGGATCCATGCATCACAATAGTAGCCGA and GSTcpc EcoRI 3′-CGGAATTCCTCTGAGTAAAAAGAAGAACAC for GST: CENPC1552–943; GSTcpc426 BamHI 5′ and GSTcpc EcoRI 3′ were used for the generation of GST:CENPC1426–943.
Mutations of three lysine residues (fragment M3) on fragment F3 was performed using a site-directed mutagenesis kit (Stratagene). The primers used were: cpCrnaMUT 5′-CTAG AAAAGATAGGGAAGAATCTGAAAAGAGGCGCT cpCrnaMUT 3′-AGCGCCTCTTTTCAGATTCTTCCCTATCTTTTCTAG.
The proteins were purified according to the manufacturer’s protocols with some modifications, passed through a PD10 column (Pharmacia), and eluted in RNA binding buffer (20 mM HEPES at pH 7.6, 100 mM KCl, 2 mM EDTA, 0.01% NP-40) and KCM buffer with 0.5% Triton X-100, for use in RNA binding and in situ “rescue” assays.
RNA binding assays
α-Satellite sequences used for RNA probe synthesis were specific for the centromeres of chromosomes 2, 4, 9, or 13/21. The resulting probes were designated α2, α4, α9, and α13/21, respectively. PCR fragments corresponding to each chromosome-specific satellite sequence were generated with relevant primer sets as described above. The sense RNA probes were produced by using T7 or Sp6 RNA polymerase (Riboprobe kit from Promega) in the presence of radioactively labeled UTP. The RNA binding assays were performed in RNA-binding buffer with radioactively labeled RNA and purified recombinant GST-CENPC1 protein. Following incubation, loading dye was added and samples were subjected to PAGE/TBE electrophoresis. Gels were then dried and exposed to Kodak film.
RNA chromatin immunoprecipitation (RNA-ChIP)
Cells were fixed, washed, and sonicated in lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors) containing RNase inhibitor, tRNA, and bovine serum albumin. ChIP was performed with the relevant antibody and captured with Protein-A Sepharose. The Sepharose was washed and eluted. The eluate fraction was de-crosslinked with high salt (200 mM NaCl) treatment at 60°C and proteinase K incubation at 50°C, followed by extraction with TRIzol:chloroform and DNA precipitation. For RNase treatment, immunoprecipitated samples were treated with RNaseA (0.2 μg/μL) or RNaseOne (0.2 μg/μL) at 37°C for 15 min, followed by DNase treatment prior to RT-PCR using the primer sets specific for the centromeres of chromosomes 4, 9, or 13/21, as follows: α9_5′start-CCTATGATGAAAAAGGTAATATCTTC; α9_3′end-CTGAAAGCGCTTAAAACGTCCGCTTG; α4_5′start-CATTCTCAGAAACTTCTTTGTGATGTG; α4_3′end-CTTCTG TCTAGTTTTTATGTGAAGATA; α13/21 for3-TAGACAGAAG CATTCTCAGAAACT; α13/21 rev4-TCCCGCTTCCAACGAAAT CCTCCAAAC.
Nuclear run-on transcription
This assay was performed using isolated HeLa nuclei as described (Poortinga et al. 2004). An equal number of nuclei, as determined by the DNA concentration, was used for each treatment. The RNA recovered from each sample was used as a probe to hybridize onto DNA arrays containing denatured DNA corresponding to housekeeping genes (GAPDH, beta-actin, and alpha-tubulin), ribosomal (18S and 5S) genes, and various α-satellite sequences (pTRA7, αXT, αR1, from chromosomes 13/14/21, 13/14, and 14/22, respectively; Lo et al. 1999). Radioactive signals were detected and quantified using a Molecular Dynamics PhosphorImager.
RNAi knockdown of CENPC1 expression levels
Human 293 cells (1.5 × 105) were subjected to RNAi knockdown using three Stealth RNAi duplexes/oligonucleotides CENPC1-85, -86, and -87, purchased from Invitrogen. These oligonucleotides have been functionally tested for knockdown in mammalian cells and prevalidated by the manufacturer to confirm their targeting specificity to human CENPC1 mRNA. As a control, a scrambled oligonucleotide was used in all RNAi knockdown experiments. The transfection of the RNAi oligonucleotides into 293 cells was performed using Lipofectamine LTX according to the manufacturer’s instructions (Invitrogen). The cells were harvested 48 h post-RNAi knockdown for RNA purification and assessment of the expression level of endogenous CENPC1 by reverse transcription and real-time PCR analysis using a cDNA reverse transcriptase kit and a SYBR-based detection real-time PCR system (Applied Biosystems). The primer sets used in the real-time PCR were: CENPC1forward-TGCCAGAAATCACATCCAAA and CENPC1reverse-GAAGGTGAGCCAACGGATAA.
The effect of RNAi knockdown of CENPC1 on cell growth was assessed by standard cell count 48 h post-transfection. The reduction in cell growth was determined by comparing the total cell numbers in the RNAi knockdown samples and those in the controls. In the rescue assays, the CENPC1-depleted cells were rescued by transfection a day after RNAi knockdown using either wild-type GFP-CENPC1 full-length construct or mutant GFP-CENPC1 full-length construct carrying mutations at K492, K497, and K498 in the NoLS domain.
Acknowledgments
We thank Bill Earnshaw for anti-CENPC1 antibody. This work was supported by the National Health and Medical Research Council of Australia, and the National Institute of General Medical Sciences and National Institute of Health.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6022807
References
- Adams R.R., Carmena M., Earnshaw W.C., Carmena M., Earnshaw W.C., Earnshaw W.C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Ainsztein A.M., Kandels-Lewis S.E., Mackay A.M., Earnshaw W.C., Kandels-Lewis S.E., Mackay A.M., Earnshaw W.C., Mackay A.M., Earnshaw W.C., Earnshaw W.C. INCENP centromere and spindle targeting: Identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Lam Y.W., Steen H., Mann M., Lamond A.I., Steen H., Mann M., Lamond A.I., Mann M., Lamond A.I., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M., Ong S.E., Lyon C.E., Lamond A.I., Mann M., Lyon C.E., Lamond A.I., Mann M., Lamond A.I., Mann M., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Burke B., Ellenberg J., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Djupedal I., Portoso M., Spahr H., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K., Portoso M., Spahr H., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K., Spahr H., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K., Gustafsson C.M., Allshire R.C., Ekwall K., Allshire R.C., Ekwall K., Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes & Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle E., Saxena A., MacDonald A., Hudson D.F., Shaffer L.G., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Saxena A., MacDonald A., Hudson D.F., Shaffer L.G., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., MacDonald A., Hudson D.F., Shaffer L.G., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Hudson D.F., Shaffer L.G., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Shaffer L.G., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Saffery R., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Cancilla M.R., Cutts S.M., Howman E., Choo K.H., Cutts S.M., Howman E., Choo K.H., Howman E., Choo K.H., Choo K.H. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet. 2000;9:187–194. doi: 10.1093/hmg/9.2.187. [DOI] [PubMed] [Google Scholar]
- Etheridge K.T., Banik S.S., Armbruster B.N., Zhu Y., Terns R.M., Terns M.P., Counter C.M., Banik S.S., Armbruster B.N., Zhu Y., Terns R.M., Terns M.P., Counter C.M., Armbruster B.N., Zhu Y., Terns R.M., Terns M.P., Counter C.M., Zhu Y., Terns R.M., Terns M.P., Counter C.M., Terns R.M., Terns M.P., Counter C.M., Terns M.P., Counter C.M., Counter C.M. The nucleolar localization domain of the catalytic subunit of human telomerase. J. Biol. Chem. 2002;277:24764–24770. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Takami Y., Nakayama T., Oshimura M., Nakayama T., Oshimura M., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Carvalho A., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C., Nigg E.A., Gerloff D.L., Earnshaw W.C., Gerloff D.L., Earnshaw W.C., Earnshaw W.C. Borealin: A novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Fowler K.J., Earle E., Hill J., Choo K.H., Fowler K.J., Earle E., Hill J., Choo K.H., Earle E., Hill J., Choo K.H., Hill J., Choo K.H., Choo K.H. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Kreil D.P., Matzke M., Matzke A.J., Matzke M., Matzke A.J., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kato H., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Urano T., Furukawa K., Murakami Y., Furukawa K., Murakami Y., Murakami Y. RNA Polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Klein U.R., Nigg E.A., Gruneberg U., Nigg E.A., Gruneberg U., Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell. 2006;17:2547–2558. doi: 10.1091/mbc.E05-12-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T., Hori T., Okada M., Fukagawa T., Okada M., Fukagawa T., Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Trinkle-Mulcahy L., Lam Y.W., Andersen J.S., Mann M., Lamond A.I., Trinkle-Mulcahy L., Lam Y.W., Andersen J.S., Mann M., Lamond A.I., Lam Y.W., Andersen J.S., Mann M., Lamond A.I., Andersen J.S., Mann M., Lamond A.I., Mann M., Lamond A.I., Lamond A.I. NOPdb: Nucleolar Proteome Database. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A.W., Liao G.C., Rocchi M., Choo K.H., Liao G.C., Rocchi M., Choo K.H., Rocchi M., Choo K.H., Choo K.H. Extreme reduction of chromosome-specific α-satellite array is unusually common in human chromosome 21. Genome Res. 1999;9:895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- Lohrum M.A., Ashcroft M., Kubbutat M.H., Vousden K.H., Ashcroft M., Kubbutat M.H., Vousden K.H., Kubbutat M.H., Vousden K.H., Vousden K.H. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Taddei A., Lachner M., Jenuwein T., Almouzni G., Lachner M., Jenuwein T., Almouzni G., Jenuwein T., Almouzni G., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Martens J.H., O’Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., O’Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Opravil S., Radolf M., Steinlein P., Jenuwein T., Radolf M., Steinlein P., Jenuwein T., Steinlein P., Jenuwein T., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau-Thuillier S., Andreassen P.R., Margolis R.L., Andreassen P.R., Margolis R.L., Margolis R.L. Colocalization of TD-60 and INCENP throughout G2 and mitosis: Evidence for their possible interaction in signalling cytokinesis. Chromosoma. 1998;107:461–470. doi: 10.1007/s004120050330. [DOI] [PubMed] [Google Scholar]
- Meder V.S., Boeglin M., de Murcia G., Schreiber V., Boeglin M., de Murcia G., Schreiber V., de Murcia G., Schreiber V., Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Guilleme M., Seeler J.S., Trouche D., Dejean A., Yaniv M., Guilleme M., Seeler J.S., Trouche D., Dejean A., Yaniv M., Seeler J.S., Trouche D., Dejean A., Yaniv M., Trouche D., Dejean A., Yaniv M., Dejean A., Yaniv M., Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A.F., Earnshaw W.C., Earnshaw W.C. Specific interaction between human kinetochore protein CENP-C and a nucleolar transcriptional regulator. J. Biol. Chem. 1996;271:18767–18774. doi: 10.1074/jbc.271.31.18767. [DOI] [PubMed] [Google Scholar]
- Politi V., Perini G., Trazzi S., Pliss A., Raska I., Earnshaw W.C., Della Valle G., Perini G., Trazzi S., Pliss A., Raska I., Earnshaw W.C., Della Valle G., Trazzi S., Pliss A., Raska I., Earnshaw W.C., Della Valle G., Pliss A., Raska I., Earnshaw W.C., Della Valle G., Raska I., Earnshaw W.C., Della Valle G., Earnshaw W.C., Della Valle G., Della Valle G. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 2002;115:2317–2327. doi: 10.1242/jcs.115.11.2317. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Hannan K.M., Snelling H., Walkley C.R., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Hannan K.M., Snelling H., Walkley C.R., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Snelling H., Walkley C.R., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Walkley C.R., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Wall M., Brandenburger Y., Palatsides M., Pearson R.B., Brandenburger Y., Palatsides M., Pearson R.B., Palatsides M., Pearson R.B., Pearson R.B., et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.A., Lens S.M., Span S.W., Vader G., Medema R.H., Kruyt F.A., Giaccone G., Lens S.M., Span S.W., Vader G., Medema R.H., Kruyt F.A., Giaccone G., Span S.W., Vader G., Medema R.H., Kruyt F.A., Giaccone G., Vader G., Medema R.H., Kruyt F.A., Giaccone G., Medema R.H., Kruyt F.A., Giaccone G., Kruyt F.A., Giaccone G., Giaccone G. Subcellular localization and nucleocytoplasmic transport of the chromosomal passenger proteins before nuclear envelope breakdown. Oncogene. 2006;25:4867–4879. doi: 10.1038/sj.onc.1209499. [DOI] [PubMed] [Google Scholar]
- Saxena A., Wong L.H., Kalitsis P., Earle E., Shaffer L.G., Choo K.H., Wong L.H., Kalitsis P., Earle E., Shaffer L.G., Choo K.H., Kalitsis P., Earle E., Shaffer L.G., Choo K.H., Earle E., Shaffer L.G., Choo K.H., Shaffer L.G., Choo K.H., Choo K.H. Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum. Mol. Genet. 2002;11:2319–2329. doi: 10.1093/hmg/11.19.2319. [DOI] [PubMed] [Google Scholar]
- Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J., Greco A., Hochstrasser D., Diaz J.J., Hochstrasser D., Diaz J.J., Diaz J.J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M., Lafontaine D.L., Lafontaine D.L. Birth of a nucleolus: The evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Topp C.N., Zhong C.X., Dawe R.K., Zhong C.X., Dawe R.K., Dawe R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A.G., Wong L., Pakusch M., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H., Wong L., Pakusch M., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H., Pakusch M., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H., Burrows F.J., Vaux D.L., Choo K.H., Vaux D.L., Choo K.H., Choo K.H. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Teng G., Grewal S.I., Martienssen R.A., Grewal S.I., Martienssen R.A., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wong J.M., Kusdra L., Collins K., Kusdra L., Collins K., Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 2002;4:731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- Wong L.H., Saffery R., Anderson M.A., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H., Saffery R., Anderson M.A., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H., Anderson M.A., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H., Stafford A.J., Fowler K.J., Choo K.H., Fowler K.J., Choo K.H., Choo K.H. Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J. Biol. Chem. 2005;280:3954–3962. doi: 10.1074/jbc.M410047200. [DOI] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C., Saitoh H., Johnson D.H., Earnshaw W.C., Johnson D.H., Earnshaw W.C., Earnshaw W.C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Hemmerich P., Grosse F., Hemmerich P., Grosse F., Grosse F. Nucleolar localization of the human telomeric repeat binding factor 2 (TRF2) J. Cell Sci. 2004;117:3935–3945. doi: 10.1242/jcs.01249. [DOI] [PubMed] [Google Scholar]