Abstract
COMMD1 (previously known as MURR1) belongs to a novel family of proteins termed the copper metabolism gene MURR1 domain (COMMD) family. The 10 COMMD family members are well conserved between vertebrates, but the functions of most of the COMMD proteins are unknown. We recently established that COMMD1 is associated with the hepatic copper overload disorder copper toxicosis in Bedlington terriers. Recent in vitro studies indicate that COMMD1 has multiple functions, including sodium transport and NF-κB signaling. To elucidate the function of Commd1 in vivo, we generated homozygous Commd1 null (Commd1−/−) mice. Commd1−/− embryos died in utero between 9.5 and 10.5 days postcoitum (dpc), their development was generally retarded, and placenta vascularization was absent. Microarray analysis identified transcriptional upregulation of hypoxia-inducible factor 1 (HIF-1) target genes in 9.5-dpc Commd1−/− embryos compared to normal embryos, a feature that was associated with increased Hif-1α stability. Consistent with these observations, COMMD1 physically associates with HIF-1α and inhibits HIF-1α stability and HIF-1 transactivation in vitro. Thus, this study identifies COMMD1 as a novel regulator of HIF-1 activity and shows that Commd1 deficiency in mice leads to embryonic lethality associated with dysregulated placenta vascularization.
COMMD1 (previously known as MURR1) is the prototype of a recently defined protein family, the copper metabolism gene MURR1 domain (COMMD) protein family (7). So far, 10 family members of largely unknown function have been identified in humans, and they share the unique COMMD of 70 to 85 amino acids. This domain appears to have an important role in protein-protein interactions and represents a novel protein-protein interaction motif. The COMMD proteins are well conserved between vertebrates, and COMMD orthologs have been found in lower organisms (7). Recent data have implicated COMMD1 in various cellular processes.
We and others established that a loss of function of the COMMD1 protein, due to a homozygous deletion encompassing exon 2 of the COMMD1 gene, is associated with autosomal recessive copper toxicosis in Bedlington terriers (8, 22, 41). This autosomal recessive disorder is characterized by inefficient excretion of copper via the bile, resulting in progressive accumulation of hepatic copper, which leads to chronic hepatitis and ultimately liver cirrhosis (17, 37, 38). COMMD1 interacts with ATP7B, the copper-transporting P-type ATPase mutated in Wilson disease, a hepatic copper overload disease in humans very similar to canine copper toxicosis (40). In addition, downregulation of COMMD1 expression by small interfering RNA (siRNA) increases cellular copper levels (6). A protein-protein interaction between COMMD1 and the X-linked inhibitor of apoptosis (XIAP) has also been identified (6). The E3 ubiquitin ligase activity of XIAP regulates the ubiquitination and proteasomal degradation of COMMD1, which subsequently influences cellular copper levels (6). Biasio et al. established that COMMD1 can also interact with several subunits of the epithelial sodium channel, resulting in inhibition of the amiloride-induced sodium current (3). Recent studies identified an association between COMMD1 and several components of the NF-κB signaling pathway (7, 13, 26). COMMD1 inhibits the NF-κB transcriptional activity that induces human immunodeficiency virus type 1 replication in resting CD4+ lymphocytes. Interestingly, all COMMD proteins can regulate NF-κB transcriptional activity, which suggests that the COMMD family members not only are structurally related but also have some functional redundancy (7, 10). These data indicate that COMMD1 is a regulator of multiple, important cellular processes, but its exact biochemical function and physiological role in vivo remain elusive.
To explore the in vivo function of COMMD1, we generated and characterized a Commd1 knockout (Commd1−/−) mouse. Our data clearly demonstrate that Commd1 has an essential role in regulating hypoxia-inducible factor 1 (HIF-1) activity during early mouse embryogenesis.
MATERIALS AND METHODS
Generation of the target vector and transgenic mice.
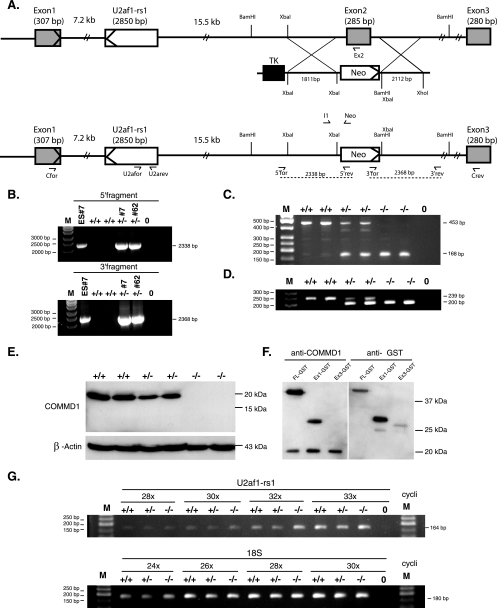
The genomic sequences that flank Commd1 exon 2 at the 5′ and 3′ ends were amplified with sense primer 5′-TCTAGACCCCAGATCAAGATACTGATTG-3′ and antisense primer 5′-TCTAGAGAGCCAGCTCTTCAGTGATCAG-3′ and with sense primer 5′-GGATCCTCTAGAGCTACATGTCTTTGCAGTTGTC-3′ and antisense primer 5′-CTCGAGCAGAACTATTCTTCACAATTGGAG-3′, respectively. The amplified products were separately cloned into the pCR2.1 TA cloning vector (Invitrogen, Breda, The Netherlands). The 5′ target sequence (1.8 kb) and the 3′ target sequence (2.1 kb) were subcloned into the target vector (pMCneo) using the XbaI restriction site for the 5′ target sequence and the BamHI and XhoI restriction sites for the 3′ target sequence (Fig. 1A). The target vector contained the neomycin (neo) and thymidine kinase (tk) genes for positive and negative selection, respectively. The XhoI-linearized targeting vector was electroporated into the 129Sv/Ev embryonic stem (ES) cell line (33). Selection for neomycin resistance and thymidine kinase was carried out, and two correctly targeted ES cell lines, cell lines 7 and 62, were injected into blastocysts collected from C57BL/6J mice according to standard procedures to generate two independent mouse lines. ES cells and F1 animals were genotyped by PCR using primers localized outside the target sequences and primers localized inside the neo gene. A 5′ fragment (2.3 kb) was amplified using sense primer 5′for (5′-CTGGTATTAAATGAGAATGCATG-3′) and antisense primer 5′rev (5′-CCTGCGGCAATCCATCTTG-3′), and a 3′ fragment (2.4 kb) was amplified using sense primer 3′for (5′-CCTCGTGCTTTACGGTATCG-3′) and antisense primer 3′rev (5′-GAGTGGTCCATTTCAATTGCTG-3′) (Fig. 1A and B). Genomic DNA isolated from tails (19), embryos, or yolk sacs (30) was used for genotyping by duplex PCR using sense primers Ex2 (5′-CAG ATA CAC AGC CCT GTT GC-3′), Neo (5′-CCT CGT GCT TTA CGG TAT CG-3′), and I1 (5′-GAG ACA ACT GCA AAG ACA TGT AGC-3′), yielding a 239-bp product for the wild-type allele and a 200-bp product for the targeted allele.
FIG. 1.
Targeted disruption of the Commd1 gene by homologous recombination. (A) Maps of the wild-type Commd1 allele, the targeting vector, and the targeted Commd1 allele. Shaded rectangles represent the three exons of the Commd1 gene. Open rectangle, the intronic gene U2af1-rs1. The orientations of the transcription of Commd1 and U2af1-rs1 are indicated by arrowheads within the first exons of the genes. The targeting vector contains the flanking genomic sequences, a neomycin gene (neo), and a thymidine kinase gene (tk). Homologous recombination is indicated by crossed lines and should result in the predicted targeted allele. Primers used for genotyping and RT-PCR are indicated by half-arrows. (B) PCR primers located outside the target sequences and primers localized inside the neo gene were used for PCR analysis to verify the correctly targeted ES cell clones. Genomic DNA was isolated from ES cell clone 7 and F1 progeny from independent lines 7 and 62. A 5′ fragment (2.3 kb) was amplified using sense primer 5′for and antisense primer 5′rev, and a 3′ fragment (2.4 kb) was amplified using sense primer 3′for and antisense primer 3′rev. (C) RT-PCR of wild-type, heterozygous, and homozygous Commd1 null embryos using primers Cfor and Crev. (D) PCR analysis of wild type (+/+), heterozygous (+/−), and homozygous Commd1 null (−/−) embryos using duplex PCR. (E) Immunoblot analysis of Commd1 and β-actin in 9.5-dpc embryos. (F) Immunoblot analysis of Commd1-GST (FL-GST), Commd1 exon 1-GST (Ex1-GST), and Commd1 exon 3-GST (Ex3-GST) in transfected HEK 293T cells using anti-Commd1 antiserum or anti-GST antibodies. (G) Semiquantitative RT-PCR analysis to determine the expression of U2af1-rs1 in relation to the expression of 18S rRNA in wild-type (+/+), heterozygous Commd1 null (+/−), and homozygous Commd1 null (−/−) 9.5-dpc embryos. The number of cycles used in this PCR is indicated (e.g., “28x” for 28 cycles). Demineralized water was used as a negative control (indicated by 0), and the size marker (M) is a 50-bp ladder.
RNA isolation and RT-PCR.
Total RNA was isolated from embryos at 9.5 days postcoitum (dpc) by using RNAStat60 (Tel-Test Inc., Friendswood, TX) according to the manufacturer's specifications. Total RNA was treated with DNase I (Invitrogen), and approximately 1 μg of total RNA was used to generate first-strand cDNA using oligo(dT) primers and Superscript SVII (Invitrogen, Breda, The Netherlands). DNase treatment and reverse transcriptase reactions were performed according to the manufacturer's specifications. Successful DNase treatment was verified by a PCR specific for genomic DNA. Reverse transcription-PCR (RT-PCR) was used to genotype the embryos by using sense primer Cfor (5′-CGCAGAACGCCTTTCACGG-3′) and antisense primer Crev (5′-TTTGCTTGACTTTAACTTCATC-3′), yielding a 453-bp product for the wild-type transcript and a 169-bp product for the mutant transcript. The expression of U2af1-rs1 in wild-type, heterozygous, and homozygous Commd1 null embryos was determined by semiquantitative PCR using sense primer U2afor (5′-TCTAATTCCCAACCAAGTTAC-3′) and antisense primer U2arev (5′-AAAACAACATGGGAAGCCAC-3′) (Fig. 1A). As an endogenous reference, the expression of 18S rRNA was determined using sense primer 18Sfor (5′-CCACATCCAAGGAAGGCAG-3′) and antisense primer 18Srev (5′-GCTGGAATTACCGCGGCTG-3′).
Copper supplementation.
Drinking water containing 6 mM CuCl2 was provided to female Commd1+/− mice starting 3 weeks before intercrossing, and copper supplementation continued through gestation and lactation. As described previously, these mice ingested approximately 50 to 100 times more copper than mice on a control diet (24).
Expression constructs, cell culture, transfections, and immunoblotting.
Sequences representing either full-length mouse Commd1 or exons 1 and 3 of mouse Commd1 were amplified from mouse liver cDNA using primers ex1for (5′-GGATCCGCCACCATGGCGGG CGATCTGGAGG-3′), ex1rev (5′-GCGGCCGCCTTGAGAAGTCCTCTCATC-3′), ex3for (5′-GGATCCGCCACCATGGAATCTGAATTTTTGTGTC-3′), and ex3rev (5′-GCGGCCGCGGCTGCCTGCATCAGCCTG-3′). PCR products were subcloned into pEBB-GST by using the BamHI and NotI restriction sites. The sequences of the constructs were verified by sequence analysis. Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, l-glutamine, and penicillin-streptomycin. Calcium phosphate precipitation was used to transfect HEK 293T cells as described previously (12). After transfection, cells were lysed in lysis buffer (1% Triton X-100, 25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 10% glycerol) supplemented with 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, protease inhibitors (Roche, Basle, Switzerland), and 10 mM dithiothreitol. Cell line extracts representing 60 μg protein were electrophoresed and then transferred to Hybond-P membranes. (These techniques are described in reference 22.) Immunoblots were probed with an anti-COMMD1 antiserum (1:2,000), an anti-glutathione S-transferase (anti-GST) antiserum (Santa Cruz, Santa Cruz, CA), or anti-β-actin monoclonal antibodies (clone AC-74; Sigma-Aldrich, Zweindrecht, The Netherlands). Normal and Commd1−/− embryos at 9.5 dpc were used for the preparation of total homogenates with the lysis buffer containing 0.4 M NaCl, 0.1% NP-40, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 400 nM NaCl. Equal amounts of protein from the embryos were transferred to Hybond-P membranes and were probed with anti-HIF-1α (NB 100-449; Novus, Huissen, The Netherlands) or anti-α-tubulin (clone DM 1A; Sigma). HIF-1α protein expression in HEK 293T cells was detected with anti-HIF-1α (clone 54; BD Bioscience, Alphen aan den Rijn, The Netherlands).
Generation of stable HEK 293T COMMD1 knockdown cells.
Plasmids encoding short hairpin RNAs were generated by cloning a target sequence specific for COMMD1 (GTCTATTGCGTCTGCAGAC) into pRETRO-SUPER as previously described (4, 5). To produce amphotropic retrovirus supernatants, Phoenix packaging cells were transfected with either empty pRETRO-SUPER or a pRETRO-SUPER construct targeting COMMD1 by using Fugene-6 (Roche, Woerden, The Netherlands) according to the manufacturer's instructions. Upon addition of 4 μg/ml Polybrene, HEK 293T cells were infected with the retrovirus supernatants and grown on a medium supplemented with 1 μg/ml puromycin (Sigma-Aldrich) to allow for monoclonal selection. Western blot analysis with an anti-COMMD1 antibody was used to quantitate the level of knockdown, typically ≥90% (P. de Bie et al., submitted for publication).
Histology and immunohistochemistry.
Embryos at 7.5 to 10.5 dpc were dissected from the decidua, and the yolk sac was removed for genotyping. The embryos were photographed and fixed for 12 h in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C. Embryos were dehydrated in ethanol and embedded in paraffin, and serial sections (thickness, 4 μm) were prepared. Sections were stained with hematoxylin and eosin. Apoptosis was detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining using the DeadEnd colorimetric TUNEL system (Promega, Leiden, The Netherlands). Incorporation of 5-bromo-2′-deoxyuridine (BrdU) was used to evaluate DNA synthesis during cell proliferation. Pregnant females received 50 mg of BrdU (Sigma)/kg of body weight by intraperitoneal injection. After 2 h, embryos were dissected, genotyped, and fixed as described above.
For the immunohistochemistry study, sections were deparaffinized, and primary antibodies against the mitosis marker phosphohistone H3 (Ser10) (06-570; Upstate, Veenendaal, The Netherlands) or against BrdU (ab6326, Abcam, Huissen, The Netherlands) were used to study cell proliferation. Immunoreactivity was visualized with the appropriate species-specific fluorescein isothiocyanate- or horseradish peroxidase-conjugated antibodies. Histochemical copper staining was performed using rhodamine.
Whole-mount in situ hybridization.
Wild-type embryos at different embryonic stages were dissected from the decidua, fixed for 12 h in 4% paraformaldehyde in PBS, washed with PBS-0.1% Tween 20, and dehydrated in methanol. Whole-mount in situ hybridization was performed as described previously (43). Commd1 antisense and sense probes were generated as described previously (44).
qRT-PCR.
Expression of BNIP3 mRNA in HEK 293T knockdown and control cell lines was determined by quantitative RT-PCR (qRT-PCR) on the ABI-7900 system (Applied Biosystems, Nieuwerkerk aan den Ijssel, The Netherlands) using SYBR PCR master mix (Applied Biosystems). Total RNA was isolated from HEK 293T cells using Trizol (Invitrogen) according to the manufacturer's specifications. The high-capacity cDNA archive kit (Applied Biosystems) was used to generate single-stranded cDNA from 1 μg of total RNA. We used primers 5′-AATATTCCCCCCAAGGAGTTCC-3′ and 5′-CTGCAGAGAATATGCCCCCTTT-3′ to amplify BNIP3. β-actin was used to normalize our data and was amplified using primers 5′-CATGTACGTTGCTATCCAGGC-3′ and 5′-CTCCTTAATGTCACGCACGAT-3′. PCR was initiated by heating for 2 min at 50°C and proceeded with a denaturing step at 94°C for 10 min, followed by 40 cycles of 94°C for 15 s and annealing and elongation for 1 min at 60°C. The results were expressed as n-fold changes in the mRNA expression level and as means ± standard errors of the means.
Luciferase reporter assay, GST-pull down assay, and coimmunoprecipitation.
For luciferase assays, HEK 293T cells were seeded in 24-well plates in triplicate for each treatment group. Cells were cotransfected with a HIF-1-dependent 5×HRE-luciferase reporter plasmid (36) and expression vectors encoding TK-Renilla luciferase, COMMD1-Flag (10), and/or HIF-1α. Twenty-four hours after transfection, cells were exposed to normoxic or hypoxic conditions (1% O2) for 24 h. A microplate luminometer (Berthold, Regensdorf, Switzerland) was used to determine the luciferase reporter activity as described by the manufacturer (Promega dual-luciferase reporter assay system). Luciferase activity was normalized for Renilla luciferase activity to correct for transfection efficiency.
HEK 293T cells were cotransfected with HIF-1α-Flag and either COMMD-GST, GST, or COMMD1. Precipitations with glutathione-Sepharose were performed as previously described (10). HIF-1α-Flag immunoprecipitation was performed using FLAG M2 agarose beads (Sigma-Aldrich) according to the manufacturer's instructions. An anti-COMMD1 antiserum was added to HEK 293T cell lysates to immunoprecipitate COMMD1; this procedure was performed as described previously, with minor changes (11). Instead of protein G-agarose beads, protein A/G-agarose (Santa Cruz, Heidelberg, Germany) was used. Protein was detected by immunoblotting for GST, HIF-1α (clone 54; BD Bioscience), COMMD1, or Flag (A8592; Sigma).
Microarray data accession numbers.
The microarray data and the complete protocols used in this study have been deposited in the MIAME (minimal information about a microarray experiment) database (in progress) at http://www.ebi.ac.uk/arrayexpress under the following accession numbers: microarray layout, A-UMCU-7; data, E-MEXP-832; protocols for embryo dissection, P-MEXP-30783; RNA isolation, P-MEXP-30781; mRNA amplification, P-MEXP-30781; cRNA labeling, P-MEXP-28902/3; generating the reference pool, P-MEXP-28722; hybridization and washing of slides, P-MEXP-28890; scanning of slides, P-MEXP-28893; data normalization, P-MEXP-28894.
RESULTS
Generation and initial characterization of Commd1-deficient mice.
To study the function of Commd1 in vivo, we generated Commd1-deficient mice by replacing exon 2 with a neo gene. As in humans and dogs, the Commd1 gene in mice consists of three exons (Fig. 1A), so this strategy mimics the in-frame exon 2 deletion observed in Bedlington terriers affected with copper toxicosis. Three of 132 ES cell clones were correctly targeted, and 2 of these clones (clones 7 and 62) (Fig. 1B) were used to generate two independent transgenic mouse lines. F1 heterozygous mice were generated by crossing the male chimeras with 129Sv/Ev females (Fig. 1B and D). Heterozygous mice were born healthy and were fertile; no apparent phenotype has been observed.
We confirmed by genomic PCR (Fig. 1B), RT-PCR, and immunoblotting (Fig. 1C and E) that exon 2 was correctly targeted. Analysis of 9.5-dpc embryos indicated that replacing exon 2 with the neo gene resulted in a truncated Commd1 transcript (Fig. 1C). After we had confirmed that Commd1 exon 2 was correctly targeted, we genotyped the mice by using a duplex genomic PCR with primers corresponding to exon 2 or the neo gene and a primer within intron 1 of the Commd1 gene (Fig. 1D). This in-frame deletion leads to a Commd1 null allele; no Commd1 protein could be detected in Commd1−/− embryos, whereas Commd1 protein was readily detected in wild-type and heterozygous embryos (Fig. 1E). We confirmed that the anti-COMMD1 antisera (22) would recognize the putative in-frame deleted protein encoded by the truncated transcript (the exon 1-exon 3 predicted protein product of 11 kDa) by immunoblotting. As shown in Fig. 1F, anti-COMMD1 antisera detected the part of the Commd1 protein encoded by Commd1 exon 1 but failed to detect amino acids 154 to 188, encoded by Commd1 exon 3. Hence, a shortened in-frame protein product, if expressed, would have been detected. These combined analyses clearly demonstrated that replacing exon 2 with a neo gene leads to a complete loss of function of the Commd1 protein, similar to that observed for Bedlington terriers affected with copper toxicosis (22, 41).
Intron 1 of the mouse Commd1 locus contains the imprinted gene U2af1-rs1 (Fig. 1A) (29). The single-exon U2af1-rs1 gene is not located within either the human or the dog COMMD1 locus. For mice, it has been shown that U2af1-rs1 is transcribed mainly from the paternal allele in the adult brain and the liver and that it may interfere with transcription of the paternal Commd1 allele (44, 46). We addressed the possibility that our targeting strategy had interfered with U2af1-rs1 expression by analyzing the expression of the intronic gene U2af1-rs1 in 9.5-dpc embryos. This analysis indicated that U2af1-rs1 mRNA expression was not affected by targeting the Commd1 allele, since no difference in U2af1-rs1 expression was observed between Commd1−/− and Commd1+/+ embryos (Fig. 1G). Recently, a novel transcript, U2mu, which is transcribed from a part of U2af1-rs1 and exons 2 and 3 of Commd1, has been described (45). The transcript of U2mu was also truncated after replacing exon 2 of the Commd1 gene. The U2mu transcript is confined to the mouse transcriptome, is in frame with exons 2 and 3 of Commd1, and encodes a predicted protein of approximately 26 kDa (45). We and others (45) performed extensive RNA expression analysis in different mouse tissues during development, which revealed that the expression of U2mu is extremely low compared to that of Commd1. Finally, although our anti-COMMD1 antisera recognize the protein encoded by Commd1 exon 2 (data not shown), no protein of 26 kDa could be detected by anti-COMMD1 antisera in normal mice. These observations strongly imply that the U2mu transcript does not encode a functionally expressed protein. Together, these data therefore suggest that our targeting of Commd1 exon 2 affects only the specific function of the Commd1 gene.
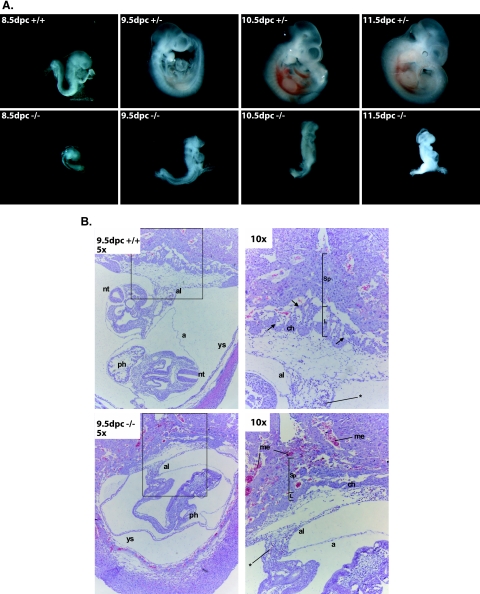
The Commd1+/− mice were intercrossed in order to produce Commd1−/− mice. Surprisingly, loss of Commd1 results in a recessive embryonic lethal phenotype, since no homozygous null mice were detected among 259 offspring (Table 1). Embryonic lethality was also observed on a C57BL/6J genetic background, indicating that this lethal phenotype is independent of the genetic background of the mouse strain (E. Burstein, University of Michigan, personal communication). To characterize the timing of the embryonic lethality, we collected embryos from timed Commd1+/− intercrosses at different stages of gestation. Commd1−/−, Commd1+/−, and Commd1+/+ embryos with the expected Mendelian ratio were recovered up to 9.5 dpc (Table 1). Only a few 10.5-dpc embryos and one Commd1−/− embryo at 11.5 dpc were recovered, but these were starting to be resorbed (Table 1; Fig. 2A), implying that Commd1−/− embryos die between 9.5 and 10.5 dpc.
TABLE 1.
Genotype analysis of offspring of Commd1+/− matings
| Age (dpc) | No. of mice with the following genotype:
|
Total no. of mice | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Weanling | 29 | 54 | 0 | 83 |
| 12.5 | 2 | 6 | 0 | 8 |
| 11.5 | 7 | 13 | 1 | 21 |
| 10.5 | 15 | 21 | 8 | 44 |
| 9.5 | 12 | 30 | 13 | 55 |
| 8.5 | 15 | 21 | 12 | 48 |
FIG. 2.
Morphological and histological analysis of wild-type and mutant embryos. (A) Different developmental stages (8.5 to 11.5 dpc) of wild-type (+/+) and heterozygous Commd1 null (+/−) embryos and homozygous Commd1 null (−/−) littermates. The embryonic development of Commd1−/− mice was delayed and was arrested at 9.5 dpc compared to that of their littermates. No axial rotation was observed for Commd1−/− embryos. (B) Hematoxylin-and-eosin staining of saggital sections of wild-type and Commd1−/− embryos in the decidua at 9.5 dpc (inset in left panel is enlarged in right panel). Besides the developmental delay, no vascularization was observed in the placenta, and increased numbers of maternal erythrocytes were observed, for Commd1−/− embryos. The allantois (al) is fused with the chorionic plate (ch). Primitive embryonic blood cells (indicated by asterisks) are present throughout the Commd1−/− embryo. a, amnion; L, labyrinth; me, maternal erythrocytes; nt, neural tube; ph, primitive heart; Sp, spongiotrophoblast; ys, yolk sac.
Phenotypes of the Commd1 null embryos.
To characterize the nature of the embryonic lethality, we studied the morphology of embryos from timed Commd1+/− intercrosses at different stages of gestation. Commd1−/− embryos were atypically small and were markedly delayed in development at 8.5 and 9.5 dpc (Fig. 2A). In spite of the developmental delay, no gross morphological abnormalities could be seen in Commd1−/− embryos at 9.5 dpc, and they were comparable in size to normal embryos at 8.5 dpc (Fig. 2A). Heart beating was observed for 9.5-dpc Commd−/− embryos as well as for normal embryos (both Commd1+/+ and Commd1+/−) at this embryonic stage, indicating that Commd1−/− embryos are still alive at 9.5 dpc. At 9.5 days of embryonic development, the wild-type and heterozygous embryos had undergone axial rotation, but this was not seen in Commd1−/− embryos (Fig. 2A).
The Commd1−/− embryos were further characterized by histochemical staining of transverse and sagittal serial sections of 9.5-dpc embryos. Although their development was markedly delayed, no gross abnormalities of the primitive organs could be seen in the Commd1−/− embryos compared to 8.5-dpc wild-type and heterozygous embryos. This was confirmed by studying the expression of the mesodermal marker Fgf8 and the somitic marker Meox1 by whole-mount in situ hybridization. No clear differences in mRNA expression of these genes were observed between 9.5-dpc Commd1−/− embryos and 8.5-dpc wild-type and heterozygous embryos (data not shown). Primitive nucleated red blood cells were seen throughout the whole Commd1−/− embryos, indicating that primitive hematopoiesis was not affected in Commd1−/− embryos (Fig. 2B). Although allantochorion fusion was observed at 9.5 dpc, the labyrinth formation was retarded and vascularization of the placenta had not started in the Commd1−/− embryos, whereas this was seen in both wild-type and heterozygous embryos (Fig. 2B). In addition, an increased number of maternal erythrocytes were seen in the placenta compared to both wild-type and heterozygous embryos (Fig. 2B).
Expression of Commd1 during normal mouse embryogenesis.
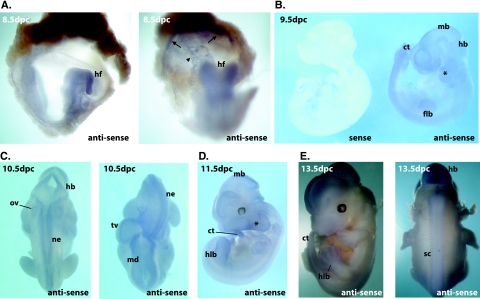
The essential role of Commd1 in mouse embryogenesis demanded a thorough analysis of the developmental expression of Commd1. We studied the expression of mouse Commd1 at different stages (8.5 to 13.5 dpc) of embryonic development by whole-mount RNA in situ hybridization (Fig. 3). At 8.5 dpc, Commd1 expression was detected throughout the embryo, with the highest expression in the headfold, allantois, and chorionic plate, but no clear expression was seen in the yolk sac (Fig. 3A). Diffuse expression of Commd1 continued during embryogenesis at stages 9.5 dpc, 10.5 dpc, and 11.5 dpc, but no expression was observed in the primitive heart (Fig. 3B to D). At later stages during development, Commd1 expression was relatively high in the neuroepithelium of the brain, the neural tube, and the epithelium of the otic vesicle (Fig. 3C and D). Expression was also clearly discernible in the caudal extremity of the tail, the limb buds, and the branchial arches. The same expression pattern was observed in 13.5-dpc embryos (Fig. 3E), but at this stage Commd1 expression was more pronounced in the interdigital regions of the hindlimb bud, the spinal cord, the caudal extremity of the tail, the developing brain, and the nose. Thus, Commd1 is ubiquitously expressed in the embryo and in the primitive placenta at 8.5 to 11.5 dpc but more distinctively at 13.5 dpc. In addition, these data clearly show that Commd1 expression is vital during the early embryonic stages of mouse development.
FIG. 3.
Expression of Commd1 during the embryonic stages 8.5 to 12.5 dpc. Whole-mount in situ hybridization was performed using an antisense probe to detect Commd1 expression, and a sense probe was used as a negative control. (A) Expression of Commd1 at 8.5 dpc was observed throughout the embryo. Higher expression was seen in the headfold (hf) and in the allantois and chorionic plate (indicated by an arrowhead and arrows, respectively). (B to E) Diffuse expression of Commd1 was also seen at 9.5, 10.5, 11.5, and 13.5 dpc but was more distinct at the neural tube, limb buds, caudal region of the tail (ct), neuroepithelium (ne), and branchial arches (*). flb, forelimb bud; hlb, hindlimb bud; md, midbrain; hb, hindbrain; tv, telencephalic vesicle; ov, otic vesicle; sc, spinal cord.
Analysis of cell proliferation and apoptosis in Commd1−/− embryos.
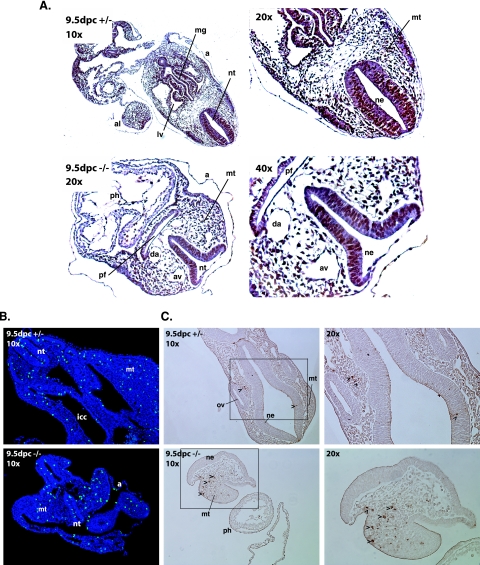
Since controlled cell proliferation and apoptosis are essential for the development of multicellular organisms, we investigated whether affected cell proliferation or increased cell death could explain the developmental delay and ultimately the arrest of development of Commd1−/− embryos. DNA replication in 9.5-dpc embryos was studied by determining BrdU incorporation in DNA. We observed no detectable difference in BrdU incorporation between Commd1-deficient embryos and either wild-type or heterozygous embryos (Fig. 4A). Cells in the G2 phase of the cell cycle were studied by determining the phosphorylation status of histone H3. At 9.5 dpc, phosphohistone H3-positive cells were clearly discerned in Commd1−/− embryos (Fig. 4B). The localization of the phosphohistone H3-positive cells (mesenchymal and epithelial cells), as well as the approximate ratios of phosphohistone H3-positive versus -negative cells, did not differ between heterozygous and wild-type embryos (Fig. 4B). Detection of TUNEL labeling was used to study cell death in 9.5-dpc embryos. An increase in the number of TUNEL-positive mesenchymal cells in the developing brain was seen for 9.5-dpc Commd1−/− embryos compared to both wild-type and heterozygous embryos (Fig. 4C), whereas no clear difference in the number of TUNEL-positive cells could be identified elsewhere in the embryos. Immunohistochemical analysis revealed almost no cleaved capsase-3-positive cells in 9.5-dpc Commd1−/− embryos or in wild-type or heterozygous embryos (data not shown). These combined data indicate that cell proliferation is not appreciably affected in 9.5-dpc Commd1−/− embryos, but the increase in the number of apoptotic cells indicates that 9.5-dpc Commd1−/− embryos are in poor vigor.
FIG. 4.
Analyses of cell proliferation and apoptosis were performed by BrdU incorporation, anti-phosphohistone H3 immunohistochemistry, and TUNEL assays of transverse sections of Commd1+/− and Commd1−/− embryos at 9.5 dpc. (A) BrdU staining; (B) anti-phosphohistone H3 staining; (C) TUNEL assay (insets enlarged in right panels). Arrowheads indicate apoptotic cells. a, amnion; al, allantois; ph, primitive heart; mt, mesenchyme tissue; nt, neural tube; ne, neural epithelium; av, anterior cardinal vein; da, dorsal aorta; pf, pharyngeal region; icc, intraembryonic coelomic cavity; mg, midgut; ov, otic vesicle.
Copper homeostasis in Commd1−/− embryos.
Since COMMD1 plays an essential role in copper homeostasis in dogs, we investigated whether embryonic copper overload could explain the lethal phenotype of Commd1−/− mice. The possibility of copper overload in Commd1−/− embryos or in the primitive placenta was analyzed using rhodamine staining of embryonic sections. This specific histochemical copper staining was used to identify copper overload in the livers of Bedlington terriers affected with copper toxicosis (18), but it did not show granular copper in 9.5-dpc Commd1−/− embryos or in the primitive placenta (data not shown). These data imply that Commd1−/− embryos do not have detectably elevated copper levels.
Previous studies have shown that severe copper deficiency leads to early embryonic death (23, 24, 27). In line with these observations, we hypothesized that inefficient copper transport from the mothers to their offspring due to the loss of Commd1 could explain the embryonic lethality of Commd1−/− embryos. In an attempt to circumvent this potential copper deficiency, heterozygous females were provided with a high-copper diet (6 mM CuCl2 supplied via the drinking water [24]) starting at 3 weeks before intercrossing and lasting through gestation and lactation. This intervention did not rescue the lethal phenotype of the Commd1−/− embryos, suggesting that copper deficiency is not the main cause for this phenotype. Alternatively, Commd1 might not be essential for copper uptake even under conditions of excessive copper intake.
Genome-wide gene expression analysis of whole embryos.
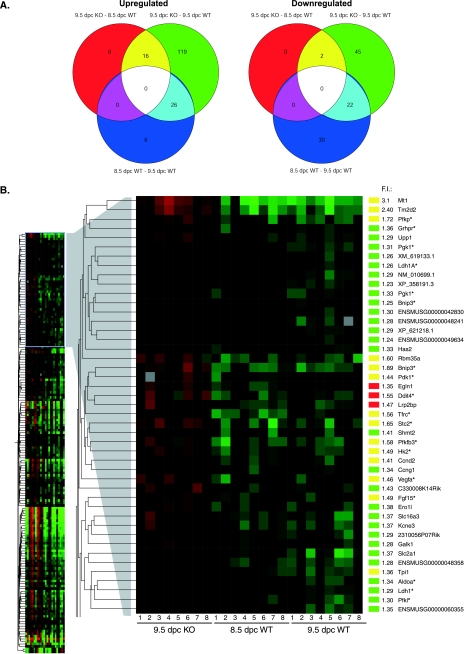
A gene expression study using microarray analysis was performed to elucidate the underlying mechanism leading to embryonic lethality in Commd1−/− embryos. Since 9.5-dpc Commd1−/− embryos are morphologically more similar to 8.5-dpc normal embryos than to 9.5-dpc normal embryos, we compared the gene expression profiles of 9.5-dpc Commd1−/− embryos with those of 8.5-dpc as well as 9.5-dpc wild-type and heterozygous embryos. Microarray analysis of variance resulted in a total of 236 genes with significantly different levels of mRNA expression (P < 0.05) in 9.5-dpc Commd1−/− embryos compared to 8.5-dpc and 9.5-dpc wild-type and heterozygous embryos (Fig. 5). As shown in the Venn diagrams, the number of differentially expressed genes for 9.5-dpc Commd1−/− embryos compared to 8.5-dpc wild-type and heterozygous embryos was lower than that for 9.5-dpc Commd1−/− embryos compared to 9.5-dpc wild-type and heterozygous embryos (Fig. 5A). Many of the same genes differentially expressed for 9.5-dpc Commd1−/− versus 9.5-dpc wild-type embryos (20%) were also differentially expressed for 8.5-dpc wild-type versus 9.5-dpc wild-type embryos (Fig. 5A). This demonstrates that the gene expression profiles of 9.5-dpc Commd1−/− embryos reflect their delay in embryonic development.
FIG. 5.
Genome-wide gene expression analysis of 9.5-dpc Commd1−/− embryos compared to 8.5- and 9.5-dpc normal embryos. (A) Venn diagrams show the numbers of genes that were significantly differentially expressed by the indicated comparison groups: 9.5-dpc Commd1−/− (KO) versus 8.5-dpc wild-type (WT) embryos, 9.5-dpc Commd1−/− versus 9.5-dpc wild-type embryos, and 8.5-dpc versus 9.5-dpc wild-type embryos. Data for upregulated and downregulated genes are given in the left and right panels, respectively. (B) Genes significantly upregulated in 9.5-dpc Commd1−/− embryos compared to each wild-type group are shown in a cluster diagram. The cluster of genes with increased mRNA expression in 9.5-dpc Commd1−/− embryos compared to both 8.5- and 9.5-dpc normal embryos is enlarged, and n-fold induction (F.I.) and gene names are given on the right. F.I. represents the mean expression of the genes in 9.5-dpc Commd1−/− embryos compared to the average expression in 8.5-dpc and 9.5-dpc normal embryos. The color bars correspond to the colors used in the Venn diagrams. Asterisks indicate the HIF-1 target genes, based on the literature.
Next, we reasoned that most information on the mechanisms of embryonic lethality would be provided by genes that were differentially expressed in Commd1−/− embryos compared with wild-type and heterozygous embryos at both 8.5 and 9.5 dpc. We identified an overlap of only 16 upregulated and 2 downregulated genes between the comparison of 9.5-dpc Commd1−/− with 9.5-dpc wild-type and heterozygous embryos and the comparison of 9.5-dpc Commd1−/− with 8.5-dpc wild-type and heterozygous embryos (Fig. 5A). Most interestingly, 8 of the 16 upregulated genes appeared to be targets of HIF-1 (Fig. 5A and B) (also see http://humgen.med.uu.nl). The mRNA expression of eight additional HIF-1 target genes was significantly higher in 9.5-dpc Commd1−/− embryos than in 9.5-dpc wild-type and heterozygous embryos, and that of two additional genes was significantly higher than in 8.5-dpc wild-type and heterozygous embryos (Fig. 5A and B) (also see http://humgen.med.uu.nl). Almost 10% of the significantly differentially expressed genes in 9.5-dpc Commd1−/− embryos have previously been shown to be induced by the transcription factor HIF-1 (1, 16, 25, 35, 42) (Table 2). These genes are involved in different cellular processes, including angiogenesis and glycolysis (Table 2). These data indicate that transcriptional regulation by HIF-1 is markedly affected in Commd1−/− embryos, which provides a possible mechanism for the embryonic lethality associated with Commd1 deficiency in mice.
TABLE 2.
Hypoxia-associated genes which are upregulated in 9.5-dpc Commd1−/− embryos compared to 8.5-dpc and 9.5-dpc normal embryos
| Function | Gene name | Protein |
|---|---|---|
| Glycolysis | Pfkfb3 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| Pfkp | Phosphofructokinase, platelet | |
| Pdk1 | Pyruvate dehydrogenase kinase, isoenzyme 1 | |
| Pgk1 | Phosphoglycerate kinase 1 | |
| Hk2 | Hexokinase 2 | |
| LdhA | Lactate dehydrogenase 1, A chain | |
| AldoA | Aldolase 1, A isoform | |
| Angiogenesis | Vegfa | Vascular endothelial growth factor A |
| Metabolism | Grhpr | Glyoxylate reductase/hydroxypyruvate reductase |
| Tf | Transferrin | |
| Tfrc | Transferrin receptor | |
| Prolyl hydroxylase | Phd2 | EGL nine homolog 1 (Caenorhabditis elegans) |
| Phd3 | EGL nine homolog 3 (C. elegans) | |
| Apoptosis | Ddit4 | DNA damage-inducible transcript 4 |
| Bnip3 | BCL2/adenovirus E1B 19-kDa-interacting protein 1, NIP3 | |
| Cytoskeleton | Krt19 | Keratin complex 1, acidic, gene 19 |
| Calcium/phosphate homeostasis | Stc2 | Stanniocalcin 2 |
| Cell-cell interaction | Lgals2 | Lectin, galactose binding, soluble 2 |
HIF-1α protein expression, activity, and interaction with COMMD1.
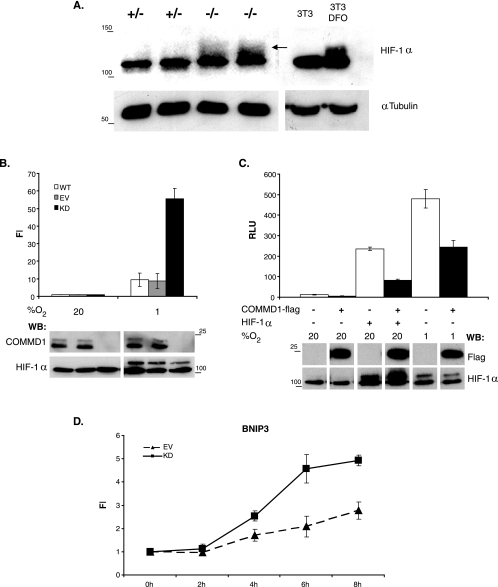
HIF-1 is a heterodimeric basic helix-loop-helix transcription factor composed of HIF-1α and HIF-1β. HIF-1α is constitutively expressed but rapidly degraded under normoxic conditions, a process that involves oxygen-dependent prolyl hydroxylation and ubiquitination. Given the observation that Hif-1 target genes were transcriptionally upregulated in Commd1−/− embryos, we investigated whether this was the result of higher Hif-1α protein levels in Commd1−/− embryos than in wild-type and heterozygous embryos. Hif-1α protein expression in 9.5-dpc normal and Commd1−/− embryos was determined by immunoblot analysis. As a control for distinguishing active Hif-1α, NIH 3T3 cells were treated with the iron chelator desferrioxamine (DFO). DFO mimics hypoxia by reducing the hydroxylation of HIF-1α via the prolyl-hydroxylases (2), resulting in a clear accumulation of active Hif-1α, which was visible as a smear, similarly to previous experiments using the same antiserum (Fig. 6A) (39). Increased active Hif-1α protein levels were also readily detected in 9.5-dpc Commd1−/− embryos compared to 9.5-dpc wild-type embryos (Fig. 6A).
FIG. 6.
Increased HIF-1α expression in Commd1−/− embryos and regulation of HIF-1-mediated transcription by COMMD1. (A) Immunoblot analysis of HIF-1α (arrow) and tubulin in 9.5-dpc embryos and in NIH 3T3 cells either left untreated or treated with DFO; the latter served as a control for distinguishing active Hif-1α. (B) HEK 293T cells with a stable knockdown of COMMD1 (KD) were cotransfected with 5×HRE-firefly luciferase reporter and Renilla luciferase control plasmids. HEK 293T cells (WT) and HEK 293T cells stably transfected with an empty siRNA vector (EV) were used as negative controls. Cells were incubated under normoxia or hypoxia. Firefly luciferase activities in the lysates were measured, corrected for Renilla luciferase activities, and expressed as n-fold induction (FI) relative to normoxic conditions. (C) HEK 293T cells were cotransfected with luciferase reporter plasmids as described for panel B, COMMD1-Flag, and/or HIF-1α and were incubated under normoxia or hypoxia. Luciferase activities in the lysates were measured and expressed as relative light units (RLU). All luciferase experiments were performed in triplicate. Error bars, standard deviations. Immunoblot analyses of HIF-1α, COMMD1, and COMMD-Flag protein expression are shown below the bar graphs. Sizes of molecular markers are given (in kilodaltons) on the left or right. WB, Western blot. (D) BNIP3 mRNA expression in HEK 293T KD and EV cells was determined at different time points during hypoxic exposure (3% O2) by qRT-PCR analysis. mRNA expression was normalized to the expression at time zero and is shown as the mean FI ± standard error of the mean for three biological replicates.
Next, we set out to investigate the relation between COMMD1 and HIF-1 activity in independent in vitro studies. To investigate whether COMMD1 deficiency leads to increased HIF-1 activity in cultured cells, reporter assays were performed with normal HEK 293T cells and with HEK 293T cells in which the expression of COMMD1 was stably knocked down to undetectable levels by RNA interference (Fig. 6B). These cells were transfected with a HIF-1-dependent reporter plasmid and were grown under normoxic or hypoxic conditions. As shown in Fig. 6B, reporter gene activity was induced by hypoxia. Consistent with our observations for Commd1-deficient embryos, COMMD1 knockdown in HEK 293T cells resulted in sixfold-higher induction of HIF-1 reporter gene activity compared to that for normal HEK 293T cells. To further corroborate these findings, overexpression of COMMD1 in HEK 293T cells resulted in marked inhibition of reporter induction under both hypoxic and normoxic conditions (Fig. 6C). To study the effect of COMMD1 deficiency on endogenous HIF-1 activity, we examined the mRNA expression of the HIF-1 target gene BNIP3 following hypoxic exposure (3% O2). BNIP3 was one of the HIF-1 target genes, which were transcriptionally induced in Commd1−/− embryos compared to wild-type embryos (Fig. 5B). In both cell lines, hypoxic exposure resulted in time-dependent increases in BNIP3 mRNA expression (Fig. 6D). Consistent with our previous observation, the level of BNIP3 mRNA expression was induced in HEK 293T COMMD1 knockdown cells relative to HEK 293T control cells.
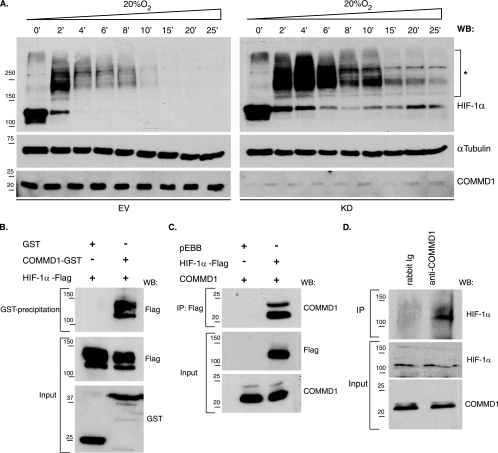
Although increased Hif-1α protein expression was observed in Commd1−/− embryos, no aberrant HIF-1α expression was seen in HEK 293T COMMD1 knockdown cells under normoxic conditions. To further investigate if COMMD1 mediates HIF-1α protein levels, we studied the stability of hypoxia-stabilized HIF-1α in HEK 293T COMMD1 knockdown and control cells. These cells were cultured for 18 h under hypoxia (1% O2) and were subsequently exposed to 20% O2 for different periods. Under hypoxia (time zero), both cell lines showed high HIF-1α expression, but the expression level decreased as soon as the cells were exposed to 20% O2. Interestingly, HIF-1α expression was almost absent in control cells after 10 min of exposure to 20% O2, whereas HIF-1α protein was still clearly observed in COMMD1 knockdown cells (Fig. 7A). Remarkably, after the cells were exposed to 20% O2 for 2 min, HIF-1α proteins of increasingly higher molecular mass were detected in both cell lines (Fig. 7A). These higher-molecular-weight proteins most probably represent ubiquitinated HIF-1α, since high oxygen levels promote the ubiquitination of HIF-1α. These data support the in vivo data showing that COMMD1 deficiency is associated with increased HIF-1α protein stability.
FIG. 7.
Increased HIF-1α protein stability in COMMD1 knockdown cells and association of COMMD1 with HIF-1α. (A) The stability of HIF-1α protein was determined in HEK 293T COMMD1 knockdown (KD) and control (EV) cells by immunoblot analysis. After the cells were cultured under hypoxic conditions (1% O2), they were exposed to 20% O2 for the indicated times. Asterisks indicate high-molecular-mass proteins detected by anti-HIF-1α. α-Tubulin expression was used as a loading control. WB, Western blot. (B) Glutathione-Sepharose precipitations using lysates of HEK 293T cells expressing HIF-1α-Flag, GST, or COMMD1-GST. (C) Flag immunoprecipitation using lysates of HEK 293T cells cotransfected with either HIF-1α-Flag or pEBB-Flag and COMMD1. (D) Endogenous COMMD1 immunoprecipitation using HEK 293T cell lysates. All precipitates were washed, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotted as indicated. “Input” indicates direct analysis of cell lysates. Ig, immunoglobulin. Sizes of molecular markers are given in kilodaltons on the left.
Next, we investigated whether the role of COMMD1 in mediating HIF-1 activity and HIF-1α stability was regulated by a physical association between COMMD1 and HIF-1α. GST-pull-down studies were performed using lysates from HEK 293T cells coexpressing COMMD1-GST and HIF-1α-Flag. HIF-1α was clearly detected in COMMD1-GST samples precipitated by glutathione-Sepharose but not in GST precipitates, indicating a specific interaction between COMMD1 and HIF-1α (Fig. 7B). Coimmunoprecipitation studies were performed using lysates from HEK 293T cells coexpressing COMMD1 and HIF-1α-Flag or untransfected cells to confirm the physical association between HIF-1α and COMMD1. COMMD1 was clearly detected in HIF-1α-Flag immunoprecipitates but not in immunoprecipitates from cells transfected with an empty vector (Fig. 7C). HIF-1α was also observed in COMMD1 immunoprecipitates but not in control immunoprecipitates, demonstrating the specificity of the interaction between COMMD1 and HIF-1α (Fig. 7D). Taken together, these in vitro data indicate that COMMD1 modulates the protein level of HIF-1α and HIF-1 activity, possibly via a COMMD1-HIF-1α protein interaction.
DISCUSSION
We and others have shown that COMMD1 is associated with copper homeostasis (8, 22, 40, 41), but recent studies have revealed that COMMD1 also has a role in NF-κB signaling, sodium transport, and XIAP signaling (3, 6, 7, 13, 15, 26). To further investigate the pleiotropic function of COMMD1 in vivo, we targeted exon 2 of the murine Commd1 gene by homologous recombination.
Extensive analyses of the targeted Commd1 locus showed that loss of Commd1 exon 2 specifically leads to Commd1 deficiency, a situation comparable to that of copper toxicosis-affected Bedlington terriers. To our surprise, Commd1−/− mice are embryonically lethal, whereas COMMD1-deficient Bedlington terriers pups are born healthy but gradually accumulate copper in their liver, leading to liver cirrhosis between the ages of 2 and 6 years (17). Although the reason for the phenotypic discrepancy between dogs and mice is currently unknown, one can speculate that there is some functional redundancy of the COMMD protein family that is species dependent. We were unable to detect any obvious copper accumulation in Commd1−/− embryos or in their placentas at 9.5 dpc. Embryonic lethality has previously been described for mice deficient in the copper transporters Ctr1 and Atp7a (23, 24, 27). The phenotypes of Commd1−/− mice differ markedly from the phenotypes of Ctr1−/− and Atp7a−/− mice, and we were unable to rescue the Commd1−/− embryonic lethality by providing a high-copper diet to the pregnant and lactating mothers. We therefore conclude that the embryonic lethal phenotype cannot be explained by a prominent role of Commd1 in copper homeostasis during mouse embryogenesis.
In this study we have uncovered a previously unidentified role of Commd1 as a mediator of HIF-1 activity during mouse embryogenesis. Gene expression profiling showed that HIF-1 target genes were induced in 9.5-dpc Commd1−/− embryos compared to 8.5-dpc and 9.5-dpc normal embryos. Interestingly, Murphy et al. suggested that hypoxia recruits metal transcription factor 1 (MTF-1) and HIF-1 to the promoter of the mouse Mt-1 gene, and this phenomenon might therefore also explain the induction of Mt-I (Fig. 5B) in 9.5-dpc Commd1−/− embryos (28). The transcriptional induction of HIF-1 target genes is associated with elevated Hif-1α protein levels in 9.5-dpc Commd1−/− embryos (Fig. 6A). HIF-1 is the master transcriptional regulator of adaptive responses to hypoxia, and previous studies have established that proper HIF-1 activity is essential for normal embryogenesis. Loss of Hif-1α leads to embryonic arrest at 9 dpc, and these embryos die at 10.5 dpc. Hif-1α−/− embryos show defects in the vascularization of the placenta and in cardiovascular and neural tube development and show increased cell death within cephalic mesenchyme (21). More importantly, increased HIF-1 activity has also been shown to lead to embryonic lethality during midgestation. This is illustrated by the phenotypes of the von Hippel-Lindau knockout mouse (Vhl−/−), lacking a HIF-1α inhibitor protein (14, 20), and the PHD2 knockout mouse (39). The embryonic lethality of both mouse models is caused by abnormalities of placental vasculogenesis due to upregulated HIF-1-mediated transcription (14). The importance of balanced HIF-1 activity for proper placental development between 8 and 10 dpc was further demonstrated by others (9, 39). They established that HIF-1 mediates the differentiation of trophoblast cells into spongiotrophoblast cells and inhibits the differentiation into labyrinthine trophoblast cells. An absence of vasculogenesis and an increased number of nonnucleated maternal erythrocytes were also observed in the placentas of Commd1−/− embryos (Fig. 2B), features very similar to what has been seen in Vhl−/− embryos. Our observation of high levels of Commd1 mRNA expression in the allantois and the chorionic plate at 8.5 dpc and recent data establishing the expression of COMMD1 in human placental villi (11) further support an essential role for Commd1 in the development of the chorioallantoic placenta. In this context it should be noted that vascularization of the placenta starts at 9.0 dpc (32), and since 9.5-dpc Commd1−/− embryos resemble the developmental stage of 8.5-dpc normal embryos, placental development in Commd1−/− embryos could be retarded rather than completely prevented. Finally, our data cannot rule out the possibility that Commd1 regulation of Hif-1 activity during mouse embryogenesis is indirect.
More evidence for the conclusion that COMMD1 mediates HIF-1 activity was provided by several independent in vitro experiments. HIF-1 luciferase reporter assays and qRT-PCR indicated that endogenous and overexpressed COMMD1 proteins directly mediate HIF-1 activity. The involvement of COMMD1 in HIF-1-mediated transcriptional regulation is corroborated by the observed protein-protein interaction between COMMD1 and HIF-1α. Interestingly, the increased HIF-1 activity in COMMD1-deficient cells compared to wild-type cells was detectable only under hypoxic conditions and not under normoxic conditions. This observation appears to be in accordance with the increased Hif-1 activity in Commd1 null embryos, since the physiological oxygen levels at 9.5 dpc are low, approximately 3% (9), suggesting that COMMD1 mediates HIF-1 activity under specific physiological conditions.
Elevated Hif-1α levels in Commd1−/− embryos and increased HIF-1α stability in COMMD1 knockdown cells suggest that COMMD1 regulates HIF-1 activity by controlling HIF-1α protein stability. The role of COMMD1 in stabilizing transcription factors has recently also been reported by others (7, 26). They demonstrated that COMMD1 mediates the protein stability of NF-κB subunits via the ubiquitin ligase complex ECSSOCS1, thereby regulating κB-mediated transcription. This may suggest that COMMD1 regulates HIF-1 activity similarly to the way it controls κB-mediated transcription, since HIF-1α protein stability is also regulated via an E3 ubiquitin ligase complex very similar to ECSSOCS1 (31).
Taken together, this study identified COMMD1 as a novel interactor and regulator of HIF-1 activity and indicates that loss of Commd1 leads to aberrant Hif-1α expression during early embryogenesis, resulting in increased mRNA expression of HIF-1 target genes. Apparently, dysregulated energy metabolism and lack of placental vascularization might explain the delay and eventual arrest of the embryonic development of Commd1−/− mice, but additional studies are needed to further delineate the role of Commd1 in the regulation of Hif-1 activity in the developing mouse embryo. The phenotypic discrepancy between COMMD1 deficiency in dog and mouse might be explained by differences between the development of the mouse placenta and that of the placentas of other mammals, even though substantial similarities exist (32). These data demonstrate for the first time that Commd1 regulates HIF-1 activity and is essential for normal mouse embryogenesis. Increased HIF-1 activity has also been associated with different diseases such as preeclampsia and cancer (reviewed in reference 34). Therefore, it will be interesting to further define the molecular mechanism by which COMMD1 regulates HIF-1 activity in different biological processes.
Acknowledgments
We thank F. Meijlink and J. Deschamps for valuable discussions and Jackie Senior for improving the manuscript. We thank Marc Vooijs (Department of Pathology, UMCU, Utrecht, The Netherlands) for reagents, advice on HIF-1α, and the use of the Ruskin INVIVO2 1000 hypoxic working station, which was financially supported by the Maurits and Anna de Kock Foundation. We acknowledge T. B. O'Brien for providing the Meox1 construct.
This work was financially supported partly by grant 40-00812-98-03106 to L.W.K. and C.W. from the Dutch Organization for Scientific Research (NWO) and partly by the Intramural Research Program of The National Human Genome Research Institute, National Institutes of Health (to B.V.D.S., A.C., and P.P.L.). A.G. was funded by the Dutch Cancer Society (UU2003-2825).
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Aprelikova, O., G. V. Chandramouli, M. Wood, J. R. Vasselli, J. Riss, J. K. Maranchie, W. M. Linehan, and J. C. Barrett. 2004. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J. Cell. Biochem. 92:491-501. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi, L., L. Tacchini, and G. Cairo. 1999. HIF-1-mediated activation of transferrin receptor gene transcription by iron chelation. Nucleic Acids Res. 27:4223-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biasio, W., T. Chang, C. J. McIntosh, and F. J. McDonald. 2004. Identification of Murr1 as a regulator of the human delta epithelial sodium channel. J. Biol. Chem. 279:5429-5434. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Burstein, E., L. Ganesh, R. D. Dick, B. van de Sluis, J. C. Wilkinson, L. W. Klomp, C. Wijmenga, G. J. Brewer, G. J. Nabel, and C. S. Duckett. 2004. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 23:244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burstein, E., J. E. Hoberg, A. S. Wilkinson, J. M. Rumble, R. A. Csomos, C. M. Komarck, G. N. Maine, J. C. Wilkinson, M. W. Mayo, and C. S. Duckett. 2005. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 280:22222-22232. [DOI] [PubMed] [Google Scholar]
- 8.Coronado, V. A., D. Damaraju, R. Kohijoki, and D. W. Cox. 2003. New haplotypes in the Bedlington terrier indicate complexity in copper toxicosis. Mamm. Genome 14:483-491. [DOI] [PubMed] [Google Scholar]
- 9.Cowden Dahl, K. D., B. H. Fryer, F. A. Mack, V. Compernolle, E. Maltepe, D. M. Adelman, P. Carmeliet, and M. C. Simon. 2005. Hypoxia-inducible factors 1α and 2α regulate trophoblast differentiation. Mol. Cell. Biol. 25:10479-10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bie, P., B. van de Sluis, E. Burstein, K. J. Duran, R. Berger, C. S. Duckett, C. Wijmenga, and L. W. Klomp. 2006. Characterization of COMMD protein-protein interactions in NF-κB signalling. Biochem. J. 398:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donadio, S., N. Alfaidy, B. De Keukeleire, J. Micoud, J. J. Feige, J. R. Challis, and M. Benharouga. 22 January 2007, posting date. Expression and localization of cellular prion and COMMD1 proteins in human placenta throughout pregnancy. Placenta. doi: 10.1016/j.placenta.2006.11.006. [DOI] [PubMed]
- 12.Duckett, C. S., R. W. Gedrich, M. C. Gilfillan, and C. B. Thompson. 1997. Induction of nuclear factor κB by the CD30 receptor is mediated by TRAF1 and TRAF2. Mol. Cell. Biol. 17:1535-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganesh, L., E. Burstein, A. Guha-Niyogi, M. K. Louder, J. R. Mascola, L. W. Klomp, C. Wijmenga, C. S. Duckett, and G. J. Nabel. 2003. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature 426:853-857. [DOI] [PubMed] [Google Scholar]
- 14.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94:9102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene, W. C. 2004. How resting T cells deMURR HIV infection. Nat. Immunol. 5:18-19. [DOI] [PubMed] [Google Scholar]
- 16.Greijer, A. E., P. van der Groep, D. Kemming, A. Shvarts, G. L. Semenza, G. A. Meijer, M. A. van de Wiel, J. A. Belien, P. J. van Diest, and E. van der Wall. 2005. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 206:291-304. [DOI] [PubMed] [Google Scholar]
- 17.Hardy, R. M., J. B. Stevens, and C. M. Stowe. 1975. Chronic progressive hepatitis in Bedlington terriers associated with elevated liver copper concentrations. Minn. Vet. 15:13-24. [Google Scholar]
- 18.Haywood, S., I. C. Fuentealba, S. J. Kemp, and J. Trafford. 2001. Copper toxicosis in the Bedlington terrier: a diagnostic dilemma. J. Small Anim. Pract. 42:181-185. [DOI] [PubMed] [Google Scholar]
- 19.Hofker, M. H., and J. van Deursen (ed.). 2002. Methods in molecular biology, vol. 209. Transgenic mouse methods and protocols. Humana Press, Totowa, NJ.
- 20.Hong, S. B., M. Furihata, M. Baba, B. Zbar, and L. S. Schmidt. 2006. Vascular defects and liver damage by the acute inactivation of the VHL gene during mouse embryogenesis. Lab. Investig. 86:664-675. [DOI] [PubMed] [Google Scholar]
- 21.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klomp, A. E., B. van de Sluis, L. W. Klomp, and C. Wijmenga. 2003. The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J. Hepatol. 39:703-709. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, Y. M., B. Zhou, D. Cosco, and J. Gitschier. 2001. The copper transporter CTR1 provides an essential function in mammalian embryonic development. Proc. Natl. Acad. Sci. USA 98:6836-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J., J. R. Prohaska, and D. J. Thiele. 2001. Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc. Natl. Acad. Sci. USA 98:6842-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. W., S. H. Bae, J. W. Jeong, S. H. Kim, and K. W. Kim. 2004. Hypoxia-inducible factor (HIF-1) alpha: its protein stability and biological functions. Exp. Mol. Med. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Maine, G. N., X. Mao, C. M. Komarck, and E. Burstein. 2007. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26:436-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer, J. F. 1998. Menkes syndrome and animal models. Am. J. Clin. Nutr. 67:1022S-1028S. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, B. J., B. G. Sato, T. P. Dalton, and K. R. Laderoute. 2005. The metal-responsive transcription factor-1 contributes to HIF-1 activation during hypoxic stress. Biochem. Biophys. Res. Commun. 337:860-867. [DOI] [PubMed] [Google Scholar]
- 29.Nabetani, A., I. Hatada, H. Morisaki, M. Oshimura, and T. Mukai. 1997. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol. Cell. Biol. 17:789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nada, S., T. Yagi, H. Takeda, T. Tokunaga, H. Nakagawa, Y. Ikawa, M. Okada, and S. Aizawa. 1993. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 73:1125-1135. [DOI] [PubMed] [Google Scholar]
- 31.Pause, A., S. Lee, R. A. Worrell, D. Y. Chen, W. H. Burgess, W. M. Linehan, and R. D. Klausner. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 94:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 33.Scheel, J. R., L. J. Garrett, D. M. Allen, T. A. Carter, L. Randolph-Moore, M. J. Gambello, F. H. Gage, A. Wynshaw-Boris, and C. Barlow. 2003. An inbred 129SvEv GFPCre transgenic mouse that deletes loxP-flanked genes in all tissues. Nucleic Acids Res. 31:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14:1983-1991. [PubMed] [Google Scholar]
- 35.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 36.Shibata, T., A. J. Giaccia, and J. M. Brown. 2000. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 7:493-498. [DOI] [PubMed] [Google Scholar]
- 37.Su, L. C., C. A. Owen, Jr., P. E. Zollman, and R. M. Hardy. 1982. A defect of biliary excretion of copper in copper-laden Bedlington terriers. Am. J. Physiol. 243:G231-G236. [DOI] [PubMed] [Google Scholar]
- 38.Su, L. C., S. Ravanshad, C. A. Owen, Jr., J. T. McCall, P. E. Zollman, and R. M. Hardy. 1982. A comparison of copper-loading disease in Bedlington terriers and Wilson's disease in humans. Am. J. Physiol. 243:G226-G230. [DOI] [PubMed] [Google Scholar]
- 39.Takeda, K., V. C. Ho, H. Takeda, L. J. Duan, A. Nagy, and G. H. Fong. 2006. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol. Cell. Biol. 26:8336-8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao, T. Y., F. Liu, L. Klomp, C. Wijmenga, and J. D. Gitlin. 2003. The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J. Biol. Chem. 278:41593-41596. [DOI] [PubMed] [Google Scholar]
- 41.van de Sluis, B., J. Rothuizen, P. L. Pearson, B. A. van Oost, and C. Wijmenga. 2002. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum. Mol. Genet. 11:165-173. [DOI] [PubMed] [Google Scholar]
- 42.Vengellur, A., B. G. Woods, H. E. Ryan, R. S. Johnson, and J. J. LaPres. 2003. Gene expression profiling of the hypoxia signaling pathway in hypoxia-inducible factor 1α null mouse embryonic fibroblasts. Gene Expr. 11:181-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vrljicak, P., D. Myburgh, A. K. Ryan, M. A. van Rooijen, C. L. Mummery, and I. R. Gupta. 2004. Smad expression during kidney development. Am. J. Physiol. Renal Physiol. 286:F625-F633. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y., K. Joh, S. Masuko, H. Yatsuki, H. Soejima, A. Nabetani, C. V. Beechey, S. Okinami, and T. Mukai. 2004. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 24:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y., K. Joh, and T. Mukai. 2002. Identification of a novel isoform of Murr1 transcript, U2mu, which is transcribed from the portions of two closely located but oppositely oriented genes. Genes Genet. Syst. 77:377-381. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Z., K. Joh, H. Yatsuki, Y. Wang, Y. Arai, H. Soejima, K. Higashimoto, T. Iwasaka, and T. Mukai. 2006. Comparative analyses of genomic imprinting and CpG island-methylation in mouse Murr1 and human MURR1 loci revealed a putative imprinting control region in mice. Gene 366:77-86. [DOI] [PubMed] [Google Scholar]